Abstract
Adult stem and progenitor cells are uniquely capable of self-renewal, and targeting this process represents a potential therapeutic opportunity. The early erythroid progenitor, burst-forming unit erythroid (BFU-E), has substantial self-renewal potential and serves as a key cell type for the treatment of anemias. However, our understanding of mechanisms underlying BFU-E self-renewal is extremely limited. Here, we found that the muscarinic acetylcholine receptor, cholinergic receptor, muscarinic 4 (CHRM4), pathway regulates BFU-E self-renewal and that pharmacological inhibition of CHRM4 corrects anemias of myelodysplastic syndrome (MDS), aging, and hemolysis. Genetic down-regulation of CHRM4 or pharmacologic inhibition of CHRM4 using the selective antagonist PD102807 promoted BFU-E self-renewal, whereas deletion of Chrm4 increased erythroid cell production under stress conditions in vivo. Moreover, muscarinic acetylcholine receptor antagonists corrected anemias in mouse models of MDS, aging, and hemolysis in vivo, extending the survival of mice with MDS relative to that of controls. The effects of muscarinic receptor antagonism on promoting expansion of BFU-Es were mediated by cyclic AMP induction of the transcription factor CREB, whose targets up-regulated key regulators of BFU-E self-renewal. On the basis of these data, we propose a model of hematopoietic progenitor self-renewal through a cholinergic-mediated “hematopoietic reflex” and identify muscarinic acetylcholine receptor antagonists as potential therapies for anemias associated with MDS, aging, and hemolysis.
INTRODUCTION
Stem and progenitor cells undergo self-renewal, which is crucial for tissue homeostasis, maintenance, and regeneration (1–4). In the hematopoietic system, the burst-forming unit erythroid (BFU-E) is the first lineage-determined erythroid progenitor, with substantial potential to undergo self-renewal to generate thousands of erythrocytes. BFU-E undergoes differentiation resulting in formation of the late erythroid progenitor, colony-forming unit erythroid (CFU-E). CFU-E generates proerythroblasts, which form erythrocytes after undergoing three to four cell divisions (5–8). Whereas survival and differentiation of CFU-Es are mainly controlled by erythropoietin (EPO), regulators of BFU-E expansion and differentiation are less well defined. EPO is mainly used to treat anemias caused by defects in EPO production, as seen in chronic kidney disease (9–13). However, many anemic patients do not have enough BFU-Es and, subsequently, not enough CFU-Es to respond to EPO treatment (12–21). An improved understanding of molecular mechanisms underlying BFU-E self-renewal is needed to treat EPO-resistant anemias and identify druggable regulators controlling this process. Because G protein–coupled receptors (GPCRs) are the largest group of pharmacologically druggable proteins, we focused on the identification of GPCRs that regulate BFU-E self-renewal.
RESULTS
Muscarinic acetylcholine receptor antagonists increase erythrocyte production by regulating BFU-E self-renewal
To identify GPCRs that regulate BFU-E self-renewal, we analyzed genome-wide gene expression profiles (14) and focused on GPCRs that are abundantly expressed in murine BFU-Es. To further narrow down our candidate list to GPCRs that are most likely to be important for regulation of BFU-E self-renewal versus differentiation, we used the fact that self-renewal and differentiation are two opposite cell fates with likely contrasting gene expression profiles. We analyzed gene expression profiles of two processes: dexamethasone-induced BFU-E self-renewal and normal BFU-E differentiation. Among the 358 druggable GPCRs examined (data file S1), six GPCRs exhibited contrasting gene expression profiles. Three GPCRs (P2ry2, Gpr124, and Calcrl) were up-regulated during BFU-E self-renewal and down-regulated during BFU-E differentiation (fig. S1, A and B), and three GPCRs (Chrm4, Fzd5, and Darc) were down-regulated during BFU-E self-renewal and up-regulated during BFU-E differentiation (fig. S1, A and C).
Using a previously established highly purified murine BFU-E (lin−Ter119−CD16/CD32−Sca-1−CD41−c-Kit+CD71/Cd24a10%low population from fetal liver) isolation and culture system (14, 15, 19), we tested chemical compounds that act as agonists of GPCRs up-regulated during BFU-E self-renewal. Conversely, for GPCRs down-regulated during BFU-E self-renewal, we tested antagonists for their capacities to promote BFU-E expansion. Of the chemical compounds tested, including purinergic receptor P2Y, G protein–coupled 2 (P2RY2) agonists PSB1114 (22) and UTPgS (23) and frizzled class receptor 5 (FZD5) antagonist Wnt antagonist III, box5 (24), oxyphenonium bromide (Fig. 1, A and B) and orphenadrine citrate (fig. S2A), two closely related muscarinic acetylcholine receptor antagonists, triggered BFU-E expansion.
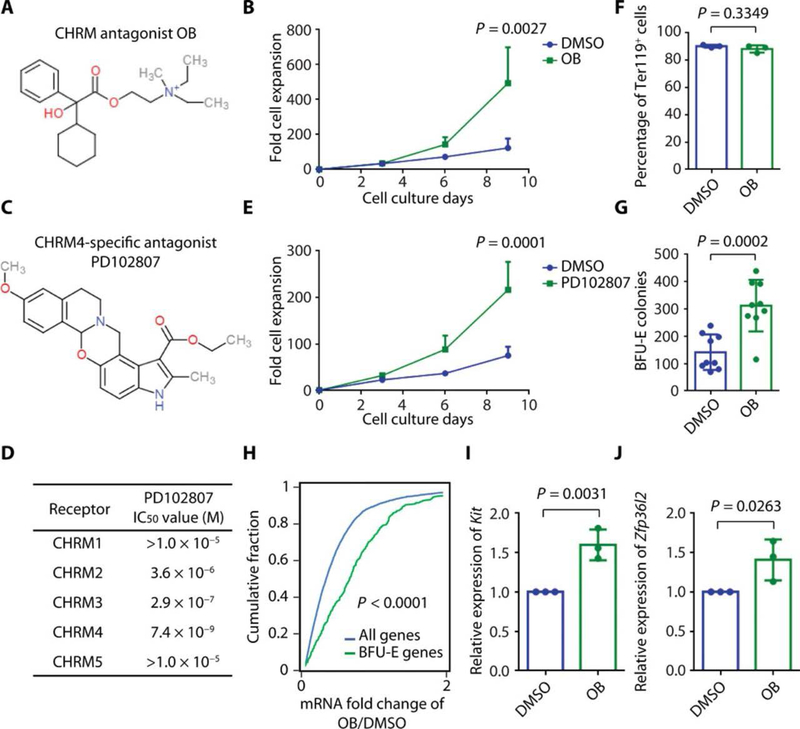
Fig. 1. Muscarinic acetylcholine receptor antagonists increase erythrocyte production by regulating BFU-E self-renewal.
(A) Chemical structure of muscarinic acetylcholine receptor antagonist oxyphenonium bromide (OB). (B) Highly purified murine BFU-Es were isolated as lin−Ter119−CD16/CD32−Sca-1−CD41−c-Kit+CD71/Cd24a10%low population from murine fetal liver (14, 15, 19) and cultured with DMSO or 100 mM OB, and cell numbers were counted from days 0 to 9. The means and SD of three measurements from distinct samples are shown. P value was calculated using two-way ANOVA (original data are in data file S3). (C) Chemical structure of CHRM4 selective antagonist PD102807. (D) IC50 values from protein binding assay for PD102807 on indicated receptors. (E) Purified murine BFUEs were cultured with DMSO or 3 μM PD102807, and cell numbers were counted from days 0 to 9. The means and SD of three measurements from distinct samples are shown. P value was calculated using two-way ANOVA analysis (original data are in data file S3). (F) Purified murine BFU-Es were cultured with DMSO or 100 μM OB, and cells were stained with anti-Ter119 antibody at the end of culture and analyzed with flow cytometry. The means and SD of the percentage of Ter119+ cells in three measurements from distinct samples are shown. P value was calculated using the one-tailed t test. (G) Purified murine BFU-Es were cultured with DMSO or 100 μM OB, and cells were plated on methylcellulose medium. BFU-E colonies were counted on day 9 of colony formation assay, and the means and SD of BFU-E colonies in nine measurements from distinct samples are shown. P value was calculated using the one-tailed t test. (H) Purified murine BFU-Es were cultured with DMSO or 100 μM OB, and RNA-seq was performed on cultured cells. The x axis represents the ratio of each gene’s expression in BFU-Es cultured with OB relative to BFU-Es cultured with DMSO. The y axis represents the cumulative fraction and is plotted as a function of the relative expression (x axis). “BFU-E genes” represent a group of 533 genes most markedly down-regulated during erythroid differentiation from the BFU-E to the CFU-E stage (data file S2) (14). “All genes” represent all the genes expressed in BFU-E, as reported previously (15). P value was calculated using the Kolmogorov-Smirnov test. (I) The means and SD of relative expression of Kit in three measurements from distinct samples extracted from RNA-seq data displayed in (H). P value was calculated using the one-tailed t test. (J) The means and SD of relative expression of Zfp36l2 in three measurements from distinct samples extracted from RNA-seq data displayed in (H). P value was calculated using the one-tailed t test.
When cultured in the presence of 100 μM oxyphenonium bromide, BFU-E underwent prolonged expansion resulting in increased production of erythrocytes (Fig. 1B). Given the fact that oxyphenonium bromide and orphenadrine citrate both inhibit the GPCR family known as cholinergic receptor, muscarinic 1–5 (CHRM1–5) (25, 26) and that CHRM4 but not other members of this family are expressed in BFU-Es (data file S1), we hypothesized that inhibition of this CHRM4 specifically may underlie the impact of these compounds on BFU-E self-renewal. In support of this notion, the CHRM4 selective antagonist PD102807 supported BFU-E expansion. Consistent with previous reports (27, 28), PD102807 selectively inhibits CHRM4 over other muscarinic acetylcholine receptors (Fig. 1, C and D, fig. S2B, and table S1). PD102807 promoted BFU-E expansion at nanomolar concentrations (Fig. 1E and fig. S2C). At day 14 of culture, most of the cultured cells underwent terminal erythroid differentiation, and there was no difference in the percentage of Ter119+ cells with or without oxyphenonium bromide or PD102807 (Fig. 1F and fig. S2, D to G). Our data further showed that oxyphenonium bromide increased formation of BFU-E colonies in colony formation assays (Fig. 1G).
Consistent with enhancement of BFU-E self-renewal, oxyphenonium bromide (Fig. 1H) and PD102807 (fig. S2H) up-regulated a gene signature characteristic of BFU-Es (data file S2) (14). This included up-regulation of Kit, a gene essential for hematopoietic stem and progenitor cell (HSPC) maintenance, and Zfp36l2, an RNA binding protein previously identified as an essential regulator for BFU-E self-renewal (15) at both mRNA (Fig. 1, I and J) and protein level (fig. S2I). PD102807 exhibited no effect on CFU-E proliferation (fig. S2, J and K). The observed effects of CHRM4 antagonists on BFU-E expansion were similar to the effects of 100 nM dexamethasone regarding BFU-E expansion (fig. S2L) and up-regulation of BFU-E gene signature (fig. S2M). In addition, CHRM4 antagonists exhibited additive but not synergistic effects with dexamethasone because there was no difference between the sum of cells generated from dexamethasone alone and CHRM4 antagonists alone and the total number of cells generated from combined dexamethasone and CHRM4 antagonist treatment (fig. S2N). This is distinct from the synergistic effects between dexamethasone and agonists of hypoxia-inducible factor 1a (HIF-1α) and peroxisome proliferator–activated receptor a (PPAR-α), where HIF-1α and PPAR-α are coregulators of the glucocorticoid receptor and function together to control the expression of their target genes (14, 19).
In the human CD34+ HSPC erythroid differentiation culture system, oxyphenonium bromide and PD102807 also expanded erythroid cells (fig. S3, A and B) and increased formation of BFU-E colonies (fig. S3, C and D). The more robust effect of CHRM4 antagonists on mouse cells is consistent with the fact that mouse BFU-E culture system contains more pure BFU-Es than human CD34+ erythroid differentiation culture system. As shown in fig. S3E, 100 purified mouse BFU-Es generated 141 BFU-E colonies, whereas 100 human CD34+ cells generated 15 BFU-E colonies. CHRM4 antagonists did not influence erythroid cell survival (fig. S3, F and G). At the end of culture, most of the cells in the CD34+ erythroid cell culture system were CD235a+ erythroid cells, and there was no difference in the percentage of CD235a+ with or without CHRM4 antagonists (fig. S3, H to M). CHRM4 antagonist also up-regulated the expression of KIT and ZFP36L2 (fig. S3N). Together, these data suggest that muscarinic acetylcholine receptor antagonists increase erythrocyte production by promoting BFU-E self-renewal.
CHRM4 negatively regulates BFU-E self-renewal
Chrm4 is the most highly expressed muscarinic acetylcholine receptor within BFU-Es (Fig. 2A), and across tissue types, erythroid precursors have the second most abundant expression of Chrm4, behind nerve tissues where a functional role of Chrm4 is established (Fig. 2B) (29, 30). Chrm4 was down-regulated during BFU-E expansion when BFU-Es were cultured with 100 nM dexamethasone (data file S1 and Fig. 2, C and D). As suggested by Western blots that detect DDK-tagged CHRM4 by both anti-DDK and anti-CHRM4 antibodies (fig. S4A), CHRM4 may undergo posttranslational modification(s) and/or RNA processing event(s) to generate two bands of differing protein size. As shown in Fig. 2 (E and F), knockdown of Chrm4 using two independent short hairpin RNAs (shRNAs) targeting Chrm4 increased production of erythroid cells (Fig. 2G) and promoted BFU-E self-renewal (Fig. 2H and fig. S4B). At the end of culture, most of the cultured cells underwent terminal erythroid differentiation (Fig. 2I), and there was no difference in cell survival with and without knockdown of Chrm4 (fig. S4C). Knockdown of Chrm4 had no influence on CFU-E proliferation (fig. S4, D and E). Evaluating for potential off-target effects of anti-Chrm4 shRNAs, we observed that human CHRM4 complementary DNA (cDNA), which was not affected by anti-mouse Chrm4 shRNA, abrogated the BFU-E expansion seen with anti-mouse Chrm4 shRNA (fig. S4, F and G). Moreover, knockdown of Chrm4 diminished the capacity of PD102807 to promote BFU-E expansion (fig. S4H), revealing the specificity of PD102807 for CHRM4. Similarly, BFU-E isolated from Chrm4−/− mice exhibited increased expansion in comparison with BFU-E isolated from wild-type mice (fig. S4I).
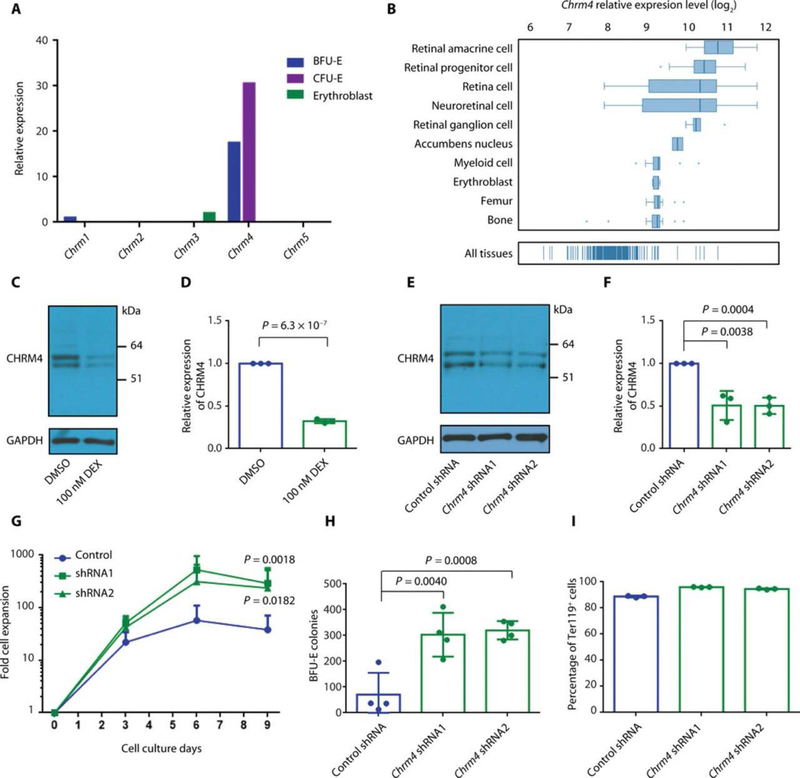
Fig. 2. CHRM4 negatively regulates BFU-E self-renewal.
(A) Relative expression of muscarinic acetylcholine receptor family members—including Chrm1, Chrm2, Chrm3, Chrm4, and Chrm5—at the BFU-E, CFU-E, and erythroblast stages of murine fetal liver (14). (B) Relative expression of Chrm4 in the indicated cell types and tissues (47). (C) Expression of CHRM4 in BFU-Es from murine fetal liver cultured in the absence or presence of 100 nM dexamethasone (DEX) was measured on day 3 of culture using Western blot. Representative results of three measurements from distinct samples are shown. Unprocessed blots are in fig. S22. (D) The means and SD of three measurements from distinct samples of expression of CHRM4 relative to GAPDH as in (C). P value was calculated using the one-tailed t test. (E) Expression of CHRM4 in BFU-Es from murine fetal liver infected with either control virus or virus encoding shRNA targeting Chrm4 was measured on day 3 of culture using Western blot. Representative results of three measurements from distinct samples are shown. Unprocessed blots are in fig. S22. (F) The means and SD of quantification of three measurements from distinct samples of expression of CHRM4 relative to GAPDH as in (E) are shown. P values were calculated using the one-tailed t test. (G) BFU-Es from murine fetal liver were infected with either control virus or virus encoding shRNA targeting Chrm4. The numbers of green fluorescent protein–positive (GFP+) cells were counted from days 0 to 9. The means and SD of five or six measurements from distinct samples are shown. P values were calculated using two-way ANOVA analysis. The original data are in data file S3. (H) BFU-Es from murine fetal liver were infected with either control virus or virus encoding shRNA targeting Chrm4. GFP+ cells were sorted and seeded for BFU-E colony formation assay. BFU-E colonies were counted on day 9, and the means and SD of four measurements from distinct samples are shown. P values were calculated using the one-tailed t test. (I) BFU-Es from murine fetal liver were infected with either control virus or virus encoding shRNA targeting Chrm4. Cells were stained with anti-Ter119 antibody at the end of culture, and GFP+ cells were gated for Ter119 expression analysis with flow cytometry. The means and SD of three measurements from distinct samples are shown.
CHRM4 is the most highly expressed muscarinic acetylcholine receptor in human erythroid progenitors (fig. S5, A and B). Consistent with results in mouse cells, knockdown of CHRM4 in human CD34+ HSPCs promoted erythroid cell expansion (fig. S5, C to E) without influence on cell survival (fig. S5F). These results suggest that CHRM4 negatively regulates BFU-E self-renewal and erythroid cell production.
CHRM4 is involved in stress erythropoiesis regulation in vivo
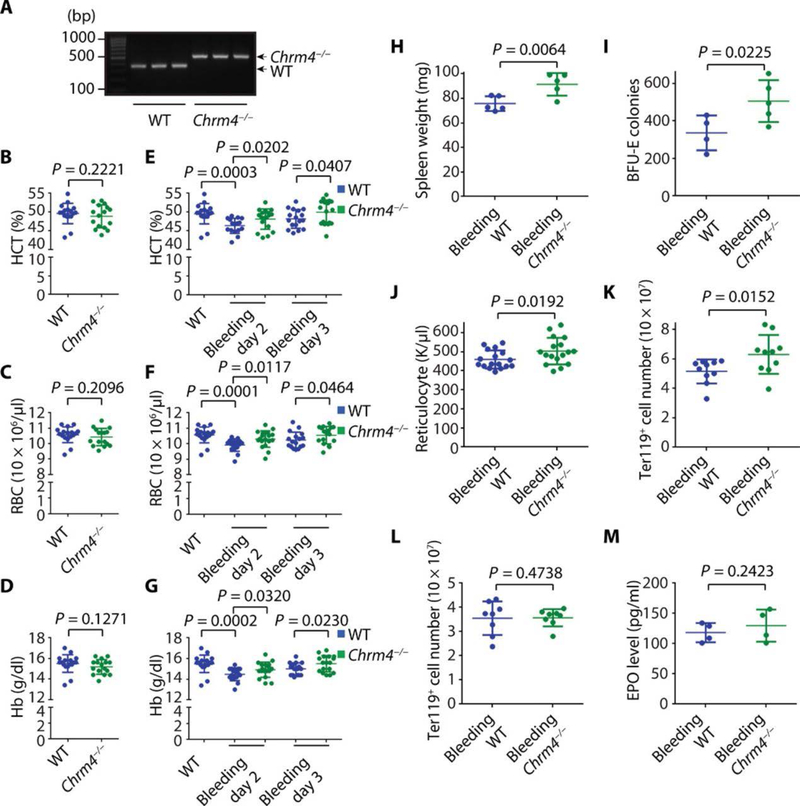
To determine whether pharmacologic inhibition of CHRM4 could be recapitulated by genetic loss of CHRM4 in vivo, we evaluated Chrm4 knockout mice (Fig. 3A) (31). Consistent with the role of CHRM4 in regulating BFU-E expansion and stress erythropoiesis, Chrm4−/− mice had no abnormalities in erythropoiesis at steady state (Fig. 3, B to D), but loss of Chrm4 increased erythroid cell production under bleeding-induced stress in vivo (Fig. 3, E to G). Loss of Chrm4 also increased splenic BFU-E and Ter119+ cells (Fig. 3, H to K), without altering bone marrow erythroid lineage cells or EPO concentrations (Fig. 3, L and M). These experiments were performed after transplantation of bone marrow from Chrm4−/− or wild-type mice into wild-type recipient mice to ensure that the effects of Chrm4 loss were hematopoietic specific.
Fig. 3. CHRM4 is involved in stress erythropoiesis regulation in vivo.
(A) Genotyping PCRs for wild-type (WT) and Chrm4−/− mice were performed. Three hundred seventy–base pair (bp) PCR product band corresponds to WT mice, and 480-bp PCR product band corresponds to Chrm4−/− mice. (B) Bone marrow cells were purified from donor WT or Chrm4−/− mice and transplanted to recipient WT mice. CBC was performed before bleeding, and the means and SD of hematocrit (HCT) values are shown (n = 17 and 16 for WT and Chrm4−/− groups, respectively). (C) The means and SD of red blood cell (RBC) values are shown (n = 17 and 16 for WT and Chrm4−/− groups, respectively). (D) The means and SD of hemoglobin (Hb) values are shown (n = 17 and 16 for WT and Chrm4−/− groups, respectively). (E) Bleeding was performed daily to induce stress erythropoiesis. CBC was performed at days 2 and 3 after the initial bleeding, and the means and SD of HCT values are shown (n = 16 and 18 for WT and Chrm4−/− groups, respectively). (F) The means and SD of RBC values are shown (n = 16 and 18 for WT and Chrm4−/− groups, respectively). (G) The means and SD of Hb values are shown (n = 16 and 18 for WT and Chrm4−/− groups, respectively). (H) The means and SD of spleen weights are shown (n = 5 and 5 for WT and Chrm4−/− groups, respectively). (I) The spleens were dissected for BFU-E colony formation assay, and the means and SD of numbers of BFU-E colony are shown (n = 4 and 5 for WT and Chrm4−/− groups, respectively). (J) The means and SD of reticulocyte counts are shown (n = 17 and 17 for WT and Chrm4−/− groups, respectively). (K) The means and SD of Ter119+ splenic cell numbers are shown (n = 10 and 10 for WT and Chrm4−/− groups, respectively). (L) The means and SD of Ter119+ bone marrow cell numbers are shown (n = 8 and 8 for WT and Chrm4−/− groups, respectively). (M) The means and SD of plasma EPO concentrations are shown (n = 4 and 4 for WT and Chrm4−/− groups, respectively). Eight-week-old female mice were used. Each dot represents one mouse. All P values were calculated using the one-tailed t test.
Muscarinic acetylcholine receptor antagonists correct anemia in vivo
To evaluate the therapeutic potential of muscarinic acetylcholine receptor antagonists in alleviating anemia in vivo, we tested muscarinic acetylcholine receptor antagonists in three disease models, including a genetically accurate myelodysplastic syndrome (MDS) mouse model, an aging mouse model, and a hemolysis mouse model. To identify the effective anti-anemia dose for these compounds in vivo, we first performed a maximal tolerance study and a pharmacodynamics study for each compound (fig. S6). We identified that the maximal tolerated doses are 50 and 100 mg/kg for oxyphenonium bromide and PD102807, respectively (fig. S6, A to C). The anti-anemia minimal effective doses were 25, 100, and 100 mg/kg for oxyphenonium bromide, PD102807, and orphenadrine citrate, respectively (fig. S6, D to I). We therefore chose 25, 100, and 100 mg/kg for oxyphenonium bromide, PD102807, and orphenadrine citrate, respectively, for further in vivo studies.
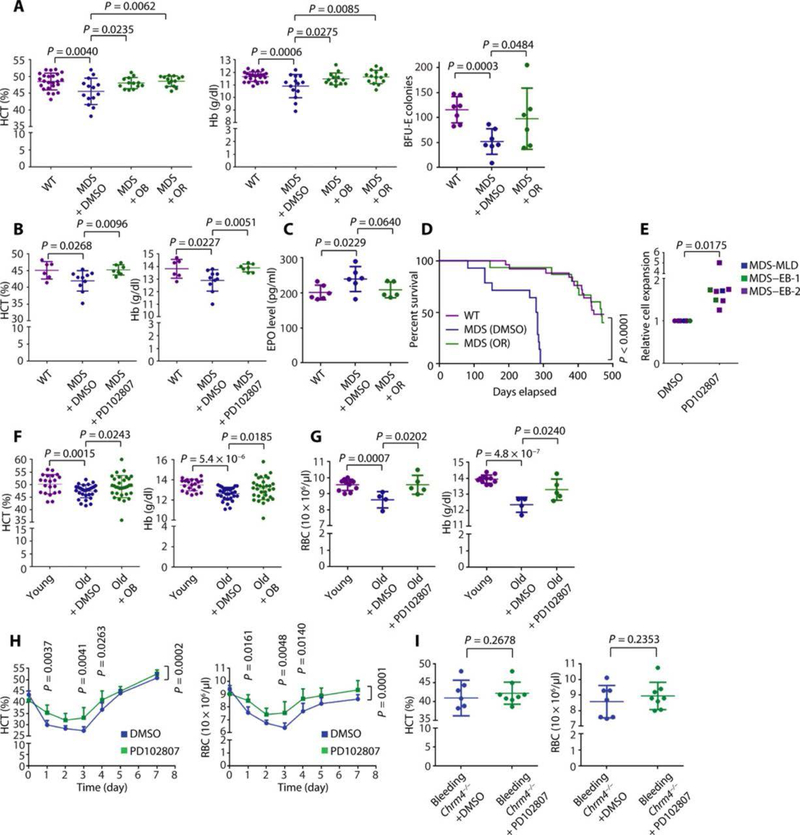
Mutations of splicing factors are the most common class of genetic abnormalities in MDS. We recently demonstrated that mutation of Srsf2 causes MDS using a conditional knock-in Mx1-Cre Srsf2P95H/WT mouse model (fig. S7A) (32–35). In this model, polyinosinic:polycytidylic acid (pIpC) treatment induces Cre recombination (fig. S7B) and anemia (fig. S7, C and D). Injection of oxyphenonium bromide, orphenadrine citrate, or PD102807 corrected anemia of MDS (Fig. 4, A and B) and promoted BFU-E self-renewal (Fig. 4A and fig. S7E). Injection of orphenadrine citrate or PD102807 had no influence on EPO concentrations (Fig. 4C and fig. S7F) or on white blood cell or platelet production (fig. S7, G to J). Treatment with orphenadrine citrate or PD102807 was well tolerated in vivo with no substantial effects on weight loss (fig. S8, A and B) or liver or kidney function (fig. S8, C to G). Orphenadrine citrate treatment extended the survival of Mx1-Cre Srsf2P95H/WT MDS disease mice to a level similar to control wild-type mice (Fig. 4D). Consistent with these favorable effects in MDS mouse models, PD102807 increased production of erythroid cells in CD34+ cells from patients with MDS (Fig. 4E, fig. S9A, and table S2). There was no difference in cell survival (fig. S9B) or differentiation (fig. S9, C and D) between cells cultured with and without PD102807, and most of the cultured cells were CD235a+ erythroid cells. Together, these data suggest that CHRM4 antagonists promote BFU-E self-renewal, increase erythrocyte production, and correct anemia of MDS.
Fig. 4. Muscarinic acetylcholine receptor antagonists correct anemia in vivo.
(A) Mx1-Cre Srsf2P95H/WT MDS mice were intraperitoneally injected with DMSO, OB (25 mg/kg), or orphenadrine citrate (OR) (100 mg/kg) daily. Mx1-Cre Srsf2WT/WT (WT) mice were used as control. CBC was performed on day 45. The means and SD of HCT and Hb are shown (n = 24, 14, 13, and 14 for WT, MDS + DMSO, MDS + OB, and MDS + OR groups, respectively). Spleens were dissected for BFU-E colony formation assay, and the means and SD of numbers of BFU-E colonies are shown (n = 7, 7, and 6 for WT, MDS + DMSO, and MDS + OR groups, respectively). P values were calculated using the one-tailed t test. (B) Mx1-Cre Srsf2P95H/WT MDS mice were orally treated with DMSO or PD102807 (100 mg/kg) daily. CBC was performed on day 15. The means and SD of HCT and Hb are shown (n = 6, 10, and 7 for WT, MDS + DMSO, and MDS + PD102807 groups, respectively). P values were calculated using the one-tailed t test. (C) The means and SD of plasma EPO concentrations are shown (n = 6, 6, and 5 for WT, MDS + DMSO, and MDS + OR groups, respectively). P values were calculated using the onetailed t test. (D) The survival curves are shown (n = 25, 14, and 15 for WT, MDS + DMSO, and MDS + OR groups, respectively). P value was calculated using the log-rank (Mantel-Cox) test. Eight-week-old female mice were used for (B) to (D). (E) CD34+ cells isolated from eight patients with MDS were cultured in erythroid differentiation culture system in the presence of DMSO or 3 nM PD102807. MDS subtype and relative cell expansion of each sample are shown. MDS–EB-½, MDS with excess blasts-½; MDS-MLD, MDS with multilineage dysplasia (48). The means and SD of relative cell expansion are shown (n = 8). P value was calculated using the one-tailed t test. (F) Aged mice (18- to 21-month-old male and female mice) were intraperitoneally injected every day with DMSO or OB (25 mg/kg). CBC was performed on day 7. Young mice (6- to 8-week-old male and female mice) were used as control. The means and SD of HCT and Hb are shown (n = 21, 31, and 32 for young, old + DMSO, and old + OB groups, respectively). P values were calculated using the one-tailed t test. (G) Aged mice (18- to 21-month-old male and female mice) were orally treated every day with DMSO or PD102807 (100 mg/kg). CBC was performed on day 7. Young mice (6- to 8-week-old male and female mice) were used as control. The means and SD of RBC count and Hb are shown (n = 11, 4, and 5 for young, old + DMSO, and old + PD102807 groups, respectively). (H) According to a previously established hemolytic anemia mouse model (19), mice were orally treated every day with DMSO or PD102807 (100 mg/kg) for 3 days (days −3 to −1) before PHZ injection. Mice were injected with PHZ (60 mg/kg) to induce hemolytic anemia on day 0. Mice were orally treated every day with DMSO or PD102807 (100 mg/kg) from days 0 to 8. CBC was performed every day. The means and 95% confidence interval of HCT and RBC over a time course are shown (n = 13 and 17 for DMSO and PD102807 groups, respectively). P values were calculated using two-way ANOVA analysis and multiple comparisons test. The original data are in data file S3. (I). Chrm4 knockout mice were orally treated with DMSO or PD102807 (100 mg/kg) under bleeding-induced stress conditions. CBC was performed, and the means and SD of HCT and RBC are shown (n = 7 and 8 for DMSO and PD102807 groups, respectively). P values were calculated using the one-tailed t test. Eight- to 10-week-old female mice were used for (H) to (I). Each dot represents one mouse.
We next evaluated CHRM4 antagonists for anemia of aging. Anemia is a common manifestation of aging and is associated with a loss of HSPC self-renewal or stem cell exhaustion, which result in bone marrow hypocellularity (36, 37). Anemia in the aging mouse model is shown in fig. S10 (A and B), and oxyphenonium bromide or PD102807 alleviated this anemia in 18- to 21-month-old aged mice (Fig. 4, F and G). Treatment with muscarinic acetylcholine receptor antagonists resulted in hemoglobin above the concentration in younger mice (8- to 10-week-old C57bl/6 mice; median hemoglobin of 13.8 g/dl) in 25% of the aged mice, whereas vehicle treatment achieved this effect in 0% of aged mice. Injection of oxyphenonium bromide had no influence on EPO concentration (fig. S10C).
In addition to impaired erythropoiesis and chronic anemia in MDS anemia (fig. S11, D to F, and Fig. 4H) and promoted BFU-E self-renewal and aging, in a phenylhydrazine (PHZ)–induced hemolysis mouse (fig. S11G). PD102807 treatment of Chrm4 knockout mice under model (fig. S11, A to C), oxyphenonium bromide or PD102807 alleviated bleeding-induced stress failed to increase erythrocyte production. This experiment further confirmed the on-target effect of PD102807 for CHRM4 (Fig. 4I). Together, these results suggest that muscarinic acetylcholine receptor antagonists represent a potential therapeutic approach for the treatment of anemia associated with MDS, aging, and hemolysis.
CHRM4 regulates genes important for the maintenance of BFU-E progenitor status
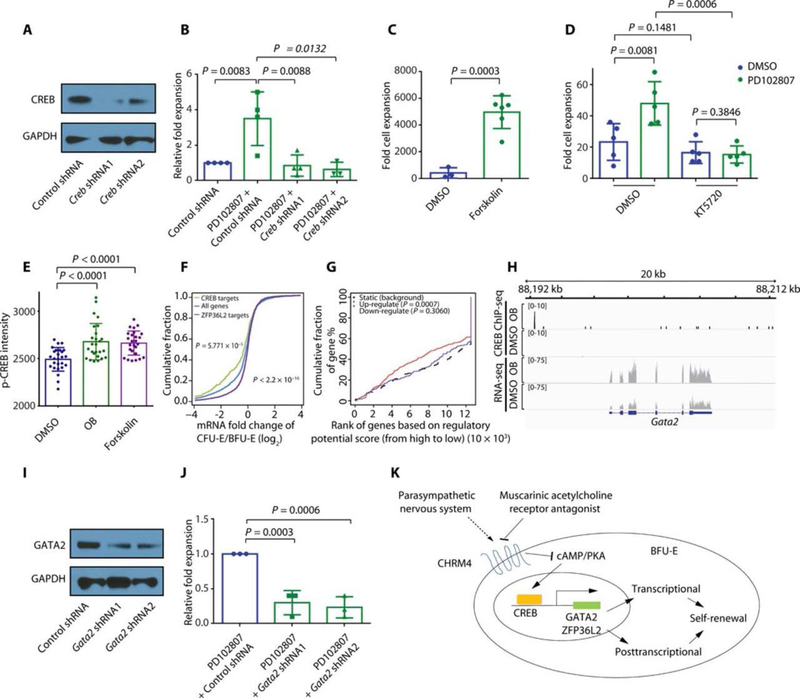
Muscarinic acetylcholine antagonists inhibit the CHRM4 pathway by raising cyclic adenosine 5′-monophosphate (cAMP) concentrations and by regulating the cAMP response element–binding protein (CREB) transcriptional program (29, 30, 38). Consistent with this, knockdown of CREB diminished the ability of CHRM4 antagonists to expand BFU-Es (Fig. 5, A and B, and fig. S12, A and B). In addition, culture of BFU-Es with the adenylate cyclase activator forskolin promoted BFU-E expansion (Fig. 5C and fig. S13, A and B), recapitulating the effect of oxyphenonium bromide on BFU-E expansion. Furthermore, culture of BFU-Es with the protein kinase A (PKA) inhibitor KT5720 (39) blocked CHRM4 antagonist–induced BFU-E expansion (Fig. 5D). In addition, cAMP increased in concentration in BFU-Es upon PD102807 or forskolin treatment relative to dimethyl sulfoxide (DMSO) (fig. S13C). Phosphorylated CREB displayed a similar increase in BFU-Es (Fig. 5E and fig. S13D). It has been shown previously that cAMP/PKA is involved in erythropoiesis regulation and cAMP concentration is not modulated by EPO (40). Our results suggest that cAMP and PKA are regulated by CHRM4.
Fig. 5. CHRM4 pathway regulates genes important for the maintenance of BFU-E progenitor status.
(A) Knockdown efficiency of anti-Creb shRNAs is shown by Western blot. Unprocessed blots are in fig. S22. (B) Murine fetal liver BFU-Es were infected with either control virus or virus encoding shRNA targeting Creb together with DMSO or 3 nM PD102807. The viral vector encodes both GFP and shRNAs. The numbers of GFP+ cells in the culture system were counted on day 9 of culture. The means and SD of four measurements from distinct samples are shown. P values were calculated using the one-tailed t test. (C) Murine fetal liver BFU-Es were cultured with DMSO or 100 μM forskolin. Total numbers of cells were counted on day 9. The means and SD of fold cell expansion of three measurements from distinct samples are shown. P value was calculated using the one-tailed t test. (D) Murine fetal liver BFU-Es were cultured with indicated compounds (3 nM PD102807 and/or 2 μM PKA inhibitor KT5720). Total numbers of cells were counted on day 9. The means and SD of fold cell expansion of five measurements from distinct samples are shown. P values were calculated using one-tailed t test. (E) Murine fetal liver BFU-Es were cultured with DMSO, 100 μM OB, or 100 μM forskolin. Immunofluorescence was performed, and p-CREB nuclear staining intensity was measured by p-CREB staining intensity within DAPI-positive area; the means and SD of 26 measurements from distinct samples are shown. P values were calculated using one-tailed t test. (F) Anti-CREB ChIP-seq was performed on BFU-Es after culture with 100 μM OB. The x axis represents the ratio of each gene’s expression in CFU-Es relative to BFU-Es, calculated as a log2 ratio. The y axis represents the cumulative fraction and is plotted as a function of the relative expression (x axis). “CREB targets” represent direct target genes of CREB in BFU-E identified by anti-CREB ChIP-seq. “ZFP36L2 targets” represent direct target genes of ZFP36L2 in BFU-E identified by anti-ZFP36L2 RIP-ChIP, as reported previously (15). All genes represent all genes expressed in BFU-E, as reported previously (15). P values were calculated using the Kolmogorov-Smirnov test. (G) Murine fetal liver BFU-Es were cultured with DMSO or 100 μM OB, and RNA-seq was performed on cultured cells. Anti-CREB ChIP-seq was performed on BFU-Es after culturing with 100 μM OB. RNA-seq results and CREB ChIP-seq results were combined for BETA analysis (46). P values were calculated using the Kolmogorov-Smirnov test. (H) Murine fetal liver BFU-Es were cultured with DMSO or 100 μM OB, and RNA-seq was performed on cultured cells. Anti-CREB ChIP-seq was performed on BFU-Es after culture with either DMSO or 100 μM OB. RNA-seq results and CREB ChIP-seq results are shown for Gata2. (I) The knockdown efficiency of mouse Gata2 shRNAs is shown using Western blots. Unprocessed blots are in fig. S22. (J) BFU-Es were infected with indicated viral shRNAs and cultured in the presence of 3 nM PD102807. Numbers of cells were counted on day 6, and the means and SD of relative fold expansion of three measurements from distinct samples are shown. (K) Muscarinic acetylcholine receptor antagonists trigger up-regulation of cAMP and CREB transcriptional programs within BFU-Es to support maintenance of BFU-E progenitor status. Key CREB transcriptional targets GATA2 and ZFP36L2 are up-regulated within BFU-Es by CHRM4 inhibition and are required for BFU-E expansion by CHRM4 inhibition.
To identify direct targets of CREB, we performed anti-CREB chromatin immunoprecipitation sequencing (ChIP-seq) on BFU-Es (fig. S14, A and B). CREB binding consensus sequence (fig. S15) and binding sites were identified (figs. S16 and S17). We previously showed that ZFP36L2 binds mRNAs of genes induced during erythroid differentiation and triggers their degradation, which, in turn, contributes to self-renewal (15). In contrast, upon treatment with CHRM4 antagonists, CREB preferentially binds near genomic loci that are highly expressed in BFU-Es (Fig. 5F) and triggers up-regulation of these genes (Fig. 5G and fig. S18, A and B). CREB targets include transcription factor Gata2 (Fig. 5H and fig. S19), which is sufficient to induce erythroid progenitor expansion (41), and Zfp36l2, which is indispensable for main-maintenance of BFU-E progenitor status (fig. S19) (15). CREB ChIP-seq peak near Gata2 contains one CREB binding motif, whereas CREB ChIP-seq peak near Zfp36l2 contains three CREB binding motifs (42). In summary, ZFP36L2 triggers degradation of genes induced during erythroid differentiation, whereas CREB increases expression of genes decreased during erythroid differentiation. In addition, Zfp36l2, GATA2, ZFP36L2, or KIT diminished CHRM4 antagonist–induced whose expression decreases during erythroid differentiation, is also mouse BFU-E expansion (Fig. 5, I to J, and fig. S20, A to D) and a target gene of CREB. We further identified that knockdown of human erythroid progenitor expansion (fig. S21, A to F). Together, these data suggest that the CHRM4-CREB pathway promotes BFU-E self-renewal by up-regulating the expression of genes important for the maintenance of BFU-E progenitor status.
DISCUSSION
Here, we found that the muscarinic acetylcholine receptor CHRM4 is a crucial regulator of BFU-E self-renewal. We showed that CHRM4 is genetically required for erythroid expansion of primary mouse BFU-Es and human CD34+ cells. We further showed that pharmacological inhibition of muscarinic acetylcholine receptor triggers expansion of both mouse and human erythroid progenitors and increased production of erythroid cells in CD34+ cells from patients with MDS. In MDS, aging, and hemolysis mouse models, muscarinic acetylcholine receptor antagonists corrected anemia in vivo.
It is important to acknowledge that there is no single ideal model of human MDS. Although we used orthogonal model systems, including the Mx1-Cre Srsf2P95H/WT mouse model and primary MDS patient samples, studies in additional models of human MDS will be essential to further understand the efficacy of CHRM4 antagonists across the diverse clinical and genetic subtypes of MDS. Moreover, given the established safety of CHRM4 antagonists in human subjects, it will also be very important to evaluate the effects of CHRM4 inhibition on erythropoiesis in patients with MDS (43). Glucocorticoids are used to treat patients with Diamond-Blackfan anemia and autoimmune hemolytic anemia, and their regulation of BFU-E expansion forms the mechanistic basis for the treatment (13–15). Thus, it will also be important to evaluate CHRM4 antagonists for the treatment of Diamond-Blackfan anemia and autoimmune hemolytic anemia.
The identification of a function of CHRM4, a receptor for neurotransmitters, in regulating BFU-E self-renewal suggests a potential connection between parasympathetic neuron activity and BFU-E self-renewal and a potential model of neural control of HSPC self-renewal through a cholinergic-mediated “hematopoietic reflex” (Fig. 5K), where CHRM4 on BFU-Es may be able to sense parasympathetic neuron activity. Parasympathetic inhibition of CHRM4 on BFU-Es results in increased activity of the downstream transcription factor CREB, which, in turn, promotes BFU-E expansion through up-regulation of critical erythroid regulators including GATA2, ZFP36L2, and KIT. These data therefore suggest an essential regulatory role of the nervous system in hematopoietic cell development and function (44, 45).
MATERIALS AND METHODS
Study design
The purpose of this study was to identify regulators of early erythroid BFU-E progenitor self-renewal and develop therapeutic strategies to treat anemia of MDS, aging, and hemolysis. On the basis of our previous publication (15) and pilot experiments, three to six replicates were used for all cell culture, mouse, and sequencing experiments. Sample size of three to six replicates was used for all cell culture work, whereas a sample size of 7 to 10 mice was used for all mouse experiments. For all mouse models used, mice were randomly assigned to control and treatment groups, with approximately equal numbers under each condition. Although treatment was not blinded, assessment of outcomes was blinded. Specific information about sample size and replication is included in figure legends.
Reagents
BFU-E culture media, StemSpan serum-free expansion medium (SFEM) (09650) and StemSpan SFEM II (09655), and methylcellulose medium, MethoCult SF M3436 (03436), were purchased from STEMCELL Technologies. Growth factors and cytokines—including recombinant murine stem cell factor (rmSCF) (250–03), recombinant murine insulin-like growth factor 1 (rmIGF-1) (250–19), recombinant human SCF (rhSCF) (300–07), and recombinant human interleukin-3 (rhIL-3) (200–03)—were purchased from PeproTech. Oxyphenonium bromide (O5501), forskolin (F3917), orphenadrine citrate (O4630), dexamethasone (D4902), and PHZ (P26252) were purchased from Sigma-Aldrich. KT 5720 (1288) was purchased from Tocris Biosciences. Anti-mouse Ter119 anaphase-promoting complex (APC) antibody (17–5921-83), anti-mouse CD71 phycoerythrin (PE) antibody (12–0711-83), and annexin V APC apoptosis detection kit (50–927-9) were purchased from eBioscience. ChIP-IT high-sensitivity kit (5340) was purchased from Active Motif. ChIPAb+ CREB-ChIP Validated Antibody and Primer Set (17–600) was purchased from EMD Millipore. Radioimmunoprecipitation assay (RIPA) lysis buffer system (sc-24948A) was purchased from Santa Cruz Biotechnology. Anti-CHRM4 antibody (TA321192) and anti-DDK antibody (TA50011) were purchased from OriGene. Anti–GAPDH (glyceraldehyde-3-phosphate dehydrogenase) antibody (ab8245), anti-CREB antibody (phospho S133; ab210124), anti-GATA2 antibody (ab22849), anti-KIT antibody (ab46758), and anti-ZFP36L2 antibody (ab70775) were purchased from Abcam. Secondary antibodies against rabbit immunoglobulin G (IgG) (111–035-003) and mouse IgG (115–035-003) were purchased from Jackson ImmunoResearch.
Mouse BFU-E culture system, CFU-E culture system, and human CD34+ cell culture system
Highly purified BFU-Es were isolated as lin−Ter-119−CD16/CD32− Sca-1−CD41−c-Kit+CD71/Cd24a10%low population from mouse embryonic day 14.5 (E14.5) fetal liver using fluorescence-activated cell sorting, as previously described (14, 15, 19). BFU-Es were cultured in StemSpan SFEM II medium containing rmSCF (100 ng/ml), EPO (2 U/ml), rmIGF-1 (40 ng/ml), and dexamethasone (1 nM), with oxyphenonium bromide (100 μM), orphenadrine citrate (1 μM), PD102807 (3 nM), KT5720 (2 μM), and forskolin (100 μM) (14, 15, 19). CFU-Es were isolated from mouse E14.5 fetal liver and cultured as previously described (5, 14, 15).
Human CD34+ HSPCs isolated from either healthy donors or patients with MDS were cultured in StemSpan SFEM medium containing rhSCF (20 ng/ml), rhIL-3 (5 ng/ml), EPO (1 U/ml), dexamethasone (1 nM), oxyphenonium bromide (200 μM), and PD102807 (3 nM) (14, 19). Bone marrow mononuclear cells were obtained from patients with MDS treated at Memorial Sloan Kettering Cancer Center (MSKCC). Written informed consent was obtained from patients for these studies according to the Helsinki convention, and this study received approval from the Institutional Review Board (IRB) at MSKCC (IRB protocol 06–107).
BFU-E colony formation, CD71 and TER119/CD235a antibody staining, apoptosis, retrovirus infection assays, and May-Grünwald-Giemsa staining
BFU-Es were cultured and placed on methylcellulose medium MethoCult SF M3436. BFU-E colonies were counted. Cultured cells were stained with CD71-PE and TER119-APC/CD235a-APC antibodies, and the percentage of CD71+ or TER119+/CD235a+ cells was measured by flow cytometry. Apoptosis rates were measured using annexin V APC and propidium iodide. For retrovirus infection, BFU-Es were infected with viruses encoding control shRNA, shRNAs targeting Chrm4, Creb, Zfp36l2, Gata2, and Kit, or human CHRM4 cDNA, followed by culture in StemSpan SFEM II medium containing rmSCF (100 ng/ml), EPO (2 U/ml), rmIGF-1 (40 ng/ml), and dexamethasone (1 nM). Human CD34+ HSPCs were infected with viral shRNAs, and 24 hours after viral infection, cells were selected by growing in culture medium containing puromycin (1 mg/ml) for 2 days (19). Murine fetal liver BFU-Es and human CD34+ HSPCs from healthy donors or patients with MDS were cultured and collected on poly-L-lysine–coated slides using Cytospin. Slides were incubated with May-Grünwald solution for 5 min, followed by phosphate-buffered saline wash. Slides were then incubated with Giemsa stain solution for 20 min. Slides were washed with water and air-dried, followed by taking images.
Animals
Animal experiments were carried out in the Cold Spring Harbor Laboratory (CSHL) Animal Shared Resource in accordance with Institutional Animal Care and Use Committee–approved procedures. Eight- to 10-week-old female C57bl/6 mice were obtained from the Jackson Laboratory. C57bl/6 female pregnant mice were used to isolate BFU-Es and CFU-Es. Eighteen- to 21-month-old male and female C57bl/6 mice were used as an aging mouse model. Eight-week-old female B6-Ly5.1/Cr mice were obtained from Charles River Laboratory as recipients for transplantation experiments.
Chrm4−/− mouse model
Bone marrow cells were harvested from Chrm4 knockout mice [Chrm4 tm1jwe (31)] and wild-type C57bl/6 mice and transplanted into lethally irradiated 8-week-old female B6-Ly5.1/Cr recipient mice via tail vein injection. Engraftment of donor cells was confirmed using flow cytometry. Mice were bled retro-orbitally daily. Peripheral blood samples were collected before and after bleeding daily for complete blood count (CBC). Spleen and bone marrow samples were collected for cell counting, colony formation assay, and flow cytometry analysis. Plasma EPO concentrations were measured using mouse EPO enzyme-linked immunosorbent assay (ELISA) kit (ab119593, Abcam). Genotyping polymerase chain reaction (PCR) primer sequences are adapted from previous report (31): primer M4A, GGAGAAGAAGGCCAAGACTCTGG; primer M4B, GGCAGTCACACATTCACTGCCTG; primer NEO-1, CAGCTCATTCCTCCCACTCATGAT.
Mx1-Cre Srsf2P95H/WT mice and aging mouse model
A total of 1 × 106 bone marrow cells were harvested from freshly dissected femora and tibiae of Mx1-Cre Srsf2WT/WT and Mx1-Cre Srsf2P95H/WT mice (32) and transplanted into lethally irradiated 8-week-old female B6-Ly5.1/Cr recipient mice via tail vein injection. pIpC was injected to induce Mx1-Cre expression. After confirming MDS disease phenotype, mice were treated with either compound or control DMSO every day. Peripheral blood samples were collected retro-orbitally for CBC using Hemavet 950. Spleens were dissected for BFU-E colony formation, and plasma EPO concentrations were measured using mouse EPO ELISA kit (ab119593, Abcam). Creatinine, aspartate transaminase, alanine transaminase, alkaline phosphatase, and total bilirubin were measured using Beckman AU680 Chemistry Analyzer.
For the aging mouse model and young mouse controls, CBC was performed on 6- to 8-week-old and 18- to 21-month-old male and female C57bl/6 mice. Eighteen- to 21-month-old mice were then intraperitoneally injected with either compound or DMSO control every day. Blood samples were collected for CBC. Spleens were dissected for BFU-E colony formation assay, and plasma EPO concentrations were measured using mouse EPO ELISA kit (ab119593, Abcam).
PHZ-induced hemolytic anemia mouse model
Eight- to 10-week-old female C57bl/6 mice, randomly distributed into compound and control treatment groups, were intraperitoneally injected with either control DMSO or compound at different doses every day. Mice were intraperitoneally injected with PHZ (60 mg/kg) to induce hemolysis. The percentages of mice dying after compound treatment at each dose were counted at 7 days after the start of treatment. Peripheral blood samples were collected retro-orbitally using heparinized capillary tubes and stored in blood collection tubes with K2-EDTA. CBC was performed using Hemavet 950. Mouse spleens were dissected and homogenized for single-cell suspension. Red blood cells were lysed by incubating the cell suspension with ammonium chloride solution. Spleen cells were then placed in methylcellulose medium MethoCult SF M3436, and BFU-E colonies were counted.
RNA sequencing
For RNA isolation, BFU-Es were cultured in StemSpan SFEM II containing SCF, EPO, IGF-1, and dexamethasone at 37°C with 100 μM oxyphenonium bromide, 3 nM PD102807, 100 nM dexamethasone, or DMSO. Cells were collected, and RNAs were isolated using RNeasy mini kit (QIAGEN). Sequencing libraries were generated using RNA sequencing (RNA-seq) sample preparation kit v2 (Illumina) and sequenced by HiSeq 2500 platform (Illumina). STAR mapper was used to map back sequence reads to the mouse genome (GRCm38, mm10). Reads were demultiplexed in line with their adapters and used for different postmapping analyses.
Chromatin immunoprecipitation sequencing
ChIP-seq was performed using ChIP-IT high-sensitivity kit. A total of 5 × 106 BFU-Es were isolated from E14.5 fetal livers of mice. BFU-Es were cultured in StemSpan SFEM II medium containing SCF, EPO, and IGF-1 at 37°C. Cells were treated with DMSO or 100 μM oxyphenonium bromide. After treatment, cells were cross-linked with 1% formaldehyde solution at room temperature with gentle shaking. Cross-linking was quenched using freshly prepared 2.5 M glycine solution. Cells were lysed using nuclei lysis buffer and sonicated for a total of 40 min using Bioruptor 2000 (Diagenode). During the whole 40-min procedure, samples were sonicated in a 30-s ON (sonication ON) and 30-s OFF (sonication OFF) manner. After each 10 min, samples were vortexed. Aliquots of samples (30 μl) were taken and reverse cross-linked using Proteinase K and ribonuclease A and then run on 1.5% agarose gel to check sonicated fragment size. Sonicated DNA samples were then placed overnight on a tube rotator at 4°C with either IgG control (10 μg) or CREB antibody (10 μg). DNA solutions were incubated with agarose G beads for 3 hours at 4°C, and then, elution and purification of immunoprecipitated DNA were performed using DNA elution and purification columns (Active Motif). To verify the specificity of the antibodies and the whole procedure, a reaction using human embryonic kidney (HEK) 293 cells was performed in parallel. After immunoprecipitation, ChIP-enriched DNAs of HEK293 cells were used to perform ChIP–quantitative PCR using c-Fos CRE ChIP primers (Active Motif), with forward primer GGCCCACGAGACCTCTGAGACA and reverse primer GCCTTGGCGCGTGTCCTAATCT. HEK293 cell line was obtained from CSHL cell bank, which authenticates and tests all the commonly used cell lines for mycoplasma using MycoAlert PLUS mycoplasma detection kit by Lonza.
Sequencing libraries were generated using TruSeq ChIP-seq sample preparation kit (Illumina). Briefly, after repairing the ends, adapters were ligated, and DNA samples with adapters were purified by collecting DNA fractions between 250 to 300 base pairs using SizeSelect 2% agarose gel electrophoresis system (Invitrogen). Purified DNA libraries were amplified using PCR amplification kit and purified using AMPure XP magnetic beads (Beckman Coulter). The size of the sequencing library was verified using high-sensitivity DNA chip in 2100 Bioanalyzer (Agilent). Sequence reads were aligned to mouse genome (GRCm38, mm10) using Bowtie 2 and demultiplexed according to their adapter sequences. MACS platform was used for peak calling. Binding and expression target analysis (BETA) was performed as previously described (46). CREB ChIP-seq peak sequences near Gata2 and Zfp36l2 gene loci were uploaded to TFBIND for CREB binding motif analysis (42), and motifs were identified as previously described (42).
Reverse transcription PCR
For reverse transcription PCR, reverse transcription was performed using SuperScript reverse transcriptase III (Invitrogen). PCR was performed using SYBR Green PCR master mix (Life Technologies) and 7900HT Real-Time PCR system (Applied Biosystems). Primer sequences are the following: c-Fos, GGCCCACGAGACCTCTGAGACA (forward primer) and GCCTTGGCGCGTGTCCTAATCT (reverse primer). Mouse: Chrm4, ATGGCGAACTTCACACCTGTC (forward primer) and CTGTCGCAATGAACACCATCT (reverse primer). Human: CHRM4, CAGCTCGGGCAATCAGTCC (forward primer) and GCCTATGATGAGATCAGCACAC (reverse primer).
Western blot
Proteins were extracted using RIPA buffer system. Protein samples were run on NuPage 4 to 12% bis-tris gel (Invitrogen), transferred to nitrocellulose membranes (Bio-Rad Laboratories), and blocked with 3% bovine serum albumin (BSA) in tris buffer saline–Tween 20 (TBST). Membranes were incubated with anti-CHRM4 rabbit polyclonal antibody (1:2000; TA321192, OriGene), anti-GAPDH mouse monoclonal antibody (1:10000), anti-ZFP36L2 antibody (1:1000; ab70775, Abcam), anti-CREB antibody (1:8000; 17–600, EMD Millipore), anti-GATA2 antibody (1:1000; ab22849, Abcam), anti-KIT antibody (1:1000; ab46758, Abcam), or anti-DDK antibody (1:2000; TA50011, OriGene) in 3% BSA overnight at 4°C. Membranes were washed six times with TBST for 5 min. Secondary antibodies against rabbit IgG (1:10000) and mouse IgG (1:10000) were incubated with membranes for 2 hours at room temperature in 3% BSA solution. Membranes were washed again six times with TBST for 5 min. Western Lightning Plus Enhanced Chemiluminescence substrate (PerkinElmer) was added to membranes for 45 s, and autoradiographs were developed. C-terminal DDK-tagged CHRM4 cell lysates were from OriGene (LY400245). HEK293T cells were transfected with plasmid encoding DDK-tagged CHRM4 or control vector, followed by 48 hours of culture, and cells were lysed in modified RIPA buffer.
cAMP assay
BFU-Es were cultured in the presence of DMSO, PD102807 (3 nM), and forskolin (10 μM) for 45 min. cAMP assay was performed using cAMP direct immunoassay kit (Abcam). BFU-Es were collected in 0.1 M HCl and incubated at room temperature for 20 min. Supernatant was collected by centrifugation for 10 min at 15,000g to remove any insoluble material. Fifty microliters of the neutralizing buffer was added to 100 μl of each sample, followed by mixing. Five microliters of the acetylating reagent mix was added to each sample, and sample tubes were vortexed and incubated for 10 min. Fifty microliters of each sample was added to the protein G–coated 96-well plate. Ten microliters of the reconstituted cAMP antibody was added to each well, followed by 1-hour incubation. Ten microliters of cAMP– horseradish peroxidase (HRP) was added to each well, followed by 1-hour incubation. After washing, 100 ml of HRP developer was added to each well, followed by 1-hour incubation. Reaction was stopped by 100 μl of 1 M HCl, and optical density at 450 nm was determined using BioTek Synergy H4 hybrid multi-mode microplate reader.
Immunofluorescence
Freshly isolated BFU-Es were treated with DMSO, 100 μM oxyphenonium bromide, or 100 μM forskolin. A total of 104 cells were collected on poly-L-lysine coated slides using Cytospin. Cells were fixed with 2% paraformaldehyde for 15 min, followed by permeabilization using 0.1% Triton X-100 for 15 min and blocking with 2% BSA solution for 30 min. Cells were incubated with phospho-CREB antibody conjugated with Alexa Fluor 405 (1:100) in 2% BSA for 1 hour. Cells were then incubated with 4′,6-diamidino-2-phenylindole (DAPI) for 5 min and mounted with mounting solution. Alex Fluor 405 maximal intensities within the DAPI-positive region were quantified using ZEISS Observer and ZEISS Efficient Navigation software.
Receptor binding assays
Receptor binding assays were performed at Eurofins Pharma Discovery Services. PD102807 at concentrations of 1 × 10−9, 1 × 10−8, 1 × 10−7, 1 × 10−6, and 1 × 10−5 M was tested in standard binding assays with radioligands and cell membranes or tissue materials with receptors. Compound binding was calculated as a percent inhibition of the binding of a radioactively labeled ligand specific for each target. Two independent binding experiments were performed for each condition. The median inhibitory concentration (IC50) values and inhibition constant (Ki) values were determined using software developed at Eurofins Pharma Discovery Services (Hill software).
For CHRM1, binding assay was performed using [3H]pirenzepine (2 nM) as a radioligand and membranes of Chinese hamster ovary (CHO) cells with human recombinant CHRM1 receptor. The incubation time was 60 min at room temperature. For CHRM2, binding assay was performed using [3H]AF-DX 384 (2 nM) as a radioligand and membranes of CHO cells with human recombinant CHRM2 receptor. Incubation time was 60 min at room temperature. For CHRM3, binding assay was performed using [3H]4-DAMP (0.2 nM) as a radioligand and membranes of CHO cells with human recombinant CHRM3 receptor. Incubation time was 60 min at room temperature. For CHRM4, the binding assay was performed using [3H]4-DAMP (0.2 nM) as a radioligand and membranes of CHO cells with human recombinant CHRM4 receptor. Incubation time was 60 min at room temperature. For CHRM5, binding assay was performed using [3H]4-DAMP (0.3 nM) as a radioligand and membranes of CHO cells with human recombinant CHRM5 receptor. Incubation time was 60 min at room temperature. For histamine H1 receptor, binding assay was performed using [3H]pyrilamine (1 nM) as a radioligand and membranes of HEK293 cells with human recombinant histamine H1 receptor. Incubation time was 60 min at room temperature. For N-methyl-D-aspartate receptor, binding assay was performed using [3H]CGP 39653 (5 nM) as a radioligand and rat cerebral cortex. Incubation time was 60 min at 4°C.
Plasmids
The following oligos were annealed and cloned into MSCV-IRES-GFP vector digested with BbsI: Chrm4 shRNA1, aaaaTCTGATGAAGCCGACATTAAgtcgacTTAATGCTCGGCTTCATCAGA; Chrm4 shRNA2, aaaaCCATCTTGTTCTGGCAGTTTGgtcgacCAAACTGCCAGAACAAGATGG; Creb shRNA1, aaaaCAATACAGCTGGCTAACAATGgtcgacCATTGTTAGCCAGCTGTATTG; Creb shRNA2, aaaaACTGATGGACAGCAGATTCTAgtcgacTAGAATCTGCTGTCCATCAGT; Gata2 shRNA1, aaaaGACGACAACCACCACCTTATGgtcgacCATAAGGTGGTGGTTGTCGTC; Gata2 shRNA2, aaaaGGGCACCTGTTGTGCAAATTGgtcgacCAATTTGCACAACAGGTGCCC; Zfp36l2 shRNA1, aaaaCCAAACACTTAGGTCTCAGATgtcgacATCTGAGACCTAAGTGTTTGG; Zfp36l2 shRNA2, aaaaGCACCACAACTCAATATGAAAgtcgacTTTCATATTGAGTTGTGGTGC; Kit shRNA1, aaaaACTTCGCCTGACCAGATTAAAgtcgacTTTAATCTGGTCAGGCGAAGT; Kit shRNA2, aaaaGACGTACGACAGGCTCATAAAgtcgacTTTATGAGCCTGTCGTACGTC. Human CHRM4 shRNAs including CHRM4 shRNA1 (NM_000741.1–1182s1c1) and CHRM4 shRNA2 (NM_000741.1270s1c1), human CHRM4 cDNA (NM_000741.2), human ZFP36L2 shRNAs including ZFP36L2 shRNA1 (NM_006887.3–2291s1c1) and ZFP36L2 shRNA2 (NM_006887.4–736s21c1), human CREB shRNAs including CREB shRNA1 (NM_004379.2–284s21c1) and CREB shRNA2 (NM_004379.2–644s21c1), human GATA2 shRNAs including GATA2 shRNA1 (NM_032638.4–1827s21c1) and GATA2 shRNA2 (NM_032638.4–1393s21c1), and human KIT shRNAs including KIT shRNA1 (NM_000222.2–2942s21c1) and KIT shRNA2 (NM_000222.2–3791s21c1) were from Sigma-Aldrich.
Statistical methods
Comparisons of amounts of a single factor, such as relative expression between cell types, were performed using pairwise one-tailed t tests. Experiments comparing two factors, such as time (cell culture days) and treatment, were analyzed using two-way analysis of variance (ANOVA). Survival outcomes were estimated by the Kaplan-Meier method and compared using the log-rank test. Comparisons of two distributions were performed using a Kolmogorov-Smirnov test. We have shown all data points where possible using dot plots and annotated these plots by showing means and SD (or confidence interval) as indicated.
Supplementary Material
Three hundred fifty-eight druggable GPCRs and their expression at three erythroid differentiation stages.
A list of BFU-E signature genes most markedly down-regulated during erythroid differentiation.
The original data of results shown as means and error bars.
Fig. S1. Gene expression analysis identifies six GPCRs with contrasting expression patterns between BFU-E differentiation and self-renewal.
Fig. S2. Muscarinic acetylcholine receptor antagonists increase erythropoiesis and BFU-E expansion in mouse BFU-E culture system.
Fig. S3. Muscarinic acetylcholine receptor antagonists induce erythroid expansion in erythroid differentiation system cultured human CD34+ cells.
Fig. S4. CHRM4 regulates erythropoiesis and BFU-E expansion in mouse BFU-E culture system.
Fig. S5. CHRM4 regulates erythroid expansion in erythroid differentiation system cultured human CD34+ cells.
Fig. S6. Maximal tolerance and pharmacodynamics studies identify maximal tolerance and effective doses of muscarinic acetylcholine receptor antagonists.
Fig. S7. Muscarinic acetylcholine receptor antagonists do not influence white blood cell and platelet production in the Mx1-Cre Srsf2P95H/WT MDS mouse model.
Fig. S8. Muscarinic acetylcholine receptor antagonists exhibit well-tolerated safety profile in the Mx1-Cre Srsf2P95H/WT MDS mouse model.
Fig. S9. Muscarinic acetylcholine receptor antagonist PD102807 promotes expansion of human CD34+ cells isolated from patients with MDS in erythroid differentiation culture system.
Fig. S10. Muscarinic acetylcholine receptor antagonists exhibit no influence on EPO concentration in an aging mouse model.
Fig. S11. Muscarinic acetylcholine receptor antagonists correct anemia in a mouse model of acute hemolysis.
Fig. S12. CREB is required for muscarinic acetylcholine receptor antagonist-induced expansion in erythroid-cultured human CD34+ cells.
Fig. S13. Muscarinic acetylcholine receptor antagonist increases cAMP and phosphorylation of CREB in BFU-E.
Fig. S14. ChIP-PCR validates CREB antibody.
Fig. S15. CREB-binding motifs are identified in BFU-E.
Fig. S16. Genomic distributions of CREB binding sites are identified in BFU-E.
Fig. S17. CREB target genes up-regulated and down-regulated by oxyphenonium bromide exhibit no binding preference.
Fig. S18. Numbers and expression changes of genes up-regulated or down-regulated by oxyphenonium bromide are identified.
Fig. S19. CREB binds to genomic loci near genes important for the maintenance of BFU-E progenitor status.
Fig. S20. ZFP36L2 and KIT function downstream of CHRM4 to regulate mouse BFU-E expansion.
Fig. S21. GATA2, ZFP36L2, and KIT function downstream of CHRM4 to regulate erythroid expansion in erythroid-cultured human CD34+ cells.
Fig. S22. Unprocessed Western blots are shown.
Fig. S23. Flow cytometry control gating plots are shown.
Table S1. IC50 and Ki values of PD102807 on indicated receptors.
Table S2. Clinical and molecular characteristics of the eight patients with MDS.
Acknowledgments:
We thank Y. Hao, M. Hammell, and Y. Jin of the bioinformatics shared resource for helping with analysis of sequencing results; B. Li for helping with cAMP assay; S. Lee for helping with Cre excision assay; P. Moody and S. D’Italia of the flow cytometry core facility for helping with cell sorting; and L. Bianco, J. Coblentz, M. Cahn, and G. Munoz of the laboratory animal resources for helping with animal studies. We thank B. Stillman and members of the Stillman Laboratory for discussions and suggestions.
Funding:
This work is supported by CSHL President’s Council, NCI–Designated Cancer Center Shared Resource grant (CA045508), NIH/NHLBI grant (U01 HL127522), CSHL–Northwell Cancer Translational Research Award, and Edward P. Evans Foundation EvansMDS Young Investigator Award. D.I. is supported by the Leukemia and Lymphoma Society. J.T. is supported by an NIH/KCI (1K08CA230319-01) award. O.A.-W. is supported by grants from the Edward P. Evans Foundation, the Henry and Marilyn Taub Foundation, the Department of Defense Bone Marrow Failure Research Program (W81XWH-12-1-0041), NIH/NHLBI (R01 HL128239), the Leukemia and Lymphoma Society, and the Pershing Square Sohn Cancer Research Alliance.
Competing interests: L.Z. holds a patent titled “Muscarinic acetylcholine receptor subtype 4 antagonists in the treatment of anemia.” O.A.-W. has served as a consultant for H3B Biomedicine, Foundation Medicine Inc., Merck, and Janssen and has received personal speaking fees from Daiichi Sankyo.
Footnotes
All other authors declare that they have no competing interests.
Data and materials availability: All deep sequencing data are available from the Gene Expression Omnibus database (GSE130107). All other data associated with this study are present in the paper or the Supplementary Materials.
REFERENCES AND NOTES
- 1.North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, Lord AM, Weber GJ, Bowman TV, Jang I-H, Grosser T, FitzGerald GA, Daley GQ, Orkin SH, Zon LI, Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 447, 1007–1011 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He S, Nakada D, Morrison SJ, Mechanisms of stem cell self-renewal. Annu. Rev. Cell Dev. Biol 25, 377–406 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Simons BD, Clevers H, Strategies for homeostatic stem cell self-renewal in adult tissues. Cell 145, 851–862 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Rossi DJ, Jamieson CHM, Weissman IL, Stems cells and the pathways to aging and cancer. Cell 132, 681–696 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Zhang L, Flygare J, Wong P, Lim B, Lodish HF, miR-191 regulates mouse erythroblast enucleation by down-regulating Riok3 and Mxi1. Genes Dev. 25, 119–124 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palis J, Primitive and definitive erythropoiesis in mammals. Front. Physiol. 5, 3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dolznig H, Kolbus A, Leberbauer C, Schmidt U, Deiner E-M, Müllner EW, Beug H, Expansion and differentiation of immature mouse and human hematopoietic progenitors. Methods Mol. Med 105, 323–344 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Harandi OF, Hedge S, Wu D-C, McKeone D, Paulson RF, Murine erythroid short-term radioprotection requires a BMP4-dependent, self-renewing population of stress erythroid progenitors. J. Clin. Invest 120, 4507–4519 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paulson RF, Targeting a new regulator of erythropoiesis to alleviate anemia. Nat. Med 20, 334–335 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Dussiot M, Maciel TT, Fricot A, Chartier C, Negre O, Veiga J, Grapton D, Paubelle E, Payen E, Beuzard Y, Leboulch P, Ribeil J-A, Arlet J-B, Coté F, Courtois G, Ginzburg YZ, Daniel TO, Chopra R, Sung V, Hermine O, Moura IC, An activin receptor IIA ligand trap corrects ineffective erythropoiesis in b-thalassemia. Nat. Med 20, 398–407 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suragani RNVS, Cadena SM, Cawley SM, Sako D, Mitchell D, Li R, Davies MV, Alexander MJ, Devine M, Loveday KS, Underwood KW, Grinberg AV, Quisel JD, Chopra R, Pearsall RS, Seehra J, Kumar R, Transforming growth factor-b superfamily ligand trap ACE-536 corrects anemia by promoting late-stage erythropoiesis. Nat. Med 20, 408–414 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Sankaran VG, Weiss MJ, Anemia: Progress in molecular mechanisms and therapies. Nat. Med 21, 221–230 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hattangadi SM, Wong P, Zhang L, Flygare J, Lodish HF, From stem cell to red cell: Regulation of erythropoiesis at multiple levels by multiple proteins, RNAs, and chromatin modifications. Blood 118, 6258–6268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flygare J, Rayon Estrada V, Shin C, Gupta S, Lodish HF, HIF1a synergizes with glucocorticoids to promote BFU-E progenitor self-renewal. Blood 117, 3435–3444 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Prak L, Rayon-Estrada V, Thiru P, Flygare J, Lim B, Lodish HF, ZFP36L2 is required for self-renewal of early burst-forming unit erythroid progenitors. Nature 499, 92–96 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bauer A, Tronche F, Wessely O, Kellendonk C, Reichardt HM, Steinlein P, Schutz G, Beug H, The glucocorticoid receptor is required for stress erythropoiesis. Genes Dev. 13, 2996–3002 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komrokji RS, Sekeres MA, List AF, Management of lower-risk myelodysplastic syndromes: The art and evidence. Curr. Hematol. Malig. Rep 6, 145–153 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Kotla V, Goel S, Nischal S, Heuck C, Vivek K, Das B, Verma A, Mechanism of action of lenalidomide in hematological malignancies. J. Hematol. Oncol 2, 36 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H-Y, Gao X, Barrasa MI, Li H, Elmes RR, Peters LL, Lodish HF, PPAR-α and glucocorticoid receptor synergize to promote erythroid progenitor self-renewal. Nature 522, 474–477 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wessely O, Deiner EM, Beug H, von Lindern M, The glucocorticoid receptor is a key regulator of the decision between self-renewal and differentiation in erythroid progenitors. EMBO J. 16, 267–280 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Migliaccio AR, Varricchio L, Concise review: Advanced cell culture models for diamond blackfan anemia and other erythroid disorders. Stem Cells 36, 172–179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Tayeb A, Qi A, Nicholas RA, Müller CE, Structural modifications of UMP, UDP, and UTP leading to subtype-selective agonists for P2Y2, P2Y4, and P2Y6 receptors. J. Med. Chem 54, 2878–2890 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Lazarowski ER, Watt WC, Stutts MJ, Boucher RC, Harden TK, Pharmacological selectivity of the cloned human P2U-purinoceptor: Potent activation by diadenosine tetraphosphate. Br. J. Pharmacol 116, 1619–1627 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenei V, Sherwood V, Howlin J, Linnskog R, Safholm A, Axelsson L, Andersson T, A t-butyloxycarbonyl-modified Wnt5a-derived hexapeptide functions as a potent antagonist of Wnt5a-dependent melanoma cell invasion. Proc. Natl. Acad. Sci. U.S.A 106, 19473–19478 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plummer AJ, Barrett WE, Rutledge R, Yonkman FF, Pharmacologic properties of antrenyl (oxyphenonium) (BA5473) diethyl(2-hydroxyethyl)methylammonium bromide a-phenyl-cyclohexaneglycolate, an anticholinergic agent. J. Pharmacol. Exp. Ther 108, 292–304 (1953). [PubMed] [Google Scholar]
- 26.Perry EK, Kilford L, Lees AJ, Burn DJ, Perry RH, Increased Alzheimer pathology in Parkinson’s disease related to antimuscarinic drugs. Ann. Neurol 54, 235–238 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Croy CH, Chan WY, Castetter AM, Watt ML, Quets AT, Felder CC, Characterization of PCS1055, a novel muscarinic M4 receptor antagonist. Eur. J. Pharmacol 782, 70–76 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Böhme TM, Augelli-Szafran CE, Hallak H, Pugsley T, Serpa K, Schwarz RD, Synthesis and pharmacology of benzoxazines as highly selective antagonists at M4 muscarinic receptors. J. Med. Chem 45, 3094–3102 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Kruse AC, Kobilka BK, Gautam D, Sexton PM, Christopoulos A, Wess J, Muscarinic acetylcholine receptors: Novel opportunities for drug development. Nat. Rev. Drug Discov 13, 549–560 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wess J, Eglen RM, Gautam D, Muscarinic acetylcholine receptors: Mutant mice provide new insights for drug development. Nat. Rev. Drug Discov 6, 721–733 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Gomeza J, Zhang L, Kostenis E, Felder C, Bymaster F, Brodkin J, Shannon H, Xia B, C.-x. Deng, J. Wess, Enhancement of D1 dopamine receptor-mediated locomotor stimulation in M4 muscarinic acetylcholine receptor knockout mice. Proc. Natl. Acad. Sci. U.S.A 96, 10483–10488 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim E, Ilagan JO, Liang Y, Daubner GM, Lee SC-W, Ramakrishnan A, Li Y, Chung YR, Micol J-B, Murphy ME, Cho H, Kim M-K, Zebari AS, Aumann S, Park CY, Buonamici S, Smith PG, Deeg HJ, Lobry C, Aifantis I, Modis Y, Allain FH-T, Halene S, Bradley RK, Abdel-Wahab O, SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell 27, 617–630 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bejar R, Stevenson KE, Caughey BA, Abdel-Wahab O, Steensma DP, Galili N, Raza A, Kantarjian H, Levine RL, Neuberg D, Garcia-Manero G, Ebert BL, Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J. Clin. Oncol 30, 3376–3382 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alderton GK, Splicing the MDS genome. Nat. Rev. Cancer 15, 393 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Obeng EA, Ebert BL, Charting the “splice” routes to MDS. Cancer Cell 27, 607–609 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Ogawa T, Kitagawa M, Hirokawa K, Age-related changes of human bone marrow: A histometric estimation of proliferative cells, apoptotic cells, T cells, B cells and macrophages. Mech. Ageing Dev 117, 57–68 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Goodell MA, Rando TA, Stem cells and healthy aging. Science 350, 1199–1204 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Shaywitz AJ, Greenberg ME, CREB: A stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem 68, 821–861 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Kase H, Iwahashi K, Nakanishi S, Matsuda Y, Yamada K, Takahashi M, Murakata C, Sato A, Kaneko M, K-252 compounds, novel and potent inhibitors of protein kinase C and cyclic nucleotide-dependent protein kinases. Biochem. Biophys. Res. Commun 142, 436–440 (1987). [DOI] [PubMed] [Google Scholar]
- 40.Boer A-K, Drayer AL, Vellenga E, cAMP/PKA-mediated regulation of erythropoiesis. Leuk. Lymphoma 44, 1893–1901 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Tsai F-Y, Orkin SH, Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood 89, 3636–3643 (1997). [PubMed] [Google Scholar]
- 42.Tsunoda T, Takagi T, Estimating transcription factor bindability on DNA. Bioinformatics 15, 622–630 (1999). [DOI] [PubMed] [Google Scholar]
- 43.Chou R, Peterson K, Helfand M, Comparative efficacy and safety of skeletal muscle relaxants for spasticity and musculoskeletal conditions: A systematic review. J. Pain Symptom Manage 28, 140–175 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Katayama Y, Battista M, Kao W-M, Hidalgo A, Peired AJ, Thomas SA, Frenette PS, Signals from the sympathetic nervous system regulate hematopoietic stem cell egress from bone marrow. Cell 124, 407–421 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Tracey KJ, The inflammatory reflex. Nature 420, 853–859 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Wang S, Sun H, Ma J, Zang C, Wang C, Wang J, Tang Q, Meyer CA, Zhang Y, Liu XS, Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat. Protoc 8, 2502–2515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P, Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics 2008, 420747 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M, Vardiman JW, The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 127, 2391–2405 (2016). [DOI] [PubMed] [Google Scholar]
- 49.An X, Schulz VP, Li J, Wu K, Liu J, Xue F, Hu J, Mohandas N, Gallagher PG, Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood 123, 3466–3477 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Three hundred fifty-eight druggable GPCRs and their expression at three erythroid differentiation stages.
A list of BFU-E signature genes most markedly down-regulated during erythroid differentiation.
The original data of results shown as means and error bars.
Fig. S1. Gene expression analysis identifies six GPCRs with contrasting expression patterns between BFU-E differentiation and self-renewal.
Fig. S2. Muscarinic acetylcholine receptor antagonists increase erythropoiesis and BFU-E expansion in mouse BFU-E culture system.
Fig. S3. Muscarinic acetylcholine receptor antagonists induce erythroid expansion in erythroid differentiation system cultured human CD34+ cells.
Fig. S4. CHRM4 regulates erythropoiesis and BFU-E expansion in mouse BFU-E culture system.
Fig. S5. CHRM4 regulates erythroid expansion in erythroid differentiation system cultured human CD34+ cells.
Fig. S6. Maximal tolerance and pharmacodynamics studies identify maximal tolerance and effective doses of muscarinic acetylcholine receptor antagonists.
Fig. S7. Muscarinic acetylcholine receptor antagonists do not influence white blood cell and platelet production in the Mx1-Cre Srsf2P95H/WT MDS mouse model.
Fig. S8. Muscarinic acetylcholine receptor antagonists exhibit well-tolerated safety profile in the Mx1-Cre Srsf2P95H/WT MDS mouse model.
Fig. S9. Muscarinic acetylcholine receptor antagonist PD102807 promotes expansion of human CD34+ cells isolated from patients with MDS in erythroid differentiation culture system.
Fig. S10. Muscarinic acetylcholine receptor antagonists exhibit no influence on EPO concentration in an aging mouse model.
Fig. S11. Muscarinic acetylcholine receptor antagonists correct anemia in a mouse model of acute hemolysis.
Fig. S12. CREB is required for muscarinic acetylcholine receptor antagonist-induced expansion in erythroid-cultured human CD34+ cells.
Fig. S13. Muscarinic acetylcholine receptor antagonist increases cAMP and phosphorylation of CREB in BFU-E.
Fig. S14. ChIP-PCR validates CREB antibody.
Fig. S15. CREB-binding motifs are identified in BFU-E.
Fig. S16. Genomic distributions of CREB binding sites are identified in BFU-E.
Fig. S17. CREB target genes up-regulated and down-regulated by oxyphenonium bromide exhibit no binding preference.
Fig. S18. Numbers and expression changes of genes up-regulated or down-regulated by oxyphenonium bromide are identified.
Fig. S19. CREB binds to genomic loci near genes important for the maintenance of BFU-E progenitor status.
Fig. S20. ZFP36L2 and KIT function downstream of CHRM4 to regulate mouse BFU-E expansion.
Fig. S21. GATA2, ZFP36L2, and KIT function downstream of CHRM4 to regulate erythroid expansion in erythroid-cultured human CD34+ cells.
Fig. S22. Unprocessed Western blots are shown.
Fig. S23. Flow cytometry control gating plots are shown.
Table S1. IC50 and Ki values of PD102807 on indicated receptors.
Table S2. Clinical and molecular characteristics of the eight patients with MDS.