Abstract
The pandemic coronavirus SARS-CoV-2 threatens public health worldwide. The viral spike protein mediates SARS-CoV-2 entry into host cells and harbors a S1/S2 cleavage site containing multiple arginine residues (multibasic) not found in closely related animal coronaviruses. However, the role of this multibasic cleavage site in SARS-CoV-2 infection is unknown. Here, we report that the cellular protease furin cleaves the spike protein at the S1/S2 site and that cleavage is essential for S-protein-mediated cell-cell fusion and entry into human lung cells. Moreover, optimizing the S1/S2 site increased cell-cell, but not virus-cell, fusion, suggesting that the corresponding viral variants might exhibit increased cell-cell spread and potentially altered virulence. Our results suggest that acquisition of a S1/S2 multibasic cleavage site was essential for SARS-CoV-2 infection of humans and identify furin as a potential target for therapeutic intervention.
Keywords: SARS-CoV-2, COVID-19, spike, cleavage, S1/S2, furin, TMPRSS2, entry, membrane fusion
Graphical Abstract
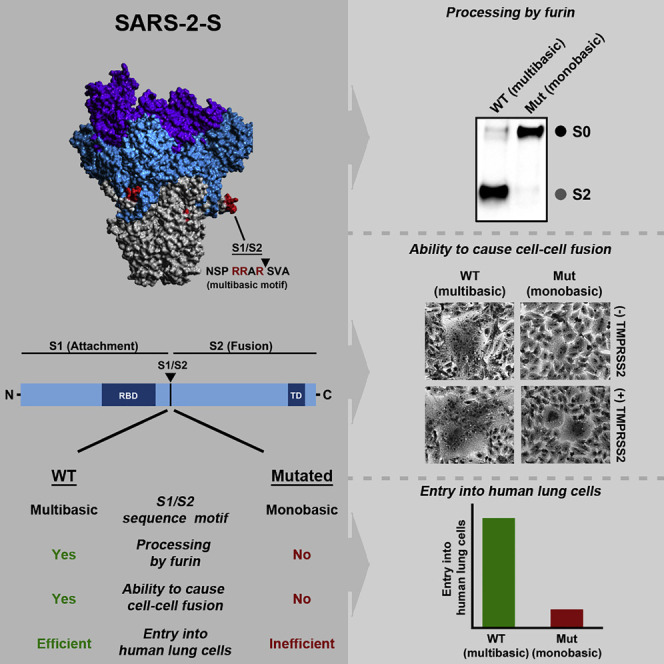
Coronavirus spike proteins are activated by host cell proteases. Hoffmann and colleagues show that the pandemic SARS-CoV-2 harbors a highly cleavable S1/S2 cleavage site not found in closely related coronaviruses. Cleavage at this site is mediated by furin and is required for viral entry into human lung cells.
Introduction
It is believed that the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, previously termed nCoV-2019) was introduced into the human population from a poorly characterized animal reservoir in late 2019 (Ge et al., 2013, Wang et al., 2020, Zhou et al., 2020b, Zhu et al., 2020). The epicenter of the subsequent SARS-CoV-2 spread was Wuhan, Hubei province, China, with more than 65,000 cases occurring in this area (WHO, 2020a). However, infections have now been detected in more than 110 countries and massive outbreaks are currently ongoing in the United States, Italy, and Spain (WHO, 2020a, WHO, 2020b). Understanding which features of SARS-CoV-2 are essential for infection of human cells should provide insights into viral transmissibility and pathogenesis and might reveal targets for intervention.
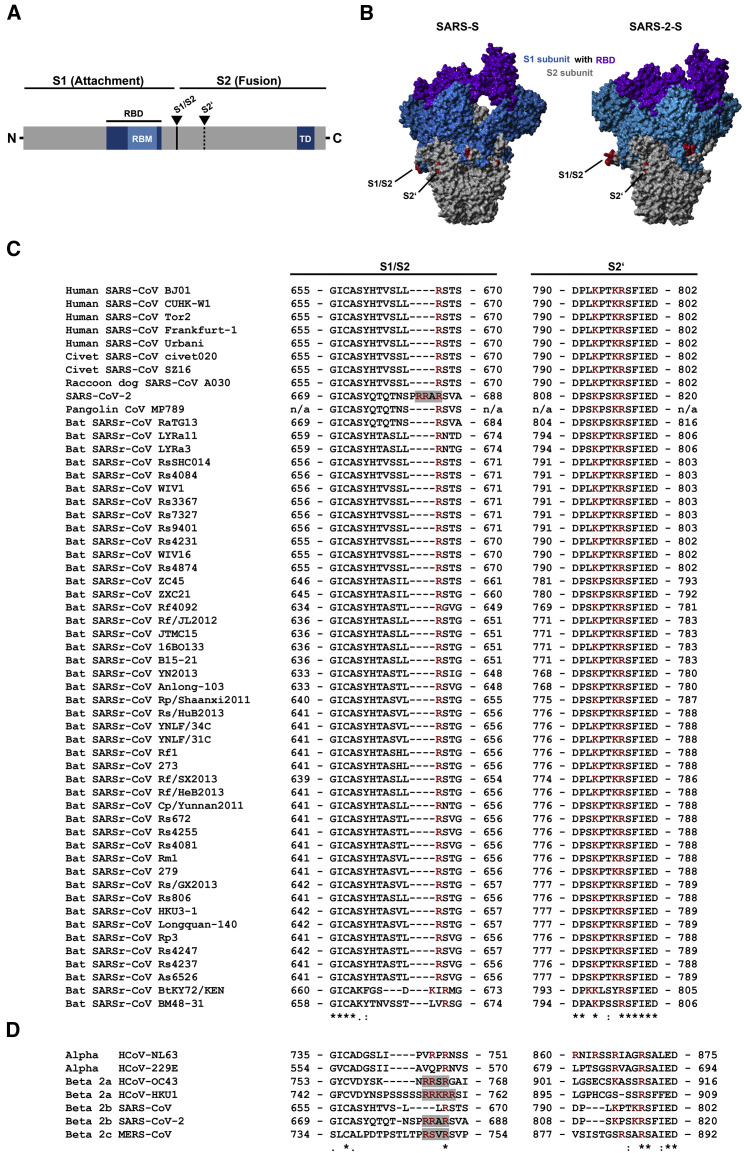
The spike protein of coronaviruses is incorporated into the viral envelope and facilitates viral entry into target cells. For this, the surface unit S1 binds to a cellular receptor while the transmembrane unit S2 facilitates fusion of the viral membrane with a cellular membrane (Hoffmann et al., 2018, Hulswit et al., 2016, Millet and Whittaker, 2018). Membrane fusion depends on S protein cleavage by host cell proteases at the S1/S2 and the S2′ site (Figure 1 A), which results in S protein activation (Hoffmann et al., 2018, Hulswit et al., 2016, Millet and Whittaker, 2018). Cleavage of the S protein can occur in the constitutive secretory pathway of infected cells or during viral entry into target cells and is essential for viral infectivity. Therefore, the responsible enzymes constitute potential targets for antiviral intervention.
Figure 1.
The Multibasic Motif at the S1/S2 Cleavage Site of SARS-2-S Is Unique among Related Group 2b Betacoronaviruses
(A) Schematic illustration of a coronavirus spike glycoprotein in which functional domains and cleavage sites are highlighted (RBD, receptor-binding domain; RBM, receptor-binding motif; TD, transmembrane domain).
(B) Protein models for SARS-S and SARS-2-S based on the PDB: 5X5B structure (Yuan et al., 2017) as a template. Colored in red are the S1/S2 and S2′ cleavage sites. Further, the S1 subunit (blue), including the RBD (purple), and the S2 subunit (gray) are depicted.
(C and D) Amino acid sequence alignment of residues around the S1/S2 and S2′ cleavage sites of group 2b betacoronaviruses found in humans, civet cats, raccoon dog, pangolin, and bats (C) or coronaviruses that are able to infect humans (D). Basic amino acid residues are highlighted in red, while gray boxes mark the presence of multibasic motifs. Numbers refer to amino acid residues (n/a, no information available). The symbol “∗” refers to amino acid residues that are conserved among all tested sequences, while the symbols “:” and “.” indicate positions with heterogeneous amino acid residues that share highly similar or similar biochemical properties.
Our previous work revealed that the activity of the cellular serine protease TMPRSS2, which activates several coronaviruses (Bertram et al., 2013, Gierer et al., 2013, Glowacka et al., 2011, Matsuyama et al., 2010, Shirato et al., 2013, Shirato et al., 2016, Shulla et al., 2011), is also required for robust SARS-CoV-2 infection of human lung cells (Hoffmann et al., 2020). However, it is conceivable that the activity of other cellular proteases is also necessary. Thus, the Middle East respiratory syndrome coronavirus spike protein (MERS-S) is activated by a two-step process: MERS-S is first cleaved by furin at the S1/S2 site in infected cells, which is required for subsequent TMPRSS2-mediated cleavage at the S2′ site (Figure 1A) during viral entry into lung cells (Kleine-Weber et al., 2018, Park et al., 2016, Millet and Whittaker, 2014). A cathepsin B/L-dependent auxiliary activation pathway is operative in many TMPRSS2− cell lines but seems not to be available in viral target cells in the lung because TMPRSS2-dependent activation of the S protein is essential for robust MERS-CoV and SARS-CoV spread and pathogenesis in the infected host (Iwata-Yoshikawa et al., 2019, Simmons et al., 2005, Zhou et al., 2015).
The S1/S2 site in SARS-CoV-2 forms an exposed loop (Figure 1B) that harbors multiple arginine residues (multibasic) (Walls et al., 2020, Wrapp et al., 2020) that are not found in SARS-CoV-related coronaviruses (SARSr-CoV) but are present in the human coronaviruses OC43, HKU1, and MERS-CoV (Figure 1C). However, the contribution of this multibasic cleavage site to SARS-CoV-2 infection of human cells is unknown and was in the focus of the present study.
Results
The Multibasic S1/S2 Site in the Spike Protein of SARS-CoV-2 Is Required for Efficient Proteolytic Cleavage of the Spike Protein
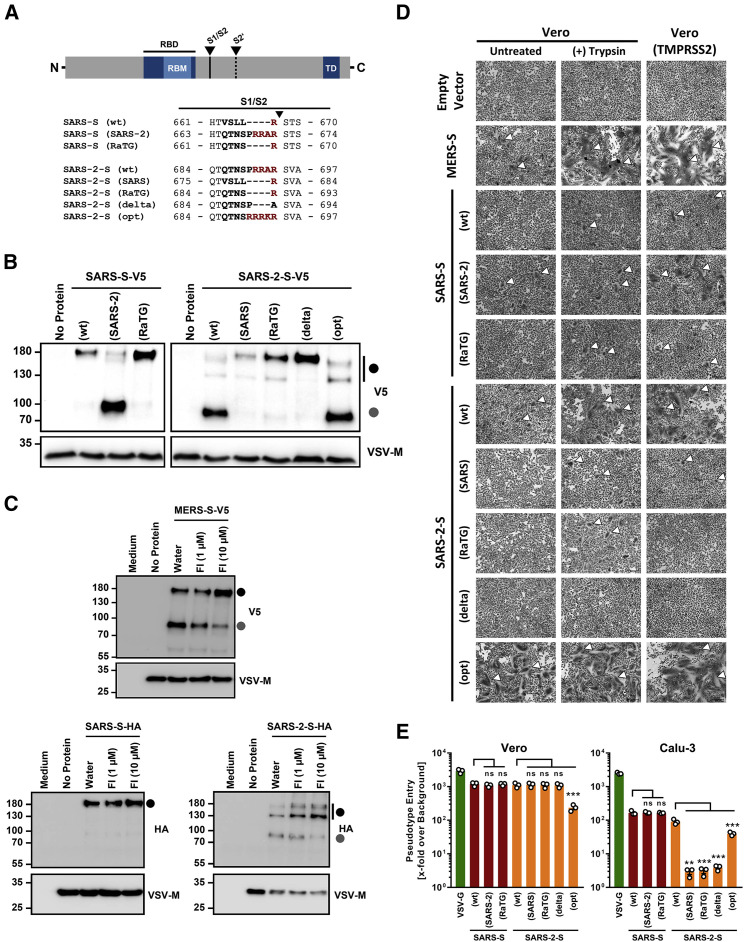
In order to address the role of the multibasic S1/S2 cleavage site in SARS-CoV-2 infection, we generated S protein mutants with altered S1/S2 cleavage sites (Figure 2 A). In particular, we exchanged the multibasic cleavage site against its monobasic counterparts present in SARS-S or RaTG13-S (Figure 2A, RaTG13 is a bat coronavirus closely related to SARS-CoV-2 [Zhou et al., 2020b]). This resulted in mutants SARS-2-S (SARS) and SARS-2-S (RaTG). Moreover, we either deleted all arginines in the S1/S2 site of SARS-2-S or inserted an additional arginine residue (jointly with an alanine to lysine exchange), giving rise to mutants SARS-2-S (delta) and SARS-2-S (opt), respectively. Finally, we introduced the S1/S2 sites of SARS-2-S and RaTG13-S into the background of SARS-S (Figure 2A), which yielded the mutants SARS-S (SARS-2) and SARS-S (RaTG).
Figure 2.
The Multibasic S1/S2 Site of SARS-2-S Is Cleaved by Furin, and Cleavage Is Required for Syncytium Formation and Entry into Human Lung Cells
(A) Overview of the SARS-S and SARS-2-S S1/S2 mutants analyzed.
(B) Analysis of furin-mediated S protein priming. Rhabdoviral particles harboring the indicated S proteins containing a C-terminal V5 tag for detection were lysed and subjected to western blot analysis. Detection of vesicular stomatitis virus matrix protein (VSV-M) served as control.
(C) Rhabdoviral particles bearing MERS-S, SARS-S, or SARS-2-S equipped with a V5 or HA epitope tag at their C terminus (or no glycoprotein at all, control) were produced in the absence or presence of furin inhibitor (FI, decanoyl-RVKR-CMK; 1 μM or 10 μM) and analyzed for S protein processing by western blot analysis. Detection of VSV-M served as control.
(D) Syncytium formation assay: Vero or Vero-TMPRSS2 cells were transfected to express the indicated S proteins (or no S protein, empty vector, control). At 24 h post transfection, cells were incubated in the presence or absence of trypsin (1 μg/mL) for an additional 24 h before they were fixed, stained with May-Gruenwald and Giemsa solution, and analyzed by bright field microscopy (scale bars, 200 μm). White arrowheads indicate syncytia. For (B)–(D), representative data from three (B and C) or four (D) independent experiments are shown.
(E) Transduction of Vero (TMPRSS2−) and Calu-3 (TMPRSS2+) cells with rhabdoviral particles bearing the indicated S proteins or vesicular stomatitis virus glycoprotein (VSV-G). At 16 h post transduction, virus-encoded firefly luciferase was quantified in cell lysates. Presented are the mean data from three independent experiments. Transduction efficiency is shown relative to that measured for particles not bearing a viral glycoprotein. Error bars indicate the standard error of the mean. Statistical significance was tested by one-way analysis of variance with Dunnett’s post test (p > 0.05, ns; ∗∗∗p ≤ 0.001).
The effects of the above described S1/S2 mutations on viral entry were examined using vesicular stomatitis virus (VSV) particles bearing S proteins because these particles are safe and adequately reflect coronavirus entry into target cells. Immunoblot of VSV particles bearing S proteins with a C-terminal antigenic tag revealed that all S proteins were readily incorporated into VSV particles. SARS-2-S WT (wild type) was efficiently cleaved at the S1/S2 site (Figure 2B), in keeping with published data (Hoffmann et al., 2020, Walls et al., 2020). Exchange of the S1/S2 site of SARS-2-S against those of SARS-S and RaTG13-S abrogated cleavage, and this effect was also seen when the multibasic motif was deleted (Figure 2B). Moreover, insertion of an additional arginine residue jointly with an alanine-to-lysine exchange at the S1/S2 site did not appreciably increase cleavability. Finally, insertion of the S1/S2 site of SARS-2-S into SARS-S increased S protein cleavability while insertion of the RaTG13 S1/S2 site did not (Figure 2B). These results indicate that the presence of several arginine residues at the S1/S2 site is required for efficient SARS-2-S proteolytic processing in human cells and also confers high cleavability to SARS-S.
Furin Cleaves the SARS-CoV-2 Spike Protein at the S1/S2 Site, and Cleavage Is Required for Efficient Cell-Cell Fusion
We next investigated which protease is required for S protein processing at the S1/S2 site. The S1/S2 motif matches the minimal furin sequence RXXR and is closely related to the furin consensus sequence RX[K/R]R. Therefore, we analyzed whether decanoyl-RVKR-CMK, a furin inhibitor, blocks SARS-2-S processing at the S1/S2 site. Decanoyl-RVKR-CMK inhibited processing of MERS-S, which is known to depend on furin (Gierer et al., 2015, Millet and Whittaker, 2014), in a concentration-dependent manner and had no effect on SARS-S expression (Figure 2C), as expected. Processing of SARS-2-S was also inhibited, indicating that furin cleaves SARS-2-S at the S1/S2 site. In order to determine whether cleavage at the S1/S2 site is required for SARS-2-S-driven cell-cell fusion, we studied S-protein-dependent formation of multinucleated giant cells (syncytia). No syncytia were observed in the absence of S protein expression while MERS-S WT expression resulted in syncytium formation, which was increased upon addition of trypsin or expression of TMPRSS2 (Figure 2D). Expression of SARS-S WT or SARS-S harboring the S1/S2 site of RaTG13-S did not induce syncytium formation in the absence of protease, but modest multikaryon formation was detected in the presence of trypsin or TMPRSS2. In contrast, SARS-S harboring the SARS-2-S S1/S2 site induced syncytia in the absence of protease, and syncytium formation was markedly increased by trypsin and, particularly, TMPRSS2. SARS-2-S expression triggered syncytium formation that was strongly increased by trypsin and TMPRSS2. Syncytium formation was clearly less prominent and required the presence of trypsin or TMPRSS2 when the SARS-2-S S1/S2 site was replaced by that of SARS-S or RaTG13-S. Moreover, deletion of the multibasic motif resulted in a spike protein that was no longer able to induce syncytium formation even in the presence of trypsin or TMPRSS2. Finally, the addition of an arginine residue to the S1/S2 site of SARS-2-S jointly with alanine-to-lysine exchange strongly increased syncytium formation, indicating that viral variants with optimized S1/S2 sites might show augmented cell-cell spread and potentially altered pathogenicity. Thus, the S1/S2 site of SARS-2-S is required for cell-cell fusion, and this process can be augmented by adding basic residues to the S1/S2 site.
Cleavage of the SARS-CoV-2 Spike Protein at the S1/S2 Site Is Required for Viral Entry into Human Lung Cells
We finally examined the importance of the S1/S2 site for S-protein-mediated virus-cell fusion. Blockade of SARS-2-S cleavage at the S1/S2 site (mutants SARS-2-S (SARS), SARS-2-S (RaTG), and SARS-2-S (delta)) abrogated entry into the TMPRSS2+ human lung cell line Calu-3 (Figure 2E), in which the cathepsin B/L-dependent S protein activation pathway is not sufficiently available (Park et al., 2016). In contrast, entry into TMPRSS2− Vero cells, which is known to be cathepsin B/L dependent, was not affected by these mutations (Figure 1E), in keeping with results reported by Walls and colleagues (Walls et al., 2020). Optimization of the S1/S2 site did not increase entry into the cell lines tested; it slightly decreased entry into both Vero and Calu-3 cells, for, at present, unclear reasons. Finally, alterations of the S1/S2 site of SARS-S did not augment entry efficiency. Collectively, these results demonstrate that a multibasic S1/S2 site is essential for SARS-2-S-driven entry into human lung cells while a monobasic site is sufficient for SARS-S.
Discussion
Our results reveal commonalities between the proteolytic activation of SARS-CoV-2 and MERS-CoV. Both viruses depend on furin-mediated pre-cleavage of their S proteins at the S1/S2 site for subsequent S protein activation by TMPRSS2 in lung cells, which fail to express robust levels of cathepsin L (Park et al., 2016). Thus, inhibitors of furin and TMPRSS2 might be considered as a treatment option for COVID-19, and a TMPRSS2 inhibitor that blocks SARS-CoV-2 infection has recently been described (Hoffmann et al., 2020). Regarding furin inhibition, it must be taken into account that furin, unlike TMPRSS2, is required for normal development (Roebroek et al., 1998). Blockade of this enzyme for prolonged time periods might thus be associated with unwanted toxic effects. In contrast, a brief treatment might be well tolerated and still associated with a therapeutic benefit (Sarac et al., 2002, Sarac et al., 2004).
For avian influenza A viruses, a multibasic cleavage site in the viral hemagglutinin protein is a central virulence factor (Luczo et al., 2015). Thus, viruses with a monobasic cleavage site are activated by TMPRSS2 or related proteases with an expression profile confined to the aerodigestive tract. As a consequence, viral replication is limited to these organs and does not result in severe disease. In contrast, viruses with a multibasic cleavage site are activated by ubiquitously expressed proprotein convertases, including furin, and can thus spread systemically and cause massive disease. In the context of coronavirus infection, S protein cleavability has been identified as a determinant of zoonotic potential (Menachery et al., 2020, Yang et al., 2014). The presence of a highly cleavable S1/S2 site in SARS-2-S may therefore not have been unexpected. However, it is noteworthy that all SARS-CoV-2-related coronaviruses of bats and pangolins identified today harbor a monobasic cleavage site (Lam et al., 2020, Li et al., 2020, Zhang et al., 2020). It will thus be interesting to determine how the multibasic motif was acquired by SARS-CoV-2, and a recent study suggested that a recombination event might have been responsible (Zhang et al., 2020, Zhou et al., 2020a).
Limitations of the Study
Our results demonstrate that the multibasic S1/S2 cleavage site is essential for SARS-2-S-driven entry into TMPRSS2+ lung cells. It will be interesting to extend these studies to primary human respiratory epithelial cells and to authentic SARS-CoV-2, which requires a reverse genetics system not available to the present study.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal anti-HA antibody produced in mouse | Sigma-Aldrich | Cat.#: H3663; RRID: AB_262051 |
| Monoclonal anti-β-actin antibody produced in mouse | Sigma-Aldrich | Cat.#: A5441; RRID: AB_476744 |
| Monoclonal anti-VSV-M (23H12) antibody | KeraFast | Cat.#: EB0011; RRID: AB_2734773 |
| Monoclonal anti-mouse, peroxidase-coupled | Dianova | Cat.#: 115-035-003; RRID: AB_10015289 |
| Anti-VSV-G antibody (I1, produced from CRL-2700 mouse hybridoma cells) | ATCC | Cat.# CRL-2700; RRID: CVCL_G654 |
| Bacterial and Virus Strains | ||
| VSV∗ΔG-FLuc | Berger Rentsch and Zimmer, 2011 | N/A |
| One Shot™ OmniMAX™ 2 T1R Chemically Competent E. coli | Thermo Fisher Scientific | Cat.#: C854003 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Lipofectamine LTX with Plus Reagent | Thermo Fisher Scientific | Cat.#: 15338100 |
| Furin inhibitor, decanoyl-RVKR-CMK | Tocris | Cat.#: 3501 |
| May-Grünwald solution | Sigma-Aldrich | Cat.#: 63590 |
| Giemsa solution | Sigma-Aldrich | Cat.#: GS500 |
| Critical Commercial Assays | ||
| Beetle-Juice Kit | PJK | Cat.#: 102511 |
| Experimental Models: Cell Lines | ||
| 293T | DSMZ | Cat.#: ACC-635; RRID: CVCL_0063 |
| Calu-3 | Laboratory of Stephan Ludwig | ATCC Cat# HTB-55; RRID: CVCL_0609 |
| Vero | Laboratory of Andrea Maisner | ATCC Cat# CRL-1586; RRID: CVCL_0574 |
| Vero-TMPRSS2 | Hoffmann et al., 2020 | N/A |
| Oligonucleotides | ||
| SARS-S (BamHI) F CTTGGATCCGCCACCATGTTTATTTTC TTATTATTTC |
Sigma-Aldrich | N/A |
| SARS-SΔ18 (XbaI) R CTTTCTAGACTACTTGCAGCAAGAA CCACAAGAGC |
Sigma-Aldrich | N/A |
| SARS-SΔ18 (-)STOP (XbaI) R CTTTCTAGACTTGCAGCAAG AACCACAAGAGC |
Sigma-Aldrich | N/A |
| SARS-2-S (BamHI) F GAATTCGGATCCGCCACCATGTTCGT GTTTCTGGTGCTGC |
Sigma-Aldrich | N/A |
| SARS-2-SΔ18 (XbaI) R AAGGCCTCTAGACTACTTGCAGCA GCTGCCACAGC |
Sigma-Aldrich | N/A |
| SARS-2-SΔ18 (-)STOP (XbaI) R AAGGCCTCTAGACTTGCA GCAGCTGCCACAGC |
Sigma-Aldrich | N/A |
| SARS-S (SARS) F CAGACAAACAGCCCCAGACGGGCCAG AAGTACTAGCCAAAAATCTATTG |
Sigma-Aldrich | N/A |
| SARS-S (SARS) R TCTGGCCCGTCTGGGGCTGTTTGTCT GTGTATGGTAACTAGCACAAATGC |
Sigma-Aldrich | N/A |
| SARS-S (RaTG) F CAGACAAACAGCAGAAGTACTAGCCA AAAATC |
Sigma-Aldrich | N/A |
| SARS-S (RaTG) R TCTGCTGTTTGTCTGTGTATGGTAACTA GCACAAATGC |
Sigma-Aldrich | N/A |
| SARS-2-S (SARS) F GTTTCTTTATTACGTTCTGTGGCCAGC CAGAGCATC |
Sigma-Aldrich | N/A |
| SARS-2-S (SARS) R ACGTAATAAAGAAACTGTCTGGTAGC TGGCACAGATG |
Sigma-Aldrich | N/A |
| SARS-2-S (RaTG) F CAGACAAACAGCAGATCTGTGGCCAGC CAGAGCATC |
Sigma-Aldrich | N/A |
| SARS-2-S (RaTG) R GCTGGCCACAGATCTGCTGTTTGTCTG TGTCTGGTAGC |
Sigma-Aldrich | N/A |
| SARS-2-S (delta) F CAAACAGCCCCGCATCTGTGGCCAGCC AGAGCATC |
Sigma-Aldrich | N/A |
| SARS-2-S (delta) R GCTGGCCACAGATGCGGGGCTGTTTGTC TGTGTCTGGTAGC |
Sigma-Aldrich | N/A |
| SARS-2-S (opt) F CGAAGACGAAAAAGATCTGTGGCCAGCCA GAGCATC |
Sigma-Aldrich | N/A |
| SARS-2-S (opt) R TCTTTTTCGTCTTCGGCTGTTTGTCTGTGT CTGG |
Sigma-Aldrich | N/A |
| pCG1 Seq F CCTGGGCAACGTGCTGGT | Sigma-Aldrich | N/A |
| pCG1 Seq R GTCAGATGCTCAAGGGGCTTCA | Sigma-Aldrich | N/A |
| SARS-S 387F TGTTATACGAGCATGTAAC | Sigma-Aldrich | N/A |
| SARS-S 790F AAGCCAACTACATTTATGC | Sigma-Aldrich | N/A |
| SARS S 1194F TGATGTAAGACAAATAGCG | Sigma-Aldrich | N/A |
| SARS S 1575F TATTAAGAACCAGTGTGTC | Sigma-Aldrich | N/A |
| SARS S 1987F GTGCTAGTTACCATACAG | Sigma-Aldrich | N/A |
| SARS S 2391F CTAAAGCCAACTAAGAGG | Sigma-Aldrich | N/A |
| SARS S 2787F TCAACTGCATTGGGCAAG | Sigma-Aldrich | N/A |
| SARS-2-S 651F CAAGATCTACAGCAAGCACACC | Sigma-Aldrich | N/A |
| SARS-2-S 1380F GTCGGCGGCAACTACAATTAC | Sigma-Aldrich | N/A |
| SARS-2-S 1992F CTGTCTGATCGGAGCCGAGCAC | Sigma-Aldrich | N/A |
| SARS-2-S 2648F TGAGATGATCGCCCAGTACAC | Sigma-Aldrich | N/A |
| SARS-2-S 3286F GCCATCTGCCACGACGGCAAAG | Sigma-Aldrich | N/A |
| pCG1-V5 F TCCCTAACCCTCTCCTCGGTCTCGATTCTACGTG AAAGCTGATCTTTTTCCCTCTGCC |
Sigma-Aldrich | N/A |
| pCG1-V5 R GACCGAGGAGAGGGTTAGGGATAGGCTTACCG CATGCCTGCAGGTTTAAACAGTCG |
Sigma-Aldrich | N/A |
| pCG1-XhoI R CTCCTCGAGTTCATAAGAGAAGAGGG | Sigma-Aldrich | N/A |
| Recombinant DNA | ||
| Plasmid: pCG1-SARS-S | Hoffmann et al., 2013 | N/A |
| Plasmid: pCG1-SARS-S-HA | Hoffmann et al., 2020 | N/A |
| Plasmid: pCG1-SARS-2-S | Hoffmann et al., 2020 | N/A |
| Plasmid: pCG1-SARS-2-S-HA | Hoffmann et al., 2020 | N/A |
| Plasmid: pCG1-SARS-SΔ18 | Hoffmann et al., 2013 | N/A |
| Plasmid: pCG1-SARS-SΔ18-V5 | This paper | N/A |
| Plasmid: pCG1-SARS-2-SΔ18 | This paper | N/A |
| Plasmid: pCG1-SARS-2-SΔ18-V5 | This paper | N/A |
| Plasmid: pCG1-SARS-SΔ18 (SARS-2) | This paper | N/A |
| Plasmid: pCG1-SARS-SΔ18-V5 (SARS-2) | This paper | N/A |
| Plasmid: pCG1-SARS-SΔ18 (RaTG) | This paper | N/A |
| Plasmid: pCG1-SARS-SΔ18-V5 (RaTG) | This paper | N/A |
| Plasmid: pCG1-SARS-2-SΔ18 (SARS) | This paper | N/A |
| Plasmid: pCG1-SARS-2-SΔ18-V5 (SARS) | This paper | N/A |
| Plasmid: pCG1-SARS-2-SΔ18 (RaTG) | This paper | N/A |
| Plasmid: pCG1-SARS-2-SΔ18-V5 (RaTG) | This paper | N/A |
| Plasmid: pCG1-SARS-2-SΔ18 (delta) | This paper | N/A |
| Plasmid: pCG1-SARS-2-SΔ18-V5 (delta) | This paper | N/A |
| Plasmid: pCG1-SARS-2-SΔ18 (opt) | This paper | N/A |
| Plasmid: pCG1-SARS-2-SΔ18-V5 (opt) | This paper | N/A |
| Plasmid: pCAGGS-MERS-S-V5 | Gierer et al., 2013 | N/A |
| Plasmid: pCAGGS-VSV-G | Brinkmann et al., 2017 | N/A |
| Plasmid: pCAGGS-DsRed | Hoffmann et al., 2013 | N/A |
| Plasmid: pCG1 | Laboratory of Roberto Cattaneo | N/A |
| Plasmid: pCG1-V5 | This paper | N/A |
| Software and Algorithms | ||
| Hidex Sense Microplate Reader Software | Hidex Deutschland Vertrieb GmbH | https://www.hidex.de/ |
| ChemoStar Imager Software (version v.0.3.23) | Intas Science Imaging Instruments GmbH | https://www.intas.de/ |
| ZEN imaging software | Carl Zeiss | https://www.zeiss.com/ |
| Clustal Omega | European Molecular Biology Laboratory – European Bioinformatics Institute (EMBL-EBI) | https://www.ebi.ac.uk/Tools/msa/clustalo/; Madeira et al., 2019 |
| Adobe Photoshop CS5 Extended (version 12.0 3 32) | Adobe | https://www.adobe.com/ |
| GraphPad Prism (version 8.3.0(538)) | GraphPad Software | https://www.graphpad.com/ |
| YASARA (version 19.1.27) | YASARA Biosciences GmbH | http://www.yasara.org/; Krieger and Vriend, 2014 |
| Microsoft Office Standard 2010 (version 14.0.7232.5000) | Microsoft Corporation | https://products.office.com/home |
| Other | ||
| Prefusion structure of SARS-CoV spike glycoprotein (5X5B) | Yuan et al., 2017 | https://www.rcsb.org/structure/5X5B |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Stefan Pöhlmann (spoehlmann@dpz.eu).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
The study did not generate unique datasets or code.
Method Details
Cell cultures
293T (human, kidney) and Vero (African green monkey, kidney) cells were cultivated in Dulbecco’s Modified Eagle Medium (PAN-Biotech) supplemented with 10% fetal bovine serum (Biochrom), 100 U/mL of penicillin and 0.1 mg/mL of streptomycin (PAN-Biotech). Vero cells that stably express human TMPRSS2 have been described previously (Hoffmann et al., 2020) and were cultivated in the presence of 10 μg/mL blasticidin (Invivogen). Calu-3 (human, lung; kindly provided by Stephan Ludwig, Westfälische Wilhelms-Universität, Muenster/Germany) cells were cultivated in Minimum Essential Medium (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (Biochrom), 100 U/mL of penicillin and 0.1 mg/mL of streptomycin (PAN-Biotech), 1x non-essential amino acid solution (from 100x stock, PAA) and 10 mM sodium pyruvate (Thermo Fisher Scientific). All cell lines were incubated at 37°C and 5% CO2 in a humidified atmosphere.
Plasmids
Expression plasmids for full-length vesicular stomatitis virus (VSV) glycoprotein (VSV-G), Middle-East respiratory syndrome coronavirus spike glycoprotein (MERS-S) containing a C-terminal V5 epitope tag, severe acute respiratory syndrome coronavirus spike glycoprotein (SARS-S) and severe acute respiratory syndrome coronavirus 2 spike glycoprotein (SARS-2-S) both equipped with a C-terminal hemagglutinin (HA) epitope tag have been described previously (Brinkmann et al., 2017, Hoffmann et al., 2020). Empty pCG1 expression vector was kindly provided by Roberto Cattaneo, Mayo Clinic, Rochester, MN/USA). Based on the SARS-S and SARS-2-S expression plasmids we cloned mutated versions with alterations at the S1/S2 cleavage site: We generated SARS-S containing the cleavage site of SARS-2-S, SARS-S (SARS-2), or BetaCoV/bat/Yunnan/RaTG13/2013 (RaTG; GISAID: EPI_ISL_402131), SARS-S (RaTG). Further, we generated SARS-2-S harboring the S1/S2 cleavage site of SARS-S, SARS-2-S (SARS) or RaTG-S, SARS-2-S (RaTG). Finally, we constructed SARS-2-S variants in which either the multibasic motif was deleted, SARS-2-S (delta), or in which the proline residue preceding the multibasic motif was mutated to arginine and the alanine residue within the minimal furin motif was changed to lysine in order to increase the basic environment at the S1/S2 site, SARS-2-S (opt). All newly cloned spike protein constructs further contained a deletion of 18 amino acids at their respective C terminus as this has been shown to improve coronavirus spike protein incorporation into VSV particles and thus transduction (Schwegmann-Weßels et al., 2009). Further, for each construct an untagged variant as well as a version containing a C-terminal V5 epitope tag was constructed.
Preparation of pseudotyped particles and transduction experiments
A previously published protocol was employed to produce VSV pseudotype particles (VSVpp) carrying foreign viral glycoproteins in their envelope (Berger Rentsch and Zimmer, 2011, Kleine-Weber et al., 2019). First, 293T cells were transfected with expression plasmid for the respective spike glycoprotein or VSV-G or empty expression vector by calcium-phosphate precipitation. At 16 h posttransfection, the cells were inoculated with VSV∗ΔG-fLuc (kindly provided by Gert Zimmer, Institute of Virology and Immunology, Mittelhäusern/Switzerland), a replication-deficient VSV vector that lacks the genetic information for VSV-G and encodes for eGFP and firefly luciferase (fLuc), at a multiplicity of infection of 3. After 1 h of incubation, the inoculum was removed and cells were washed with phosphate-buffered saline (PBS) before medium containing anti-VSV-G antibody (I1, mouse hybridoma supernatant from CRL-2700; ATCC) was added to all cells except for those expressing VSV-G (here, medium without antibody was added). Cells were further incubated for 16 h, before the VSVpp containing supernatants were harvested, freed from cellular debris by centrifugation and used for experiments.
For transduction, target cells were grown in 96-well plates until they reached 50%–80% confluency. The culture supernatant was removed by aspiration and 100 μl/well of the respective pseudotype were added (quadruplicate samples). At 16 h posttransduction, culture supernatants were aspirated and cells lysed in 1x cell culture lysis reagent (prepared from 5x stock, Promega) for 20 min at room temperature. The lysates were then transferred to white, opaque-walled 96-well plates and luciferase activity was quantified by measuring luminescence upon addition of a substrate (PJK) using a Hidex Sense plate luminometer (Hidex).
Western blot analysis
For the analysis of S protein processing, we subjected VSVpp harboring V5- or HA-tagged S proteins to SDS-PAGE and western blot analysis. For this, we loaded 1 mL VSVpp onto 50 μl of a 20% (w/v) sucrose cushion and performed high-speed centrifugation (25.000 g for 120 min at 4°C). Next, we removed 1 mL of supernatant, added 50 μl of 2x SDS-sample buffer and incubated the samples for 15 min at 96°C. Thereafter, the samples were subjected to SDS-PAGE and protein transfer to nitrocellulose membranes by western blot. The membranes were subsequently blocked in 5% skim milk solution (PBS containing 0.05% Tween-20 [PBS-T] and 5% skim milk powder) for 1 h at room temperature. The blots were then incubated over night at 4°C with primary antibody solution (all antibodies were diluted in PBS-T containing 5% skim milk; mouse anti-HA tag [Sigma-Aldrich, H3663, 1:2,500], mouse anti-V5 tag [Thermo Fisher Scientific, R960-25, 1:2,500] or VSV matrix protein [Kerafast, EB0011, 1:2,500]). Following this incubation, the blots were washed 3x with PBS-T before they were incubated for 1 h at room temperature with peroxidase-coupled goat anti-mouse antibody (Dianova, 115-035-003, 1:10,000). Finally, the blots were again washed and imaged. For this, an in house-prepared enhanced chemiluminescent solution (0.1 M Tris-HCl [pH 8.6], 250 μg/mL luminol, 1 mg/mL para-hydroxycoumaric acid, 0.3% H2O2) and the ChemoCam imaging system along with the ChemoStar Professional software (Intas Science Imaging Instruments GmbH) were used.
Syncytium formation assay
Vero or Vero-TMPRSS2 cells were grown on coverslips seeded in 24-well plates and transfected with S protein expression plasmids (1 μg/well) using Lipofectamine 2000 LTX with Plus reagent (Thermo Fisher Scientific) and OptiMEM medium (GIBCO). After 6 h the transfection solutions were aspirated and the cells further incubated for 24 h in standard culture medium. Next, the medium was changed to serum free medium ± 1 μg/mL bovine trypsin (Sigma-Aldrich) and the cells were incubated for additional 24 h. Then, the cells were washed with PBS, fixed with 4% paraformaldehyde solution for 20 min at room temperature, washed again, air-dried and incubated for 30 min with May-Gruenwald solution (Sigma-Aldrich). Thereafter, the cells were washed three times with deionized water, air-dried and incubated for 30 min with 1:10 diluted Giemsa solution (Sigma-Aldrich). After an additional washing interval with deionized water, the samples were air-dried and analyzed by bright-field microscopy using a Zeiss LSM800 confocal laser scanning microscope and the ZEN imaging software (both from Zeiss).
Sequence analysis and protein models
Sequence alignments were performed using the Clustal Omega online tool (https://www.ebi.ac.uk/Tools/msa/clustalo/). Protein models were designed using the YASARA software (http://www.yasara.org/index.html). For the generation of the SARS-2-S protein model the protein sequence was first modeled on a SARS-S template (PDB: 5X5B, (Yuan et al., 2017)) using the SWISS-MODEL online tool (https://swissmodel.expasy.org/). The following sequences information were obtained from National Center for Biotechnology Information (NCBI) database: SARS-CoV BJ01 (GenBank: AY278488.2), SARS-CoV CUHK-W1 (GenBank: AY278554.2), SARS-CoV Frankfurt-1 (GenBank: AY291315.1), SARS-CoV Tor2 (GenBank: CS050815.1), SARS-CoV Urbani (GenBank: AY278741.1), civet SARS-CoV SZ16 (GenBank: AY304488.1), civet SARS-CoV civet020 (GenBank: AY572038.1), raccoon dog SARS-CoV A030 (GenBank: AY687357.1), bat SARSr-CoV BtKY72/KEN (GenBank: KY352407.1), bat SARSr-CoV BM48-31/BGR/2008 (GenBank: GU190215.1), bat SARSr-CoV Rs4231 (GenBank: KY417146.1), bat SARSr-CoV WIV16 (GenBank: KT444582.1), bat SARSr-CoV Rs4874 (GenBank: KY417150.1), bat SARSr-CoV SL-CoVZC45 (GenBank: MG772933.1), bat SARSr-CoV SL-CoVZXC21 (GenBank: MG772934.1), bat SARSr-CoV LYRa11 (GenBank: KF569996.1), bat SARSr-CoV LYRa3 (GenBank: KF569997.1), bat SARSr-CoV WIV1 (GenBank: KF367457.1), bat SARSr-CoV RsSHC014 (GenBank: KC881005.1), bat SARSr-CoV Rs3367 (GenBank: KC881006.1), bat SARSr-CoV Cp/Yunnan2011 (GenBank: JX993988.1), bat SARSr-CoV Rp/Shaanxi2011 (GenBank: JX993987.1), bat SARSr-CoV HKU3-1 (GenBank: DQ022305.2), bat SARSr-CoV Rm1 (GenBank: DQ412043.1), bat SARSr-CoV Rp3 (GenBank: DQ071615.1), bat SARSr-CoV Rf1 (GenBank: DQ412042.1), bat SARSr-CoV 279 (GenBank: DQ648857.1), bat SARSr-CoV 273 (GenBank: DQ648856.1), bat SARSr-CoV YN2013 (GenBank: KJ473816.1), bat SARSr-CoV Rs/HuB2013 (GenBank: KJ473814.1), bat SARSr-CoV Rs/GX2013 (GenBank: KJ473815.1), bat SARSr-CoV Rf/SX2013 (GenBank: KJ473813.1), bat SARSr-CoV Rf/JL2012 (GenBank: KJ473811.1), bat SARSr-CoV Rf/HeB2013 (GenBank: KJ473812.1), bat SARSr-CoV YNLF/34C (GenBank: KP886809.1), bat SARSr-CoV YNLF/31C (GenBank: KP886808.1), bat SARSr-CoV Rs672 (GenBank: FJ588686.1), bat SARSr-CoV Rs7327 (GenBank: KY417151.1), bat SARSr-CoV Rs4084 (GenBank: KY417144.1), bat SARSr-CoV Rs9401 (GenBank: KY417152.1), bat SARSr-CoV Rs4247 (GenBank: KY417148.1), bat SARSr-CoV Rs4255 (GenBank: KY417149.1), bat SARSr-CoV Rs4081 (GenBank: KY417143.1), bat SARSr-CoV Rs4237 (GenBank: KY417147.1), bat SARSr-CoV As6526 (GenBank: KY417142.1), bat SARSr-CoV Rf4092 (GenBank: KY417145.1), bat SARSr-CoV Longquan-140 (GenBank: KF294457.1), bat SARSr-CoV Rs806 (GenBank: FJ588692.1), bat SARSr-CoV Anlong-103 (GenBank: KY770858.1), bat SARSr-CoV JTMC15 (GenBank: KU182964.1), bat SARSr-CoV 16BO133 (GenBank: KY938558.1), bat SARSr-CoV B15-21 (GenBank: KU528591.1), pangolin coronavirus MP789 (GenBank: MT084071.1). In addition the following sequences information were obtained from the Global Initiative on Sharing All Influenza Data (GISAID) database: SARS-CoV-2 (GISAID: EPI_ISL_404895), bat SARSr-CoV RaTG13 (GISAID: EPI_ISL_402131).
Quantification and Statistical Analysis
If not stated otherwise, statistical significance was tested by one-way analysis of variance with Dunnet’s posttest (GraphPad Prism 7.03). Only p values of 0.05 or lower were considered statistically significant (p > 0.05 [ns, not significant], p ≤ 0.05 [∗], p ≤ 0.01 [∗∗], p ≤ 0.001 [∗∗∗]). For all statistical analyses, the GraphPad Prism 7 software package was used (GraphPad Software).
Acknowledgments
We thank Inga Nehlmeier for technical assistance. We gratefully acknowledge the authors and the originating and submitting laboratories for their sequence and metadata shared through GISAID, on which this research is based. This work was supported by the Bundesministerium für Bildung und Forschung (BMBF, RAPID Consortium, 01KI1723D to S.P.). We further like to thank Andrea Maisner and Stephan Ludwig for providing the Vero cells and Calu-3 cells, respectively.
Author Contributions
Conceptualization, M.H. and S.P.; Formal Analysis, M.H. and S.P.; Investigation, M.H. and H.K.-W; Writing – Original Draft, M.H. and S.P.; Writing – Review & Editing, all authors; Funding Acquisition, S.P.
Declaration of Interest
The authors declare not competing interests
Published: May 1, 2020
References
- Berger Rentsch M., Zimmer G. A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon. PLoS ONE. 2011;6:e25858. doi: 10.1371/journal.pone.0025858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Dijkman R., Habjan M., Heurich A., Gierer S., Glowacka I., Welsch K., Winkler M., Schneider H., Hofmann-Winkler H., et al. TMPRSS2 activates the human coronavirus 229E for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J. Virol. 2013;87:6150–6160. doi: 10.1128/JVI.03372-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann C., Hoffmann M., Lübke A., Nehlmeier I., Krämer-Kühl A., Winkler M., Pöhlmann S. The glycoprotein of vesicular stomatitis virus promotes release of virus-like particles from tetherin-positive cells. PLoS ONE. 2017;12:e0189073. doi: 10.1371/journal.pone.0189073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierer S., Bertram S., Kaup F., Wrensch F., Heurich A., Krämer-Kühl A., Welsch K., Winkler M., Meyer B., Drosten C., et al. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J. Virol. 2013;87:5502–5511. doi: 10.1128/JVI.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierer S., Müller M.A., Heurich A., Ritz D., Springstein B.L., Karsten C.B., Schendzielorz A., Gnirß K., Drosten C., Pöhlmann S. Inhibition of proprotein convertases abrogates processing of the middle eastern respiratory syndrome coronavirus spike protein in infected cells but does not reduce viral infectivity. J. Infect. Dis. 2015;211:889–897. doi: 10.1093/infdis/jiu407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Müller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Hofmann-Winkler H., Pöhlmann S. In: Activation of Viruses by Host Proteases. Böttcher-Friebertshäuser E., Garten W., Klenk H., editors. Springer; 2018. Priming Time: How Cellular Proteases Arm Coronavirus Spike Proteins; pp. 71–98. [Google Scholar]
- Hoffmann M., Müller M.A., Drexler J.F., Glende J., Erdt M., Gützkow T., Losemann C., Binger T., Deng H., Schwegmann-Weßels C., et al. Differential sensitivity of bat cells to infection by enveloped RNA viruses: coronaviruses, paramyxoviruses, filoviruses, and influenza viruses. PLoS ONE. 2013;8:e72942. doi: 10.1371/journal.pone.0072942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulswit R.J., de Haan C.A., Bosch B.J. Coronavirus Spike Protein and Tropism Changes. Adv. Virus Res. 2016;96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., Nagata N. TMPRSS2 Contributes to Virus Spread and Immunopathology in the Airways of Murine Models after Coronavirus Infection. J. Virol. 2019;93:93. doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Weber H., Elzayat M.T., Hoffmann M., Pöhlmann S. Functional analysis of potential cleavage sites in the MERS-coronavirus spike protein. Sci. Rep. 2018;8:16597. doi: 10.1038/s41598-018-34859-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Weber H., Elzayat M.T., Wang L., Graham B.S., Müller M.A., Drosten C., Pöhlmann S., Hoffmann M. Mutations in the Spike Protein of Middle East Respiratory Syndrome Coronavirus Transmitted in Korea Increase Resistance to Antibody-Mediated Neutralization. J. Virol. 2019;93:93. doi: 10.1128/JVI.01381-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger E., Vriend G. YASARA View - molecular graphics for all devices - from smartphones to workstations. Bioinformatics. 2014;30:2981–2982. doi: 10.1093/bioinformatics/btu426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T., Shum M.H., Zhu H.C., Tong Y.G., Ni X.B., Liao Y.S., Wei W., Cheung W.Y., Li W.J., Li L.F., et al. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020 doi: 10.1038/s41586-020-2169-0. Published online March 26, 2020. [DOI] [PubMed] [Google Scholar]
- Li X., Zai J., Zhao Q., Nie Q., Li Y., Foley B.T., Chaillon A. Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. J. Med. Virol. 2020 doi: 10.1002/jmv.25731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luczo J.M., Stambas J., Durr P.A., Michalski W.P., Bingham J. Molecular pathogenesis of H5 highly pathogenic avian influenza: the role of the haemagglutinin cleavage site motif. Rev. Med. Virol. 2015;25:406–430. doi: 10.1002/rmv.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D., Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47(W1):W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Dinnon K.H., 3rd, Yount B.L., Jr., McAnarney E.T., Gralinski L.E., Hale A., Graham R.L., Scobey T., Anthony S.J., Wang L., et al. Trypsin Treatment Unlocks Barrier for Zoonotic Bat Coronaviruses Infection. J. Virol. 2020;94:e01774-19. doi: 10.1128/JVI.01774-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. USA. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Physiological and molecular triggers for SARS-CoV membrane fusion and entry into host cells. Virology. 2018;517:3–8. doi: 10.1016/j.virol.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.E., Li K., Barlan A., Fehr A.R., Perlman S., McCray P.B., Jr., Gallagher T. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc. Natl. Acad. Sci. USA. 2016;113:12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roebroek A.J., Umans L., Pauli I.G., Robertson E.J., van Leuven F., Van de Ven W.J., Constam D.B. Failure of ventral closure and axial rotation in embryos lacking the proprotein convertase Furin. Development. 1998;125:4863–4876. doi: 10.1242/dev.125.24.4863. [DOI] [PubMed] [Google Scholar]
- Sarac M.S., Cameron A., Lindberg I. The furin inhibitor hexa-D-arginine blocks the activation of Pseudomonas aeruginosa exotoxin A in vivo. Infect. Immun. 2002;70:7136–7139. doi: 10.1128/IAI.70.12.7136-7139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarac M.S., Peinado J.R., Leppla S.H., Lindberg I. Protection against anthrax toxemia by hexa-D-arginine in vitro and in vivo. Infect. Immun. 2004;72:602–605. doi: 10.1128/IAI.72.1.602-605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwegmann-Weßels C., Glende J., Ren X., Qu X., Deng H., Enjuanes L., Herrler G. Comparison of vesicular stomatitis virus pseudotyped with the S proteins from a porcine and a human coronavirus. J. Gen. Virol. 2009;90:1724–1729. doi: 10.1099/vir.0.009704-0. [DOI] [PubMed] [Google Scholar]
- Shirato K., Kawase M., Matsuyama S. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J. Virol. 2013;87:12552–12561. doi: 10.1128/JVI.01890-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Kanou K., Kawase M., Matsuyama S. Clinical Isolates of Human Coronavirus 229E Bypass the Endosome for Cell Entry. J. Virol. 2016;91:91. doi: 10.1128/JVI.01387-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Novel Coronavirus(2019-nCoV) Situation Report 52. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200312-sitrep-52-covid-19.pdf?sfvrsn=e2bfc9c0_4
- WHO Coronavirus disease 2019 (COVID-19) Situation Report – 81. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200410-sitrep-81-covid-19.pdf?sfvrsn=ca96eb84_2
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Du L., Liu C., Wang L., Ma C., Tang J., Baric R.S., Jiang S., Li F. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc. Natl. Acad. Sci. USA. 2014;111:12516–12521. doi: 10.1073/pnas.1405889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y., et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Wu Q., Zhang Z. Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr. Biol. 2020;30:1346–1351.e2. doi: 10.1016/j.cub.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Vedantham P., Lu K., Agudelo J., Carrion R., Jr., Nunneley J.W., Barnard D., Pöhlmann S., McKerrow J.H., Renslo A.R., Simmons G. Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res. 2015;116:76–84. doi: 10.1016/j.antiviral.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Chen X., Hu T., Li J., Song H., Liu Y., Wang P., Liu D., Yang J., Holmes E.C., et al. A novel bat coronavirus reveals natural insertions at the S1/S2 cleavage site of the Spike protein and a possible recombinant origin of HCoV-19. bioRxiv. 2020 doi: 10.1101/2020.03.02.974139. [DOI] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. China Novel Coronavirus Investigating and Research Team A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The study did not generate unique datasets or code.