SUMMARY
The mammalian heart is incapable of regenerating a sufficient number of cardiomyocytes to ameliorate the loss of contractile muscle after acute myocardial injury. Several reports have demonstrated that mononucleated cardiomyocytes are more responsive than are binucleated cardiomyocytes to proproliferative stimuli. We have developed a strategy to isolate and characterize highly enriched populations of mononucleated and binucleated cardiomyocytes at various times of development. Our results suggest that an E2f/Rb transcriptional network is central to the divergence of these two populations and that remnants of the differences acquired during the neonatal period remain in adult cardiomyocytes. Moreover, inducing binucleation by genetically blocking the ability of cardiomyocytes to complete cytokinesis leads to a reduction in E2f target gene expression, directly linking the E2f pathway with nucleation. These data identify key molecular differences between mononucleated and binucleated mammalian cardiomyocytes that can be used to leverage cardiomyocyte proliferation for promoting injury repair in the heart.
Graphical Abstract
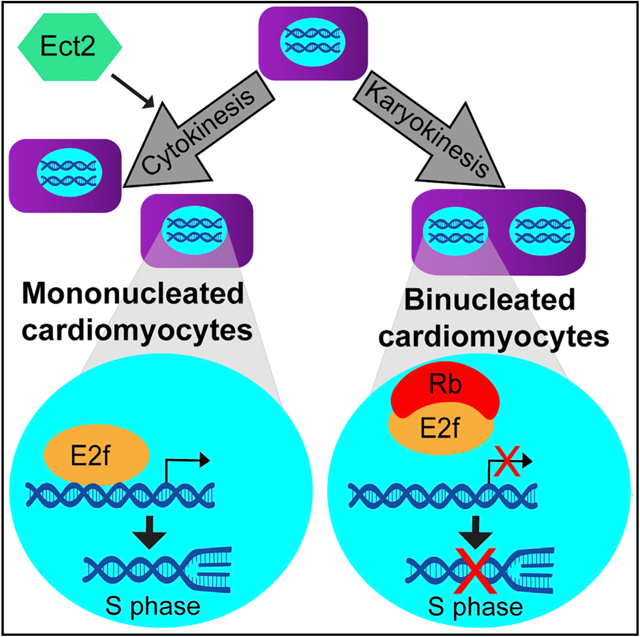
In Brief
Windmueller et al. develop a strategy to separate mononucleated and binucleated cardiomyocytes and examine transcriptional differences acquired as the two subsets diverge during the neonatal period. Binucleation is associated with silencing of proliferation genes and upregulation of maturation genes. Genetic loss of Ect2 links binucleation to the Rb/E2f pathway.
INTRODUCTION
The mammalian heart has traditionally been considered a terminally differentiated organ, incapable of regeneration. Acute injury to the heart results in the loss of functional cardiomyocytes (CMs), and in response to this, the heart undergoes a maladaptive remodeling process, including hypertrophy and fibrotic scar formation, which can result in heart failure. While the adult mammalian heart does not appear to exhibit significant regeneration, a number of lower vertebrate species, including zebrafish, are able to regenerate myocardium (Oberpriller and Oberpriller, 1974; Poss et al., 2002). Furthermore, the murine heart displays regenerative capacity during the embryonic and early postnatal stages, although this ability is largely lost by 1 week after birth (Porrello et al., 2011). There is also evidence that a low level of CM turnover does occur over the lifetime in both the mouse and the human heart (Bergmann et al., 2009; Mollova et al., 2013). In all of these contexts, new CMs are thought to arise from the proliferation of pre-existing CMs (Jopling et al., 2010; Senyo et al., 2013).
The majority of CMs in the adult rodent heart are either binucleated (BiNucs) or polyploid. However, mounting evidence suggests that the smaller population of CMs that are mononucleated (MoNucs) and diploid may exhibit a greater potential to proliferate and to contribute to cardiac regeneration. When the adult murine heart is treated with a proliferative stimulus in vivo, a higher percentage of MoNucs than BiNucs re-enter the cell cycle (Bersell et al., 2009; Kühn et al., 2007). Furthermore, natural variation in the percentage of diploid MoNucs in the heart across different mouse strains is positively correlated with the number of cells that re-enter the cell cycle after injury and with functional repair of the heart after injury (Patterson et al., 2017). The zebrafish heart, which retains a lifelong ability to regenerate, contains almost solely MoNucs under physiological conditions. However, the induction of polyploidy and binucleation via expression of a dominant-negative Ect2 impairs the regenerative ability of the zebrafish heart (Gonzalez-Rosa et al., 2018). Despite these intriguing observations, there have been no studies directly comparing MoNucs and BiNucs in the mammalian heart, and the molecular basis for their distinct proliferative responses remains unclear.
During early murine development, almost all CMs are MoNucs, which correlates with ongoing proliferation to drive organ growth. However, during the neonatal maturation period, the number of BiNuc CMs increases rapidly, which correlates with the exit of CMs from the cell cycle. This is accompanied by other changes that include increased organization of the sarcomere structure and a switch from utilization of glycolytic metabolism to oxidative phosphorylation for energy production. To address whether the developmental divergence of MoNucs and BiNucs contributes to their differential response to proliferative stimuli, we developed a strategy to separate MoNuc and BiNuc CMs using fluorescence-activated cell sorting (FACS) and assessed the transcriptional differences between these subsets of CMs across late development and in the adult. These data revealed that during the neonatal period, binucleation is associated with E2f transcription factor-mediated silencing of genes that permit the onset of DNA synthesis. Loss of retinoblastoma (Rb) family member expression led to increased E2f target gene expression with no change in CM nucleation. In contrast, the inhibition of cytokinesis through cardiomyocyte-specific genetic loss of Ect2 expression resulted in an increased ratio of BiNuc CMs with a commensurate decrease in an E2f-related proliferation pathway. Our study highlights a strategy for isolating MoNucs and BiNucs and suggests that the decreased competency for proliferation in BiNucs is due to an E2f/Rb pathway that is regulated via binucleation.
RESULTS
MoNuc and BiNuc CMs Can Be Separated by FACS
We sought to characterize transcriptional differences between MoNuc and BiNuc CMs during maturation from late embryonic development through adulthood. Single-cell suspensions were prepared from hearts at the indicated time points to be sorted by FACS (Figure 1A). A wide nozzle was used to accommodate the large size of mature CMs. A wide nozzle also subjects the CMs to a lower shear stress than is typical for FACS (Shapiro, 1985). Ventricular CMs were lineage labeled with Mlc2vcre: R26REYFP and nuclei were stained with Vybrant DyeCycle DNA dye. The intensity of the fluorescent signal emitted by this dye is proportional to DNA quantity. The dye is used to assess DNA content and ploidy of nuclei by FACS (Patterson et al., 2017). Cells were gated on enhanced yellow fluorescent protein (EYFP) presence and then determined to be MoNucs or BiNucs by a strategy that combines the use of DyeCycle dye with the concept behind doublet discrimination (Figures 1B–1E). Doublet discrimination is used in FACS analysis to exclude instances in which two cells are stuck together and characterized as a single event. Doublet discrimination takes into account the width (time) and the height (voltage intensity) of the signal produced and assumes that two cells will produce a signal of greater width than a single cell, while the height of the signal will remain the same. We used a similar metric to distinguish BiNucs from MoNucs. Nucleation was gated on the height and width of the fluorescent signal produced by the 405-nm laser excitation of DyeCycle dye (Figures 1C–1E). The nuclei of BiNucs produced a signal of equal intensity to that produced by diploid MoNucs but of greater width. MoNucs with increased voltage intensity, presumed to be polyploid or undergoing DNA synthesis, were excluded. Since the size and shape of CMs change over maturation, each time point required tailored gating parameters. These parameters were empirically fine-tuned to maximize enrichment by sorting cells onto microscope slides for the visualization of nucleation (Figures 1F–1K). The relative percentages of MoNucs and BiNucs collected at each time point faithfully reflect the developmental switch to BiNucs, being the predominant CM subtype, and are in agreement with the reported percentages of MoNucs and BiNucs over CM maturation (Ikenishi et al., 2012). This methodology resulted in >70% enrichment of either MoNucs or BiNucs at the embryonic day 18.5 (E18.5) time point and >90% enrichment at the postnatal day 7 (P7) and adult time points (Figures 1L–1N). We quantified the viability of sorted CMs by propidium iodide (PI) staining. At each time point, >97% of cells remained negative for PI after being sorted (Figures S1A and S1B). Finally, we expanded the application of our FACS strategy to separate adult MoNucs and BiNucs without the requirement of a lineage marker (Figure 1O). We reasoned that a long, rod-shaped CM would produce a wider light scatter than a smaller, round, non-myocyte, similar to our gating for two nuclei versus one. To quantify the ability of this strategy to properly identify CMs, we gated lineage-traced cells on the width of side scatter and then backgated on YFP fluorescence, showing that 99.1% of cells identified are CMs by their YFP expression (Figures 1P and 1Q). We were able to separate MoNucs and BiNucs by their distinct light scatter with equal success rates using this strategy (Figures S1C and S1D). The techniques described here allowed us to isolate highly enriched populations of lineage labeled MoNuc and BiNuc CMs from E18.5, P7, and adult mice to assess changes in gene expression during CM maturation.
Figure 1. Mononucleated and Binucleated Cardiomyocytes Can Be Separated by FACS.
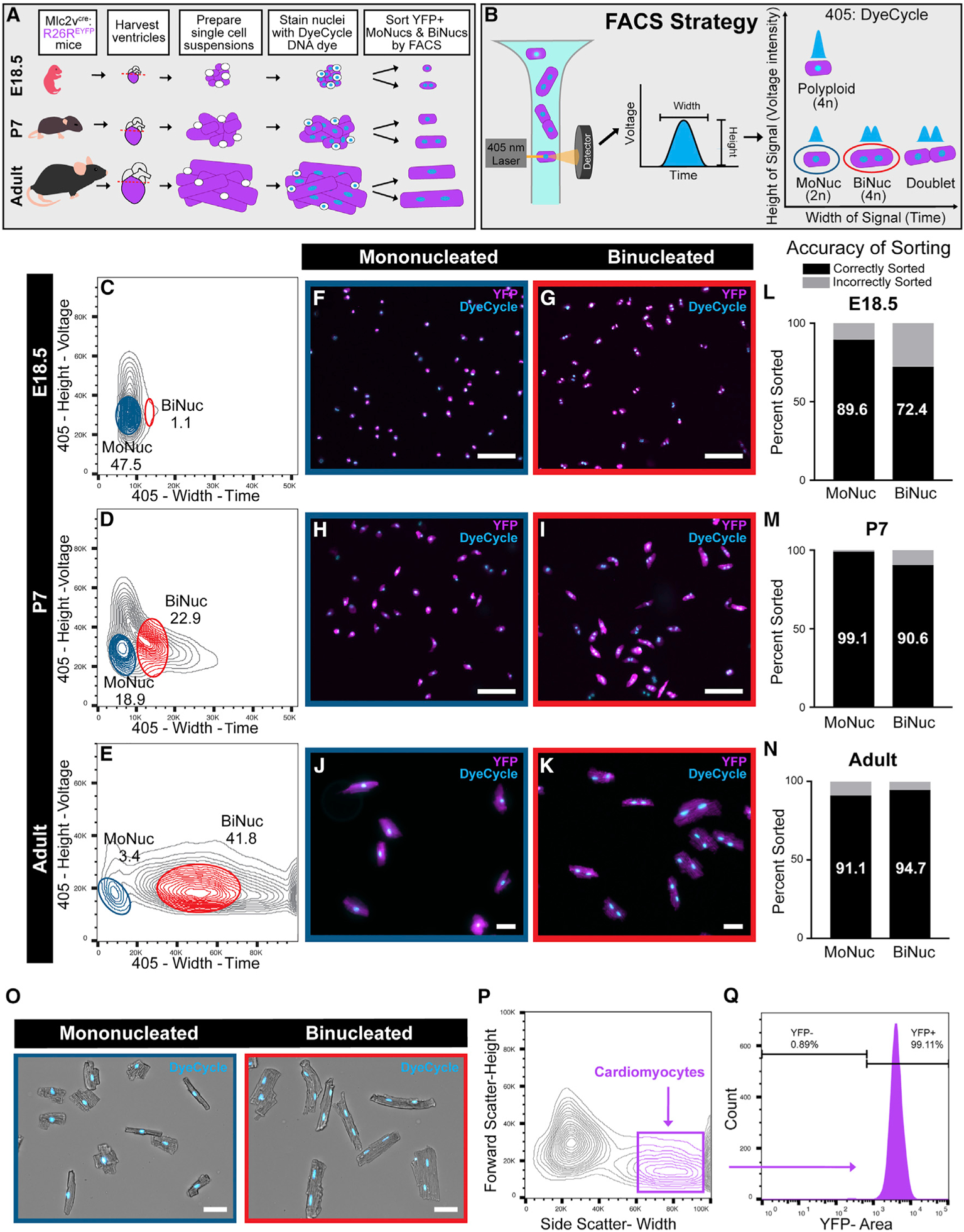
(A) Schematic of experimental design to collect mononucleated (MoNucs) and binucleated (BiNucs) cardiomyocytes (CMs) for RNA-sequencing (RNA-seq) analysis. Single-cell suspensions are isolated from the hearts of Mlc2vcre:R26REYFP mice at age E18.5, P7, and adult. Nuclei are stained with Vybrant DyeCycle DNA dye. CMs are identified by FACS as YFP+ and then sorted by nucleation.
(B) Schematic of sorting strategy for separating MoNucs and BiNucs. As nuclei of CMs pass the 405-nm laser, DyeCycle DNA dye is activated and produces a signal of which the width is proportional to the length of time that the nuclei travel through the laser. Two nuclei (BiNucs) take longer to travel past the laser than one nucleus (MoNucs) and produce two overlapping signals, which are interpreted as a single signal of greater width. The height of the signal produced is proportional to DNA content per nucleus, allowing the separate gating of diploid and polyploid cells. MoNucs and BiNucs cluster separately on a plot of width versus height of the 405 signal.
(C–E) Representative flow cytometry plots of the width versus height of the 405 channel. Gates used to sort E18.5 (C), P7 (D), and adult (E) MoNucs and BiNucs are shown. The mean percentages of total YFP+ cells that fall inside MoNuc or BiNuc gates are indicated. n = 3 animals for each time point.
(F–K) Cropped images of live Mlc2vcre:R26REYFP MoNucs (F, H, and J) and BiNucs (G, I, and K) from E18.5, P7, and adult mice sorted onto slides according to gates shown in (C)–(E). Scale bar, 50 μm.
(L–N) Percentages of E18.5, P7, and adult MoNucs and BiNucs correctly and incorrectly sorted into indicated groups as determined by slide images taken during sorts. More than 100 cells were counted for each group.
(O) Adult CD-1 MoNuc and BiNuc CMs sorted onto slides with expanded FACS strategy that allows CMs to be sorted without a lineage marker. Scale bar, 50 μm.
(P) FACS plot showing expanded FACS strategy.
(Q) Success of expanded FACS strategy tested on Mlc2vcre:R26REYFP CMs. Histogram shows that >99% of cells gated as CMs are YFP+.
See also Figure S1.
Binucleation Is Accompanied by a Switch from a Proliferation- to a Maturation-Associated Gene Expression Program
To characterize transcriptional differences between MoNucs and BiNucs over the neonatal maturation period, we sorted MoNuc and BiNuc CM populations and performed RNA sequencing (RNA-seq) analysis. The principal-component analysis (PCA) shows that at each time point, MoNucs and BiNucs are transcriptionally distinct with the primary and secondary components accounting for 76.2% and 9.6%, respectively, of the variance across all of the samples (Figure 2A). At E18.5, BiNucs were enriched for genes involved in cell division, including several genes known to be involved in cytokinesis (Figures S2A and S2B). These data suggest that at this time point, CMs identified as BiNucs may still have been undergoing, or attempting to undergo, cytokinesis. The RNA-seq data revealed that MoNucs and BiNucs exhibit the most statistically significant differences in gene expression at P7. To further characterize the set of genes differentially expressed between P7 MoNucs and BiNucs, we examined their expression levels across all of the samples (Figure 2B). We observed that the expression of this set of genes also changes significantly between the embryonic and adult time points. The genes upregulated in P7 MoNucs tend to be more highly expressed in both embryonic CM populations compared to adult CMs, whereas the genes upregulated in P7 BiNucs tend to be enriched in both adult CM populations versus embryonic CMs. Gene set enrichment analysis (GSEA) showed that P7 MoNucs are enriched for genes involved in the cell cycle, especially those that are E2f targets and those that are involved in DNA synthesis (Figures 2C–2E). In contrast, P7 BiNucs are enriched for genes involved in CM maturation processes, including sarcomere organization and a switch to fatty acid oxidative metabolism (Figures 2C, 2F, and 2G). These data indicate that during the postnatal period, CM binucleation is accompanied by the termination of the fetal gene expression program, including genes required for proliferation, in exchange for the activation of the mature CM gene expression program.
Figure 2. Binucleation Is Accompanied by a Switch from a Proliferation-Associated Gene Expression Program to One Associated with Maturation.
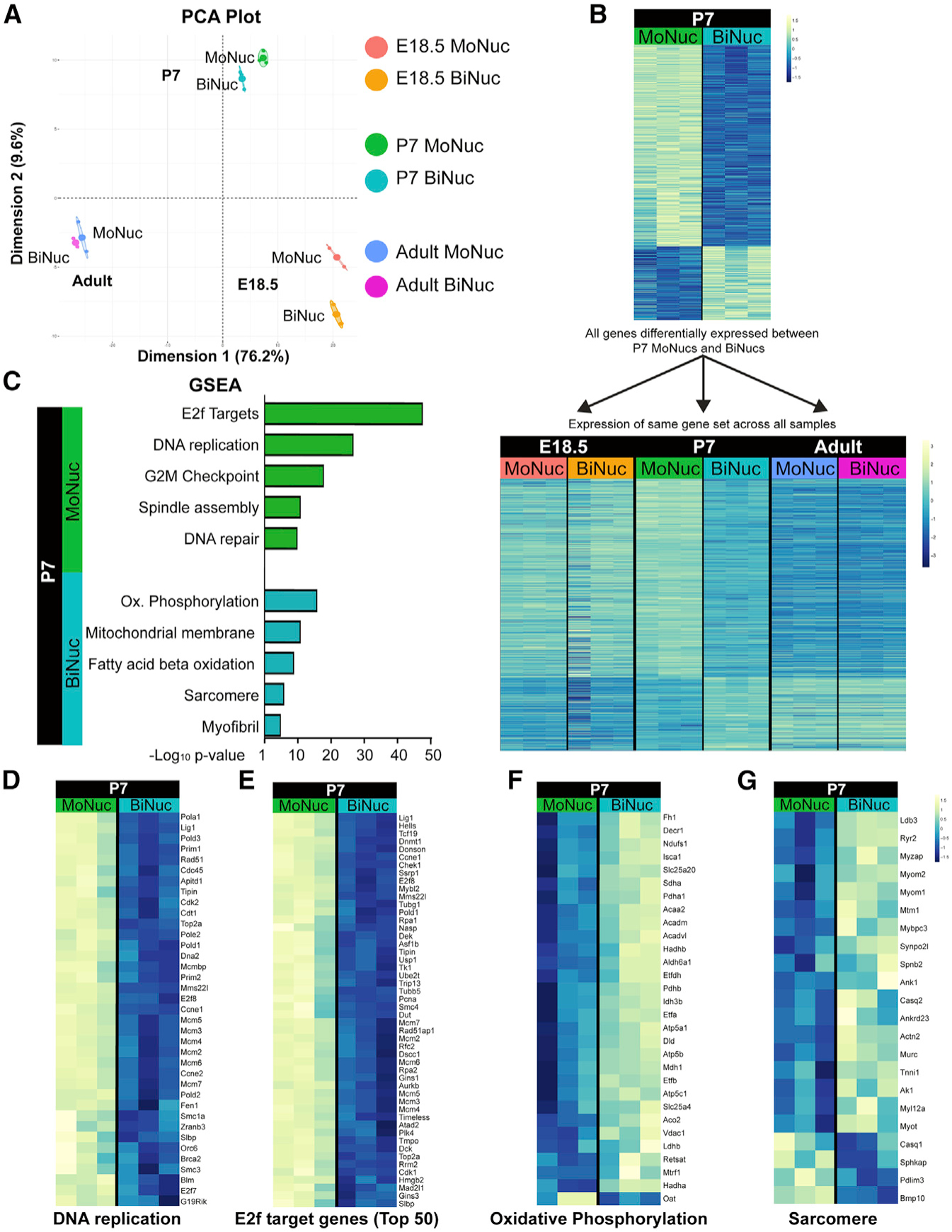
(A) Principal component analysis (PCA) from RNA-seq of MoNucs and BiNucs from E18.5, P7, and adult hearts shows that at each time point, MoNucs and BiNucs are transcriptionally distinct. n = 3 animals per time point.
(B) Heatmaps of genes with significant differences (FDR < 0.05) in expression between P7 MoNucs and BiNucs. Gene expression values are represented at P7 only (top) and at all time points (bottom). Bottom heatmap reveals that the gene expression profile of P7 BiNucs but not P7 MoNucs bears a resemblance to the profiles of adult samples. The difference in intensity between top and bottom heatmaps is due to the different scales used.
(C) Representative categories from gene set enrichment analysis (GSEA) of differential gene expression between P7 MoNucs and BiNucs. P7 MoNucs are enriched for genes involved in the cell cycle, while P7 BiNucs are enriched for genes involved in cardiomyocyte maturation. Analysis was done using Camera.
(D–G) Heatmaps comparing expression between P7 MoNucs and BiNucs of gene sets included in (C). E2f target gene heatmap shows top 50 differentially expressed genes from the set of 199 genes.
See also Figure S2.
BiNuc CMs at P7 Turn Off E2f Target Gene Expression Required for G1/S Phase Transition
Transcriptome analysis revealed that at P7, binucleation is accompanied by the termination of a gene expression program closely associated with regulating the onset of proliferation. To gain insight into which transcription factors may be involved in regulating the apparent switch in transcription program, we analyzed the promoters of differentially expressed genes at P7 for the enrichment of DNA-binding sites and associated transcription factors. This analysis revealed that binucleation leads to the downregulation of targets of the E2f transcription factor family, whereas genes upregulated in P7 BiNucs are targets of the Ppara, Esrra, Mef2, and Myod transcription factors (Figure 3A). The E2f transcription factor family is known to control the onset of DNA synthesis (Nevins, 1992). E2f1 has also been implicated in repressing the expression of genes involved in oxidative phosphorylation, including several of the genes upregulated in P7 BiNucs (Blanchet et al., 2011). It has been suggested that this dual control by E2f creates a regulatory switch between proliferation and metabolism. E2f transcription factor activity is regulated by Rb (Johnson et al., 1993; Lundberg and Weinberg, 1998). Rb-mediated silencing of E2f target genes is involved in the irreversible exit of the cell cycle during senescence, leaving cells unable to respond to proliferative stimuli (Narita et al., 2003). Our RNA-seq data show that 54% of the top 50 downregulated genes in P7 BiNucs are targets of E2f that are known to regulate the G1/S transition and onset of DNA synthesis (Figures 3B and 3C). This downregulation occurs most dramatically between P7 MoNucs and BiNucs (Figure 3D). In addition to their target genes, 4 of the E2f family members, E2f1, E2f2, E2f7, and E2f8, were strongly downregulated in P7 BiNucs (Figure S3A). These data demonstrate that DNA synthesis and onset of the cell cycle are dramatically downregulated in association with binucleation in P7 CMs through the silencing of E2f target genes.
Figure 3. BiNuc CMs at P7 Turn Off E3f Target Gene Expression Required for G1/S Phase Transition and S Phase.
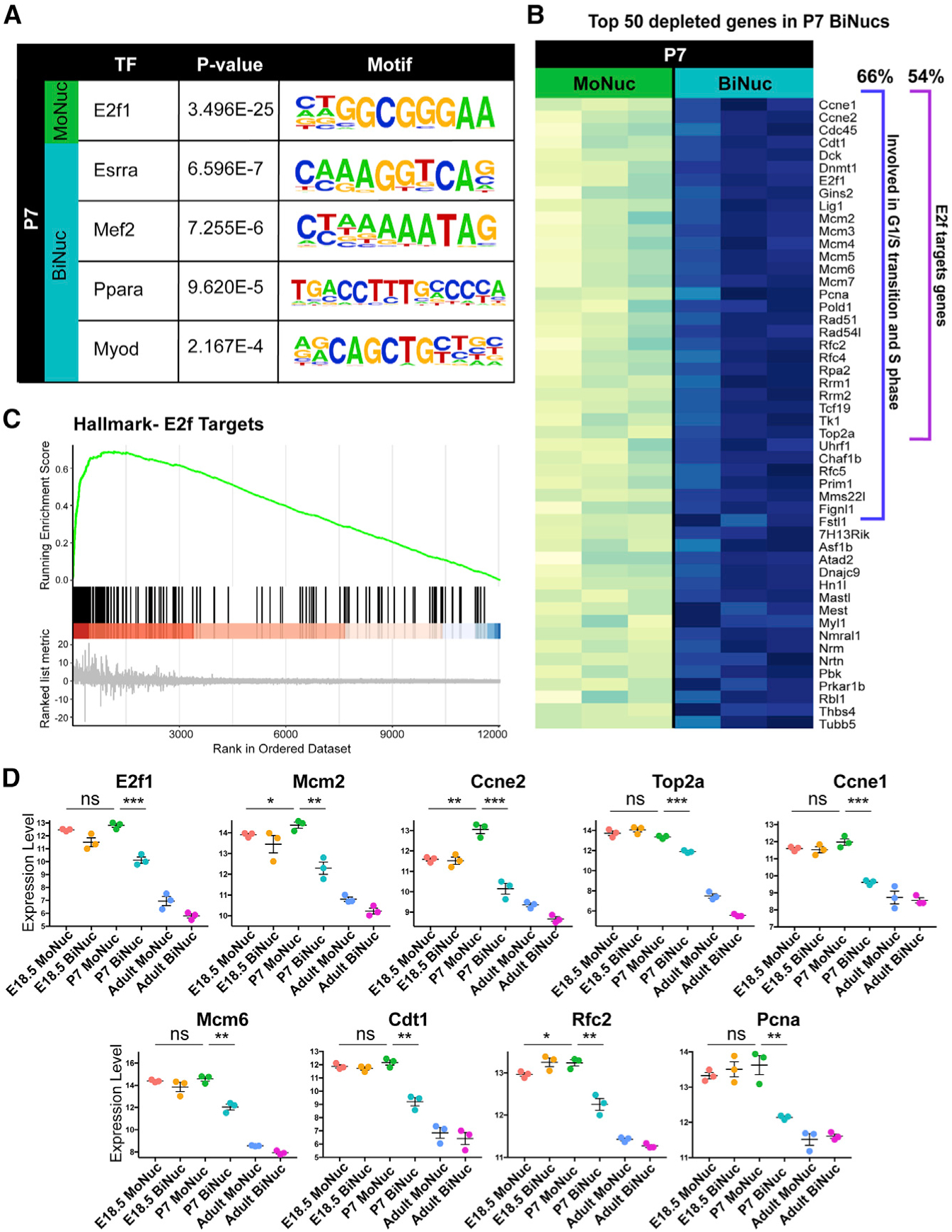
(A) Enriched transcription factor motifs within promoters of genes differentially expressed between P7 MoNucs and BiNucs reveal that E2f target genes are the most highly enriched. Analysis was performed using the ToppGene suite.
(B) Analysis of the top 50 differentially expressed genes between P7 MoNucs and BiNucs reveals that 66% (33/50) are involved in G1/S transition and S phase of the cell cycle; 27 of the 50 (54%) are E2f target genes.
(C) GSEA plot of E2f target genes differentially expressed between P7 MoNucs and BiNucs.
(D) E2f target gene expression decreases specifically between P7 MoNucs and BiNucs, but not between E18.5 and P7 MoNucs. The expression values of indicated E2f target genes measured by RNA-seq are shown across all of the samples. The significance of differences in gene expression between E18.5 and P7 MoNucs and between P7 MoNucs and BiNucs are indicated. Data are reported as mean ± SEM. n = 3 animals per time point. p values determined by Student’s t test. ns, not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
See also Figure S3.
Adult MoNuc and BiNuc CMs Retain Differences Established during Neonatal Maturation
To determine whether differences between P7 MoNucs and BiNucs remained in adult CMs, we compared the transcriptional profiles of MoNucs and BiNucs from adult hearts. Transcriptionally, these CM subsets are quite similar with only 3 differentially expressed genes with a false discovery rate (FDR) < 0.05 (Figure S4A). However, two of these genes, Necdin and Cenpf, are known to be involved in the regulation of E2f/Rb signaling (Papadimou et al., 2005; Taniura et al., 1998). A total of 456 genes had a non-adjusted p < 0.05 (Figure 4A). GSEA of these genes revealed the enrichment of categories related to those that are differentially expressed between P7 MoNucs and BiNucs (Figure 4B). While adult MoNucs are enriched for genes involved in DNA synthesis and G2 phase of the cell cycle, adult BiNucs are enriched for genes involved in fatty acid oxidation. Furthermore, several of the most highly downregulated E2f target genes in the P7 BiNucs remain downregulated in the adult BiNucs (Figure 4C). These data show that adult MoNucs and BiNucs retain remnants of transcriptional differences established during the neonatal maturation period.
Figure 4. Adult MoNuc and BiNuc CMs Retain Differences Established during Neonatal Maturation.
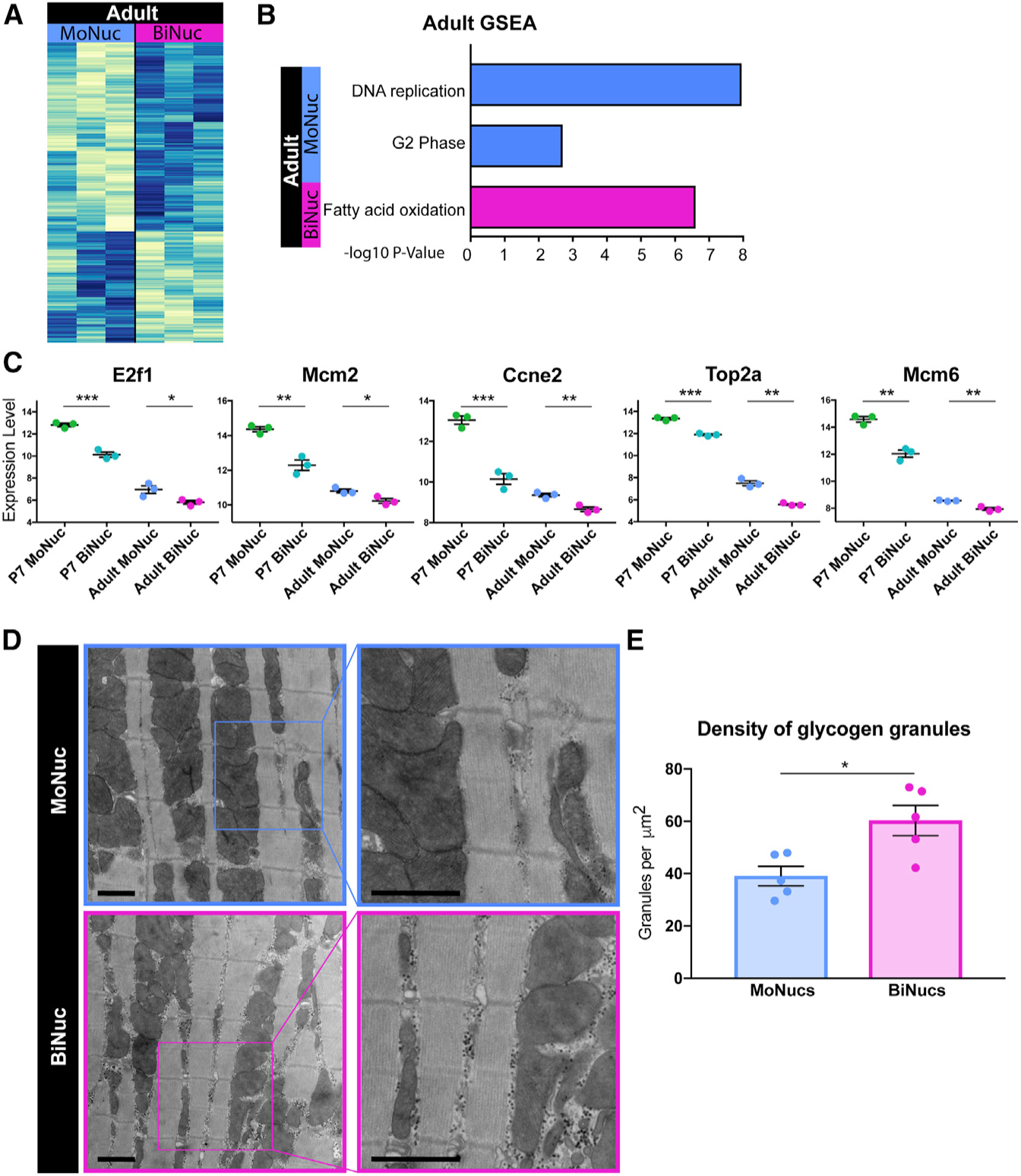
(A) Heatmap of gene expression differences with p < 0.05 between adult MoNucs and BiNucs.
(B) Representative categories from GSEA of differential gene expression between Adult MoNucs and BiNucs reveals overlap with categories enriched between P7 MoNucs and BiNucs. Analysis was done using Camera.
(C) A subset of E2f target genes most differentially expressed between P7 MoNucs and BiNucs remain differentially expressed between adult MoNucs and BiNucs. Expression values of these genes measured by RNA-seq are shown in P7 and adult samples. The significance of differences in gene expression between P7 MoNucs and BiNucs and between adult MoNucs and BiNucs are indicated. Data are reported as mean ± SEM. n = 3 animals per time point. p values are determined by Student’s t test. ns, not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
(D) Electron micrographs of sorted adult MoNucs and BiNucs. Scale bar, 1 μm.
(E) Quantification of glycogen granule density in non-sarcomeric cytoplasmic space visualized by electron microscopy. The significance of difference between adult MoNucs and BiNucs is indicated. Data are reported as mean ± SEM. n = 5 cells per sample. p values are determined by Student’s t test. ns, not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
See also Figure S4.
To further examine differences between adult MoNucs and BiNucs, we performed transmission electron microscopy studies on sorted CMs. We observed that adult BiNucs have a significantly higher density and buildup of glycogen granules surrounding the mitochondria than do MoNucs (Figures 4D and 4E). Glycogen granules are a stored energy source that is depleted when used to drive glycolytic metabolism (Prats et al., 2018; Schneider et al., 2014). This observation suggests that metabolic differences exist between adult MoNucs and BiNucs. One possible explanation for this observation is that diploid adult MoNucs may use glycolytic metabolism more readily than BiNucs and are therefore not able to maintain stores of excess glycogen. This would be consistent with the switch away from glycolytic metabolism that occurs during CM maturation and the oxidative metabolism gene expression profile that we observed to be enriched in BiNucs at the P7 and adult stages. These data further support the concept that adult MoNucs and BiNucs retain remnants of differences established during the neonatal maturation period.
Rb Is Required for Downregulation of E2f Target Genes
Previous work has shown that Rb recruits HP1-γ to H3K9me3 marks at a subset of E2f target genes to stably repress their expression during CM maturation (Sdek et al., 2011). Therefore, we sought to determine whether Rb is required for the repression of E2f target genes that we saw in P7 BiNucs. We used a triple Rb family knockout (TKO) mouse model, Rbflox/flox;p130flox/flox;p107−/− (Viatour et al., 2008) to determine the role of the Rb family in CM binucleation. CMs were isolated from P1 mice and infected with either adenoviral-Cre/GFP (TKO) or adenoviral-GFP (control) in vitro (Figure 5A). qPCR revealed strong downregulation of the Rb1 transcript in TKO CMs (Figure 5B). qPCR revealed that TKO CMs strongly upregulated E2f target genes, whose expression is decreased in P7 BiNucs versus MoNucs (Figure 5E). In contrast, non-E2f targets were upregulated to a lesser degree in TKO CMs (Figures 5E and 5F). This suggests that Rb is required for the downregulation of E2f target genes in neonatal CMs. We then quantified the percentage of GFP+ MoNucs and BiNucs in both control and TKO samples to determine whether Rb activity regulates the onset of binucleation. There was no difference in the ratio of BiNucs to MoNucs between control and TKO CMs (Figures 5C and 5D). This suggests that Rb-mediated changes in E2F target expression are not responsible for the onset of binucleation.
Figure 5. Rb Acts Downstream of Binucleation and Is Required for Downregulation of E2f Target Genes.
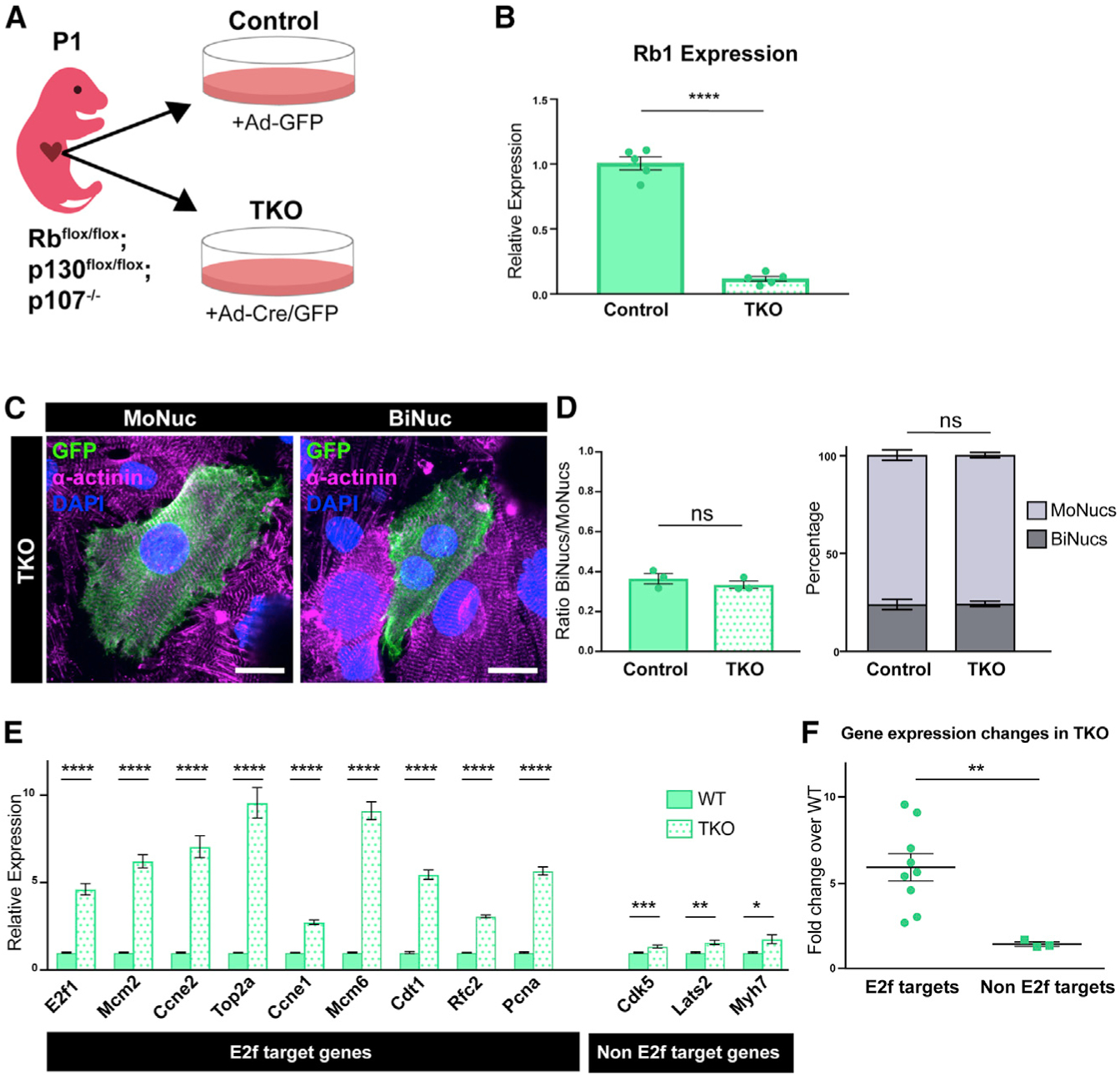
(A) Schematic of experimental design of in vitro Rb family KO experiments. CMs were isolated from Rbflox/flox;p130flox/flox;p107−/− mice at P1 and infected with adenovirus containing either Cre and GFP (Ad-Cre/GFP) or GFP alone (Ad-GFP). Cells infected with Ad-Cre/GFP are denoted as triple knockout (TKO) samples, and cells infected with Ad-GFP are denoted as control samples.
(B) Expression values of the Rb transcript in TKO samples compared to control samples by qPCR. Data are reported as mean ± SEM. n = 5 animals.
(C) Confocal images of fixed TKO MoNuc and BiNuc CMs stained with antibodies against a-actinin and GFP. Scale bar, 20 μm.
(D) Quantification of the ratio of BiNucs to MoNucs and percentages of MoNucs and BiNucs in GFP+ control and TKO CMs fixed and stained with antibodies against α-actinin and GFP. n = 3 animals. Data are reported as mean ± SEM. p values are determined by Student’s t test.
(E) Gene expression differences between control and TKO samples of E2f target genes and non-E2f target genes by qPCR. Data are reported as mean ± SEM. n = 5 animals. p values are determined by Student’s t test.
(F) Comparison of mean fold change expression of TKO samples over control samples of E2f target genes versus non-E2f target genes. p values are determined by Student’s t test. ns, not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Binucleation Directly Results in the Downregulation of E2f Target Genes
Our data show that the downregulation of E2f target genes, a process that requires Rb, occurs in BiNucs at P7, but is not responsible for inducing binucleation. To determine whether these transcriptional changes instead occur as a direct result of binucleation, we used a mouse model in which we induced increased binucleation of CMs through a CM-specific genetic deletion of Ect2, a gene required for cytokinesis (Cook et al., 2011). We generated an Ect2flox/flox allele and crossed this into both the Nkx2.5cre and Mlc2vcre lines to inactivate Ect2 at different time points of CM development. Nkx2.5cre:Ect2flox/flox mice did not survive beyond E10.5 (Figures 6A, 6C, and 6D). At E10.5, Nkx2.5cre:Ect2flox/flox mice had altered myocardial morphology and underwent a significant decrease in ventricular Ect2 expression (Figures 6B–6D). E10.5 Nkx2.5cre:Ect2flox/flox mutant hearts primarily comprised BiNucs, while control hearts comprised almost exclusively MoNucs. The mean ratio of BiNucs to MoNucs increased from 0.026 ± 0.01 in control hearts to 3.716 ± 0.07 in mutant hearts (Figures 6E and 6F). Mlc2vcre:Ect2flox/flox mutants are viable, likely due to the later onset and mosaic nature of Cre-mediated recombination driven by the Mlc2vcre line during cardiac development (Gillers et al., 2015; Guimarães-Camboa et al., 2015; Figure 7A). During neonatal development, there is a rapid increase in binucleated CMs between P4 and P7 (Soonpaa et al., 1996). Therefore, we examined Mlc2vcre:Ect2flox/flox mutants at P3, at which time they exhibited a 61.8% decrease in Ect2 expression compared to controls (Figure 7B). Mlc2vcre:Ect2flox/flox mutants had an increase in the mean ratio of BiNucs to MoNucs at P3 of 0.901 ± 0.08 compared to 0.339 ± 0.02 in control mice (Figures 7C and 7D). This result agrees with recent work by Liu et al. (2019), which also showed that the loss of Ect2 results in an increase in BiNucs during the neonatal period and that increased expression of Ect2 leads to increased MoNucs. qPCR revealed a significant decrease in the expression of E2f target genes in Mlc2vcre: Ect2flox/flox mutants compared to controls, but no significant change in genes that are not E2f targets (Figure 7E). These data suggest that Rb-mediated downregulation of E2f target genes occurs as a direct result of the binucleation process in CMs. To examine whether these events impair the regenerative ability of the neonatal heart, we performed left anterior descending (LAD) ligation to induce a myocardial infarction in Mlc2vcre:Ect2flox/flox mutants and control mice at P1. When injured at this developmental time point, the murine heart is still able to mount a substantial regenerative response (Porrello et al., 2011). However, the higher percentage of BiNucs that we observed in Mlc2vcre:Ect2flox/flox mutants at P3 would be expected to impair the regenerative capacity of the heart. We observed that by P8, Mlc2vcre:Ect2flox/flox mutants exhibited a higher percentage of scar tissue compared to control mice (Figures 7F and 7G). Whether this is due to reduced proliferation, decreased CM survival, or other mechanisms, these data support a model whereby binucleation directly results in decreased competency for neonatal cardiac regeneration.
Figure 6. Loss of Ect2 Results in Binucleation of CMs.
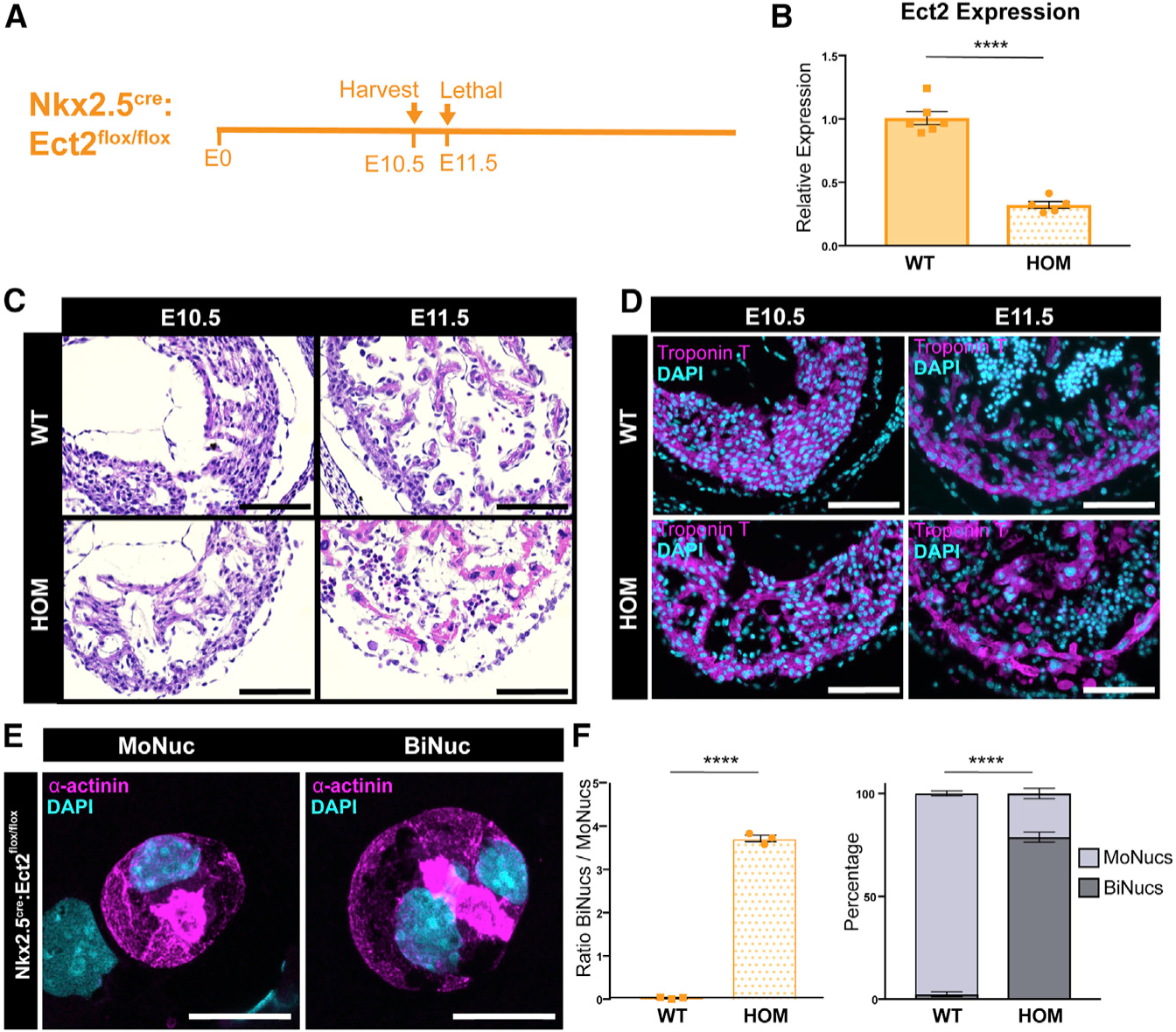
(A) Time course of Nkx2.5cre:Ect2flox/flox KO.
(B) Expression values of the Ect2 transcript in Nkx2.5cre:Ect2flox/flox (HOM; homozygous knockout) samples compared to Ect2flox/flox and Ect2flox/+ (wild-type [WT]) samples at E10.5 by qPCR. Data are reported as mean ± SEM. n = 5–6 animals per group.
(C and D) Representative tissue sections of WT and HOM hearts at E10.5 and E11.5 stained with H&E (C) or an antibody against cardiac troponin T (D) reveals that the Nkx2.5cre:Ect2flox/flox allele is lethal by E11.5. Scale bar, 100 μm. (D) Representative images of a MoNuc and BiNuc present in E10.5 HOM samples. Single-cell suspensions were cytospun and stained with an antibody against α-actinin. Scale bar, 20 μm.
(E) Quantification of the ratio of BiNucs to MoNucs and percentages of MoNucs and BiNucs in E10.5 WT and HOM samples. Data are reported as mean ± SEM. n = 3 animals per group. p values are determined by Student’s t test. ns, not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Figure 7. Binucleation Results in Downregulation of E2f Target Genes and Impairment of Regenerative Potential.
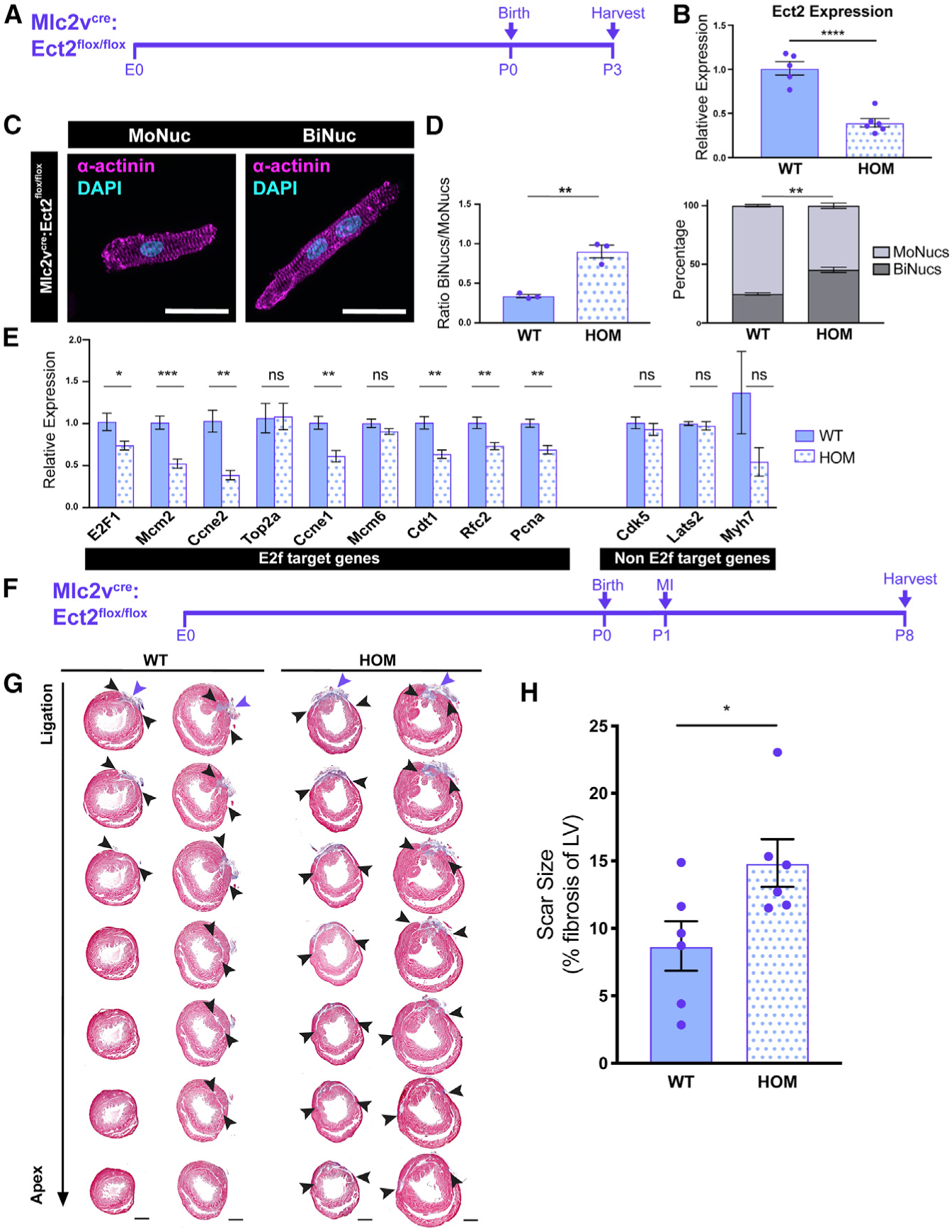
(A) Time course of Mlc2vcre:Ect2flox/flox KO.
(B) Expression values of the Ect2 transcript in Mlc2vcre:Ect2flox/flox (HOM) samples compared to Ect2flox/flox and Ect2flox/+ (WT) samples at P3 by qPCR. Data are reported as mean ± SEM. n = 5–6 animals per group.
(C) Images of P3 HOM MoNuc and BiNuc CMs from fixed single-cell suspensions stained with anti-α-actinin. Scale bar, 20 μm.
(D) Quantification of the ratio of BiNucs to MoNucs and percentages of MoNucs and BiNucs in P3 WT and HOM samples. Data are reported as mean ± SEM. n = 3 animals per group.
(E) Gene expression differences between WT and HOM samples of E2f target genes and non-E2f target genes. Data are reported as mean ± SEM. n = 5–6 animals per group. p values are determined by Student’s t test. ns, not significant.
(F) Schematic showing study design of myocardial infarction (MI) by ligation of the left anterior descending (LAD) artery in Mlc2vcre:Ect2flox/flox mutant and control mice at P1.
(G) Masson’s trichrome-stained heart sections from the ligation site toward the apex 7 days after injury. n = 6 per group.
(H) Quantification of the percentage of left ventricular myocardium containing scar tissue at P8. Suture location is indicated with the purple arrow. Scar boundaries are indicated with black arrows. Scale bar, 500 μm. Data are reported as mean ± SEM. n = 6 animals per group. p values are determined by Mann-Whitney U test. ns, not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
DISCUSSION
A lack of effective therapeutic strategies to treat the acutely injured heart has led scientists to search for a means to replace CMs that have been lost due to injury. One compelling approach would be to develop therapies that activate endogenous CM proliferation to regenerate damaged tissue, similar to what occurs in lower vertebrates. While previous work has suggested a correlation between CM nucleation and the ability to re-enter the cell cycle, with MoNucs being more receptive to proliferative stimuli, the molecular basis for this difference has remained elusive due to the technical hurdles in isolating and independently characterizing MoNucs and BiNucs. Our method for isolating these two different CM populations, combined with analysis of their unique transcriptional profile, reveals that MoNucs exhibit an E2f-related gene signature that emerges during the early postnatal period and is retained into adulthood. These studies provide a methodological advancement that has allowed us to begin to characterize the molecular differences in MoNucs and BiNucs. Such information may eventually be leveraged in manipulating MoNucs to enhance cardiac regeneration.
The MoNuc and BiNuc CM subsets diverge during a neonatal maturation period, at which time CMs exit the cell cycle and undergo terminal differentiation. Our data revealed that MoNucs and BiNucs are most distinct during the neonatal period, as these subsets begin to diverge. At P7, binucleation was associated with the termination of a proliferation-associated gene expression program in exchange for a mature CM gene expression program. While this programmatic switch has long been associated with the neonatal period, our findings show that by P7 only BiNucs have begun to undergo this shift, while MoNucs remain transcriptionally immature. While our data show that MoNucs eventually assume a similar mature gene expression program, they appear to do so at a later time and not as a result of binucleation. These findings suggest that the mechanism that couples binucleation to cell-cycle exit in BiNucs also contributes to the increased loss of proliferative competency observed in this CM subpopulation.
E2f target genes make up a large portion of the most dramatically downregulated genes in P7 BiNucs, some of which remain downregulated in adult BiNucs. The E2f transcription factor family is known to regulate the expression of genes required to initiate DNA synthesis (Johnson et al., 1993). While CMs continue to undergo hypertrophic growth after the neonatal maturation period, they do not enter into S phase under physiological conditions. E2f activity is known to be regulated by Rb, and this interaction can have temporary and long-term control over entry into S phase (Harrington et al., 1998; Lundberg and Weinberg, 1998; Nevins, 1992). While the phosphorylation state of Rb dictates temporary repression of S phase entry during the cell cycle, other post-translational modifications to Rb regulate its ability to irreversibly terminate the cell cycle (Kwon et al., 2017; Markham et al., 2006; Saddic et al., 2010).
Previous work has also shown that Rb family members are required for the cell-cycle exit of CMs during the postnatal period (MacLellan et al., 2005). This offers an intriguing potential mechanism by which binucleation could be coupled to irreversible cell-cycle exit.
While the loss of Rb family members did not result in a difference in the ratio of BiNucs to MoNucs, it did result in the increased expression of E2f target genes depleted in P7 BiNucs. This suggests that in neonatal CMs, Rb is required for downregulating E2f target gene expression and does not itself regulate the onset of binucleation. By altering the ratio of BiNucs to MoNucs during the maturation process with a CM-specific Ect2 KO mouse model, we demonstrated that increased binucleation led to decreased expression of E2f target genes. These data support the hypothesis that silencing E2f target genes is a direct result of binucleation.
In senescent cells, the regulation of irreversible cell-cycle exit by Rb includes the recruitment of chromatin-modifying proteins that alter the architecture of E2f target genes (Blais et al., 2007; Frolov and Dyson, 2004). CpG methylation changes and histone modifications to E2f target genes are also known to occur during the process of CM maturation (Gilsbach et al., 2018; Sdek et al., 2011). One possible explanation for our results is that in response to binucleation, Rb may initiate chromatin modifications at E2f target genes that ultimately contribute to the difference in proliferative competency between BiNucs and MoNucs. This possibility is in line with recent work showing that E2f transcription factor networks are among those that undergo extensive chromatin remodeling during the postnatal period and therefore fail to reactivate in adult CMs after injury (Quaife-Ryan et al., 2017). Our work provides a potential mechanism whereby these changes are initiated.
While a majority of adult murine CMs are binucleated, the majority of adult CMs in the human heart are thought to be mononucleated and polyploid (Bergmann et al., 2011; Brodsky et al., 1980). Therefore, the applicability of our findings to the human heart remains to be determined. However, the processes resulting in binucleation and polyploidy share similarities and both can result from failed cell division, whether at the karyokinesis stage or the cytokinesis stage. Karyokinesis and cytokinesis are closely coupled processes and are orchestrated by a shared group of molecular regulators (Jeyaprakash et al., 2007; McCollum, 2004). Therefore, it is possible that whether destined to become polyploid or binucleated, human and mouse CMs share similar molecular mechanisms responsible for, and in response to, the failure of CMs to complete cell division.
Our results suggest that binucleation may act as a switch during the neonatal period to shut down a proliferative gene expression program in exchange for the expression of genes required for CM maturation. This possibility is further supported by the recent work of Hirose et al. (2019), which shows that increasing the percentage of diploid MoNucs by the expression of a dominant-negative thyroid hormone receptor-a is associated with the upregulation of E2f signaling and the downregulation of oxidative phosphorylation gene expression. We observed that adult MoNucs and BiNucs retain the remnants of some of the differences established during the neonatal period, including differences in E2f target gene expression. Our technique to isolate MoNucs and BiNucs should allow for further studies into the differences between these CM subpopulations and whether these differences can be leveraged to more effectively promote cardiac regeneration.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Edward E. Morrisey (emorrise@pennmedicine.upenn.edu). This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
R26REYFP (Jackson Labs; cat# 006148) (Srinivas et al., 2001), ROSA26FLPe (Jackson Labs; cat# 009086) (Farley et al., 2000), Nkx2.5cre (Jackson Labs; cat# 024637) (Stanley et al., 2002), and Mlc2vcre(Jackson Labs; cat# 029465) (Chen et al., 1998) mice were purchased from Jackson Labs and have been previously described. To generate the Ect2flox/flox allele, cryopreserved sperm from the Ect2tm1a(EUCOMM)Wtsi mouse line was purchased from the Institut Clinique de la Souris (Illkirch, France). The mouse line was derived through in-vitro fertilization by the Transgenic and Chimeric Mouse Facility at the University of Pennsylvania. The resulting mouse was then bred with the ROSA26FLPe allele to remove the neomycin cassette and generate a conditional allele with LoxP sites flanking exon 13. For cardiomyocyte sorting experiments, Adult (8–13 weeks), P7, or E18.5 Mlc2vcre/+:R26REYFP/+ offspring were generated by breeding female R26REYFP/EYFP mice with Mlc2vcre/+ males. For Ect2 experiments, Ect2flox/flox mice were bred to either Mlc2vcre/+ or Nkx2.5cre/cre to generate mice heterozygous for both alleles. Resulting mice were then bred with Ect2flox/+ mice to generate litters used in experiments. Genotyping was performed with the primers in Table S1. A mix of male and female age-matched and litter-matched healthy mice were used for all experiments. No animals were used for more than one experiment. Animals were housed in breeding pairs or with sex-matched littermates in the small animal vivarium at the Smilow Center for Translational Research at the Perelman School of Medicine and maintained in a 12-h light, 12-h dark cycle with unlimited access to food and water. All animal procedures were approved by the University of Pennsylvania Institutional Animal Care and Use Committee under protocol #806345.
METHOD DETAILS
Isolation of Cardiomyocytes for FACS
Isolation of Adult Cardiomyocytes
Cardiomyocytes were isolated from mice aged 8–13 weeks using a protocol modified from methods previously described (Judd et al., 2016; Robison et al., 2016; Tian et al., 2015). Buffers were prepared on the day of isolation as follows. Cell Isolation Buffer (CIB): 130 mM NaCl (Sigma; S6191), 1 mM Lactic acid (Sigma; L1750), 5.4 mM KCl (Sigma; P3911), 25 mM HEPES (Bioworld; 0820000–3), 0.50 mM MgCl2 (Sigma; M9272), 0.33 mM NaH2PO4 (Sigma; S9638), 22 mM Glucose (Sigma; 16301), 20 mM Creatine Monohydrate (GNC; 350526), brought to pH 7.4 with 10M NaOH. 10X CIB stock excluding Glucose, Creatine, and NaOH was previously prepared and frozen in aliquots; Digestion buffer: CIB with 60 U/mL Collagenase Type II (Worthington; LS004176) and 0.025 mg/mL Protease Type XIV (Sigma; P5147); Dissociation Buffer: CIB with 1 mg/mL BSA (Omnipor; 2960). Mice were injected with 100 USP units Heparin (Abraxis; 401586B) 25 minutes prior to removal of heart. Mice were terminally anesthetized with Isoflurane by placement in an induction chamber containing 4% isoflurane followed by additional 2% isolflurane inhaled via nose cone for maintenance. After achieving deep anesthesia, as confirmed from lack of eye blink and toe pinch reflex, the chest was opened via bilateral thoracotomy and the heart was removed and placed in ice cold CIB. Extraneous connective tissue was removed and the heart was cannulated via the aorta, secured with 5–0 silk suture, and mounted on a Langendorf apparatus. The heart was then perfused with CIB for 3 min at a temperature of 37°C and a constant flow rate of 3 ml/min, followed by perfusion with Digestion Buffer for 14 minutes. The ventricles were then removed from the Langendorf apparatus and placed in Dissociation Buffer at a temperature of 37°C and gently teased apart with forceps. Tissue was then triturated with a transfer pipette and the cell suspension was passed through a 100 μm cell strainer. To begin gradually adding back calcium, an equal volume of Dissociation Buffer supplemented with 200 μM Ca2+ was then added dropwise to the cell suspension. Cardiomyocytes were enriched by centrifugation at 300 RPM for 3 min and resuspended in Dissociation Buffer supplemented with 250 μM Ca2+. Cardiomyocytes were then allowed to settle by gravity sedimentation for 20 minutes at 37°C and resuspended in 4 mL Dissociation Buffer with 500 μM Ca2+. Vybrant DyeCycle Violet (Invitrogen; V35003) was added to the cell suspension at 1 μL/mL and cells were incubated at 37°C for 30 minutes prior to FACS.
Isolation of P7 Cardiomyocytes
Buffers were prepared on the day of isolation as follows. Cell Isolation Buffer (CIB) was the same as used for isolation of adult cardiomyocytes; P7 Digestion Buffer: CIB with 506 U/mL Collagenase Type II and 0.52 mg/mL Trypsin (Fisher; J63993, 1:250); P7 Dissociation Buffer: CIB with 506 U/mL Collagenase Type II and 0.52 mg/mL Trypsin, 10% Horse Serum (GIBCO; 16050), 5% Fetal Bovine Serum (GIBCO; 10437); P7 Sort Buffer: CIB with 0.66% Horse Serum, and 0.33% Fetal Bovine Serum. Hearts were removed from 7 day old mice and placed in ice cold CIB. YFP+ hearts were identified and used for cell isolation. Under a dissection microscope, atria and extraneous tissue was removed and ventricles were cut into fourths. Each heart was then placed in 2 mL P7 Digestion Buffer and incubated for 30 minutes at 37°C with rocking. Each heart was then transferred to 2 mL P7 Dissociation Buffer supplemented with 60 μM Ca2+ where it was triturated with a transfer pipette followed by a 1000 μL pipette tip. Cell suspensions were then combined and passed through a 70 μm cell strainer. Cell suspension was then centrifuged twice for 5 minutes at 500 RPM and resuspended first in P7 Dissociation Buffer supplemented with 140 μM Ca2+, then in 4 mL P7 Sort Buffer supplemented with 240 μM Ca2+. Vybrant DyeCycle Violet was added to the cell suspension at 1 μL/mL and cells were incubated at 37°C for 30 minutes prior to FACS.
Isolation of E18.5 Cardiomyocytes
Buffers were prepared on the day of isolation as follows. E18.5 Digestion Buffer: HBSS (GIBCO; 14170) with 10 mM HEPES (Invitrogen; 15630), 0.54 mM EDTA (Invitrogen; 15575) and 2 mg/mL Trypsin; E18.5 Dissociation Buffer: HBSS with 10mM HEPES, 10% Horse Serum, 5% Fetal Bovine Serum; E18.5 Sort Buffer: Opti-MEM (GIBCO; 31985) with 0.66% Horse Serum, and 0.33% Fetal Bovine Serum. Time pregnant females were euthanized by CO2 inhalation. Embryos were removed and hearts were excised and placed in ice cold HBSS. YFP+ hearts were identified and used for cell isolation. Under a dissection microscope, atria and extraneous tissue was removed and ventricles were splayed open. Each heart was then placed in 2 mL E18.5 Digestion Buffer and incubated for 10 minutes at 37°C with rocking. Each heart was then transferred in a drop of Digestion buffer to a well of a 12 well plate and incubated for 30 minutes. 2 mL E18.5 Dissociation Buffer was then added to each well and the heart was triturated with a 1000 μL pipette tip. Cell suspensions were then combined and passed through a 70 μm cell strainer. Cell suspension was then centrifuged for 5 minutes at 500 RPM and resuspended in 4 mL E18.5 Sort Buffer. Vybrant DyeCycle Violet was added to the cell suspension at 1 μL/mL and cells were incubated at 37°C for 30 minutes prior to FACS.
Sorting Mononucleated and Binucleated Cardiomyocytes by FACS
Cell sorting was performed on a MoFlo Astrios EQ (Beckman Coulter) at room temperature. Embryonic and neonatal cardiomyocytes were sorted using a 100 um nozzle while adult cardiomyocytes were sorted using a 150 um nozzle with a maximum pressure difference of 1. Cardiomyocytes from Mlc2vCre:R26REYFP mice were gated on YFP presence with a 488 nm laser. Nucleation was gated on height and width of signal from a 405 nm laser as described above. Prior to collection, 50–100 cells within each gate were sorted onto a slide and imaged with the DAPI, GFP, and transmitted light channels of an EVOS FL Auto imaging system microscope (ThermoFisher; AMAFD1000) to ensure correct separation of mononucleated and binucleated cells. If necessary, gates were adjusted until the targeted cells were correctly identified. For RNA, an equal number of mononucleated and binucleated cardiomyocytes were sorted into Trizol-LS (LifeTech; 10296028).
RNA Isolation for RNA-seq and qPCR
RNA was isolated by phenol-chloroform extraction followed by precipitation with 75% Ethanol. RNA was then purified using the RNeasy micro Kit (QIAGEN; 74004) as follows. Solution containing precipitated RNA was applied to an RNeasy MinElute Spin Column and washes were done according to the manufacturer’s protocol. RNA was then eluted in 20 μL H2O. RNA to be sequenced was tested for integrity via Bioanalyzer using the RNA 6000 Pico Kit (Agilent; 5067). RNA for qPCR was quantified using a Nanodrop One (Thermo) and cDNA was generated using the Superscript IV First-Strand Synthesis System (ThermoFisher; 18090050). Quantitative Real-Time PCR was performed on a QuantStudio 7 Flex with Power SYBR Green PCR Mastermix (Applied Biosystems; 4367659) with the primers listed in Table S2.
RNA Sequencing and Analysis
Library prep was conducted using Illumina truSeq stranded mRNA kit and Clontech SMARTer RNA-seq amplification kit. Fastq files were assessed for quality control using the FastQC program. Fastq files were aligned against the mouse reference genome (mm9) using the STAR aligner (Dobin et al., 2013). Duplicate reads were flag using the MarkDuplicates program from Picard tools. Per gene read counts for Ensembl (v67) gene annotations were computed using the R package with duplicate reads removed. Gene counts represented as counts per million (CPM) were first nominalized using TMM method in the edgeR R package and genes with 25% of samples with a CPM < 1 were removed and deemed low expressed. The data were transformed using the VOOM function from the limma R package (Law et al., 2014). Differential gene expression was performed as a paired analysis between mono-nuclear and bi-nuclear samples using limma. To perform the pairwise test the subject correlation was first computed using duplicate Correlation() function, the linear model was then fit using the subject as a blocking variable and adjusting for the subject correlation. Given the small sample size of the experiment, we employed the empirical Bayes procedure as implemented in limma to adjust the linear fit and calculate p values. p values were adjusted for multiple comparisons using Benjamini-Hochberg procedure. Heatmaps and PCA plots were generated in R. Gene Ontology enrichment analysis was performed using the ToppGene Suite (https://toppgene.cchmc.org/) (Chen et al., 2009).
Surgical Procedures
To induce MI in neonatal mice, we permanently ligated the LAD artery on postnatal day 1 (P1) as previously described (Leach et al., 2017). To minimize stress and reduce the likelihood of the pups being rejected by the nursing dam, only half of the litter was removed prior to commencing surgery. Pups were housed in an incubation chamber to provide warmth throughout and after the surgical procedure. The dam was housed away from the operating table during the procedure. P1 mice were anesthetized by cooling on ice. Once the appropriate plane of anesthesia is reached, the neonate was removed from the ice for the surgical procedure. Appropriate anesthesia was recognized by lack of breathing, movement, and reflexes. The pup was placed atop a firm operating surface in the supine position. Forelimbs and tail were held in position using masking tape. A dissecting microscope with a fibro-optic light was used for visualization throughout the surgery. A lateral skin incision was then made on the left side of the chest, just below the insertion of the left forelimb, using fine scissors and forceps. Blunt dissection was used to expose the ribs, and a lateral thoracotomy was performed at the fourth intercostal space by blunt dissection of the intercostal muscles following skin incision. The left anterior descending coronary artery (LAD) was identified under the microscope. 8–0 nylon suture was then sutured across the LAD and permanently ligated. The visualization of the whitish color of left ventricular apex assured the effective ligation of LAD. Following the LAD ligation, the thoracotomy was closed by re-approximating the ribs using 6–0 non-absorbable suture. The skin incision was then closed using Vetbond, a skin adhesive glue. The entire procedure took less than 10 minutes per mouse. The instruments were sterilized between procedures using a hot bead sterilizer. After surgery, pups were brought back to body temperature by placing them in a 33C incubator for 20–30 minutes.
In Vitro Neonatal Cardiomyocyte Culture Experiments
Cardiomyocytes were isolated from Rbflox/flox;p130flox/flox;p107−/− pups at postnatal day 1. Buffers and protocol were as described for isolating E18.5 cardiomyocytes with the following modifications. Rinse buffer: DMEM with 10% Horse Serum, 5% Fetal Bovine Serum, 1% Pen/Strep. High Serum Culture Media: Opti-MEM with 10% Horse Serum, 5% Fetal Bovine Serum, 1% Pen/Strep. Low Serum Culture Media: Opti-MEM with 0.7% Horse Serum, 0.3% Fetal Bovine Serum, 1% Pen/Strep. Each heart was placed in 2 mL Digestion Buffer and incubated overnight at 4°C with rocking instead of 10 minutes at 37°C. Following dissociation, cell suspensions were washed with Rinse Media then resuspended in High Serum Culture media and passed through a 70 μm cell strainer. Fibroblasts were depleted by plating cell suspensions for 2 hours on cell culture plastic. Cell suspensions were then collected and cells counted with a hemocytometer. 75,000 cells in High Serum Culture Media were plated in per well of a 48 well plate, precoated overnight with 100 μg/mL laminin. The following day, each well of cells was treated with 5×10^6 PFU of adenovirus expressing either GFP (Ad-GFP) (Vector Biolabs; 1060) or both Cre recombinase and GFP (Ad-Cre-GFP) (Vector Biolabs; 1700). The morning after adenoviral infection, media was changed to Low Serum Culture Media for 24 hours to inhibit growth of fibroblasts. 36 hours after adenoviral infection, media was changed back to High Serum culture media and cells were cultured for an additional 36 hours. 72 hours after adenoviral infection, cells were harvested in Trizol for RNA as described above or fixed for staining to quantify nucleation.
Staining
Single cell suspensions of cardiomyocytes were fixed in 4% PFA and permeabilized in PBS with 0.5% Triton X-100. SEA BLOCK blocking buffer (ThermoFisher; 37527) was used for blocking, diluting antibodies, and washes. Cells were incubated with primary antibody overnight at 4°C followed by secondary antibody for 1 hour at room temperature. Cells were then incubated with DAPI, pelleted, resuspended in ProLong Diamond Antifade Mountant (ThermoFisher; P36961) and mounted on slides. Images were acquired with the Nikon Eclipse Ni-E microscope and mononucleated and binucleated cells were counted using Fiji Software. For Histology, tissues were fixed in 4% paraformaldehyde, dehydrated through a series of ethanol washes, and embedded in paraffin. Tissue was sectioned at 6 μm intervals and stained with either hematoxylin and eosin, Masson’s trichrome, or the following antibodies: Sarcomeric α-Actinin (Sigma; A7811), Troponin T-C (Santa Cruz; sc-8121), GFP (Abcam; ab6673).
Electron Microscopy
Cardiomyocytes for electron microscopic examination were sorted directly into fixative containing 2.5% glutaraldehyde, 2.0% paraformaldehyde in 0.1M sodium cacodylate buffer, pH 7.4, overnight at 4°C. After subsequent buffer washes, the samples were post-fixed in 2.0% osmium tetroxide for 1 hour at room temperature, and rinsed in DH2O prior to en bloc staining with 2% uranyl acetate. After dehydration through a graded ethanol series, the tissue was infiltrated and embedded in EMbed-812 (Electron Microscopy Sciences, Fort Washington, PA). Thin sections were stained with uranyl acetate and lead citrate and examined with a JEOL 1010 electron microscope fitted with a Hamamatsu digital camera and AMT Advantage image capture software.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of Scar Size
For each animal, scar size was measured in Masson’s trichrome–stained heart sections. Every 10th section from the site of the suture to the apex was analyzed. Scar size was measured as the percent fibrosis of the left ventricle myocardium using MIQuant, an automated image segmentation open source code for MATLAB (Nascimento et al., 2011). The average of two independent measurements of scar size are reported. The initial measurement was not obtained blind, to ensure reproducibility a second investigator measured scar size blind to genotype.
Statistical Analysis
Data are reported as Mean ± SEM. Statistical analysis was performed in Prism 7 for Mac. Unless otherwise specified, an unpaired Student’s t test was used to compare two experimental groups. For injury studies a Mann-Whitney U test was used. Data were considered significant if p < 0.05.
DATA AND CODE AVAILABILITY
The sequencing data in this manuscript is available through the accession number GEO: GSE140851
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Troponin T-C | Santa Cruz | Cat#sc-8121; RRID:AB_2287642 |
| GFP | Abcam | Cat#ab6673; RRID:AB_305643 |
| Sarcomeric a-Actinin | Sigma | Cat#A7811; RRID:AB_476766 |
| Bacterial and Virus Strains | ||
| Cre Recombinase Adenovirus (Ad-Cre-GFP) | Vector Biolabs | Cat#1700 |
| eGFP Adenovirus (Ad-GFP) | Vector Biolabs | Cat#1060 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Vybrant DyeCycle Violet | Invitrogen | Cat#V35003 |
| Critical Commercial Assays | ||
| Superscript IV First-Strand Synthesis System | ThermoFisher | Cat#18090050 |
| Power SYBR Green PCR Mastermix | Applied Biosystems | Cat#4367659 |
| RNeasy micro Kit | QIAGEN | Cat#74004 |
| RNA 6000 Pico Kit | Agilent | Cat#5067 |
| Deposited Data | ||
| RNA-Sequencing | N/A | GEO: GSE140851 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Nkx2.5cre | Jackson Labs | Cat#024637 |
| Mouse: ROSA26FLPe | Jackson Labs | Cat#009086 |
| Mouse: Mlc2vcre | Jackson Labs | Cat#029465 |
| Mouse: Ect2fl/fl: Ect2tm1a(EUCOMM)Wtsi | Institut Clinique de la Souris | MGI:4433090; RRID:IMSR_EM:09964 |
| Mouse: R26REYFP | Jackson Labs | Cat#006148 |
| Oligonucleotides | ||
| Genotyping Primers | See Table S1 | N/A |
| qPCR Primers | See Table S2 | N/A |
| Software and Algorithms | ||
| Prism 7.0 | Graphpad | GraphPad Prism, RRID:SCR_002798 |
| Fiji | ImageJ | Fiji, RRID:SCR_002285 |
| R 3.2 | R Project | R Project for Statistical Computing, RRID:SCR_001905 |
| Picard Tools | Broad Institute | Picard, RRID:SCR_006525 |
| GAGE | PMCID: PMC2696452, Bioconductor | GAGE, RRID:SCR_017067 |
| NMF | R package | https://cran.r-project.org/web/packages/NMF/index.html |
| FactoMineR | R package | FactoMineR, RRID:SCR_014602 |
| STAR 2.5 | PMCID: PMC3530905 | https://github.com/alexdobin/STAR |
| Limma | PMCID: PMC4402510, Bioconductor | https://bioconductor.org/packages/release/bioc/html/limma.html |
| MIQuant | PMCID: PMC3184116 | https://paginas.fe.up.pt/~quelhas/MIQuant/MIQuant.zip |
| ToppGene Suite | PMCID: PMC2703978 | ToppGene Suite, RRID:SCR_005726 |
Highlights.
A FACS-based strategy separates mononucleated and binucleated cardiomyocytes
Mononucleated and binucleated cardiomyocytes are transcriptionally distinct
Binucleation results in the silencing of E2f transcriptional targets
Increased binucleation due to the loss of Ect2 impairs regenerative potential
ACKNOWLEDGMENTS
We would like to thank F. Tuluc, J. Murray, L. Wu, J. Lora, A. Albertus, and G. Suplinskas at the Flow Cytometry Core Laboratory at Children’s Hospital of Philadelphia for assistance with flow cytometry; the Cell and Developmental Biology Microscopy Core at the University of Pennsylvania for confocal microscopy services; the Electron Microscopy Resource Laboratory at the University of Pennsylvania for electron microscopy services; and the Histology and Gene Expression Laboratory at the University of Pennsylvania Cardiovascular Institute for histology services. These studies were supported by funding from the NIH (U01-HL134745, to E.E.M., and T32-GM007229 and T32-HL007954, to R.W.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.02.034.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabé-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, et al. (2009). Evidence for cardiomyocyte renewal in humans. Science 324, 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann O, Zdunek S, Alkass K, Druid H, Bernard S, and Frisén J (2011). Identification of cardiomyocyte nuclei and assessment of ploidy for the analysis of cell turnover. Exp. Cell Res 317, 188–194. [DOI] [PubMed] [Google Scholar]
- Bersell K, Arab S, Haring B, and Kühn B (2009). Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell 138, 257–270. [DOI] [PubMed] [Google Scholar]
- Blais A, van Oevelen CJ, Margueron R, Acosta-Alvear D, and Dynlacht BD (2007). Retinoblastoma tumor suppressor protein-dependent methylation of histone H3 lysine 27 is associated with irreversible cell cycle exit. J. Cell Biol 179, 1399–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchet E, Annicotte JS, Lagarrigue S, Aguilar V, Clapé C, Chavey C, Fritz V, Casas F, Apparailly F, Auwerx J, and Fajas L (2011). E2F transcription factor-1 regulates oxidative metabolism. Nat. Cell Biol 13, 1146–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky WY, Arefyeva AM, and Uryvaeva IV (1980). Mitotic polyploidization of mouse heart myocytes during the first postnatal week. Cell Tissue Res. 210, 133–144. [DOI] [PubMed] [Google Scholar]
- Chen J, Kubalak SW, Minamisawa S, Price RL, Becker KD, Hickey R, Ross J Jr., and Chien KR (1998). Selective requirement of myosin light chain 2v in embryonic heart function. J. Biol. Chem 273, 1252–1256. [DOI] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, and Jegga AG (2009). ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37, W305–W311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DR, Solski PA, Bultman SJ, Kauselmann G, Schoor M, Kuehn R, Friedman LS, Cowley DO, Van Dyke T, Yeh JJ, et al. (2011). The ect2 rho Guanine nucleotide exchange factor is essential for early mouse development and normal cell cytokinesis and migration. Genes Cancer 2, 932–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, and Dymecki SM (2000). Widespread recombinase expression using FLPeR (flipper) mice. Genesis 28, 106–110. [PubMed] [Google Scholar]
- Frolov MV, and Dyson NJ (2004). Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J. Cell Sci 117, 2173–2181. [DOI] [PubMed] [Google Scholar]
- Gillers BS, Chiplunkar A, Aly H, Valenta T, Basler K, Christoffels VM, Efimov IR, Boukens BJ, and Rentschler S (2015). Canonical wnt signaling regulates atrioventricular junction programming and electrophysiological properties. Circ. Res 116, 398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilsbach R, Schwaderer M, Preissl S, Grüning BA, Kranzhö fer D, Schneider P, Nührenberg TG, Mulero-Navarro S, Weichenhan D, Braun C, et al. (2018). Distinct epigenetic programs regulate cardiac myocyte development and disease in the human heart in vivo. Nat. Commun 9, 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rosa JM, Sharpe M, Field D, Soonpaa MH, Field LJ, Burns CE, and Burns CG (2018). Myocardial Polyploidization Creates a Barrier to Heart Regeneration in Zebrafish. Dev. Cell 44, 433–446.e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães-Camboa N, Stowe J, Aneas I, Sakabe N, Cattaneo P, Henderson L, Kilberg MS, Johnson RS, Chen J, McCulloch AD, et al. (2015). HIF1α Represses Cell Stress Pathways to Allow Proliferation of Hypoxic Fetal Cardiomyocytes. Dev. Cell 33, 507–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington EA, Bruce JL, Harlow E, and Dyson N (1998). pRB plays an essential role in cell cycle arrest induced by DNA damage. Proc. Natl. Acad. Sci. USA 95, 11945–11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose K, Payumo AY, Cutie S, Hoang A, Zhang H, Guyot R, Lunn D, Bigley RB, Yu H, Wang J, et al. (2019). Evidence for hormonal control of heart regenerative capacity during endothermy acquisition. Science 364, 184–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenishi A, Okayama H, Iwamoto N, Yoshitome S, Tane S, Nakamura K, Obayashi T, Hayashi T, and Takeuchi T (2012). Cell cycle regulation in mouse heart during embryonic and postnatal stages. Dev. Growth Differ 54, 731–738. [DOI] [PubMed] [Google Scholar]
- Jeyaprakash AA, Klein UR, Lindner D, Ebert J, Nigg EA, and Conti E (2007). Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell 131, 271–285. [DOI] [PubMed] [Google Scholar]
- Johnson DG, Schwarz JK, Cress WD, and Nevins JR (1993). Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature 365, 349–352. [DOI] [PubMed] [Google Scholar]
- Jopling C, Sleep E, Raya M, Martí M, Raya A, and Izpisúa Belmonte JC (2010). Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464, 606–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd J, Lovas J, and Huang GN (2016). Isolation, Culture and Transduction of Adult Mouse Cardiomyocytes. J. Vis. Exp (114) 10.3791/54012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn B, del Monte F, Hajjar RJ, Chang YS, Lebeche D, Arab S, and Keating MT (2007). Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat. Med 13, 962–969. [DOI] [PubMed] [Google Scholar]
- Kwon JS, Everetts NJ, Wang X, Wang W, Della Croce K, Xing J, and Yao G (2017). Controlling Depth of Cellular Quiescence by an Rb-E2F Network Switch. Cell Rep. 20, 3223–3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, and Smyth GK (2014). voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach JP, Heallen T, Zhang M, Rahmani M, Morikawa Y, Hill MC, Segura A, Willerson JT, and Martin JF (2017). Hippo pathway deficiency reverses systolic heart failure after infarction. Nature 550, 260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhang CH, Ammanamanchi N, Suresh S, Lewarchik C, Rao K, Uys GM, Han L, Abrial M, Yimlamai D, et al. (2019). Control of cytokinesis by β-adrenergic receptors indicates an approach for regulating cardiomyocyte endowment. Sci. Transl. Med 11, eaaw6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg AS, and Weinberg RA (1998). Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol. Cell. Biol 18, 753–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLellan WR, Garcia A, Oh H, Frenkel P, Jordan MC, Roos KP, and Schneider MD (2005). Overlapping roles of pocket proteins in the myocardium are unmasked by germ line deletion of p130 plus heart-specific deletion of Rb. Mol. Cell. Biol 25, 2486–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham D, Munro S, Soloway J, O’Connor DP, and La Thangue NB (2006). DNA-damage-responsive acetylation of pRb regulates binding to E2F-1. EMBO Rep. 7, 192–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollum D (2004). Cytokinesis: the central spindle takes center stage. Curr. Biol 14, R953–R955. [DOI] [PubMed] [Google Scholar]
- Mollova M, Bersell K, Walsh S, Savla J, Das LT, Park SY, Silberstein LE, Dos Remedios CG, Graham D, Colan S, and Kühn B (2013). Cardiomyocyte proliferation contributes to heart growth in young humans. Proc. Natl. Acad. Sci. USA 110, 1446–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Nũnez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, and Lowe SW (2003). Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113, 703–716. [DOI] [PubMed] [Google Scholar]
- Nascimento DS, Valente M, Esteves T, de Pina Mde.F., Guedes JG, Freire A, Quelhas P, and Pinto-do-Ó P (2011). MIQuant–semi-automation of infarct size assessment in models of cardiac ischemic injury. PLoS One 6, e25045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins JR (1992). E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science 258, 424–429. [DOI] [PubMed] [Google Scholar]
- Oberpriller JO, and Oberpriller JC (1974). Response of the adult newt ventricle to injury. J. Exp. Zool 187, 249–253. [DOI] [PubMed] [Google Scholar]
- Papadimou E, Ménard C, Grey C, and Pucéat M (2005). Interplay between the retinoblastoma protein and LEK1 specifies stem cells toward the cardiac lineage. EMBO J. 24, 1750–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson M, Barske L, Van Handel B, Rau CD, Gan P, Sharma A, Parikh S, Denholtz M, Huang Y, Yamaguchi Y, et al. (2017). Frequency of mononuclear diploid cardiomyocytes underlies natural variation in heart regeneration. Nat. Genet 49, 1346–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, and Sadek HA (2011). Transient regenerative potential of the neonatal mouse heart. Science 331, 1078–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD, Wilson LG, and Keating MT (2002). Heart regeneration in zebrafish. Science 298, 2188–2190. [DOI] [PubMed] [Google Scholar]
- Prats C, Graham TE, and Shearer J (2018). The dynamic life of the glycogen granule. J. Biol. Chem 293, 7089–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaife-Ryan GA, Sim CB, Ziemann M, Kaspi A, Rafehi H, Ramialison M, El-Osta A, Hudson JE, and Porrello ER (2017). Multicellular Transcriptional Analysis of Mammalian Heart Regeneration. Circulation 136, 1123–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison P, Caporizzo MA, Ahmadzadeh H, Bogush AI, Chen CY, Margulies KB, Shenoy VB, and Prosser BL (2016). Detyrosinated microtubules buckle and bear load in contracting cardiomyocytes. Science 352, aaf0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saddic LA, West LE, Aslanian A, Yates JR 3rd, Rubin SM, Gozani O, and Sage J (2010). Methylation of the retinoblastoma tumor suppressor by SMYD2. J. Biol. Chem 285, 37733–37740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JL, Suh Y, and Cuervo AM (2014). Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab. 20, 417–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sdek P, Zhao P, Wang Y, Huang CJ, Ko CY, Butler PC, Weiss JN, and Maclellan WR (2011). Rb and p130 control cell cycle gene silencing to maintain the postmitotic phenotype in cardiac myocytes. J. Cell Biol 194, 407–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, Wu TD, Guerquin-Kern JL, Lechene CP, and Lee RT (2013). Mammalian heart renewal by pre-existing cardiomyocytes. Nature 493, 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro HM (1985). Practical Flow Cytometry (Alan R. Liss).
- Soonpaa MH, Kim KK, Pajak L, Franklin M, and Field LJ (1996). Cardiomyocyte DNA synthesis and binucleation during murine development. Am. J. Physiol 271, H2183–H2189. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, and Costantini F (2001). Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley EG, Biben C, Elefanty A, Barnett L, Koentgen F, Robb L, and Harvey RP (2002). Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3′UTR-ires-Cre allele of the homeobox gene Nkx2–5. Int. J. Dev. Biol 46, 431–439. [PubMed] [Google Scholar]
- Taniura H, Taniguchi N, Hara M, and Yoshikawa K (1998). Necdin, a postmitotic neuron-specific growth suppressor, interacts with viral transforming proteins and cellular transcription factor E2F1. J. Biol. Chem 273, 720–728. [DOI] [PubMed] [Google Scholar]
- Tian Y, Liu Y, Wang T, Zhou N, Kong J, Chen L, Snitow M, Morley M, Li D, Petrenko N, et al. (2015). A microRNA-Hippo pathway that promotes cardiomyocyte proliferation and cardiac regeneration in mice. Sci. Transl. Med 7, 279ra38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viatour P, Somervaille TC, Venkatasubrahmanyam S, Kogan S, McLaughlin ME, Weissman IL, Butte AJ, Passegué E, and Sage J (2008). Hematopoietic stem cell quiescence is maintained by compound contributions of the retinoblastoma gene family. Cell Stem Cell 3, 416–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data in this manuscript is available through the accession number GEO: GSE140851


