Abstract
Objectives:
Chemotherapy remains a cornerstone treatment in non-small cell lung cancer either in combination with checkpoint inhibitors or as subsequent therapy. Identifying molecular predictors of response allows for optimal treatment selection. We performed genomic analysis on tumor samples of patients treated with carboplatin and nab-paclitaxel as part of a phase II trial to evaluate the prognostic and predictive value of mutations in DNA repair pathway in patients treated with this regimen.
Materials and methods:
Next-generation sequencing libraries were produced using a capture-based targeted panel covering the coding exons of 278 genes on patients treated on clinical trial NCT00729612. Overall survival (OS) and progression-free survival (PFS) were assessed as part of the clinical outcomes and correlated with mutation analysis.
Results:
Of 63 patients enrolled, 25 patients had sufficient and acceptable DNA isolated from archival tumor samples for targeted sequencing. The most commonly altered pathways included DNA repair (DR) including Fanconi anemia and homologous recombination, JAK-STAT signaling, IGF-1, mTOR, and MAPK-ERK. Four patients with mutations in homologous recombination mutations had a shorter PFS (hazard ratio [HR] = 4.54, 95% CI 1.2, 17.1, p = 0.026) and OS (HR = 6.3, 95% CI 1.8, 21.3, p = 0.003).
Conclusion:
In this analysis of patients with predominantly squamous cell non-small cell lung cancer treated with carboplatin and nab-paclitaxel in a phase II trial, patients with mutations in homologous recombination pathways had shorter overall and progression-free survival. Validation on additional datasets of patients treated with platinum-based chemotherapy and immunotherapy combinations is warranted.
Keywords: Non-small cell lung cancer, DNA repair, Homologous recombination, Biomarker, Chemotherapy
1. Introduction
Cytotoxic chemotherapy remains an important treatment for patients with non-small cell lung cancer (NSCLC) either alone or in combination with immunotherapy. Studies demonstrating the efficacy of first line treatment with immune checkpoint inhibitors (ICI) alone and in combination with chemotherapy have led to new first-line treatment options for patients with NSCLC [1–3]. Carboplatin, pembrolizumab and either paclitaxel or nab-paclitaxel are now standard first line options for patients with lung squamous cell carcinoma (LSCC) [4]. With multiple treatment options now available, patient selection will be key to guide clinical decision making.
A better understanding of the genomic landscape of NSCLC has led to novel treatments for patients with select targetable mutations (i.e. EGFR, ALK, ROS, and BRAF among others), and in many cases targeted therapy offers improved response rates compared to chemotherapy [5–7]. Targeted therapies have so far shown clinical benefit primarily in patients with lung adenocarcinoma where driver mutations are more common than in patients with LSCC [8]. Mutations in TP53 represent the most common alteration in LSCC; mutations in CDKN2A, PIK3CA, KEAP1, MLL2, NFE2L2, and RB1 are also frequently observed [8,9]. Mutations in MLL2, CDH8, NFE2L2 or RB1 have been associated with a worse prognosis for patients with early stage LSCC [9]. Mutations in NFE2L2 and KEAP1 have also been associated with resistance to chemotherapy and targeted therapies as well as radiation therapy [10,11]. For patients whose tumors do not harbor actionable driver mutations, platinum-based chemotherapy remains a backbone of treatment with or without ICI [12]. For these patients, PD-L1 and tumor mutational burden (TMB) have been the primary biomarkers to guide therapy [2,13]. However, these have been predominantly studied in patients receiving ICI alone, and not in the context of chemotherapy-ICI combinations. There is an ongoing need to develop predictive biomarkers for chemotherapy to enhance patient selection for novel combination therapies.
The mechanism by which platinum agents exert antineoplastic effect is largely through the creation of stable DNA adducts, leading to DNA damage and cell death [14]. This has led to DNA repair mechanisms being evaluated as a possible predictive biomarker for platinum-based therapy [15,16]. Understanding the role of DNA repair pathway mutations in response to platinum based chemotherapy is vital to determine optimal treatment approaches for patients with NSCLC. In this study we retrospectively performed next-generation DNA-sequencing on NSCLC patients who received first-line chemotherapy with carboplatin and nanoparticle albumin-bound (nab)-paclitaxel on an NCCN-sponsored clinical trial to evaluate the predictive value of DNA repair pathway mutations in this patient population.
2. Material and methods
Clinical trial NCT00729612 (NCCN A-08) was a phase II trial of carboplatin and nab-paclitaxel in 63 patients with advanced or metastatic NSCLC conducted at The Ohio State University with the support of the National Comprehensive Cancer Network (NCCN) Oncology Research Program. The methods and results of the clinical study have been reported previously [17]. Briefly, treatment consisted of nab-paclitaxel (300 mg/m2 for the first 40 patients; dose reduced to 260 mg/m2 in the remaining 23 patients) and carboplatin area under the curve (AUC) 6 administered as an intravenous infusion on day 1 of a 21-day cycle. The study was conducted between September 2008 and December 2011. Eligible patients included previously untreated patients with contraindications to bevacizumab therapy, including squamous histology, hemoptysis, thromboembolic disease, requirement for therapeutic anticoagulation, or cavitary lung lesions. Major inclusion criteria included measurable disease, ECOG performance status of 0–2, and adequate organ function. Relevant exclusion criteria included recent major surgery, baseline peripheral neuropathy, or previous chemotherapy. All patients were consented as part of the trial to have their initial diagnostic specimen (i.e pretreatment specimen) evaluated for research purposes. The study was approved by the Institutional Review Board and conducted in accordance with Good Clinical Practice Guidelines.
2.1. DNA sequencing
DNA was purified from archival tumor tissue collected from patients consented to the trial (NCT00729612). Next-generation sequencing libraries were produced using a capture-based targeted panel covering the coding exons of 278 genes and libraries were sequenced to a 215X average depth of coverage [18]. Raw sequence reads were processed and aligned to human reference genome using the Genome Analysis Toolkit (GATK) workflow [19]. Ensembl Variant Effect Predictor (VEP) was used to annotate and determine functional consequences of tumor specific variants [20].
2.2. Statistical analysis
NCT00729612 utilized a 2-stage Simon model. The primary endpoint was response rate measured by RECIST. All clinical data was collected as part of the study including objective response rate (ORR), overall survival (OS) and progression-free survival (PFS). These parameters were correlated with mutation analysis. Mutation pathways were grouped according to prior studies [21,22]. Clinical response to treatment was compared between patients whose best response to treatment was partial response versus those with stable disease or progressive disease as best response per RECIST criteria [23]. Kaplan–Meier method and log-rank tests were used to estimate and test the differences of probabilities in OS and PFS between groups. Hazard ratios were estimated by COX regression model. Analyses were performed for all patients with genomic data (genomic evaluable population, GEP) as well as for only squamous cell carcinoma patients within GEP.
3. Results
A total of 63 patients (mean age: 63 yrs) were accrued to the study, with 54 patients evaluable for response. The majority of patients (n = 48) had squamous cell NSCLC, 42 were male (66.7%), and 57 (90%) were current or former smokers (Supplemental Table 1). As reported previously, the overall response rate was 38% (24 partial responses and no complete responses), with median PFS of 5 months and median OS of 9.7 months [17].
3.1. Targeted DNA sequencing
Sufficient DNA sequencing data was available for 25 patients (the genomic evaluable population, GEP). Two patients came off trial for clinical progression before re-staging scans, and another 3 patients were non-evaluable for RECIST response but had clinical disease progression. Four (16%) patients had no known pathogenic mutations detected. The most frequent mutations were in TP53, occurring in 13 patients (52%). Other mutations detected included PRKDC in 4 (16%) patients; MAP3K1 and EPHA5 (3 patients each, 12%); CREBBP, FANCM, NOTCH3, CSF3R, PTEN, BLM, LRP1B, FBXW7, RAD50 (2 patients each, 8%). The most represented mutated pathways included DNA repair (DR) pathways such as the Fanconi anemia and homologous recombination repair pathways, JAK-STAT signaling pathway, IGF-1 pathway, and MAPK-ERK pathway (Table 1 and Supplemental Table 2).
Table 1.
Partial response rates by mutation pathways among 23 evaluable patients. Fewer partial responses were observed in patients with DNA repair pathway mutations compared to those without (11% vs 50%, p=0.069, Fisher’s Exact test).
| Pathway | Genes Included | Response rate (n,%) in patients with specified mutations | Response rate (n,%) in patients without specified mutations | P-value |
|---|---|---|---|---|
| DNA Repair | ATR, BLM, BRCA2, ERCC2, FANCA, FANCM, MSH6, PROC, RAD50 | 1/9 (11%) | 7/14 (50%) | P = 0.069 |
| JAK-STAT | BTK, CSF3R, JAKI, JAK2, MAP3K1, MAP3K13, MAP3K5, MAP3K9, NOTCH2, NOTCH3, NOTCH4, RACI | 2/10 (20%) | 6/13 (46%) | P = 0.195 |
| IGF-I | CHUK, IGFIR, IGF2R, JAKI, JAK2, PIK3C2G, PIK3CA, PIK3CG | 0/4 (0%) | 8/19 (42%) | P = 0.154 |
| MAPK-ERK | EPHA5, EPHA6, EPHBI, EPHB6, ERBB4, FGF7, FGFR4, FLTI, FLT3, GRIN2A, JAKI, JAK2, MAP3K1, MAP3K13, MAP3K5, MAP3K9, RBI, VEGFA | 2/9 (22%) | 6/14 (43%) | P = 0.290 |
| TP53 | 5/11 (45%) | 3/12 (25%) | P = 0.929 |
3.2. Survival analysis by alterations in non-DNA repair related pathways
When evaluating gene mutations by pathway (Table 1), no pathway alterations were significantly associated with either OS or PFS. For example, 11 patients’ tumors with MAPK-ERK pathway mutations had median OS 17.7 compared to median OS 9.6 months for 14 patients’ tumors without MAPK-ERK mutations (HR 0.69, 95% CI 0.3, 1.6, p = 0.378). Five patients whose tumors had mutations in IGF-1 pathway had median OS of 14.6 months compared to 13.2 months in 20 those without IGF-1 mutations (HR 0.72, 95% CI 0.24, 2.2, p = 0.5635). Eleven patients with tumor mutations in JAK-STAT pathway had median OS 14.6 months compared to median 13.2 months for 14 of those without STAT mutations (HR 0.8, 95% CI 0.35, 1.8, p = 0.609). Furthermore, alterations in JAK-STAT, IGF-1, or MAPK-ERK were not associated with significant differences in response rates.
3.3. DNA repair pathway mutations
In addition to TP53 which is important in mediating DNA repair and genomic integrity, other mutations in DNA repair genes detected included ATR, BLM, BRCA2, ERCC2, FANCA, FANCM, MSH6, PRKDC, and RAD50 (Table 1). Interestingly, patients whose tumors harbored DNA repair pathway mutations had a lower rate of partial response to treatment by RECIST (11% vs 50%), although this did not meet statistical significance (Table 1, p = 0.069). More specifically, in 18 patients with squamous cell carcinoma evaluable for a response, mutations in the DNA repair pathway (p = 0.382) and homologous recombination pathway (BLM, RAD50, BRCA2; p = 0.446) were not associated with objective response to treatment (Fig. 1). Of the four patients with homologous recombination mutations, there were no partial responses observed, and one patient had clinical progression during the first cycle prior to staging scans and was non-evaluable for radiographic response. The remaining three patients had progressive (n = 1) or stable disease (n = 2) as their best response. Thus, these homologous recombination alterations may be contributing to the trend in differences in response rate between tumors with and without DNA repair pathway mutations (Fig. 1).
Fig. 1.
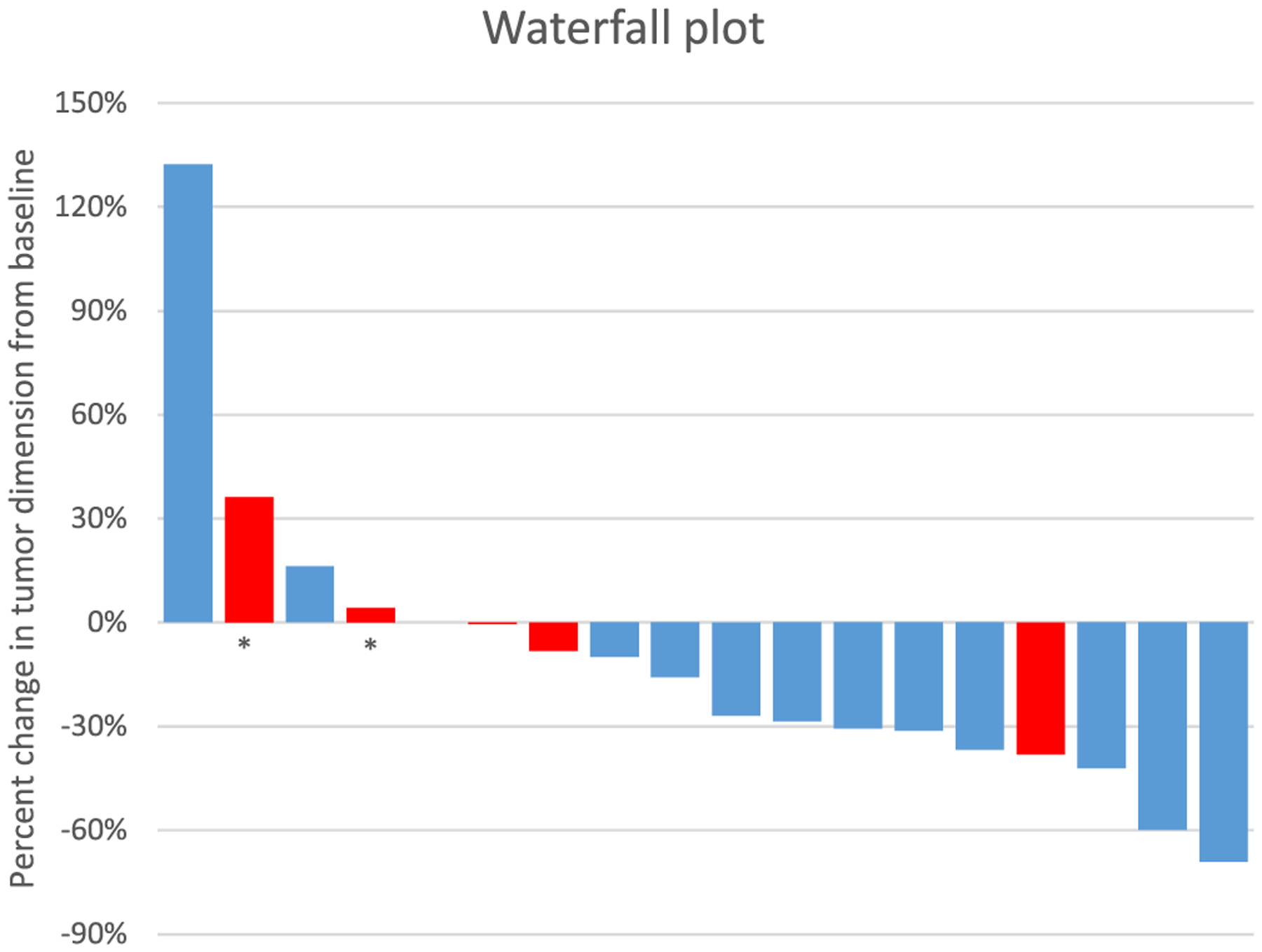
Waterfall plot: Maximum percent change from baseline in the sum of the diameters of target lesions in 18 evaluable patients with squamous cell histology, with patients’ tumors harboring DNA repair mutations present indicated in red. Patients whose tumors possess DNA repair mutations appeared to have less clinical benefit from carboplatin and nab-paclitaxel, although this did not meet statistical significance. Red bars indicate DNA repair mutations are present, asterisk indicates homologous repair mutations.
Overall, within the GEP, there was no difference in PFS or OS in 10 patients with DNA repair mutations (Fig. 2A, C), and there was no difference in PFS or OS in three patients with mutations in the FA pathway. However, four patients whose tumors bear homologous recombination mutations had significantly shorter PFS than patients without homologous recombination mutations (HR 4.5, 95% CI 1.2, 17.1, p = 0.026, Fig. 2B). In addition, these patients exhibited significantly shorter OS (HR 6.3, 95% CI 1.8, 21.3, p = 0.003, Fig. 2D). Two patients with RAD50 mutations had significantly worse OS (p = 0.003). For those patients within the GEP with squamous cell histology, homologous recombination mutations remained significantly associated with OS (HR 4.2, 95% CI 1.1, 16.1, p = 0.035, Fig. 3D) although the association with PFS was weaker (HR 4.2, 95% CI 1.0, 17.9, p = 0.05, Fig. 3B). Finally, an analysis of response and survival based on the presence or absence of TP53 alterations revealed no change in response rate (Table 1) or overall survival (Fig. 3).
Fig. 2.

Kaplan Meier curves for progression free survival (A, B) and overall survival (C, D) within the 25 patient GEP. No statistically significant difference in PFS (A) or OS (C) was observed in patients with tumors possessing mutations in DNA repair pathways. However, patients whose tumors have mutations in the homologous recombination repair pathway had a shorter PFS (B, HR = 4.54, 95% CI 1.2, 17.1, p = 0.026; log-rank p = 0.014), and OS (D, HR = 6.3, 95% CI 1.8, 21.3, p = 0.003; log-rank p = 0.0008).
Fig. 3.

Kaplan Meier curves for progression free survival (A, B) and overall survival (C, D) for only those patients within the GEP with squamous cell histology. No statistically significant difference in PFS (A) or OS (C) was observed in patients with tumors possessing mutations in DNA repair pathways. However, patients whose tumors have mutations in the homologous recombination repair pathway had a shorter OS (D, HR 4.2, 95% CI 1.1, 16.1, p = 0.035; log-rank p = 0.022) although the association with PFS was weaker (B, HR 4.2, 95% CI 1.0, 17.9, p = 0.05, log-rank p = 0.031).
4. Discussion
Chemotherapy remains a cornerstone of the treatment for patients with advanced NSCLC. For patients with LSCC, treatment with carboplatin and nab-paclitaxel has been shown to have increase rate of response compared to carboplatin and paclitaxel, and improvement in survival has been reported in some patient populations [24]. Recently, the addition of immune-checkpoint inhibitors (ICI) to chemotherapy in the first line treatment demonstrated improvement in survival for patients with NSCLC of all histologies compared to chemotherapy alone [1]. Platinum based chemotherapy in combination with ICI is now an approved first line treatment for patients with metastatic NSCLC regardless of histology [1,4]. There remains an urgent need for reliable predictive biomarkers for both chemotherapy and immune-directed therapies. Tumor expression of PD-L1 and TMB have emerged as potential biomarkers for ICI treatment [2,13,25,26]. However, both are imperfect biomarkers due to inter-assay variability and tumor heterogeneity for PD-L1 [27,28] and the lack of standardization for TMB calculation and reporting [13,25]. The role of these biomarkers for patients treated with combination chemotherapy and ICI remains unclear. In this study, we retrospectively evaluated tumor samples from patients treated with carboplatin and nab-paclitaxel in a phase 2 trial to evaluate possible predictive biomarkers. We found that patients whose tumors have mutations in homologous recombination DNA repair pathways had shorter PFS and OS than those without homologous recombination mutations, as well as a trend toward lower response rate. Mutations in other pathways including JAK-STAT, MAPK-ERK, and IGF-1 were not associated with survival or response to treatment.
Aberrations in the genes involved in DNA repair can lead to the development of cancer, and ineffective or defective DNA repair has long been evaluated as a biomarker or target for anti-neoplastic therapy [29]. Mutations leading to defective mismatch repair enzymes results in microsatellite instability which is associated with a response to ICI [30]. In NSCLC, DNA repair pathways have been evaluated as a predictor of clinical benefit from platinum-based chemotherapy. The DNA repair gene excision repair cross-complementation group-1 (ERCC1), a component of the nucleotide excision repair (NER) pathway, has been studied as a predictive biomarker for platinum-based chemotherapy in NSCLC [15,16]. However, encouraging findings of the predictive utility of ERCC1 protein expression were not replicated in subsequent studies, possibly due to differing isoforms of ERCC1 and limitations of the antibodies used for testing [31,32]. DNA repair pathway mutations have been shown to play a role in response to platinum-based chemotherapy in NSCLC including RAD50 [33,34]. Mutations in DNA repair pathways have also been associated with increased neo-antigen load and T-cell infiltration [21], as well as with overall tumor mutational burden in several cancers [35], including NSCLC [36]. These studies support further investigation of DNA repair pathways as a predictor of response to treatment.
In our study, mutations in homologous recombination genes were associated with poor survival and a trend toward lower response rate. It is well established that mutations in germline homologous recombination genes such as BRCA1 and BRCA2 are associated with the development with several cancers including most notably breast and ovarian cancer [37,38]. Treatment with polyadenosine diphosphateribose (PARP) inhibitors is associated with dramatic clinical benefit in both breast [39] and ovarian cancer [40], and several PARP inhibitors are now approved for select patients with these mutations. RAD50 is a key component of the Mre11/Rad50/Nbs1 (MRN) complex which is required for recognition of double-strand breaks, and has also been shown to be involved in cell cycle regulation [41].
Expression of RAD50 protein has also been associated with resistance to radiotherapy in patients with NSCLC [42]. Disruption of the MRN complex due to mutant RAD50 led to increased tumor sensitivity to cisplatin in a murine squamous cell xenograft model [33]. RAD51 protein expression has also been shown to be associated with a worse prognosis in NSCLC [43]. In our study, the two RAD50 mutations identified included R1198G and E605Q. To our knowledge, the association between mutations in HR repair genes including RAD50 being associated with decreased response to treatment, PFS, and OS with carboplatin and nab-paclitaxel combination therapy in patients with NSCLC has not previously been reported.
The rationale for why mutations in homologous recombination genes are associated with worse outcomes is not clear. Most mutations observed in these genes are predicted to be disruptive, leading to loss of function. Thus, decreased tumor cell DNA repair capacity could be hypothesized to lead to increased tumor cell death and better response to DNA damaging therapy with platinum-based combination chemotherapy. On the other hand, defects in homologous recombination might lead to compensatory activation of other repair pathways such as non-homologous end-joining or nucleotide excision repair that ultimately lead to tumors that are “primed” for DNA repair in the face of DNA damaging chemotherapy. Unfortunately, the activation of these repair pathways (and repair capacity) is not able to be reliably measured by DNA sequencing, and are best assessed using functional DNA repair assays in tumor cell lines or fresh tumor tissue. In addition, while activation can sometimes be inferred by immunohistochemistry of phosphorylated DNA repair proteins, sufficient archival tissue did not exist to perform these studies.
There are several important limitations in our study, including the small sample size and the lack of sufficient tissue to perform DNA sequencing on many of the patients enrolled in the clinical trial. The filtering of genetic calls by likely deleterious impact and mutation allelic frequency may also have led to the omission of possibly relevant mutations. These findings in a subset of patients treated with carboplatin and nab-paclitaxel combination therapy may be used to guide studies of patients treated with this regimen in combination with ICI. Lastly, the fact that we observed differences in PFS and OS but not response rate in patients with homologous recombination mutations raises the possibility of these mutations being prognostic but not specifically predictive of response to platinum-based treatment. OS may also be impacted by second and third line treatments which may limit the generalizability of these findings.
5. Conclusion
In the era of first line chemo-immunotherapy for patients with metastatic NSCLC, predictive and reliable biomarkers for response to treatment are more important than ever. In a clinical study of patients with metastatic NSCLC treated with first-line carboplatin and nab-paclitaxel with predominantly squamous cell histology, patients with mutations in homologous recombination mutations had shorter overall and progression-free survival. Patients with mutations in DNA repair genes had a trend toward lower response rate. These findings should be evaluated in other cohorts of patients treated with platinum combination chemotherapy and patients treated with combination chemo-immunotherapy to evaluate the predictive value of these mutations.
Supplementary Material
Funding
Clinical study NCT00729612 was approved and funded in part by the National Comprehensive Cancer Network (NCCN) Oncology Research Program from general research support provided by Celgene.
Abbreviations:
- AUC
area under the curve
- DR
DNA repair
- ICI
immune-checkpoint inhibitor
- LSCC
lung squamous cell carcinoma
- NSCLC
non-small cell lung cancer
- OS
overall survival
- PD-L1
programmed death-ligand 1
- PFS
progression free survival
- TMB
tumor mutational burden
Footnotes
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.lungcan.2019.06.017.
References
- [1].Gandhi L, Rodriguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F, Domine M, Clingan P, Hochmair MJ, Powell SF, Cheng SY, Bischoff HG, Peled N, Grossi F, Jennens RR, Reck M, Hui R, Garon EB, Boyer M, Rubio-Viqueira B, Novello S, Kurata T, Gray JE, Vida J, Wei Z, Yang J, Raftopoulos H, Pietanza MC, Garassino MC, Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer, N. Engl. J. Med 378 (22) (2018) 2078–2092. [DOI] [PubMed] [Google Scholar]
- [2].Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A, Gottfried M, Peled N, Tafreshi A, Cuffe S, O’Brien M, Rao S, Hotta K, Leiby MA, Lubiniecki GM, Shentu Y, Rangwala R, Brahmer JR, Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer, N. Engl. J. Med 375 (19) (2016) 1823–1833. [DOI] [PubMed] [Google Scholar]
- [3].Socinski MA, Jotte RM, Cappuzzo F, Orlandi F, Stroyakovskiy D, Nogami N, Rodríguez-Abreu D, Moro-Sibilot D, Thomas CA, Barlesi F, Finley G, Kelsch C, Lee A, Coleman S, Deng Y, Shen Y, Kowanetz M, Lopez-Chavez A, Sandler A, Reck M, Atezolizumab for first-line treatment of metastatic nonsquamous NSCLC, N. Engl. J. Med 378 (24) (2018) 2288–2301 [DOI] [PubMed] [Google Scholar]
- [4].Paz-Ares L, Luft A, Vicente D, Tafreshi A, Gümüş M, Mazières J, Hermes B, Çay Şenler F, Csőszi T, Fülöp A, Rodríguez-Cid J, Wilson J, Sugawara S, Kato T, Lee KH, Cheng Y, Novello S, Halmos B, Li X, Lubiniecki GM, Piperdi B, Kowalski DM, Pembrolizumab plus chemotherapy for squamous non–small-cell lung cancer, N. Engl. J. Med 379 (21) (2018) 2040–2051. [DOI] [PubMed] [Google Scholar]
- [5].Soria J-C, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, Dechaphunkul A, Imamura F, Nogami N, Kurata T, Okamoto I, Zhou C, Cho BC, Cheng Y, Cho EK, Voon PJ, Planchard D, Su W-C, Gray JE, Lee SM, Hodge R, Marotti M, Rukazenkov Y, Ramalingam SS, Osimertinib in untreated EGFR-mutated advanced non–small-cell lung cancer, N. Engl. J. Med 378 (2) (2018) 113–125. [DOI] [PubMed] [Google Scholar]
- [6].Shaw AT, Kim D-W, Nakagawa K, Seto T, Crinó L, Ahn M-J, De Pas T, Besse B, Solomon BJ, Blackhall F, Wu Y-L, Thomas M, O’Byrne KJ, Moro-Sibilot D, Camidge DR, Mok T, Hirsh V, Riely GJ, Iyer S, Tassell V, Polli A, Wilner KD, Jänne PA, Crizotinib versus chemotherapy in advanced ALK-positive lung cancer, N. Engl. J. Med 368 (25) (2013) 2385–2394. [DOI] [PubMed] [Google Scholar]
- [7].Peters S, Camidge DR, Shaw AT, Gadgeel S, Ahn JS, Kim D-W, Ou S-HI, Pérol M, Dziadziuszko R, Rosell R, Zeaiter A, Mitry E, Golding S, Balas B, Noe J, Morcos PN, Mok T, Alectinib versus Crizotinib in untreated ALK-positive non–small-cell lung cancer, N. Engl. J. Med 377 (9) (2017) 829–838. [DOI] [PubMed] [Google Scholar]
- [8].Comprehensive genomic characterization of squamous cell lung cancers, Nature 489 (7417) (2012) 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Choi M, Kadara H, Zhang J, Moran C, Kalhor N, Rimm D, Kaftan E, Schlessinger J, Lynch TJ, Swisher S, Chow C, Parra ER, Fujimoto J, Rodriguez-Canales J, Wistuba II, Lee J, Townsend JP, Gaffney SG, Zhao Z, Behrens C, Gibbons DL, Heymach J, Lifton RP, Herbst RS, Kim K, Mutation profiles in early-stage lung squamous cell carcinoma with clinical follow-up and correlation with markers of immune function, Ann. Oncol 28 (1) (2016) 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Frank R, Scheffler M, Merkelbach-Bruse S, Ihle MA, Kron A, Rauer M, Ueckeroth F, Konig K, Michels S, Fischer R, Eisert A, Fassunke J, Heydt C, Serke M, Ko YD, Gerigk U, Geist T, Kaminsky B, Heukamp LC, Clement-Ziza M, Buttner R, Wolf J, Clinical and pathological characteristics of KEAP1- and NFE2L2-mutated non-small cell lung carcinoma (NSCLC), Clin. Cancer Res 24 (13) (2018) 3087–3096. [DOI] [PubMed] [Google Scholar]
- [11].Jeong Y, Hoang NT, Lovejoy A, Stehr H, Newman AM, Gentles AJ, Kong W, Truong D, Martin S, Chaudhuri A, Heiser D, Zhou L, Say C, Carter JN, Hiniker SM, Loo BW Jr, West RB, Beachy P, Alizadeh AA, Diehn M, Role of KEAP1/NRF2 and TP53 mutations in lung squamous cell carcinoma development and radiation resistance, Cancer Discov. 7 (1) (2017) 86–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ettinger DS, Aisner DL, Wood DE, Akerley W, Bauman J, Chang JY, Chirieac LR, D’Amico TA, Dilling TJ, Dobelbower M, Govindan R, Gubens MA, Hennon M, Horn L, Lackner RP, Lanuti M, Leal TA, Lilenbaum R, Lin J, Loo BW Jr, Martins R, Otterson GA, Patel SP, Reckamp K, Riely GJ, Schild SE, Shapiro TA, Stevenson J, Swanson SJ, Tauer K, Yang SC, Gregory K, Hughes M, NCCN guidelines insights: non-small cell lung cancer, version 5.2018, J. Compr. Canc. Netw 16 (7) (2018) 807–821. [DOI] [PubMed] [Google Scholar]
- [13].Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, Miller ML, Rekhtman N, Moreira AL, Ibrahim F, Bruggeman C, Gasmi B, Zappasodi R, Maeda Y, Sander C, Garon EB, Merghoub T, Wolchok JD, Schumacher TN, Chan TA, Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer, Science (New York, N.Y.) 348 (6230) (2015) 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bonanno L, Favaretto A, Rosell R, Platinum drugs and DNA repair mechanisms in lung cancer, Anticancer Res. 34 (1) (2014) 493–501. [PubMed] [Google Scholar]
- [15].Lord RV, Brabender J, Gandara D, Alberola V, Camps C, Domine M, Cardenal F, Sanchez JM, Gumerlock PH, Taron M, Sanchez JJ, Danenberg KD, Danenberg PV, Rosell R, Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer, Clin. Cancer Res 8 (7) (2002) 2286–2291. [PubMed] [Google Scholar]
- [16].Shin J, Lee KS, Yoh KA, Cho HJ, Choi MK, Kim SH, Kim YJ, Chung JH, Cho S, Kim K, Jheon S, Yoon HI, Lee JH, Lee CT, Lee JS, Prognostic impact of DNA repair protein expression in non-small cell lung cancers treated with platinum-based chemotherapy and subsequent curative lung resection, Oncology 95 (1) (2018) 20–30. [DOI] [PubMed] [Google Scholar]
- [17].Bertino EM, Williams TM, Nana-Sinkam SP, Shilo K, Chatterjee M, Mo X, Rahmani M, Phillips GS, Villalona-Calero MA, Otterson GA, Stromal caveolin-1 is associated with response and survival in a phase II trial of nab-paclitaxel with carboplatin for advanced NSCLC patients, Clin. Lung Cancer 16 (6) (2015) 466–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Samorodnitsky E, Datta J, Jewell BM, Hagopian R, Miya J, Wing MR, Damodaran S, Lippus JM, Reeser JW, Bhatt D, Timmers CD, Roychowdhury S, Comparison of custom capture for targeted next-generation DNA sequencing, J. Mol. Diagn 17 (1) (2015) 64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, Banks E, Garimella KV, Altshuler D, Gabriel S, DePristo MA, From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline, Curr. Protoc. Bioinformatics 43 (11.10) (2013) 1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F, Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor, Bioinformatics (Oxford, England) 26 (16) (2010) 2069–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chae YK, Anker JF, Bais P, Namburi S, Giles FJ, Chuang JH, Mutations in DNA repair genes are associated with increased neo-antigen load and activated T cell infiltration in lung adenocarcinoma, Oncotarget 9 (8) (2018) 7949–7960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lok BH, Carley AC, Tchang B, Powell SN, RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination, Oncogene 32 (30) (2013) 3552–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, Kaplan R, Lacombe D, Verweij J, New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1), Eur. J. Cancer (Oxford, England: 1990) 45 (2) (2009) 228–247. [DOI] [PubMed] [Google Scholar]
- [24].Socinski MA, Bondarenko I, Karaseva NA, Makhson AM, Vynnychenko I, Okamoto I, Hon JK, Hirsh V, Bhar P, Zhang H, Iglesias JL, Renschler MF, Weekly nab-paclitaxel in combination with carboplatin versus solvent-based paclitaxel plus carboplatin as first-line therapy in patients with advanced non-small-cell lung cancer: final results of a phase III trial, J. Clin. Oncol 30 (17) (2012) 2055–2062. [DOI] [PubMed] [Google Scholar]
- [25].Carbone DP, Reck M, Paz-Ares L, Creelan B, Horn L, Steins M, Felip E, van den Heuvel MM, Ciuleanu T-E, Badin F, Ready N, Hiltermann TJN, Nair S, Juergens R, Peters S, Minenza E, Wrangle JM, Rodriguez-Abreu D, Borghaei H, Blumenschein GR, Villaruz LC, Havel L, Krejci J, Corral Jaime J, Chang H, Geese WJ, Bhagavatheeswaran P, Chen AC, Socinski MA, First-line nivolumab in stage IV or recurrent non–small-cell lung cancer, N. Engl. J. Med 376 (25) (2017) 2415–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hellmann MD, Ciuleanu T-E, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C, Minenza E, Linardou H, Burgers S, Salman P, Borghaei H, Ramalingam SS, Brahmer J, Reck M, O’Byrne KJ, Geese WJ, Green G, Chang H, Szustakowski J, Bhagavatheeswaran P, Healey D, Fu Y, Nathan F, Paz-Ares L, Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden, N. Engl. J. Med 378 (22) (2018) 2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hirsch FR, McElhinny A, Stanforth D, Ranger-Moore J, Jansson M, Kulangara K, Richardson W, Towne P, Hanks D, Vennapusa B, Mistry A, Kalamegham R, Averbuch S, Novotny J, Rubin E, Emancipator K, McCaffery I, Williams JA, Walker J, Longshore J, Tsao MS, Kerr KM, PD-L1 immunohistochemistry assays for lung cancer: results from phase 1 of the blueprint PD-L1 IHC assay comparison project, J. Thorac. Oncol 12 (2) (2017) 208–222. [DOI] [PubMed] [Google Scholar]
- [28].McLaughlin J, Han G, Schalper KA, Carvajal-Hausdorf D, Pelekanou V, Rehman J, Velcheti V, Herbst R, LoRusso P, Rimm DL, Quantitative assessment of the heterogeneity of PD-L1 expression in non-small-cell lung cancer, JAMA Oncol. 2 (1) (2016) 46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Curtin NJ, DNA repair dysregulation from cancer driver to therapeutic target, Nat. Rev. Cancer 12 (12) (2012) 801–817. [DOI] [PubMed] [Google Scholar]
- [30].Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, Biedrzycki B, Donehower RC, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Duffy SM, Goldberg RM, de la Chapelle A, Koshiji M, Bhaijee F, Huebner T, Hruban RH, Wood LD, Cuka N, Pardoll DM, Papadopoulos N, Kinzler KW, Zhou S, Cornish TC, Taube JM, Anders RA, Eshleman JR, Vogelstein B, Diaz LA, PD-1 blockade in tumors with mismatch-repair deficiency, N. Engl. J. Med 372 (26) (2015) 2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Friboulet L, Olaussen KA, Pignon J-P, Shepherd FA, Tsao M-S, Graziano S,Kratzke R, Douillard J-Y, Seymour L, Pirker R, Filipits M, André F, Solary E, Ponsonnailles F, Robin A, Stoclin A, Dorvault N, Commo F, Adam J, Vanhecke E, Saulnier P, Thomale J, Le Chevalier T, Dunant A, Rousseau V, Le Teuff G, Brambilla E, Soria J-C, ERCC1 isoform expression and DNA repair in non–small-cell lung cancer, N. Engl. J. Med 368 (12) (2013) 1101–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bepler G, Williams C, Schell MJ, Chen W, Zheng Z, Simon G, Gadgeel S, Zhao X, Schreiber F, Brahmer J, Chiappori A, Tanvetyanon T, Pinder-Schenck M, Gray J, Haura E, Antonia S, Fischer JR, Randomized international phase III trial of ERCC1 and RRM1 expression-based chemotherapy versus gemcitabine/carboplatin in advanced non-small-cell lung cancer, J. Clin. Oncol 31 (19) (2013) 2404–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Abuzeid WM, Jiang X, Shi G, Wang H, Paulson D, Araki K, Jungreis D, Carney J, O’Malley BW Jr, Li D, Molecular disruption of RAD50 sensitizes human tumor cells to cisplatin-based chemotherapy, J. Clin. Invest 119 (7) (2009) 1974–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Al-Ahmadie H, Iyer G, Hohl M, Asthana S, Inagaki A, Schultz N, Hanrahan AJ, Scott SN, Brannon AR, McDermott GC, Pirun M, Ostrovnaya I, Kim P, Socci ND, Viale A, Schwartz GK, Reuter V, Bochner BH, Rosenberg JE, Bajorin DF, Berger MF, Petrini JH, Solit DB, Taylor BS, Synthetic lethality in ATM-deficient RAD50-mutant tumors underlies outlier response to cancer therapy, Cancer Discov. 4 (9) (2014) 1014–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Campbell BB, Light N, Fabrizio D, Zatzman M, Fuligni F, de Borja R, Davidson S, Edwards M, Elvin JA, Hodel KP, Zahurancik WJ, Suo Z, Lipman T, Wimmer K, Kratz CP, Bowers DC, Laetsch TW, Dunn GP, Johanns TM, Grimmer MR, Smirnov IV, Larouche V, Samuel D, Bronsema A, Osborn M, Stearns D, Raman P, Cole KA, Storm PB, Yalon M, Opocher E, Mason G, Thomas GA, Sabel M, George B, Ziegler DS, Lindhorst S, Issai VM, Constantini S, Toledano H, Elhasid R, Farah R, Dvir R, Dirks P, Huang A, Galati MA, Chung J, Ramaswamy V, Irwin MS, Aronson M, Durno C, Taylor MD, Rechavi G, Maris JM, Bouffet E, Hawkins C, Costello JF, Meyn MS, Pursell ZF, Malkin D, Tabori U, Shlien A, Comprehensive analysis of hypermutation in human cancer, Cell 171 (5) (2017) 1042–1056 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chae YK, Davis AA, Raparia K, Agte S, Pan A, Mohindra N, Villaflor V, Giles F, Association of tumor mutational burden with DNA repair mutations and response to anti-PD-1/PD-L1 therapy in non-small-cell lung cancer, Clin. Lung Cancer (2018). [DOI] [PubMed] [Google Scholar]
- [37].Capoluongo E, Ellison G, Lopez-Guerrero JA, Penault-Llorca F, Ligtenberg MJL, Banerjee S, Singer C, Friedman E, Markiefka B, Schirmacher P, Buttner R, van Asperen CJ, Ray-Coquard I, Endris V, Kamel-Reid S, Percival N, Bryce J, Rothlisberger B, Soong R, de Castro DG, Guidance statement on BRCA1/2 tumor testing in ovarian cancer patients, Semin. Oncol 44 (3) (2017) 187–197. [DOI] [PubMed] [Google Scholar]
- [38].Stuckey AR, Onstad MA, Hereditary breast cancer: an update on risk assessment and genetic testing in 2015, Am. J. Obstet. Gynecol 213 (2) (2015) 161–165. [DOI] [PubMed] [Google Scholar]
- [39].Litton JK, Rugo HS, Ettl J, Hurvitz SA, Goncalves A, Lee KH, Fehrenbacher L, Yerushalmi R, Mina LA, Martin M, Roche H, Im YH, Quek RGW, Markova D, Tudor IC, Hannah AL, Eiermann W, Blum JL, Talazoparib in patients with advanced breast cancer and a germline BRCA mutation, N. Engl. J. Med 379 (8) (2018) 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mirza MR, Monk BJ, Herrstedt J, Oza AM, Mahner S, Redondo A, Fabbro M, Ledermann JA, Lorusso D, Vergote I, Ben-Baruch NE, Marth C, Madry R, Christensen RD, Berek JS, Dorum A, Tinker AV, du Bois A, Gonzalez-Martin A, Follana P, Benigno B, Rosenberg P, Gilbert L, Rimel BJ, Buscema J, Balser JP, Agarwal S, Matulonis UA, Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer, N. Engl. J. Med 375 (22) (2016) 2154–2164. [DOI] [PubMed] [Google Scholar]
- [41].Kostyrko K, Bosshard S, Urban Z, Mermod N, A role for homologous recombination proteins in cell cycle regulation, Cell Cycle 14 (17) (2015) 2853–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wang Y, Gudikote J, Giri U, Yan J, Deng W, Ye R, Jiang W, Li N, Hobbs BP, Wang J, Swisher SG, Fujimoto J, Wistuba II, Komaki R, Heymach JV, Lin SH, RAD50 expression is associated with poor clinical outcomes after radiotherapy for resected non-small cell lung cancer, Clin. Cancer Res 24 (2) (2018) 341–350. [DOI] [PubMed] [Google Scholar]
- [43].Chen X, Qian D, Cheng J, Guan Y, Zhang B, Ding X, Zeng J, Chen X, Er P, Zhang F, Zhao N, Chen X, Zhao L, Yuan Z, Pang Q, Wang P, High expression of Rad51c predicts poor prognostic outcome and induces cell resistance to cisplatin and radiation in non-small cell lung cancer, Tumour Biol. 37 (10) (2016) 13489–13498. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


