Abstract
The need for portable and reproducible genomics analysis pipelines is growing globally as well as in Africa, especially with the growth of collaborative projects like the Human Health and Heredity in Africa Consortium (H3Africa). The Pan-African H3Africa Bioinformatics Network (H3ABioNet) recognized the need for portable, reproducible pipelines adapted to heterogeneous compute environments, and for the nurturing of technical expertise in workflow languages and containerization technologies. To address this need, in 2016 H3ABioNet arranged its first Cloud Computing and Reproducible Workflows Hackathon, with the purpose of building key genomics analysis pipelines able to run on heterogeneous computing environments and meeting the needs of H3Africa research projects. This paper describes the preparations for this hackathon and reflects upon the lessons learned about its impact on building the technical and scientific expertise of African researchers. The workflows developed were made publicly available in GitHub repositories and deposited as container images on quay.io.
Keywords: Bioinformatics, hackathon, workflow, reproducible, pipeline, capacity building
Introduction
As an inherently interdisciplinary science, bioinformatics depends upon complementary expertise from biomedical scientists, statisticians and computer scientists 1. This opportunity for collaborative projects also creates a need for avenues to exchange knowledge 1. Hackathons, along with codefests and sprints, are emerging as an efficient mean for driving successful projects 2. They can be in the form of science hackathons that aim to derive research plans and scientific write up 3, community-driven software development 4, and data hackathons or datathons 5. In addition to the scientific and technical outcomes, these intensive and focused activities offer necessary skills development and networking opportunities to young and early career scientists.
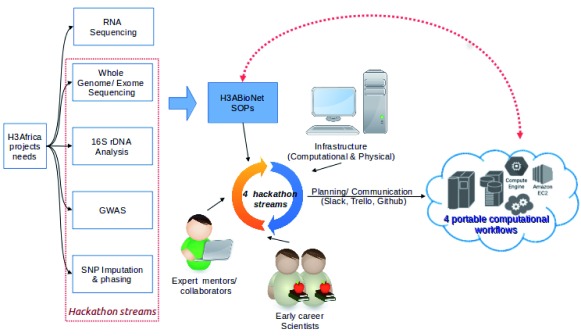
On the African continent, there is generally limited access to such events. However, with the growing capacity for Africans to generate genomic data, the need to analyze these data locally by African scientists, is also growing. H3ABioNet 6, the Bioinformatics Network within the H3Africa initiative 7, has invested in capacity building via different approaches 8. The H3ABioNet Cloud Computing hackathon was a natural extension of the network’s efforts in developing Standard Operating Procedures (SOPs) via its Network Accreditation Task Force (NATF) 9; aimed at building and assessing capacity in genomic analysis. This also follows other efforts by the H3ABioNet Infrastructure Working Group (ISWG) towards setting up infrastructure at various H3ABioNet Nodes at the hardware, software, networking, and personnel level. The H3ABioNet Cloud Computing hackathon, therefore, provided an excellent opportunity to assess the computational skills capacity development of the network through training, learning and adoption of novel technologies ( Figure 1). These technologies included workflow languages for reproducible science, containerization of software, and creation of computational products that can be used in heterogeneous computing environments encountered by African and international scientists in the form of standalone servers, cloud allocations and High Performance Computing (HPC) resources.
Figure 1. Planning and execution of the H3ABioNet Cloud computing hackathon.
(SOPs: Standard Operating Procedures).
In this paper, we discuss the organization of the H3ABioNet Cloud Computing hackathon, the interactions between the participants, and the lessons learnt. A paper describing the technical aspects of the pipelines developed has been in preparation (unpublished paper- Baichoo et. al. 2018 1), whereas the code and pipelines themselves have been made publicly available via H3ABioNet’s GitHub page in the following repositories: ( h3agatk, h3abionet16S, h3agwas and chipimputation) as well as container images hosted on Quay.io.
H3ABioNet Cloud Computing Hackathon Activities
Prior to the H3ABioNet Cloud Computing hackathon, H3ABioNet, via its Infrastructure Working Group (ISWG), formed a Cloud Computing task force to investigate cloud computing technologies, to familiarize H3ABioNet members with current cloud implementations and gauge their suitability for H3Africa data analyses. The H3ABioNet Cloud Computing hackathon was one of the first deliverables of this task force, with the specific objective to test and implement the use of four analysis workflows that can be ported on multiple compute platforms. Figure 1 shows this hackathon within the broader H3Africa context and provides a broad overview of the planning and execution of this activity, with details in the following subsections.
Pre-hackathon preparations
The computational pipelines put forward for development during the H3ABioNet Cloud Computing hackathon were identified based on the data being generated by different H3Africa projects and the SOPs used for the H3ABioNet Node Accreditation exercises. Reproducibility and portability were also identified as key features for the workflows, due to the heterogeneous computational platforms available in Africa. H3ABioNet Nodes that used or helped develop current H3ABioNet workflows and SOPs were part of the planning team, as well as other nodes that had technically strong scientists who were willing to extend their skills.
In the course of planning for the H3ABioNet Cloud Computing hackathon, two technical areas were identified where additional expertise was required. These were containerization technology such as Docker, and the writing of genomic pipelines in popularly used workflow languages and newly emerging community-standards like Nextflow 10 and the Common Workflow Language (CWL) 11, respectively. While expertise for Nextflow already existed within the network, two collaborators from outside of Africa were interested to join the project given their expertise in running genomic pipelines in cloud environments, containerization of code 12 and developing CWL 11. They subsequently joined the planning and participated in the hackathon.
The H3ABioNet Cloud Computing hackathon was announced on the internal H3ABioNet consortium mailing list as a call for interested applicants and in some cases, individuals were invited based on their specific expertise. Most of the participants selected were early career scientists with strong computational skills, an understanding of genomic pipelines and willingness to work in teams. The pipelines for the Cloud Hackathon where divided into four “streams”: 1) Stream A: variant calling from whole genome sequencing (WGS) and whole exome sequencing (WES) data ( https://github.com/h3abionet/h3agatk), 2) Stream B: 16S rDNA Diversity Analysis ( https://github.com/h3abionet/h3abionet16S), 3) Stream C: Genome Wide-association studies (Illumina array data) ( https://github.com/h3abionet/h3agwas) and 4) Stream D: SNP Imputation and phasing using different reference panels ( https://github.com/h3abionet/chipimputation). Successful applicants were given a choice to select a project stream based on their skills and interest or if unsure, assigned to a specific stream. Streams A and B decided to use CWL for their pipeline development, whereas Streams C and D opted to use Nextflow Language, due to their prior experience using Nextflow.
Vital in setting up the teams for each of the streams was that each team had a balanced composition. This included bioinformaticians with strong computational skills to create the Docker containers and implement the workflow languages, knowledge in the specific genomic analyses and computational tools required, and strong system administration skills to assist with the installation of numerous software components. We also included bioinformaticians with experience in running the workflows or components of the workflows, and software developers who could assist with creating Docker containers, troubleshoot and implement workflow languages.
To maximize the learning experience, upon selection, participants were given prerequisite tutorials and materials (Github, Nextflow, CWL, Docker and the SOPs) to go through. Communication and planning infrastructure in the form of Slack channels and Trello boards were created beforehand with all the participants added in order to allow them to brainstorm and share ideas with team members before the hackathon began ( Table 1). Fortnightly planning meetings were held starting from 3 months in advance in order for hackathon participants to get involved in planning their proposed tools and to get to know one another and develop a working rapport before the start of the hackathon.
Table 1. Communication channels used for the hackathon.
| Channel | Link | Purpose |
|---|---|---|
| Mailing list | - | Group wide announcements and
communications |
| Mconf | https://mconf.sanren.ac.za/ | Online meetings |
| Slack | https://slack.com/ | Inner group discussions and chat |
| Trello | https://trello.com/ | Plan goals and activities, and track
progress |
| GitHub | https://github.com/ | Code repository and version control |
The hackathon ran in August 2016 and was hosted at the University of Pretoria Bioinformatics and Computational Biology Unit in South Africa. The choice of the hackathon venue was based on availability of Unix/Linux desktop machines with the facility for sudo/root access enabling participants to install software and deploy Docker containers for testing. Besides the local machines, participants also had access to cloud computing platforms such as Azure and Amazon, Nebula (made available by the National Center for Supercomputing Applications, University of Illinois at Urbana-Champaign), and the African Research Cloud (through a collaboration with the University of Cape Town eResearch initiative). After the hackathon, more testing was also done on EGI Federated Cloud resources (through an agreement contract with the University of Khartoum).
Hackathon week activities
The initial day of the H3ABioNet Cloud Computing hackathon was dedicated to introductions, expectations by the participants and practical tutorials covering the use of CWL, Nextflow and creation of Docker containers to ensure all participants had the same basic level of knowledge. The teams had a breakout session where overall milestones for the streams during the hackathon week were refined, tasks were identified and assigned to team members and Trello boards updated with the specific tasks. Each stream reported back on their progress and overall work plan for the coming hackathon days. For the remaining days of the hackathon, participants were split into their respective streams to work on developing and containerizing their pipelines as well as creating the related documentation. To ensure a successful hackathon with concrete outcomes, the streams spent the first 30 minutes of each hackathon day reviewing their prior progress and updating their Trello boards and reporting to the group what they will be working on. At the end of the day each stream provided a progress report to the whole group on what they had achieved, what they struggled with and what they will be working on. The start and end of day reporting proved useful as it allowed groups that had encountered and solved an issue to share the implemented solution with another stream, and for different streams to work together to solve any shared issues encountered, thus speeding up the development of the pipelines. Area experts and collaborators would switch between the streams to provide necessary technical expertise.
Communication during the hackathon was facilitated by Slack integration with Trello (for tasks managements and progress tracking) and code developed was pushed to GitHub (for live code integration). Table 1 lists the various communication media used during the hackathon. Some groups also utilized Google docs for documenting their progress prior to migrating documentation into github README files. Remote participation in the hackathon was facilitated through the MConf conference system. One stream had a participant with very strong coding skills working remotely from the US; who managed to make progress on the corresponding workflow when the other group members were not working due to the big time difference between the USA and South Africa (SA). This ensured continuous development on the workflow when the team in SA would clock off and provide a to-do list which was accomplished by the participant from the USA. Noticeable during the hackathon was the team spirit created and the increasingly later end time for the days with most days ending at 8:30 pm as participants continued working after the different streams provided their daily reports. All participants wished for an extra day or two to complete their pipelines.
Post-hackathon feedback and actions
After the week-long hackathon at the University of Pretoria, members of each stream continued working on their respective pipelines communicating via Slack and Trello. Meetings were held over MConf every two weeks to report on the progress of each pipeline. Upon completion, each group handed their pipeline to other groups to test on different platforms to avoid any bias in implementation and improve the documentation and hence facilitate the ease of use of the four pipelines developed.
Discussion
The H3ABioNet Cloud Computing Hackathon was aimed at producing portable, cloud-deployable Docker containers for a variety of bioinformatics workflows including variant calling, 16S rDNA diversity analysis, quality control, genotype calling, and imputation and phasing for genome-wide association studies. Dockerization provides a method to package and manage software, tools and workflows within a portable environment/container, similar to virtualization but with a smaller computing overhead compared to virtualization. Docker containers can easily be developed and deployed on computing environments (especially cloud based infrastructure) and can be used by a variety of groups to ensure reproducible analysis using the same tools, software versions, and workflows used.
The novelty of the H3ABioNet Cloud Computing Hackathon was that all the participants selected were involved in the latter stages of the planning and the setting of some of the outcomes for the hackathon. Critical recommendations during the hackathon planning meetings were that the resulting Docker containers and pipelines developed should be compatible with heterogeneous African research compute environments with portability and good documentation being key. This is especially important considering the fact that access to Cloud computing environments within Africa is still in its infancy. Hence, it was decided that development and testing of the pipelines should occur on a single machine, with the ability to be ported to a cluster or an HPC environment, and ultimately tested and deployed on cloud-based platforms (Amazon, Microsoft Azure, EGI FedCloud, IBM Bluemix, and the new African Research Cloud initiative).
Lessons learnt and concluding remarks
The opportunity to link people physically and focus solely on one project has been highly effective in providing the main outline and proof of concept outputs. However, once people were back home, continuing the tasks has been a challenge. Clearly defining the roles and commitment of all the participants in the papers reporting the results should encourage them to complete the work, and increase their accountability.
The communication and management tools used for this hackathon were important as these tools facilitated interaction between and across team members and enabled the participants to continue to work in a structured manner once back at their respective institutions, despite time zones differences.
The H3ABioNet Cloud Computing Hackathon has been an important milestone for the Network as it brought together people with various skills to work on focused projects. It signalled the shift from capacity building to utilizing the capacity developed in order to tackle problems specific to the heterogeneous African computing environments, as defined and implemented by the mostly African participants.
As software packages and computing environments evolve with varying build cycles and new bioinformatics tools become available, we envision that hackathons to keep these pipelines current, adopt new technology implementations such as Singularity, and develop new workflows such as for RNA-Seq analysis will occur. The pipelines developed during the H3ABioNet Cloud Computing hackathon will be used for training and data analyses for intermediate level bioinformatics workshops, and for scientific collaborations requiring bioinformatics expertise for data analysis such as with the H3Africa genotyping chip and GWAS analyses. Future H3ABioNet hackathons would also provide an opportunity to utilize the skills of trained bioinformaticians at intermediate and advanced levels, who would not otherwise attend bioinformatics training workshops, to come together to derive practical solutions that are of benefit to the African and wider scientific community.
Data and software availability
All data underlying the results are available as part of the article and no additional source data are required.
The four pipelines are available publicly via H3ABioNet’s github organization page https://github.com/h3abionet in the following repositories: (h3agwas, chipimputation, h3agatk and h3abionet16S) as well as container images on quay.io at https://quay.io/organization/h3abionet_org.
All code is available under MIT license, except for the h3agtk pipeline which is available under Apache license 2.0
Note
1Full author list for unpublished study: Shakuntala Baichoo, Yassine Souilmi, Sumir Panji, Gerrit Botha, Ayton Meintjes, Scott Hazelhurst, Hocine Bendou, Eugene de Beste, Phelelani T. Mpangase, Oussema Souiai, Mustafa Alghali, Long Yi, Brian D. O’Connor, Michael Crusoe, Don Armstrong, Shaun Aron, Fourie Joubert, Azza E. Ahmed, Mamana Mbiyavanga, Peter van Heusden, Lerato E. Magosi, Jennie Zermeno, Liudmila Sergeevna Mainzer, Faisal M. Fadlelmola, C. Victor Jongeneel and Nicola Mulder
Acknowledgments
We acknowledge the advice and help from Ananyo Choudhury from Sydney Brenner Institute for Molecular Bioscience, University of the Witwatersrand, Johannesburg, South Africa.
Funding Statement
H3ABioNet is supported by the National Institutes of Health Common Fund [U41HG006941]. H3ABioNet is an initiative of the Human Health and Heredity in Africa Consortium (H3Africa) programme of the African Academy of Science (AAS. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 3 approved with reservations]
References
- 1. Yanai I, Chmielnicki E: Computational biologists: moving to the driver’s seat. Genome Biol. 2017;18(1):223. 10.1186/s13059-017-1357-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Möller S, Afgan E, Banck M, et al. : Community-driven development for computational biology at Sprints, Hackathons and Codefests. BMC Bioinformatics. 2014;15 Suppl 14:S7. 10.1186/1471-2105-15-S14-S7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Groen D, Calderhead B: Science hackathons for developing interdisciplinary research and collaborations. eLife. 2015;4:e09944. 10.7554/eLife.09944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Crusoe MR, Brown CT: Channeling Community Contributions to Scientific Software: A sprint Experience. J Open Res Softw. 2016;4(1): pii: e27. 10.5334/jors.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aboab J, Celi LA, Charlton P, et al. : A “datathon” model to support cross-disciplinary collaboration. Sci Transl Med. 2016;8(333):333ps8. 10.1126/scitranslmed.aad9072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mulder NJ, Adebiyi E, Alami R, et al. : H3ABioNet, a sustainable pan-African bioinformatics network for human heredity and health in Africa. Genome Res. 2016;26(2):271–7. 10.1101/gr.196295.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. H3Africa Consortium, Rotimi C, Abayomi A, et al. : Research capacity. Enabling the genomic revolution in Africa. Science. 2014;344(6190):1346–8. 10.1126/science.1251546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aron S, Gurwitz K, Panji S, et al. : H3abionet: developing sustainable bioinformatics capacity in africa. EMBnet j. 2017;23:e886 10.14806/ej.23.0.886 [DOI] [Google Scholar]
- 9. Jongeneel CV, Achinike-Oduaran O, Adebiyi E, et al. : Assessing computational genomics skills: Our experience in the H3ABioNet African bioinformatics network. PLoS Comput Biol. 2017;13(6):e1005419. 10.1371/journal.pcbi.1005419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peter A, Crusoe MR, Nebojša T, et al. : Common Workflow Language, v1.0. 2016. 10.6084/m9.figshare.3115156.v2 [DOI] [Google Scholar]
- 11. Di Tommaso P, Chatzou M, Floden EW, et al. : Nextflow enables reproducible computational workflows. Nat Biotechnol. 2017;35(4):316–9. 10.1038/nbt.3820 [DOI] [PubMed] [Google Scholar]
- 12. O’Connor BD, Yuen D, Chung V, et al. : The Dockstore: enabling modular, community-focused sharing of Docker-based genomics tools and workflows. [version 1; referees: 2 approved]. F1000Res. 2017;6:52. 10.12688/f1000research.10137.1 [DOI] [PMC free article] [PubMed] [Google Scholar]