Abstract
CRISPR/Cas9–based gene knockouts (KOs) enable precise perturbation of target gene function in human cells, which is ideally assessed in an unbiased fashion by molecular omics readouts. Typically, this requires the lengthy process of isolating KO subclones. We show here that KO subclones are phenotypically heterogenous, regardless of the guide RNA used. We present an experimental strategy that avoids subcloning and achieves fast and efficient gene silencing on cell pools, based on the synergistic combination of two guide RNAs mapping at close (40–300 bp) genomic proximity. Our strategy results in better predictable indel generation with a low allelic heterogeneity, concomitant with low or undetectable residual target protein expression, as determined by MS3 mass spectrometry proteomics. Our method is compatible with nondividing primary cells and can also be used to study essential genes. It enables the generation of high confidence omics data which solely reflect the phenotype of the target ablation.
Introduction
The discovery of the bacterial defense CRISPR-Cas system1 and its adaptation to silence mammalian genes has been a revolutionary step forward in the use of gene editing as a broad and easy-to-use laboratory research tool to silence (CRISPR knockout [KO]),2–4 mutate,5,6 repress/interfere (CRISPRi)7,8 or activate (CRISPRa)9–11 targeted genes. The most commonly used system, CRISPR-Cas9 is based on an endonuclease (Cas9 from Streptococcus pyogenes) that recognizes a three nucleotide motif NGG on DNA called protospacer adjacent motif (PAM) immediately 3′ of a 20 nucleotide sequence that is complementary to a small guide CRISPR (crRNA), itself binding to another small RNA (trans-activating crRNA [tracrRNA]) with a specific secondary structure. Cas9 binds the RNA dimer and is selectively recruited to the 20 bp PAM sequence to cut blunt the double-stranded DNA, usually 3 bp (sometimes 4 bp) 5′ of the PAM motif. The DNA is cleaved and repaired repeatedly via the nonhomologous end-joining pathway until errors occur resulting in small insertions or deletions (indels).12 Cas9 mutants have been engineered to increase its selectivity to minimize off-target editing,13–15 to broaden its PAM site recognition motifs,16,17 to change the endonuclease activity towards nickase (Cas9 D10A),4 to enhance editing specificity,18 or for complete loss of enzymatic activity (dCas9) to be used as a specific shuttle molecule under the form of a fusion protein with transcriptional repressor (CRISPRi)7,8 or activator (CRISPRa)9–11 domains. It is now widely accepted that guide RNA (gRNA) off-targets depend on the amount and duration of Cas9 expression in a cell, leading users to prefer the introduction of highly active recombinant Cas9 bound to crRNA–tracrRNA duplex or to a single gRNA (sgRNA) resulting from the fusion of the crRNA and tracrRNA, forming the ribonucleoprotein (RNP) complex that gets quickly degraded inside the cell, to the use of constitutive expression of Cas9 via plasmids.15,19 Several studies have painstakingly dissected the 20 nucleotide gRNA sequence to define its ideal length and composition20–25 resulting into the development of webtools to design gRNAs. Although specificity of the gRNAs can be enhanced by increasing the number of mismatches to the closest homologous sequence, their efficiency remains hard to predict. Historically, most studies were based on CRISPR-Cas9 editing of immortalized cell lines with subsequent isolation of KO subclones, typically by dilution cloning. These KO cell lines are relatively straightforward to generate, however at the expense of time and resources. Nonetheless, this strategy cannot be easily adapted to induced pluripotent stem cells, which are difficult to subclone, and is even less suitable for primary cells that usually do not divide. In these cases a successful gene knockout strategy becomes critically dependent on the identification of highly efficient gRNAs that achieve efficient editing of the majority of alleles in a pool of cells. Frequently this turns out to be challenging, in particular for genes for which there are limited possibilities of designing suitable gRNA sequences. Checking the gene editing result only at the DNA sequence level is not enough to ensure disappearance of the targeted protein since cells can rescue some protein functionality via exon skipping or secondary translation initiation sites.26 Similarly, antibody-based detection of residual protein levels may miss newly generated splice variants or truncations and are not always quantitative.
We have developed a CRISPR-Cas9 approach to generate, within a few days, KO cell pools using two gRNAs hybridizing at close proximity on the target DNA sequence that synergize to edit close to 100% of targeted alleles. Combined with an adapted mass spectrometry protocol (MS3) to quantify the remaining protein target, this represents an efficient approach to study protein functions in diverse model systems (cell lines, induced pluripotent stem cells, primary cells).
Materials and Methods
Design and synthesis of gRNAs
All gRNAs were designed using the Benchling web tool with at least three mismatches for NGG PAM sites and at least two mismatches for NAG PAM sites (human genome GRch38). Unless specified otherwise, all gRNAs were synthesized in vitro as sgRNAs from DNA oligonucleotides purchased from Sigma using the TranscriptAid kit and purified with the Gene jet RNA clean-up kit (both from Thermo) according to the manufacturer's instructions. Synthetic sgRNAs, crRNAs and tracrRNA were purchased from IDT. All gRNA sequences are provided in Supplementary Table S1.
HepG2 cell culturing
HepG2 cells (ATCC HB-8065) were grown in modified Eagle's medium (MEM) supplemented with 10% fetal calf serum at 37°C in presence of 5% CO2 in a humidified incubator. Cells were detached with Accutase (Gibco) and mechanically dissociated by pipetting through a 100 μL plastic tip prior to seeding or electroporation.
Human CD4+ T cells
Human CD4+ T cells from peripheral blood were purchased from Biotrend (Stemexpress). Cells were sourced ethically, and their research use was in accord with the terms of the informed consents under an institutional review board/ethical committee protocol. Cells were thawed and grown in modified RPMI27 supplemented with 10% fetal calf serum at 37°C in presence of 5% CO2 in a humidified incubator. For the gene editing on activated T cells, CD4+ cells were activated 3 days with Transact (anti-CD3/anti-CD28) at 1/500 and interleukin 2 at 20 IU/mL (both from Miltenyi).
Gene editing of HepG2 and primary T cells
Electroporations were performed with the nucleofector 4D-X (Lonza) following the manufacturer's instructions. Buffer SF and program EH-100 were used with HepG2 cells and buffer P3 and program EH-115 with the T cells. Cell numbers and the origin and amounts of gRNAs and Cas9 used are listed in Supplementary Table S2. Information concerning the oligonucleotide sequences of the PCR primers, the size of the PCR fragments and the distance between the Cas9 sites is shown in Supplementary Table S1.
Generation of HepG2 KO clones
Electroporated cells were plated into MEM with 50% conditioned MEM (MEM medium that has been incubated 20–30 h on low density HepG2) supplemented with 4 mM pyridoxal for pyridoxal kinase (PDXK) gene-edited cells. KO clones were generated by cell dilution in 96 well plates. After ∼14 days, wells were checked under the microscope and single clones were isolated and grown further.
Gene editing analysis
KO indels were analyzed by Sanger sequencing (Sequiserve) following PCR amplification using AmpliTaq 360 Gold (Thermo) with an elongation time of 30 to 50 s. PCR oligos were designed so that the resulting deletion between the two Cas9 sites would not exceed 45% of the length of the wild-type fragment (with the exception of EPHX2). Sequences of PCR oligos are listed in Supplementary Table S1. Amplified fragments were purified using the MinElute PCR kit (Qiagen). Sequencing data were analyzed using TIDE or ICE (Synthego) webtools.
The expected additive gene editing effect of the two gRNAs [%GE(A)] was calculated as: %GE(D) + [100 − %GE(D)] × [%GE(H)/100]. The synergistic benefit was calculated as the difference between the percentage of gene editing obtained with the synergistic tandem gRNA combination and the calculated percentage of gene editing of the gRNA combination if the effect of the two gRNAs would only be additive (%GE(T) − %GE(A)) [GE, gene editing; D, driver gRNA; H, helper gRNA; T, tandem (synergistic) gRNA combination; A, calculated additive gRNA combination].
Liquid chromatography–tandem mass spectrometry and data analyses
See Supplementary Material and Methods for a detailed description of these analyses.
Results
Target KO subclones exhibit divergent proteome signatures
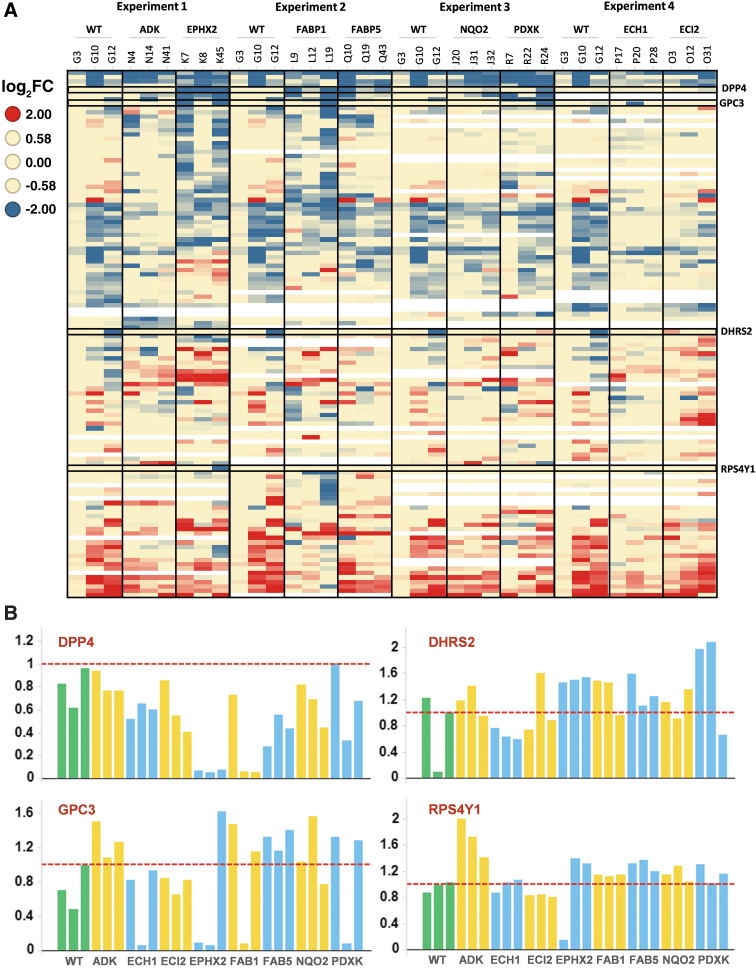
We used CRISPR-Cas9 gene editing technology to generate KO cell clones for several genes encoding metabolic enzymes (ADK, ECH1, ECI2, EPHX2, NQO2, PDXK) or lipid transporters (FABP1, FABP5) in the hepatocarcinoma cell line HepG2. KO clones were generated by electroporating a pre-associated RNP complex (recombinant Cas9 and in vitro synthetized sgRNA) and were isolated via dilution cloning. The presence of an indel leading to disruption of the coding frame in all alleles was verified by Sanger DNA sequencing. Three different clones per target were then grown in parallel and harvested at similar cell confluency (70–90%) to characterize their entire proteome by comparing it to that of clones derived from cells mock-electroporated with Cas9 without sgRNAs. The proteome signature (>5,000 proteins quantified by tandem mass spectrometry based on ≥2 unique peptides) was analyzed to assess the depletion of the protein target (Supplementary Fig. S1). A heatmap displaying the proteins with a high fold change when compared with a control clone but not considered significantly regulated showed a strong heterogeneity amongst the three KO clones for the same target originated from the same electroporation (Fig. 1A). Notably this heterogeneity was also visible for the mock-electroporated clones. A few proteins, such as glypican 3 (GPC3) and dipeptidyl-peptidase 4 (DPP4), show strong differential expression in several clones irrespective of the targeted gene and of the sgRNA used (Fig. 1A and B). In other cases, strong downregulation is observed only in a single clone, like for ribosomal subunit RSP4Y1 in EPHX2 KO clone 1 or DHRS2 in wild-type control clone 2. This heterogeneity is therefore unlikely to be due to off-target effects of the sgRNA used. It could be caused by genetic or epigenetic heterogeneity of the HepG2 cell pool at the time of the electroporation, or from a genetic or epigenetic stress response induced by electroporation and dilution cloning. The cell subclones have undergone around 20 divisions before sampling for proteomics analysis and had the time to adapt to the consequences of the gene editing. This clonal heterogeneity is not specific for HepG2 cells, as we have observed it in other cell lines (like the THP-1 monocytic cell line) and clonal diversity was recently reported as the major cause of confounding variation in drug response screens across MCF7 and other cell lines28.
FIG. 1.
Clones knocked out for same target display heterogenous proteomes. Three HepG2 knockout (KO) clones have been generated for each of the eight targets using CRISPR-Cas9 gene editing. (A) Clone heterogeneity is visualized in a heatmap using proteins that in the statistical analysis have a high fold-change compared with wildtype (WT) clones [abs(log2) ≥ 1.15] and high variation between clones (adjusted p-value >0.05). Protein up- and downregulation is clearly different among three clones knocked out for the same target confirming clone heterogeneity. Clones are indicated via their identification codes. (B) Bar chart displaying relative (to control, WT) mass spectrometry (MS) quantification of the four selected proteins from A). Data show that expression is strongly reduced in some clones, independent of the gene targeted for gene editing. FAB1, FABP1; FAB5, FABP5.
Tandem gRNA CRISPR on cell pools is a fast and efficient strategy to create KO indels
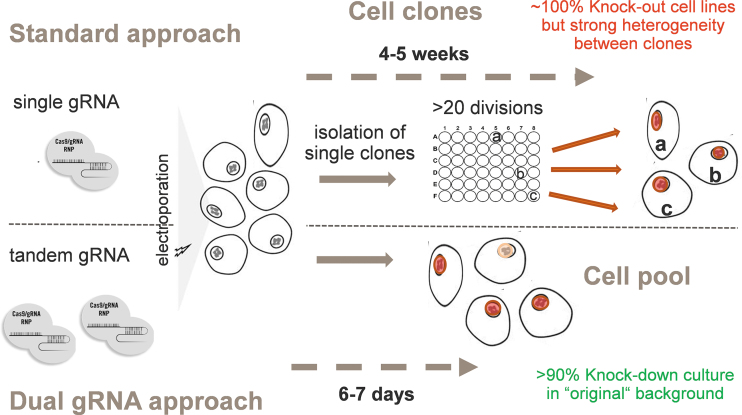
To circumvent subclone heterogeneity and cell adaptation issues, it is critical to rapidly generate—within a few days—KO cell pools. These pools would ideally contain a clear majority of KO cells and only few cells that would have retained at least one wild-type allele (Fig. 2). To reach this aim, there was a clear requirement for highly efficient gRNAs to create a frameshift indel in the majority (ideally close to 100%) of alleles. As the target genes are often present as two or three copies per HepG2 cell, suboptimal editing would leave at least one wild-type allele. Very few human genes display haploinsufficiency, and thus a residual wild-type allele would usually be sufficient to express the encoded protein at, or close to, wild-type levels.
FIG. 2.
Clonal versus cell pool gene editing strategies. Cartoon comparing the standard, painstaking protocol to generate KO clones with the versatile and fast tandem gRNA approach that produces a whole cell population, nearly entirely KO for the chosen target within days.
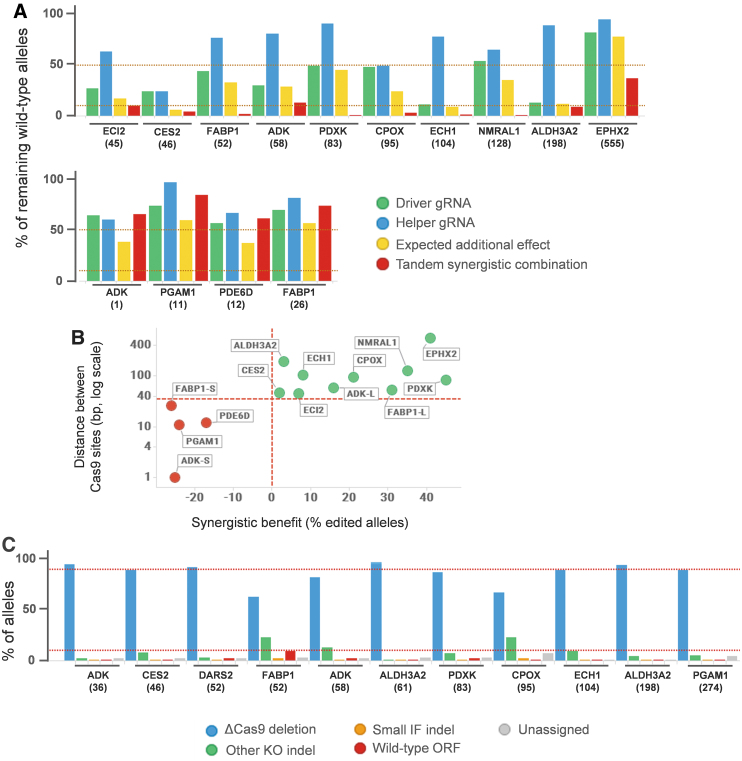
We performed a series of studies comparing the efficacy of RNP complexes comprising a single gRNA or combinations of two gRNAs. We generally observed that combining two gRNAs which recognize sequences at close proximity on the target DNA leads to a high indel rate, in many cases much higher than expected from each gRNA. We hypothesized that the close proximity of the two RNA complexes has a synergistic effect translating into an increased indel rate. To validate this observation, we compared side by side the efficacy of RNP complexes formed with each of the two gRNAs separately or in combination to knock out the alleles for 12 targets (14 gRNA combinations). When combined, only half of the amount of each gRNA was used to maintain the same amount of active RNP complexes and the stoichiometry between gRNA and recombinant Cas9. Suboptimal experimental conditions were chosen to better assess the synergistic effect. The percentage of remaining wild-type alleles was monitored by Sanger sequencing and quantified by the Synthego ICE webtool (Fig. 3A). PCR were designed to minimize bias towards amplifying the shorter fragment containing the deletion (see “Materials and Methods”). Under these conditions, indel rates above 90% are achieved using two gRNAs that alone, in twice the amount compared with the combination, give poor indel rates (in the range of 10 to 60%). The combination of the two gRNAs gives higher gene editing rates than what would be expected if the effect of the two gRNAs would only be additive. Synergy appears to work at best when the two Cas9 cuts are 40 to 300 bp apart and is completely lost when the Cas9 cuts are less than 35 bp apart. In this latter situation, the calculated synergistic benefit (difference between the proportion of gene edited alleles after tandem synergistic gRNA combination and after a gRNA combination with a simple additive effect, as calculated in the methods section; Fig. 3B) becomes negative, reflecting a possible hindrance between RNP complexes. On the other hand, synergy may still be observed beyond a distance of 300 bp, as shown for EPHX2 KO (Fig. 3A and B) for which two gRNAs with Cas9 cutting sites distant 555 bp from each other display very low indel efficiency when used alone (above 80% of remaining wild-type alleles) but reach less than 40% of wild-type alleles when used in combination. Hence, when the aim is to reach close to 100% alleles modified, our results suggest that it would be recommended to not exceed a distance of 300 bp between the two Cas9 cutting sites. A larger scale study would, however, be needed to dissect more precisely the benefits of the synergistic effect in relation to the distance between the Cas9 sites.
FIG. 3.
Synergistic effect of tandem guide RNA (gRNA) combinations. (A) Bar chart displaying the percentage of remaining wild-type alleles in CRISPR/Cas9 gene editing (GE) experiments in HepG2 cells using either each gRNA alone—the most efficient guide (driver [D]), the second guide (helper [H]), or the synergistic tandem combination (T)—for 12 targets (14 combinations). An expected additive gRNA combination has been calculated (see Methods) and is also displayed. Distance between the two Cas9 sites is indicated inside brackets. No synergistic effect is observed when the two sites are too close. Horizontal bars: 50% and 90%. (B) Scatter plot showing the synergistic benefit (calculated as the difference between the %GE obtained with the tandem synergistic approach and the %GE that would be expected if the effect of the two gRNAs would only be additive) in relation to the distance in between the two Cas9 sites. Green dots, positive synergistic benefit; red dots, negative synergistic benefit. ADK-S, 1 bp in between Cas9 sites; ADK-L, 58 bp; FABP-S, 26 bp; FABP-L, 52 bp. Horizontal bar, 35 bp. (C) Bar chart showing the relative quantification of the different gene editing outcomes. Under these optimal conditions almost all alleles are gene edited, with a large majority carrying the deletion resulting from both Cas9 cuts. IF, in frame; ORF, open reading frame. Horizontal bars, 10% and 90%.
Optimized gRNA design for tandem gRNA-CRISPR approach
Our tandem gRNA CRISPR strategy has several advantages over the generation of KO cell clones: (i) Clone isolation is a lengthy, work intensive process whereas KO in cell pools is fast and versatile. (ii) KO clones display genetic heterogeneity, whereas cell pools mitigate cell-to-cell differences. (iii) The tandem gRNA protocol does not require the tedious process of finding highly efficient gRNAs. A combination between a main (“driver”) gRNA with medium efficiency (>30% indel rate) and a second (“helper”) gRNA with at least some measurable efficiency (>10%) is often enough to reach close to 100% indel rate. Because lower efficacy is tolerated, this strategy allows to design gRNAs with more stringent specificity criteria (e.g., more mismatches to next homologous sequence, see “Material and Methods”), which minimize further the risk of gRNA off-target effects. (iv) The tandem approach leads to higher homogeneity in the gene editing outcome since the deletion in between the two Cas9 site is highly favored, which also increases the chances to get a full KO. Whereas the driver gRNA is designed within the exon, the helper gRNA may target an intron sequence, leading then to the skipping of the whole exon thereby increasing the probability to get a full gene KO, especially if the splicing exclusion results in a disruption of the open reading frame (ORF). However, it should be noted that in this case the helper gRNA should be significantly less efficient than the driver gRNA in generating mutations, since indels generated only in the intronic sequence would almost always result in the expression of the wild-type protein. Following those guidelines, Figure 3C summarizes data obtained for 11 different sgRNA combinations. In all cases the gene editing rate is very high with, often, less than 2% of alleles with wild-type ORF. For FABP1 and PDXK, most of the remaining alleles with wild-type ORF are due to indels located within the targeted intron. In all cases, the deletion between the two Cas9 sites represents 60–95% of gene edited alleles predicting with good confidence a functional KO of the targeted gene.
Quantification of residual target expression is necessary to monitor efficiency of target KO
In a previous study,26 we have demonstrated that many clones predicted to have a full gene KO after DNA sequencing, still express significant levels of the encoded protein. These results can be explained either by a splicing event which removes the edited exon or by the expression of a protein fragment generated 5′of the indel site or through a new translation initiation methionine. These truncated forms of the target protein may retain some functionality.26 It is therefore essential to make sure that the protein target becomes depleted in the gene edited cell pools. This can be achieved by measuring the residual target levels using an MS3 protocol that allows a precise quantification of protein depletion. Table 1 shows that the lowest residual protein levels are often reached from day 7 (D7) onwards following electroporation (labelled as D0). In all examples listed, residual target levels were quantified from 18% down to 1% of wild-type target levels. These quantification values include a background linked to the MS method used that is estimated to be between 1% and 10%. To better quantify this signal background for each target, we included in the same MS measurement extracts from clones KO for that target (when available). Table 1 shows that the residual target quantification in the KO clone extracts is in the range of 3–9%, which should solely reflect the MS background. When this value is deduced from the quantification of the residual target levels from the cell pools at D7 after electroporation, we come to a value between 2 (PDXK) and 12% (ALDH3A2, ECH1) of wild-type levels. No background reference from KO clones is available for CES2 and PGAM1, but the residual target quantification measured in the KO cell pools is already quite low (9 and 1% respectively). The protein levels usually remain stable from Day 7 onwards, as seen for the metabolic enzyme PGAM1 (still only 1% at D17). In a few cases, however, the small fraction of cells that still expresses wild-type alleles increases with time because the target gene impacts cell proliferation, as in the case of the aspartate tRNA ligase DARS2. The lowest residual protein level was quantified at 17% at D7 (Table 1); cells start to die after ∼2 weeks of culture and at D17, cells still carrying a wild-type allele become enriched, as shown by the increased levels of quantified DARS2 protein levels (38%). This enrichment is confirmed by the Sanger sequencing of the DARS2 allele population over time, showing an increase of the wild-type allele fraction from 0.7% (D4) to 8.6% (D17) (Supplementary Fig. S2). Overall, these data show that very low residual target protein levels can be achieved with the tandem pool strategy due to a very small fraction of remaining wild-type alleles.
Table 1.
Mass spectrometry protein quantification of residual targets of gRNA combinations from Fig. 3C
| Target | Exon targeted | ΔCas9 sites (bp) | Guide RNA location | MS target quantification % of WT target level (days after electroporation) |
||||
|---|---|---|---|---|---|---|---|---|
| D5 | D6 | D7 | D17 | KO clone | ||||
| ADK | exon 5 | 58 | Ex5 - Ex5 | 20 | 12 | 7 | 3 | |
| ADK | exon 9 | 36 | Ex9 - In9 | 13 | 15 | 10 | 5 | |
| ALDH3A2 | exon 4 | 198 | In3 - Ex4 | 23 | 17 | 17 | 5 | |
| ALDH3A2 | exon 4 | 61 | Ex4 - Ex4 | 24 | 13 | 10 | 2 | |
| CES2 | exon 2 | 46 | Ex2 - Ex2 | 23 | 15 | 9 | ||
| CPOX | exon 1 | 95 | Ex1 - Ex1 | 22 | 17 | 16 | ||
| ECH1 | exon 2 | 104 | Ex2 - Ex2 | 66 | 30 | 18 | 6 | |
| FABP1 | exon 1 | 52 | Ex1 - In1 | 26 | 20 | 16 | 5 | |
| PDXK | exon 4 | 83 | Ex4 - In4 | 7 | 11 | 9 | ||
| PGAM1 | exon 2 | 274 | Ex2 -In2 | 6 | 1 | 1 | ||
| DARS2 | exon 4 | 52 | In3 - Ex4 | 26 | 17 | 38 | ||
Targeted genes and exons and location (exon or intron) of the targeted sequences and the distance between both Cas9 sites are listed, as well as the relative residual mass spectrometry (MS) protein quantification at days 5, 6, 7, and 17 after the electroporation. Note that the MS background accounts for ∼1% to 10% of total quantification and can be better resolved by comparing with the quantification of the protein sample coming from a respective target knock-out clone (KO column). Quantification is usually lowest from D7 on, but for essential genes like DARS2, longer culture leads to an enrichment of cells carrying a nondefective allele.
In vitro synthesized sgRNA induces a strong but transient antiviral interferon response
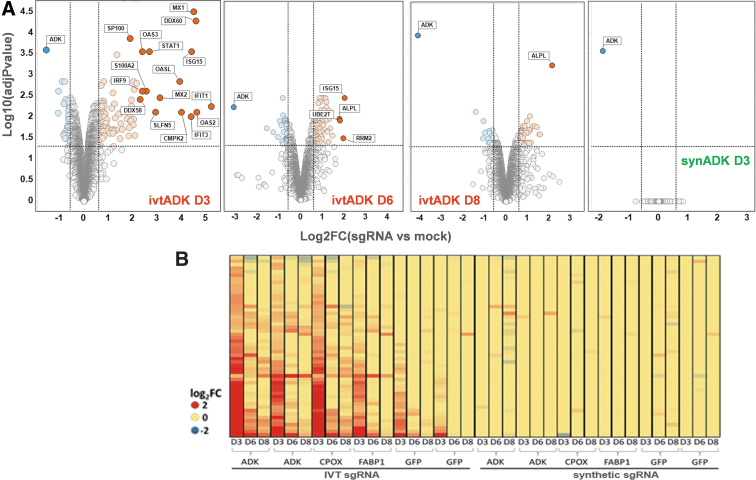
It has been previously reported that sgRNAs synthesized in vitro are cytotoxic. These sgRNAs are phosphorylated in 5′ and are hence recognized as viral RNA29,30. Since the recognition of foreign oligonucleotides activates several cellular pathways which may modify the proteome - and thus confounds our analysis -, we aimed at monitoring the extent and duration of the cellular response to electroporation of in vitro synthesized sgRNAs at the protein level using unbiased MS whole proteome profiling. We used sgRNAs directed against three cellular targets: ADK, FABP1, and CPOX as well as a sgRNA targeting GFP. These sgRNAs were generated either by in vitro transcription of double-stranded DNA oligonucleotides (and were therefore phosphorylated in 5′) or purchased as synthetic sgRNAs (not phosphorylated). Cells were harvested at day 3 (D3), day 6 (D6) and day 8 (D8) after electroporation and proteomes were compared with mock-electroporated HepG2 cells. In vitro synthesized ADK sgRNAs induced a strong antiviral response (Fig. 4A); in particular, several proteins linked to the interferon pathway were strongly upregulated at D3 when compared with the mock electroporated cells (Table 2). Such an effect is also seen with all in vitro synthesized sgRNAs (Fig. 4B) although the intensity of response may depend on the sequence and/or batch preparation of the sgRNA. This antiviral response is however transient: the number and level of upregulated proteins belonging to the interferon pathway are lower at D6 compared with D3 and these effects are barely detectable at D8. As a milder effect is induced by the control GFP sgRNA which should not target any site in the genome, it cannot be excluded that the double strand breaks generated during the DNA cutting and repairing process exacerbate the antiviral response. By comparison, no significant response was detected when using the synthetic sgRNAs that are not phosphorylated (Fig. 4A and B). So, synthetic sgRNAs are much more tolerated by the cells and should be preferentially used, especially when generating omics datasets or when using primary cells that sense foreign oligonucleotides (as T cells).
FIG. 4.
In vitro transcribed sgRNAs induce a strong but transient antiviral response (HepG2). (A) Volcano plots displaying proteins significantly regulated (red, upregulated; blue, downregulated) in ADK gene-edited cell pools compared with mock electroporated cells using either in vitro transcribed sgRNAs (ivtADK; at D3, D6, and D8 after electroporation) or synthetic sgRNA (synADK at D3). Data show the strong but transient antiviral response induced by IVT sgRNAs but not by synthetic sgRNAs. Significance is determined by the FC (|log2(FC)| > 0.58; vertical lines) and adjusted p-values (p < 0.05; horizontal line). (B) Heatmap displaying all proteins identified that are significantly regulated at D3 when using IVT ADK sgRNA (when compared with mock electroporated cells). ADK and GFP experiments have been performed twice as true biological duplicates. Antiviral response is not observed when using synthetic guides. FC, fold change.
Table 2.
Gene ontology enrichment of cells transfected with in vitro transcribed single guide RNA shows a strong antiviral response
| Term | Count | % | p-Value | Fold enrichment | Benjamini | FDR |
|---|---|---|---|---|---|---|
| GO:0060337, type I interferon signaling pathway | 18 | 18.18 | 5.33 × 10−24 | 39.50 | 3.87 × 10−21 | 8.08 × 10−21 |
| GO:0051607, defense response to virus | 22 | 22.22 | 1.10 × 10−22 | 21.04 | 4.00 × 10−20 | 1.67 × 10−19 |
| GO:0009615, response to virus | 17 | 17. 17 | 1.24 × 10−17 | 21.87 | 3.01 × 10−15 | 1.89 × 10−14 |
| GO:0060333, interferon gamma—mediated signaling pathway | 14 | 14.14 | 7.74 × 10−17 | 31.65 | 2.02 × 10−14 | 1.67 × 10−13 |
| GO:0045071, negative regulation of viral genome replication | 10 | 10.1 | 2.25 × 10−11 | 28.70 | 3.27 × 10−9 | 3.42 × 10−8 |
| GO:0045087, innate immune response | 14 | 14.14 | 1.26 × 10−8 | 8.03 | 1.53 × 10−6 | 1.91 × 10−5 |
| GO:0006955, immune response | 9 | 9.09 | 5.25 × 10−6 | 9.07 | 5.44 × 10−4 | 7.95 × 10−3 |
| GO:0032480, negative regulation of type I interferon production | 6 | 6.06 | 1.29 × 10−5 | 18.65 | 1.17 × 10−3 | 1.95 × 10−2 |
Gene ontology analysis was performed on the dataset corresponding to the ADK KO samples generated with the in vitro transcribed (IVT) sgRNAs and harvested 3 days after electroporation. GO analysis shows strong enrichment of terms related to viral infection and interferon pathway activation.
Tandem gRNA CRISPR enables highly efficient gene editing in primary cells
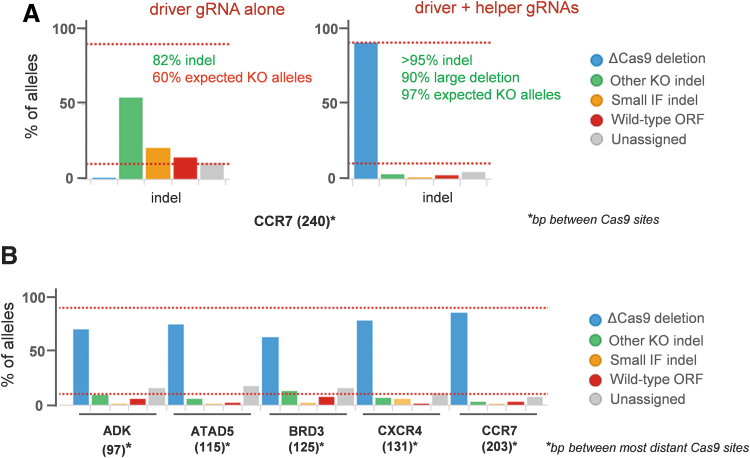
To assess the performance of our method in primary cells, we chose to knock out the CCR7 chemokine receptor in nonactivated primary human CD4+ T cells using a pair of synthetic gRNAs (crRNA + tracrRNA) as RNP complexes. DNA sequencing data showed that when used alone, the driver gRNA still leaves ∼12% of wild-type alleles; in addition, about 21% of alleles carry short deletions corresponding to the loss of one or two amino acids that may still result into the expression of a functional receptor and thus may not generate any detectable phenotype (Fig. 5A). However, when associated with a helper gRNA located in a neighboring intron, 90% of alleles carried the deletion in between the two Cas9 sites (238–240 bp) and only ∼2% of wild-type alleles could be quantified. This shows that the deletion between the gRNA binding sites is also greatly favored in primary cells, increasing the probability of generating a genuine KO cell population. This strategy based on combining several gRNAs produces highly efficient gene editing also in anti-CD3/anti-CD28 activated CD4+ T cells (Fig. 5B).
FIG. 5.
Multiple gRNA approach in primary T cells. (A) KO of CCR7 chemokine receptor in nonactivated primary T cells. When used alone, the driver guide is not performant enough, with only ∼60% of alleles carrying indels disrupting the ORF. In combination with a helper guide located in an intron, nearly all alleles carry the deletion resulting from the Cas9 digest, leading to a performant and homogenous gene editing. (B) Efficient KO of several targets using gRNA triplets in activated CD4+ T cells. Large deletions are highly favored, leading to a relatively homogenous gene editing outcome. IF: in frame; ORF: open reading frame. Horizontal bars: 10% and 90%.
Tandem gRNA CRISPR KO enables rapid identification of KO phenotypes
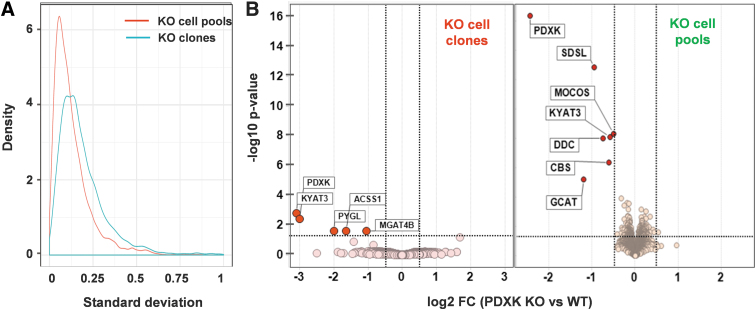
We compared the phenotypic proteomics data generated in KO clones with those generated via the tandem gRNA strategy of KO cell pools. We assessed the proteomics signatures of three KO cell clones for the pyridoxal kinase PDXK (Fig. 1) to the proteomics data generated on cells pools using synthetic sgRNA pairs from three biological replicates (generated at different dates). PDXK converts pyridoxal (vitamin B6) into pyridoxal phosphate, a cofactor for several metabolic enzymes. The standard deviation between replicates clearly shows that the cell pool dataset shows less variation around the mean than the dataset from the three PDXK KO clones (Fig. 6A), confirming the heterogeneity of these latter (similar comparisons for three other targets are displayed in Supplementary Fig. S3). Unbiased whole proteome analysis identified four proteins significantly (adjP <0.05) downregulated in the PDXK KO clones (in addition to PDXK itself) when compared with clones generated from mock electroporated HepG2 cells (Fig. 6B, left panel). These downregulated proteins include two metabolic enzymes which use pyridoxal-phosphate as cofactor: KYAT3 (kynurenine-oxoglutarate transaminase 3) and PYGL (liver glycogen phosphorylase). From the corresponding dataset generated by using tandem gRNAs in cell pools, 6 proteins were identified as significantly downregulated in addition to PDXK (Fig. 6B, right panel); all 6 are pyridoxal-phosphate dependent metabolic enzymes: KYAT3, SDSL (serine dehydratase-like), DDC (aromatic -L-amino-acid decarboxylase), GCAT (2-amino-3-ketobutyrate coenzyme A ligase), MOCOS (molybdenum cofactor sulfurase), and CBS (cystathionine beta-synthase). PYGL was quantified but not significantly downregulated. In particular, the strong similarity between the three biological replicates resulted into very low p-values, increasing confidence into the identification of proteins specifically affected by PDXK silencing. Two additional enzymes (ACSS1 and MGAT4B) are significantly downregulated in the clonal dataset but not in the cell pool dataset. As they do not use pyridoxal phosphate as cofactor and are not a priori linked to pyridoxal metabolism, these proteins may be false negative possibly resulting from the clonal heterogeneity. In summary, our approach using KO cell pools generated with the tandem gRNAs leads to more clearly identifiable phenotypes by proteomic analysis.
FIG. 6.
Proteomics phenotype of PDXK KO cells. Comparison between HepG2 clones (average of three clones) and HepG2 cell pools (average of three biological replicates) KO for PDXK. (A) Graphs comparing the standard deviation (normalized log10 of sum ion area) of MS protein quantification for samples for KO clones (blue) and KO cell pools (red). The cell pool dataset is much tighter than the clonal one. (B) Side-by-side comparison of volcano plots generated from PDXK KO clones or from PDXK KO cell pools. FC is calculated by comparison with mock-electroporated HepG2 cells. All five proteins (in addition to PDXK) that are identified as significantly down-regulated (log2(FC) <-0.5; P < 0.05) in the cell pool experiments are enzymes using pyridoxal-P as cofactor. Only two out of four proteins significantly downregulated in the KO clone experiment are enzyme binding pyridoxal-P; in addition, adjusted p-values are much higher than for the cell pool experiment.
Discussion
Gene KO by CRISPR-Cas9 has become a commodity but the most commonly used procedure of generating KO clones is time and resource intensive and there is no general agreement in the field regarding the most efficient gRNA design. In addition, the subcloning procedure is not amenable to silencing essential genes nor to primary cells that most often do not efficiently proliferate in culture. Current CRISPR-Cas9 strategies using cell pools are unsatisfactory since they require either extremely efficacious gRNAs or the use of vector-based systems coupled with drug selection. The first requirement is quite challenging in general and in particular for genes for which a limited number of specific gRNAs can be designed. The second bears the risk of increasing off-target effects which confound downstream studies. Our tandem gRNA CRISPR strategy on cell pools benefits from extremely efficient gene editing (most often >95% of edited alleles), the use of simple tools (not optimized gRNAs), versatility and speed. It takes advantage of the synergy occurring when two gRNAs target sequences that are located at close proximity (ideally within 40–300 bp), resulting in a strong increase in the indel rate. It has been previously reported that the binding of an efficient Cas9 RNP complex (containing spCas9) allows, by altering the local chromatin context, the nearby recruitment of a low-efficiency Cas9 RNP complex (fnCas9, from francisella novicida) that would not have alone any activity.31 We hypothesize that the binding of the most efficient (driver) RNP complex in a similar way helps recruiting a less efficient (helper) complex in its vicinity. The tandem approach also leads to a more predictable and more homogenous gene editing since the deletion in between the two Cas9 cuts is strongly favored (representing in most cases 60 to 95% of allele editing). These relatively small deletions (<300 bp) generate minimal genome perturbation when compared with approaches using a pair of gRNAs targeting sequences located in different exons many kilobases apart, resulting in deletion of large intragenic regions, in less predictable editing outcome and in possible interference with gene regulatory sequences.32–34 This strategy is amenable to the study of essential genes, as the cells can be used for biological assay or to derive omics datasets quite early after the CRISPR process (i.e., prior to the induction of cell death). It is also applicable to primary cells, especially when using synthetic gRNAs that are less cytotoxic since they do not induce a strong antiviral response. Using a pair of gRNAs might even reduce the frequency of double-strand DNA breaks, because when Cas9 cuts simultaneously at both sites, the ligation of both ends is favored, breaking the “DNA cut and repair” futile cycle12 that is observed when using a single gRNA. MS quantification of the residual levels of the cognate protein is required to exclude that truncated versions of the protein are expressed following an early stop codon or exon skipping which might confound the analysis and interpretation of the KO phenotype. Moreover, in KO cell pools there might be less successful attempts to rescue the gene KO, since the sample generation is much faster, leaving little time for the cells to adapt to the KO situation.
Conclusion
We have designed an efficient and versatile approach to generate CRISPR-Cas9 KO cell pools within a few days with highly predictable gene editing outcome. This method is well suitable to derive omics datasets for target characterization and validation and is amenable to CRISPR array screening.
Supplementary Material
Acknowledgments
Authors would like to thank the tissue culture and mass spectrometry Cellzome teams for their technical expertise.
Gérard Joberty participated in study design, performed experiments, analyzed data, and participated in writing the manuscript; Maria Fälth-Savitski participated in study design, analyzed data, and participated in writing the manuscript; Marcel Paulmann performed experiments; Markus Bösche performed mass spectrometry analyses; Carola Doce analyzed data; Aaron T. Cheng, Gerard Drewes, and Paola Grandi participated in study design and writing the manuscript. All co-authors reviewed and approved of the manuscript prior to submission. The manuscript has been submitted solely to this journal and is not published, in press, or submitted elsewhere.
Author Disclosure Statement
All authors are employees and some are shareholders of GlaxoSmithKline, which funded the work.
Funding Information
Funding was received from GlaxoSmithKline.
Supplementary Material
References
- 1. Barrangou R, Fremaux C, Deveau H. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007;315:1709–1712. DOI: 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- 2. Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012;337:816–821. DOI: 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mali P, Yang L, Esvalt KM, et al. RNA-guided human genome engineering via Cas9. Science 2013;339:823–826. DOI: 10.1126/science [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819–823. DOI: 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu Y, Liang D, Wang Y, et al. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell 2013;13:659–662. DOI: 10.1016/j.stem.2013.10.016 [DOI] [PubMed] [Google Scholar]
- 6. Platt RJ, Chen S, Zhou Y, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 2014;159:440–455. DOI: 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Qi LS, Larson MH, Gilbert LA, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013;152:1173–1183. DOI: 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gilbert LA, Larson MH, Morsut L, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013;154:442–451. DOI: 10.1016/j.cell.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gilbert LA, Horlbeck MA, Adamson B, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 2014;159:647–661. DOI: 10.1016/j.cell.2014.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Konermann S, Brigham MD, Trevino AE, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2015;517:583–588. DOI: 10.1038/nature14136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zalatan JG, Lee ME, Almeida R, et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 2015;160:339–350. DOI: 10.1016/j.cell.2014.11.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mladenov E, Iliakis G. Induction and repair of DNA double strand breaks: the increasing spectrum of non-homologous end joining pathways. Mutat Res 2011;711:61–72. DOI: 10.1016/j.mrfmmm.2011.02.005 [DOI] [PubMed] [Google Scholar]
- 13. Kleinstiver BP, Pattanayak V, Prew MS, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016. 529:490–495. DOI: 10.1038/nature16526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen JS, Dagdas YV, Kleinstiver BP, et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 2017;550:407–410. DOI: 10.1038/nature24268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vakulskas CA, Dever DP, Rettig GR, et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat Med 2018;24:1216–1224. DOI: 10.1038/s41591-018-0137-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kleinstiver BP, Prew MS, Tsai SQ, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015;523:481–485. DOI: 10.1038/nature14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu JJ, Orlova N, Oakes BL, et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature 2019;566:218–223. DOI: 10.1038/s41586-019-0908-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ran FA, Hsu PD, Lin CY, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013;154:1380–1389. DOI: 10.1016/j.cell.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim S, Kim D, Cho SW, et al. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 2014;24:1012–1019. DOI: 10.1101/gr.171322.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Doench JG, Hartenian E, Graham DB, et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol 2014;32:1262–1267. DOI: 10.1038/nbt.3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tsai SQ, Zheng Z, Nguyen NT, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 2015;33:187–197. DOI: 10.1038/nbt.3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Doench JG, Fusi N, Sullender M, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 2016;34:184–191. DOI: 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Allen F, Crepaldi L, Alsinet C, et al. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat Biotechnol 2018; Nov 27. [Epub ahead of print] DOI: 10.1038/nbt.4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chakrabarti AM, Henser-Brownhill T, Monserrat J, et al. Target-specific precision of CRISPR-mediated genome editing. Mol Cell 2019;73:699–713.e6. DOI: 10.1016/j.molcel.2018.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang D, Zhang C, Wang B, et al. Optimized CRISPR guide RNA design for two high-fidelity Cas9 variants by deep learning. Nat Commun 2019;10:4284 DOI: 10.1038/s41467-019-12281-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smits AH, Ziebell F, Joberty G, et al. Biological plasticity rescues target activity in CRISPR knock outs. Nat Methods 2019;16:1087–1093. DOI: 10.1038/s41592-019-0614-5 [DOI] [PubMed] [Google Scholar]
- 27. Oh SA, Seki A & Rutz S. Ribonucleoprotein transfection for CRISPR/Cas9-mediated gene knockout in primary T cells. Curr Protoc Immunol 2019;124:e69 DOI: 10.1002/cpim.69 [DOI] [PubMed] [Google Scholar]
- 28. Ben-David U, Siranosian B, Ha G, et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature 2018;560:325–330. DOI: 10.1038/s41586-018-0409-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim S, Koo T, Jee HG, et al. CRISPR RNAs trigger innate immune responses in human cells. Genome Res. 2018; Feb 22. [Epub ahead of [print]. DOI: 10.1101/gr.231936.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wienert B, Shin J, Zelin E, et al. In vitro-transcribed guide RNAs trigger an innate immune response via the RIG-I pathway. PLoS Biol 2018;16:e2005840 DOI: 10.1371/journal.pbio.2005840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen F, Ding X, Feng Y, et al. Targeted activation of diverse CRISPR-Cas systems for mammalian genome editing via proximal CRISPR targeting. Nat Commun 2017;8:14958 DOI: 10.1038/ncomms14958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Canver MC, Bauer DE, Dass A, et al. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J Biol Chem 2014;289:21312–21324. DOI: 10.1074/jbc.M114.564625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Giuliano CJ, Lin A, Girish V, et al. Generating single cell-derived knockout clones in mammalian cells with CRISPR/Cas9. Curr Protoc Mol Biol 2019;128:e100 DOI: 10.1002/cpmb.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zheng Q, Cai X, Tan MH, et al. Precise gene deletion and replacement using the CRISPR/Cas9 system in human cells. BioTechniques 2014;57:115–124. DOI: 10.2144/000114196 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.