Abstract
Bemisia tabaci cryptic species Middle East-Asia Minor I (MEAM1) is a serious agricultural polyphagous insect pest and vector of numerous plant viruses, causing major worldwide economic losses. B. tabaci control is limited by lack of robust gene editing tools. Gene editing is difficult in B. tabaci due to small embryos that are technically challenging to inject and which have high mortality post injection. We developed a CRISPR-Cas9 gene editing protocol based on injection of vitellogenic adult females rather than embryos (“ReMOT Control”). We identified an ovary-targeting peptide ligand (“BtKV”) that, when fused to Cas9 and injected into adult females, transduced the ribonucleoprotein complex to the germline, resulting in efficient, heritable editing of the offspring genome. In contrast to embryo injection, adult injection is easy and does not require specialized equipment. Development of easy-to-use gene editing protocols for B. tabaci will allow researchers to apply the power of reverse genetic approaches to this species and will lead to novel control methods for this devastating pest insect.
Introduction
Bemisia tabaci cryptic species Middle East-Asia Minor I (MEAM1) is a widely distributed invasive agricultural pest that causes the economic loss of billions of dollars in crop damages worldwide.1,2 B. tabaci is polyphagous, with a broad host range. This insect feeds on plant phloem sap through its life cycle using piercing-sucking mouthparts and can cause direct damage to plants. The honeydew excretions from whiteflies promote fungus growth that can reduce photosynthesis and crop yields. Finally, B. tabaci is an important vector of numerous plant viruses that affect economically critical crop species. B. tabaci is the only known vector of begomoviruses—a family of plant viruses known to cause plant diseases and adversely affect crop yield.2 Current control methods for B. tabaci are insecticides and limited use of biological control.3,4 Plant-mediated RNAi targeting begomoviruses or the whitefly vector has shown some promise in laboratory settings but has not been translated to field applications.5–8 Although RNAi can be effective at reducing gene transcription, efficacy can be highly variable, depending on the gene and tissue of interest. Thus far, the lack of tools to manipulate whitefly genetically hinders the screening of potential genetic targets that can be used to design agricultural control strategies.
The economic importance of B. tabaci demands new methods to control this devastating pest insect. Gene editing, using CRISPR-Cas9, has formed the foundation for novel control strategies for insect vectors of human diseases9–13 and plant diseases,14 but a lack of gene editing techniques for B. tabaci is a significant barrier to the application of gene editing for basic biological studies and control of this insect. Arthropod gene editing by CRISPR-Cas9 is usually performed by injecting gene editing materials into pre-blastoderm embryos, but the exceedingly small size of B. tabaci embryos (0.2 mm) and high mortality of injected eggs makes this technically challenging. A method has recently been developed called “ReMOT Control” (Receptor-Mediated Ovary Transduction of Cargo), which circumvents the need to inject embryos, using instead a small ovary-targeting peptide to transduce the Cas9 ribonucleoprotein complex (RNP; the Cas9 protein complexed with a guide RNA) directly into the developing ovaries upon injection into the hemolymph of adult female insects. ReMOT Control has been shown to edit the germline of the mosquitoes Aedes aegypti and Anopheles stephensi efficiently.9,10
Here, we develop an efficient ReMOT Control CRISPR-Cas9-based adult injection protocol for gene editing in B. tabaci. The development of robust gene editing methodologies for this species opens the power of CRISPR and reverse genetic approaches to study the biology and to develop new control strategies for this important economic pest.
Methods
B. tabaci colony maintenance
A colony of B. tabaci MEAM1 (originally collected from a poinsettia plant in Ithaca, NY) was maintained in a growth chamber set at 28°C ± 2°C, 14L/10D photoperiod, and 50% relative humidity in 63.5 cm × 63.5 cm × 63.5 cm cages with organza access sleeves. Insects were maintained on organic soybean (var. Viking; Johnny's Selected Seeds), organic radish (var. Cherry Belle; Burpee Seeds and Plants), and/or gerbera daisy (var. Garvinea; Burpee Seeds and Plants). Plants were grown in a separate growth chamber using the same parameters as the insect growth chamber. Plants were grown in 15 cm pots using enriched potting media with liquid fertilizer (Peter's Excel Cal-Mag 15-5-15) applied twice a month.
Plasmid construction and protein expression
Based on a predicted 24 aa vitellogenin binding sequence from the crustacean Macrobrachiumrosen bergii,15 we identified the homologous sequence from multiple B. tabaci vitellogenin genes (XP_018897090, XP_018912902, XP_018897089), and from the mosquitoes Ae. aegypti (AAA994861.1) and An. gambiae (AAF82131.1). We identified a lysine conserved across B. tabaci sequences and a fully conserved valine residue that defined the ends of the targeting sequence (“BtKV”; Supplementary Fig. S1). pET28A-BtKV-Cas9 was constructed from pET28-P2C-Cas910 and a gblock (IDT) of BtKV-mCherry. pET28A-P2C-Cas9 was digested with restriction enzymes BamHI and SalI. A gblock of BtKV fused to mCherry was synthesized and inserted into the digested pET28-P2C-Cas9 using In-Fusion Cloning (Takara Bio). Sanger sequencing was used to verify the sequence. pET28A-BtKV-mCherry-Cas9 was further digested with either SalI or XhoI to remove mCherry or Cas9, respectively, gel purified, and re-ligated using T4 ligase to obtain the plasmids pET28A-BtKV-Cas9 or pET28A-BtKV-mCh. Plasmids were transformed into BL21(DE3) cells for BtKV-Cas9/mCh expression following standard protein expression protocols. P2C-Cas9 and Cas9 were expressed using plasmids, as previously described.10 Briefly, a 5 mL starter culture grown overnight at 37°C with shaking at 225 rpm was used to inoculate 50 mL Terrific Broth (Invitrogen) at a 1:100 dilution. When the culture reached an OD600 of 0.4–0.8, it was induced with isopropyl-β-D-1-thiogalactopyranoside (IPTG) at a concentration of 500 μM at 20°C overnight. Cultures were pelleted, re-suspended in lysis buffer (20 mM Tris-HCl, pH 8.0, 300 mM NaCl, 20 mM imidazole) and sonicated. Cell debris was pelleted, and the supernatant was collected and used for immobilized metal affinity chromatography using Ni-NTA agarose beads (Qiagen) and agitated for at least 2 h at 4°C. Subsequently, the beads were separated in a chromatography column and washed with lysis buffer allowing gravity flow. Proteins were eluted (20 mM Tris-HCl, pH 8.0, 300 mM NaCl, 250 mM imidazole) and dialyzed in 20 mM Tris-HCl, pH 8.0, 300 mM KCl, 500 μM phenylmethylsulfonyl fluoride. Proteins were concentrated using an Amicon Ultra 0.5 mL centrifugal filter device with a cutoff of 100 kDa (Millipore Sigma).
Generating guide RNAs
Single guide RNA (sgRNA) were generated following the protocols found here.11 The 20 nucleotide guide sequences were designed against exon 2, exon 3, and exon 5 of the B. tabaci white gene (NW_017550151, region 408016-472564) both manually and by CRISPOR.16 Primers and guide RNA sequences are listed in Supplementary Table S1.
Injection protocol for B. tabaci
For initial experiments, B. tabaci adults of unknown age and mating status were aspirated from the colony cage into two-dram screw cap vials, placed in ice to anesthetize the insects, sexed, and transferred to a standard plain glass slide with double-sided tape on a chill table. For later experiments, we established a synchronous colony so that we could control for the age of injected insects (see below). For localization of BtKV fusion protein into the embryos, female B. tabaci were injected in the abdomen with BtKV-mCherry fusion protein (3 mg/mL) or phosphate-buffered saline (PBS) control using quartz capillary needles. Samples were collected 24 h post injection, and ovaries were dissected onto a concavity microscope slide with a glass coverslip and imaged by epifluorescence microscopy.
To generate mutants, the injection mixture consisted of the RNP complex of BtKV-Cas9 (3 mg/mL) with a mix of all sgRNAs (3 mg/mL) at a 1:3 molar ratio, and a third of the volume of saponin (4, 8, or 16 μg/mL) as an endosome escape reagent (or buffer as a control).9,10 Five sgRNA multiplexed were used at a concentration of 250 ng/μL each and 1.25 μg/μL in total. Two groups of females were collected: (1) females of unknown age and unknown mated status were collected at random, and (2) females <24 h post emergence (hpe) were collected to reduce their chances of having already developed eggs.17,18 The females were injected and placed on a piece of soybean leaflet in a Petri dish with a moist paper towel wrapped around the stem and another paper towel on the bottom plate to prolong the life of the leaflet. Water and liquid fertilizer were added as needed. Females were removed from the leaflet 2 weeks post injection. The Petri dishes were incubated at 28°C, 16L/8D cycle, in a humidified Heratherm IMH750-S reach-in chamber, and the progeny of injected females screened visually for altered eye color in nymphs or adults using an Olympus SZX7 stereomicroscope. Mutations in insects showing altered eye color were confirmed by polymerase chain reaction (PCR) using PHIRE Tissue Direct PCR Master Mix (Thermo Fisher Scientific), the amplicon was cloned into pJET1.2, and clones were submitted for Sanger sequencing.
Heritability crosses
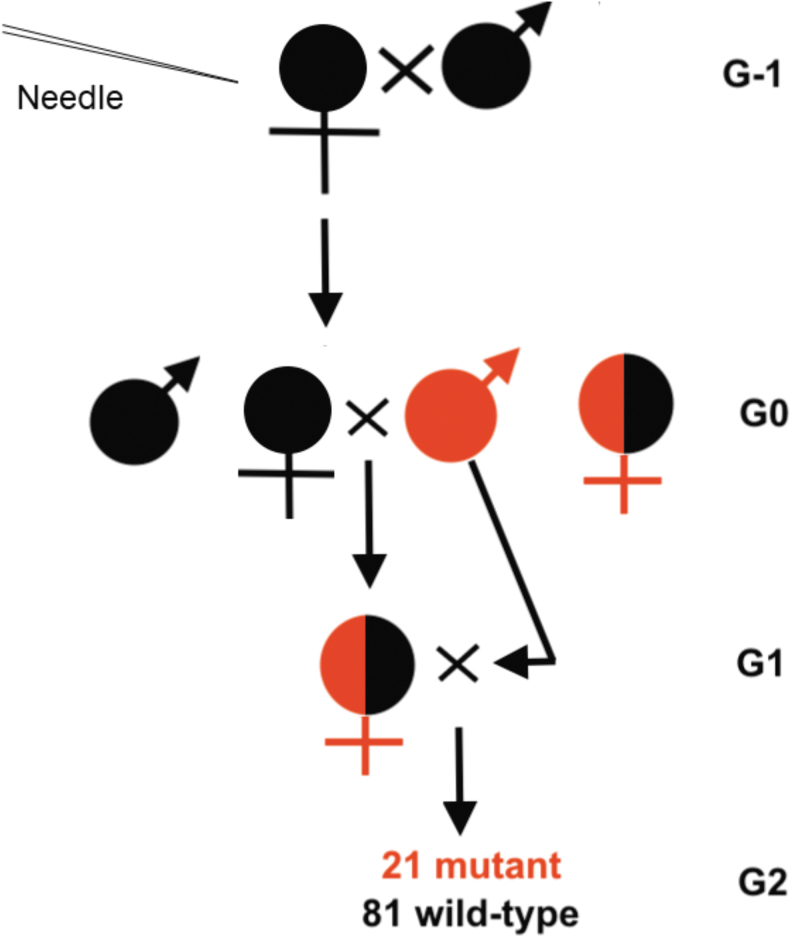
Crosses of mutant males and wild-type females were performed (Fig. 4) to demonstrate heritability of the mutant allele and phenotype. In brief, G0 mutant males resulting from injection of G1 females were interbred with wild-type G0 females. Resulting G1s were sexed and separated approximately 24 hpe. G1 females were backcrossed to a G0 mutant male to generate G2 offspring. Insects were screened for eye color in the nymphal stage.
FIG. 4.
Crossing scheme to identify heritability. Injected females (G1) can produce wild-type males and females (black), mutant males (red), and/or hemizygous females (red/black) in G0. The resulting mutant males were crossed with the female siblings to generate the G1. The G1 hemizygous females were backcrossed with G0 mutant males to generate G2 offspring, which were scored as juveniles for white or wild-type eyes.
Homology modeling of mutated proteins
A homology model of one in-frame insertion mutant (21 bp) was generated using the Swiss Model homology server (swissmodel.expasy.org). The structure of the B. tabaci white gene has not been empirically solved. So, a model of the wild-type protein was first generated against the database using the server, the result of which was used as template to model the 21 bp insertion mutant.
Results
Identification and validation of the BtKV ovary targeting ligand
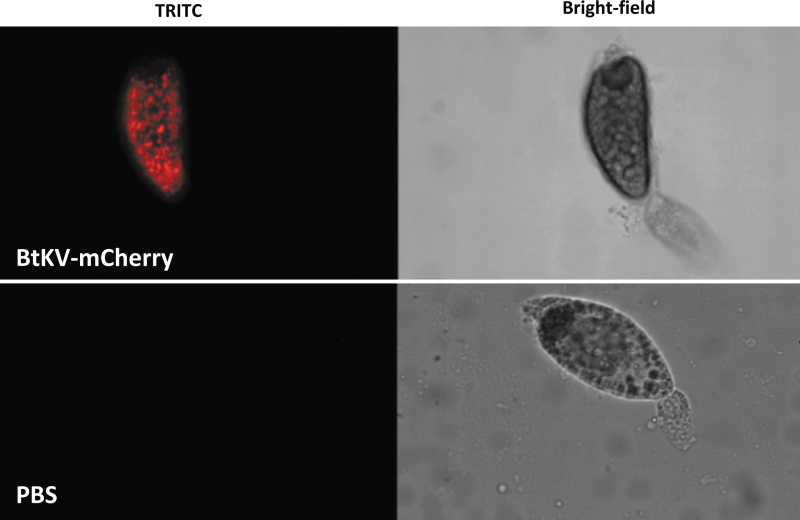
While the P2C ligand works to target cargo to the ovaries of multiple mosquito species,9,10 it did not function robustly in B. tabaci (Table 1). We therefore designed a new targeting peptide suitable for this insect species based on its endogenous vitellogenin protein (“BtKV”; KPYGVYKTMEDSV; see Methods). B. tabaci oocyte development is asynchronous with stage I, II, III, and IV at 11 days or more post eclosion where oocytes are always developing. Developmental phase II oocytes have the highest amount of vitellogenin uptake via endocytosis and nutrient cord.19 After injection of BtKV-mCherry into reproductive adult B. tabaci females, ovaries were dissected after 24 h and visualized for red fluorescence. We observed red fluorescence in the developing oocytes from females injected with BtKV-mCherry fusion protein but not PBS-injected controls (Fig. 1).
Table 1.
CRISPR editing efficiency in Bemisia tabaci
| Injection | Saponin (μg/mL) | Females injected (n) | Sampling method | Offspring screened (n) | Offspring edited (n) | KO/injected (%) | KO/offspring (%) | KO/offspring (%) sex ratio corrected |
|---|---|---|---|---|---|---|---|---|
| Cas9 | 0 | 31 | Random | 70 | 0 | 0 | 0 | 0 |
| Cas9 | 4 | 28 | Random | 218 | 0 | 0 | 0 | 0 |
| P2C-Cas9 | 0 | 20 | Random | 54 | 0 | 0 | 0 | 0 |
| P2C-Cas9 | 4 | 13 | Random | 74 | 0 | 0 | 0 | 0 |
| BtKV-Cas9 | 0 | 42 | Random | 961 | 1 | 2.38% | 0.10% | 0.39% |
| BtKV-Cas9 | 4 | 47 | Random | 261 | 2 | 4.26% | 0.77% | 2.84% |
| BtKV-Cas9 | 4 | 47 | Random | 189 | 1 | 2.13% | 0.53% | 1.96% |
| BtKV-Cas9 | 4 | 30 | Random | 314 | 1 | 3.33% | 0.32% | 1.18% |
| BtKV-Cas9 | 4 | 28 | Random | 180 | 1 | 3.57% | 0.56% | 2.06% |
| BtKV-Cas9 | 8 | 31 | Random | 378 | 0 | 0 | 0 | 0 |
| BtKV-Cas9 | 8 | 29 | Random | 144 | 0 | 0 | 0 | 0 |
| BtKV-Cas9 | 16 | 34 | Random | 121 | 0 | 0 | 0 | 0 |
| BtKV-Cas9 | 16 | 22 | Random | 284 | 0 | 0 | 0 | 0 |
| BtKV-Cas9 | 16 | 29 | Random | 73 | 0 | 0 | 0 | 0 |
| BtKV-Cas9 | 0 | 14 | <24 hpe | 226 | 0 | 0 | 0 | 0 |
| BtKV-Cas9 | 0 | 21 | <24 hpe | 118 | 15 | 71.43% | 12.71% | 20.72%a |
| BtKV-Cas9 | 4 | 15 | <24 hpe | 96 | 0 | 0 | 0 | 0 |
| BtKV-Cas9 | 4 | 26 | <24 hpe | 243 | 2 | 7.69% | 0.82% | 1.03%a |
Sex ratio corrected based on ratio of emerged males and females in the injected group.
KO, knockout; hpe, hours post emergence.
FIG. 1.
Targeting of molecular cargo to Bemisia tabaci ovaries. The BtKV peptide targets molecular cargo (mCherry-Cas9 fusion protein) into B. tabaci ovaries and is visible in dissected mature oocytes 24 h post injection (top). No signal is visible in control ovaries (bottom).
Knockout of the B. tabaci white gene by ReMOT Control
For proof of principle gene editing experiments, we chose the white gene, which is an ABC transporter protein responsible for transport of ommochrome pigments into the eyes. Null mutants for this gene have white eyes in multiple taxa.20–22 Cas9 proteins were complexed with five sgRNAs targeting exons 2, 3, and 5 of the gene (Supplementary Table S1). We used saponin as an endosome escape reagent, as it was functional in Aedes aegypti and Anopheles gambiae ReMOT Control editing experiments.9,10
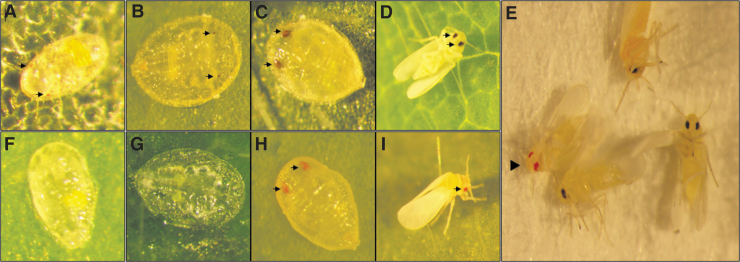
Cas9 RNP injections with no targeting peptide or with P2C as the targeting peptide, with or without saponin, did not result in any visibly edited offspring (Table 1). For injections using BtKV as the targeting peptide, white-eye nymphal offspring were recovered from 7/14 independent replicate injection experiments (Fig. 2 and Table 1). The data showed that mutants were only recovered in treatments without saponin or the lowest concentration of saponin; higher concentrations of saponin were inhibitory (Table 1). Mutants were recovered in 7/9 independent injection experiments using ≤4 μg/mL saponin. Finally, females <24 hpe with or without saponin generated mutants with higher efficiency compared to females of unknown ages (Table 1). Early nymphal instar mutants had white (colorless) eyes, but during development, the color changed to a bright orange-red color (Fig. 2) compared to the dark red-brown color of the wild type.
FIG. 2.
Wild-type and mutant B. tabaci. Wild-type from (A) second, (B) third, and (C) fourth instar nymphs and (D) adults. (E) Mutant and wild-type in the same image. (F–I) Matched-stage mutant individuals resulting from ReMOT Control editing of the B. tabaci white gene. Black arrows point to eye(s) of wild-type and mutants. Black arrowhead points to mutant adult.
Due to the haplodiploid nature of B. tabaci, we are much more likely to observe mutant males, as they are haploid. While we cannot determine the sex of mutants that did not survive to adulthood, all mutants that did make it to the adult stage were male. We assume that mutants identified in the immature stage were male as well. This can bias actual estimates of editing efficiency if the sex ratio is not 50:50. B. tabaci females can actively control the sex ratio of their offspring by controlling which eggs are fertilized, and published estimates of the sex ratio range from 25% to 75% female.23 We examined the sex ratio in our colony nine replicate times by random sampling and found it to be highly female biased, with estimates ranging from 65% to 85% female, and an average value of 73% female (Supplementary Table S2). When adjusted for sex ratio bias, we calculated an editing rate in male offspring within a range of 0.39–20.72% (Table 1), depending on whether young females were injected. When females <24 hpe were injected, editing rates in the offspring were >20% in some replicates—high enough for lineage screening by PCR to identify mutant individuals in the case of a gene without an obvious morphological phenotype. Across all replicates using ≤4 μg/mL saponin, injection of approximately 12 females was enough to obtain at least one edited offspring—an editing efficiency almost twofold greater than observed in mosquitoes.9,10
Analysis of mutants
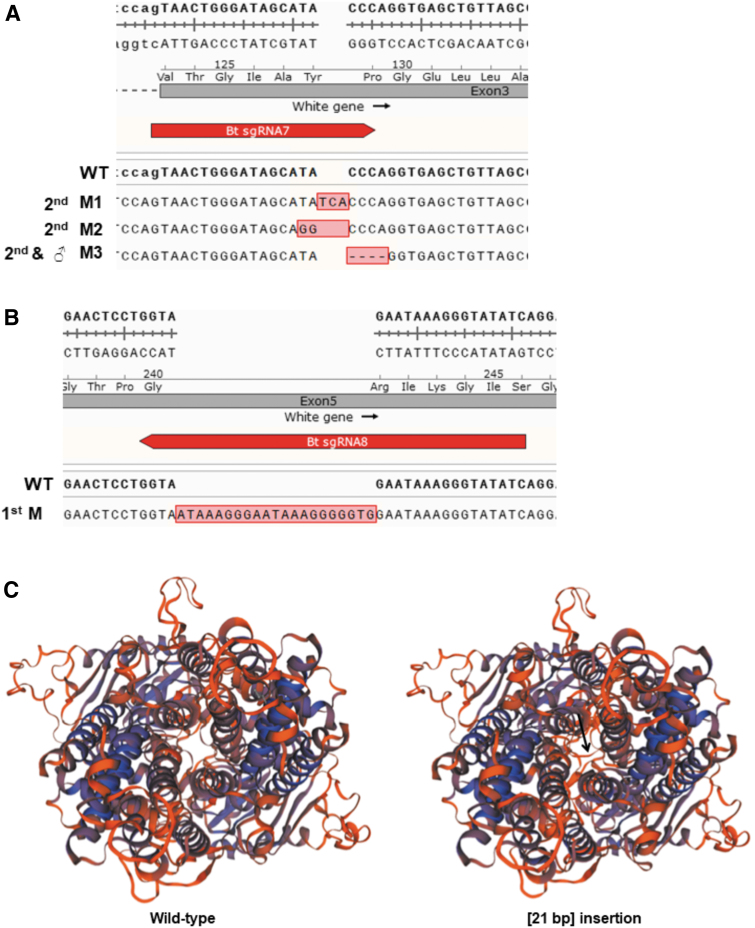
From the 23 mutant offspring visually identified, we randomly selected eight for molecular characterization and were able to obtain DNA of sufficient quality for sequencing from five offspring (three white-eye 1st or 2nd nymphal instars and two orange eye male adults). PCR of housekeeping genes was unsuccessful for the other three individuals, indicating degraded DNA. We validated gene knockout by PCR and sequencing of the product of the targeted gene locus in these five individuals. The sequencing produced multiple peaks for one individual, and so the PCR product was cloned into pJET1.2, and nine clones were sequenced. This individual (although phenotypically white eye) appeared to be a genetic mosaic, where we detected wild-type alleles and two insertion alleles (a three nucleotide insertion and a two nucleotide substitution). Another white-eye juvenile had a 21 bp in-frame insertion (insertion mutant; 21 bp). Homology modeling of this mutation predicted that it caused a loop to be cast across the pore region of the transporter, likely blocking the transport function of the gene by steric interference (Fig. 3C). The final white-eye juvenile had a 4 bp deletion in exon 3, causing a frame shift of the white gene. Similarly, the two orange-red eye males had the same 4 bp deletion.
FIG. 3.
Mutant alleles and homology modeling of the 21 b insertion. (A) The mutant alleles (M1: 3 nt insertion; M2: 2 nt substitution; M3: 4 nt deletion) identified at exon 3 were compared to the wild-type sequence. 2nd and ♂ indicates the instar/life stage the mutation was identified in (2 adult males shared this mutation). (B) The 21 bp insertion (M) at exon 5 in a first nymphal instar. (C) Predicted 3D structure of the wild-type B. tabaci white protein (left) compared to the structure of the in-frame 21 bp insertion mutant (right). The insertion is predicted to cause a loop (arrow) across the channel pore, sterically hindering function of the transporter, leading to a null mutation.
Heritability of generated mutations
To demonstrate that the mutations generated by ReMOT Control were heritable in B. tabaci, we performed a cross (Fig. 4) between male and female offspring of injected females. The males used in the cross were visibly mutant (white-eye as juvenile, orange-red eye as adults). The females were phenotypically wild type. All 90 G1 offspring had wild-type eye color, which indicates that the parents consisted of a mutant male and wild-type females. Subsequently, G1 females were backcrossed with the same G0 mutant male. The expected ratio of mutant to wild-type offspring resulting from a cross between a haploid mutant male to a hemizygous mutant female is 50:50. We observed 22 mutants and 81 wild-type offspring, suggesting a deviation from the expected ratio (χ2 = 17.84, p < 0.0001), Nevertheless, the data demonstrate that the mutation was heritable and thus the germline was edited.
Discussion
In this report, we demonstrated heritable CRISPR-Cas9 gene editing in B. tabaci using ReMOT Control by changing the ovary targeting ligand from P2C to BtKV. P2C was derived from the yolk protein of Drosophila melanogaster and was used for gene editing in Aedes aegypti10 and Anopheles stephensi.9 In our experiments, P2C did not generate knockouts in B. tabaci. Thus, we identified a new 13 aa ligand from the native vitellogenin protein of B. tabaci to target the RNP for ReMOT Control. This resulted in high efficiency gene editing. In mosquitoes, use of an endosomal escape reagent is critical to the success of ReMOT Control,9,10 and saponin was shown to be highly effective.9 However, in B. tabaci, we found a chemical endosomal escape reagent was unnecessary, and in fact saponin was detrimental to the process. This result demonstrates that ReMOT Control must be independently optimized for each new species, as parameters that are successful in one system may not work in others.
ReMOT Control transduces Cas9 RNP into the ovaries of females. So, it cannot edit the paternal derived chromosome until after the egg has been fertilized. Thus, the maternal chromosome is edited more efficiently than the paternal chromosome.9,10 B. tabaci is haplodiploid, where females are diploid and haploid males result from unfertilized eggs. Because ReMOT Control preferentially edits the maternal chromosome, obtained white-eye nymphal instars were likely haploid males that developed from an unfertilized egg with a single edited chromosome. Haplodiploidy is an advantage for ReMOT Control because mutations in the female germline can be immediately recognized in the haploid male offspring. Consistent with this, all G0 mutant adults with orange-red eyes were males. To exploit the haplodiploidy feature effectively, we obtained females <24 hpe to limit pre-injection egg development, as well as potentially limiting the chances of mating, thus favoring male bias offspring, making it easier to screen for the visually noticeable phenotype.
Our cross experiment demonstrated that mutations generated by ReMOT Control are in the germline and can be passed down to offspring by heredity, rather than just editing the somatic tissues. Mutations were inherited in deviation of expected Mendelian ratios, possibly due to differential hatching of mutant versus wild-type eggs.
We observed white-eye nymphs from first to early fourth instar and an unexpected orange-red eye phenotype in the mutant late fourth instar nymphs and adults. During the late fourth instar nymphal stage, the eyes of the pharate adult become diffused, heavily pigmented, and distinct,24 which means that the noticeable change in eye color during the late stages of the fourth nymphal instar are the adult eyes. The change in pigmentation in B. tabaci is different from the brown leafhopper Nilaparvata lugens, another hemimetabolous insect. Mutation of the white gene in N. lugens yielded a white ocelli and a light red pigmented eye where the eye pigments were consistent throughout the life stages.14 Both ommochrome (brown/black) and pteridine (red/orange/yellow) pigments contribute to eye color in insects.25 We speculate that the B. tabaci white gene is responsible for transportation of the brown ommochrome pigments into the eye. With it mutated, the red pteridine pigments become visible, explaining the shift in eye color of mutant insects, although further research is required to confirm or refute this hypothesis.
Conclusion
We have shown in this work that ReMOT Control allows easy and efficient CRISPR editing in B. tabaci without the need to inject embryos. The eggs of B. tabaci have a pedicel that is embedded into leaf tissue and acts a conduit for water and solute absorption into the eggs for successful embryonic development and hatching.26 This character, along with the exceeding small size of the embryos (∼0.2 mm × 0.01 mm) and high mortality of injected eggs presents significant challenges for the success of embryo injections. ReMOT Control removes these constraints and significantly expands the ability for any laboratory to apply CRISPR techniques to whitefly research, which will greatly accelerate molecular biology research on this organism and lead to the development of novel control strategies for this economically devastating pest insect. Furthermore, this technology can be applied readily to other whitefly species and to related insect groups, for example psyllids and mealybugs, which also constitute major agricultural pests.
Supplementary Material
Acknowledgments
We thank Dr. Angela Douglas for helpful advice regarding whitefly rearing and experiments, Abdulla M.A.J. Alghawas for assistance with laboratory procedures, and Dr. Blake Bextine for facilitating our initial investigations in this area of research.
Author Disclosure Statement
J.L.R. has filed for patent protection on the ReMOT Control technology. No competing financial interests exist for the remaining authors.
Funding Information
This work was funded by NSF/BIO grant 1645331, NIH/NIAID grant R21AI111175, USDA/NIFA grant 2014-10320, USDA Hatch funds (Accession #1010032; Project #PEN04608), and a grant with the Pennsylvania Department of Health using Tobacco Settlement Funds to J.L.R.
Supplementary Material
References
- 1. Inoue-Nagata AK, Lima MF, Gilbertson RL. A review of geminivirus diseases in vegetables and other crops in Brazil: current status and approaches for management. Hortic Bras 2016;34:8–18. DOI: 10.1590/S0102-053620160000100002 [DOI] [Google Scholar]
- 2. Czosnek H, Hariton-Shalev A, Sobol I, et al. The incredible journey of Begomoviruses in their whitefly vector. Viruses 2017;9:273 DOI: 10.3390/v9100273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Faria M, Wraight SP. Biological control of Bemisia tabaci with fungi. Crop Prot 2001;20:767–778. DOI: 10.1016/S0261-2194(01)00110-7 [DOI] [Google Scholar]
- 4. Gerling D, Alomar Ò, Arnò J. Biological control of Bemisia tabaci using predators and parasitoids. Crop Prot 2001;20:779–799. DOI: 10.1016/S0261-2194(01)00111-9 [DOI] [Google Scholar]
- 5. Kanakala S, Kontsedalov S, Lebedev G, et al. Plant-mediated silencing of the whitefly Bemisia tabaci cyclophilin B and heat shock protein 70 impairs insect development and virus transmission. Front Physiol 2019;10:557 DOI: 10.3389/fphys.2019.00557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luo Y, Chen Q, Luan J, et al. Towards an understanding of the molecular basis of effective RNAi against a global insect pest, the whitefly Bemisia tabaci. Insect Biochem Mol Biol 2017;88:21–29. DOI: 10.1016/j.ibmb.2017.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Malik HJ, Raza A, Amin I, et al. RNAi-mediated mortality of the whitefly through transgenic expression of double-stranded RNA homologous to acetylcholinesterase and ecdysone receptor in tobacco plants. Sci Rep 2016;6:38469 DOI: 10.1038/srep38469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Raza A, Malik HJ, Shafiq M, et al. RNA interference based approach to down regulate osmoregulators of whitefly (Bemisia tabaci): potential technology for the control of whitefly. PLoS One 2016;11:e0153883 DOI: 10.1371/journal.pone.0153883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Macias VM, McKeand S, Chaverra-Rodriguez D, et al. Cas9-mediated gene-editing in the malaria mosquito Anopheles stephensi by ReMOT Control. G3 (Bethesda) 2020. Mar 2 [Epub ahead of print]; DOI: 10.1534/g3.120.401133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chaverra-Rodriguez D, Macias VM, Hughes GL, et al. Targeted delivery of CRISPR-Cas9 ribonucleoprotein into arthropod ovaries for heritable germline gene editing. Nat Commun 2018;9:3008 DOI: 10.1038/s41467-018-05425-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kistler KE, Vosshall LB, Matthews B J. Genome engineering with CRISPR-Cas9 in the mosquito Aedes aegypti. Cell Rep 2015;11:51–60. DOI: 10.1016/j.celrep.2015.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dong Y, Simões ML, Marois E, et al. CRISPR/Cas9-mediated gene knockout of Anopheles gambiae FREP1 suppresses malaria parasite infection. PLoS Pathog 2018;14:e1006898 DOI: 10.1371/journal.ppat.1006898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kyrou K, Hammond AM, Galizi R, et al. A CRISPR-Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol 2018;36:1062–1066. DOI: 10.1038/nbt.4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xue W.-H, Xu N, Yuan XB, et al. CRISPR/Cas9-mediated knockout of two eye pigmentation genes in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Insect Biochem Mol Biol 2018;93:19–26. DOI: 10.1016/j.ibmb.2017.12.003 [DOI] [PubMed] [Google Scholar]
- 15. Roth Z, Wells S, Aflalo ED, et al. Identification of receptor-interacting regions of vitellogenin within evolutionarily conserved β-sheet structures by using a peptide array. Chembiochem 2013;14:1116–1122. DOI: 10.1002/cbic.201300152 [DOI] [PubMed] [Google Scholar]
- 16. Haeussler M, Schönig K, Eckert H, et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol 2016;17:148 DOI: 10.1186/s13059-016-1012-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li T-Y, Vinson SB, Gerling D. Courtship and mating behavior of Bemisia tabaci (Homoptera: Aleyrodidae). Environ Entomol 1989;18:800–806. DOI: 10.1093/ee/18.5.800 [DOI] [Google Scholar]
- 18. Guo J-Y, Dong S-Z, Yang X-L, et al. Enhanced vitellogenesis in a whitefly via feeding on a begomovirus-infected plant. PLoS One 2012;7:e43567 DOI: 10.1371/journal.pone.0043567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guo J-Y, Wan F-H, Ye G-Y. Oogenesis in the Bemisia tabaci MEAM1 species complex. Micron 2016;83:1–10. DOI: 10.1016/j.micron.2016.01.003 [DOI] [PubMed] [Google Scholar]
- 20. Perera OP, Little NS, Pierce CA. CRISPR/Cas9 mediated high efficiency knockout of the eye color gene Vermillion in Helicoverpa zea (Boddie). PLoS One 2018;13:e0197567 DOI: 10.1371/journal.pone.0197567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mackenzie SM, Brooker MR, Gill TR, et al. Mutations in the white gene of Drosophila melanogaster affecting ABC transporters that determine eye colouration. Biochim Biophys Acta 1999;1419:173–185. DOI: 10.1016/S0005-2736(99)00064-4 [DOI] [PubMed] [Google Scholar]
- 22. Bai X, Zeng T, Ni XY, et al. CRISPR/Cas9-mediated knockout of the eye pigmentation gene white leads to alterations in colour of head spots in the oriental fruit fly, Bactrocera dorsalis. Insect Mol Biol 2019;28:837–849. DOI: 10.1111/imb.12592 [DOI] [PubMed] [Google Scholar]
- 23. Horowitz AR, Gerling D. Seasonal variation of sex ratio in Bemisia tabaci on cotton in Israel. Environ Entomol 1992;21:556–559. DOI: 10.1093/ee/21.3.556 [DOI] [Google Scholar]
- 24. Gelman DB, Blackburn MB, Hu JS, et al. The nymphal-adult molt of the silverleaf whitefly (Bemisia argentifolii): timing, regulation, and progress. Arch Insect Biochem Physiol 2002;51:67–79. DOI: 10.1002/arch.10051 [DOI] [PubMed] [Google Scholar]
- 25. Rasgon JL, Scott TW. Crimson: a novel sex-linked eye color mutant of Culex pipiens L. (Diptera: Culicidae). J Med Entomol 2004;41:385–391. DOI: 10.1603/0022-2585-41.3.385 [DOI] [PubMed] [Google Scholar]
- 26. Buckner JS, Freeman TP, Ruud RL, et al. Characterization and functions of the whitefly egg pedicel. Arch Insect Biochem Physiol 2002;49:22–33. DOI: 10.1002/arch.10006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.