ABSTRACT
Salt stress is one of the environmental factors that negatively affect plant growth and development. We have previously reported a putative C3HC4 zinc-finger ubiquitin E3 ligase (AtPPRT1) negatively regulates Abscisic acid (ABA) and drought stress response. According to previous studies, the accumulation of ABA in plants can further regulate the salt stress response. Therefore, in this study, we further analyzed whether AtPPRT1 negatively regulates the salt stress response. The results showed that AtPPRT1 expression was induced by salt stress. Furthermore, under salt stress, the β-glucuronidase (GUS) gene driven by the AtPPRT1 promoter has shown increased activity in the hypocotyl and petioles of Arabidopsis seedlings. Additionally, seedlings of the T-DNA insertion mutant atpprt1 showed significant growth advantage under salt stress, whereas overexpressing AtPPRT1 (OE lines) in Arabidopsis seedlings displayed hypersensitive under salt stress. Etiolated atpprt1 seedlings also demonstrated significantly elongated hypocotyl lengths in salt stress. The elevated or reduced salt tolerance of atpprt1 and AtPPRT1 overexpressing lines was confirmed by the changes in chlorophyll content and 3,3ʹ-Diaminobenzidine (DAB) staining. The above data suggest that AtPPRT1 has a negative effect on salt tolerance in Arabidopsis seedlings.
KEYWORDS: Arabidopsis, AtPPRT1, salt tolerance
1. Introduction
Salt stress is considered to be one of the most severe threats to agricultural yield: it has a negative effect on the growth and development of plants, thereby dramatically reducing the yield of crops. Thus, for a long time, studies have focused on the mechanisms that might enhance the salt tolerance of plants and improve the yield of crops under salt stress.
The salty environment can decrease the water potential in the soil outside the plant, which reduces the plant’s ability to absorb water. Reduced water absorption capacity exposes plant roots to osmotic stress and eventually leads to plants’ premature senescence and death.1 Plant hormones, such as the ABA, vary in their response to salt and osmotic stress.2,3 ABA often acts as a signal in the initial regulation process of plants to regulate the salt stress response.4,5 Many reports have further revealed that exogenous ABA can promote the expression of salt-responsive genes, thereby enhancing the salt tolerance of plants.6
The ubiquitination pathway is a common protein modification mechanism that helps to better control the half-life, localization and physiological activity of plant proteins. Ubiquitination in plants requires the involvement of E1 ubiquitin-activating enzyme, E2 ubiquitin-binding enzyme and E3 ubiquitin ligase. Compared with E1 and E2s, E3s are the most diverse and abundant enzymes in the ubiquitin cascade, during which they explicitly recognize proteins and then ubiquitinate them.7 The RING finger E3 ligase is one of the earliest discovered and well-studied ubiquitin ligases, many of which are involved in the regulation of a variety of plant responses to abiotic stress. It was found that AT1G68820 contains a C3HC4-type RING domain at the C-terminus.8 Therefore AtPPRT1, which is encoded by At1G68820, is a putative C3HC4 zinc-finger ubiquitin E3 ligase. A recent study showed that OsSIRH2-14, a rice (Oryza sativa) really interesting new gene (RING) H2‐type E3 ligase, could mediate the ubiquitination of an HKT‐type Na+ transporter (OsHKT2;1), resulting in the degradation of OsHKT2;1 by the 26S proteasome and thereby enhancing the salt tolerance of rice under high salt stress.9 Additionally, the pepper (Capsicum annuum) Drought Induced RING-type E3 ligase 1 (CaDIR1) plays a negative regulatory role in the plant response to ABA and drought stress in peppers,10 while the RING finger E3 ligase SpRing in wild tomato species positively regulates seed germination and salt tolerance in seedlings of transformant Arabidopsis lines.11 A previous study showed that the ABA-Related RING-type E3 ligase (AtARRE) acted as a negative regulator in response to ABA signaling in Arabidopsis thaliana.12 Overall, these findings prove that E3 ubiquitin ligase plays a crucial role in the regulation of abiotic stress response in plants.
In our previous study, we found that AtPPRT1 plays a negative regulatory role in response to ABA and drought stress.8 To confirm whether AtPPRT1 has a negative effect on salt resistance in Arabidopsis, in this study, we characterized the function of AtPPRT1 in the salt stress response. The results indicate that the transcriptional expression of AtPPRT1 is induced by salt stress, and AtPPRT1 plays a negative regulatory role in the salt stress response in Arabidopsis seedlings.
2. Materials and methods
2.1. Plant materials and growth conditions
The background of Arabidopsis thaliana used in this study was Columbia (Col-0). The T-DNA insertion mutant atpprt1 was obtained from the Arabidopsis Biological Resource Center (ABRC; http://abrc.osu.edu/). We also selected two independent AtPPRT1 overexpressing lines, referred to as OE2 and OE10. The seeds were placed in water, vernalized for 3 days under storage conditions at 4°C, and then sterilized with NaClO for 15 min in a sterile environment. The sterilized seeds were sown on 1/2 MS medium plates consisting of 1% (w/v) sucrose and 0.65% (w/v) agar. All experimental materials were incubated in a greenhouse at 23°C with a photoperiod of 16 hours light/8 hours dark and relative humidity of 70%.
2.2. Promoter and regulatory element analysis
Plant cis-acting regulatory elements (PlantCARE) is a database that contains information on plant cis-acting elements, enhancers and repressors. This database was used to retrieve information related to the putative AtPPRT1 promoter.
2.3. Phenotypic analysis
For germination rates assays and seedlings with green cotyledon assays, 150 seeds from each line were sown on the identical MS medium plates with different concentrations of NaCl (0, 100 mM, 120 mM and 140 mM). Then, the germination rates of all lines were recorded during germination, and cotyledon greening rates were calculated 7 days after 120 mM and 140 mM NaCl treatment.
Seeds were seeded on 1/2 MS medium plates and cultured vertically in a greenhouse. After 2 days, 30 seedlings of each line sharing similar root lengths were transferred to new 1/2 MS medium plates supplemented with different concentrations of NaCl (0, 120 mM,150 mM) and 300 mM mannitol. For root elongation measurements, the lengths of all roots under 120 mM NaCl and 300 mM mannitol were measured after 7 days of vertical culture. A separate set of seedlings were subjected to 120 mM NaCl was used for chlorophyll, fresh weight and first leaf pair span estimation. Then, the survival rates of all lines were recorded 7 days after 150 mM NaCl treatment and the survival seedlings of each line were selected for 3,3ʹ-Diaminobenzidine (DAB) staining.
For hypocotyl elongation assays, seeds were sowed on 1/2 MS medium plates and incubated for 7 days in the dark. At the same time, etiolated seedlings that were treated for 2 days in the dark were transferred to a 1/2 MS plate containing 120 mM NaCl and incubated for another 5 days in the dark. The hypocotyl lengths under non-stress and salt stress conditions were measured and counted.
2.4. GUS staining
The β-glucuronidase (GUS) gene was used as a reporter gene, and the pCAMBIA1301 plasmid was used as an effector plasmid to obtain positively transformed plants. We selected 3-day-old (#21), 7-day-old seedlings (#5, #8, #21), etiolated seedlings (#21) and 4-week-old transformed plants with flowers and leaves (#21) as experimental materials. The GUS staining assay was applied while materials were treated with and without 120 mM NaCl for 4.5 hours. The GUS staining solution (Real-Times Company, Detroit, Michigan, USA) was added until the materials were completely immersed. The staining was carried out for 3–4 hours under light-shielding conditions at 37°C until the test material was stained with stable blue color. Then, the blue-stained plant material was decolored with 70% ethanol. Finally, the GUS staining activity of different experimental materials was observed under the microscope.
2.5. Chlorophyll estimation
For chlorophyll estimation, 20 mg of seedlings were homogenized in extraction buffer (containing ethanol, acetone and H2O in a ratio of 5: 5: 1). The absorbance of the clear supernatant was measured at 663 and 645 nm. Total chlorophyll contents were estimated according to previously reported methods.13
2.6. DAB staining
The survival seedlings of Col-0, atpprt1, OE2 and OE10 after 150 mM NaCl treatment were selected for DAB staining. The DAB staining solution (1 mg/ml) was added until the materials were completely immersed. After 5 hours of staining under light-shielding conditions, seedlings were boiled in 70% ethanol for 15 minutes. Finally, the DAB staining of different lines was observed under the microscope.
2.7. Real-time qRT-Pcr
The differently treated experimental materials were frozen in liquid nitrogen and subjected to total RNA extraction according to previously reported methods.14 For qRT-PCR analysis, the TB GREEN Premix Ex Taq kit (Takara) was used in the Applied Biosystems 7500 real-time PCR system as required, and ACTIN2 was used as an internal reference to analyze the transcription levels of the genes of interest (primers are shown in Supplementary Table S1).
3. Results
3.1. Analysis of cis-acting elements of the atpprt1 promoter and transcriptional expression of AtPPRT1induced by abiotic stresses
In order to study the AtPPRT1 response to salt stress, we analyzed the potential promoter region in both the sense and antisense directions within 3 kbp upstream of the transcription start site according to the plant cis-acting regulatory element database (PlantCARE).15 According to previous reports, the ABRE binding site motifs,16–18 G-box promoter motif19 and MYB binding-site motif20 are all involved in the salt stress response. Under abiotic stress, transcription factors bind to transcription factor binding sites in the cis-acting elements of stress response genes and initiate their transcriptional expression, thereby regulating the plant stress response. In this study, AtPPRT1 was induced by a variety of abiotic stress and participated in salt stress responses, which were highly correlated with the functional analysis of these cis-acting elements (Figure 1(a)).
Figure 1.
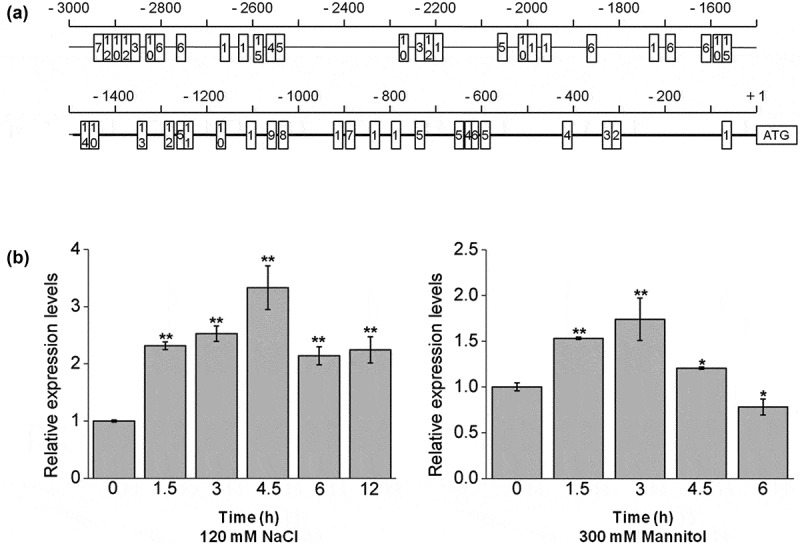
Bioinformatics analysis of the AtPPRT1 promoter and the transcriptional expression levels of AtPPRT1 under abiotic stresses. (a) The region of 3 kbp upstream of the AtPPRT1 gene was analyzed for the cis-acting elements according to the plant cis-acting regulatory element database (PlantCARE). The promoter region of 1492 bp used in this study is shown in bold line. The numbers in the boxes indicate different binding sites. 1, MYC binding-site motif; 2, ACE binding-site motif; 3, ABRE binding-site motif; 4, O2 site motif; 5, BOX4 promoter motif; 6, ARE binding-site motif; 7, LTR binding-site motif; 8, AE-box promoter motif; 9, W-box promoter motif; 10, MYB binding-site motif; 11, TGA-element; 12, G-Box promoter motif; 13, MBS binding-site motif; 14, P-box promoter motif; 15, TCA-element. (b) the qRT-PCR analysis of AtPPRT1 transcriptional expression levels induced by 120 mM NaCl and 300 mM mannitol. These experiments were repeated three times with similar results. Error bars represent ± SD (n = 3, *p < .05, **p < .01, t-test).
Ten-day-old wild-type Arabidopsis seedlings were adopted as experimental materials, and the transcriptional expression levels of AtPPRT1 for different treatment durations were analyzed. The transcriptional expression levels of AtPPRT1 first increased and then decreased under these abiotic stresses. The transcriptional expression of AtPPRT1 peaked at 4.5 h during 120 mM NaCl treatment and at 3 h during 300 mM mannitol treatment (Figure 1(b)). The results indicate that the transcriptional expression of AtPPRT1 was induced by salt and osmotic stress.
3.2. GUS activity in proAtPPRT1:: GUS transformant lines
The upstream region (~1.5 kbp) of the AtPPRT1 ATG start codon fused with the GUS gene was constructed and transformed into the wild-type plants. Three independent T3 transgenic lines were histochemically analyzed by GUS staining.
Three-day-old and seven-day-old transformed plants’ seedlings as well as the flowers and leaves of mature plants of ProAtPPRT1:: GUS was dyed with GUS staining solution and then discolored with 70% ethanol. The GUS activity was detected in cotyledons and hypocotyl of 3-day-old seedlings (Figure 2(d)). With the seedling growth, GUS activity was found mainly in the vascular bundle tissue of leaves, leaf petioles, and hypocotyls (Figure 2(a)). Under control conditions, GUS activity was detected in cotyledons of etiolated seedlings (Figure 2(e)). However, the expression level of AtPPRT1 changed after the samples were treated with 120 mM NaCl for 4.5 hours. For example, GUS was more active in the petiole of 7-day-old seedlings, hypocotyl of 3-day-old and 7-day-old seedlings under salt stress (Figure 2(a,d)). At the cell elongation stage, strong GUS activity was detected in vascular bundle tissue of hypocotyls under salt stress (Figure 2(e)). These results suggest that the promoter activity was up-regulated at these sites (Figure 2(a, d,e)).
Figure 2.
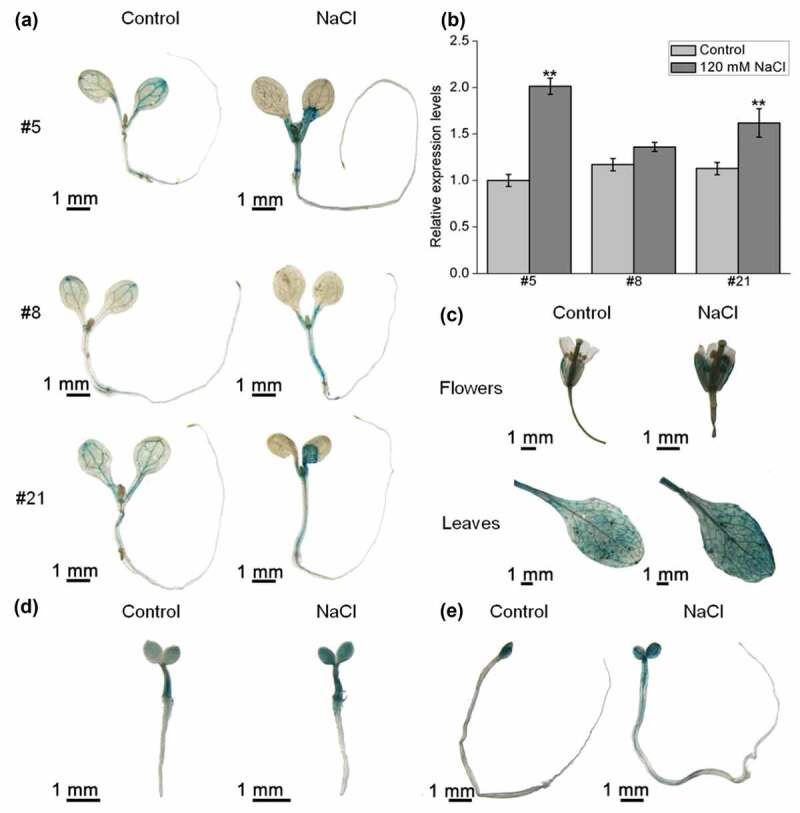
GUS activity in different tissues and its transcriptional expression levels in ProAtPPRT1:: GUS transformant lines. (a) 7-day-old and (d) 3-day-old homozygous transformed plants’ seedlings and (e) etiolated seedlings were used for GUS staining under non-stress conditions and 120 mM NaCl treatment; (b) the transcriptional expression levels of the GUS gene were analyzed by qRT-PCR. These experiments were repeated three times with similar results. Error bars represent ± SD (n = 3, *p < .05, **p < .01, t-test); (c) the flowers and leaves of 4-week-old ProAtPPRT1:: GUS transformed plants (#21) were used for GUS staining under non-stress conditions and 120 mM NaCl treatment. Scale bar = 1 mm. #5, #8 and #21 indicate different transformant lines.
The total RNA was extracted from three independent positive transformant homozygous lines of 7-day-old ProAtPPRT1:: GUS transformed plants’ seedlings treated with or without salt stress. The transcriptional expression levels of the GUS gene were analyzed by qRT-PCR according to the reference.14 The results show that the transcriptional expression levels of the GUS gene in all three lines increased after salt stress-treated, especially in transformant lines #5 and #21. The qRT-PCR results were consistent with the previous histochemical staining results (Figure 2(b)).
In the reproductive stage (4-week-old), strong GUS activity was detected in mature leaves, as well as some reproductive organs, such as calyxes and filaments. After salt stress-treated, the promoter activity of AtPPRT1 was slightly enhanced in mature leaves, calyxes, filaments and styles (Figure 2(c)).
The above results indicate that the expression pattern of AtPPRT1 varies in different growth phases under salt stress compared with non-stress conditions (Figures 1, 2).
3.3. AtPPRT1 acts as a negative regulator in the plant response to salt stress in Arabidopsis seedlings
The germination rates and cotyledon greening rates of all lines treated with different salt concentrations were analyzed to confirm whether AtPPRT1 is involved in salt stress. The results show that during seed germination, there was no significant difference between Col-0, atpprt1 and OE lines on MS plates. However, when MS medium plates were supplemented with different concentrations of NaCl (100 mM, 120 mM and 140 mM), the germination rates of atpprt1 were faster than Col-0 and OE lines (Figure 3(a,b)). The rates of seedlings with green cotyledons of atpprt1 was higher than Col-0, while OE lines were lower than Col-0 under 120 mM and 140 mM NaCl (Figure 3(c)). In addition, the atpprt1 seedlings also demonstrate significantly elongated root lengths when treated with 120mM NaCl (Figure 4(a,b)).
Figure 3.
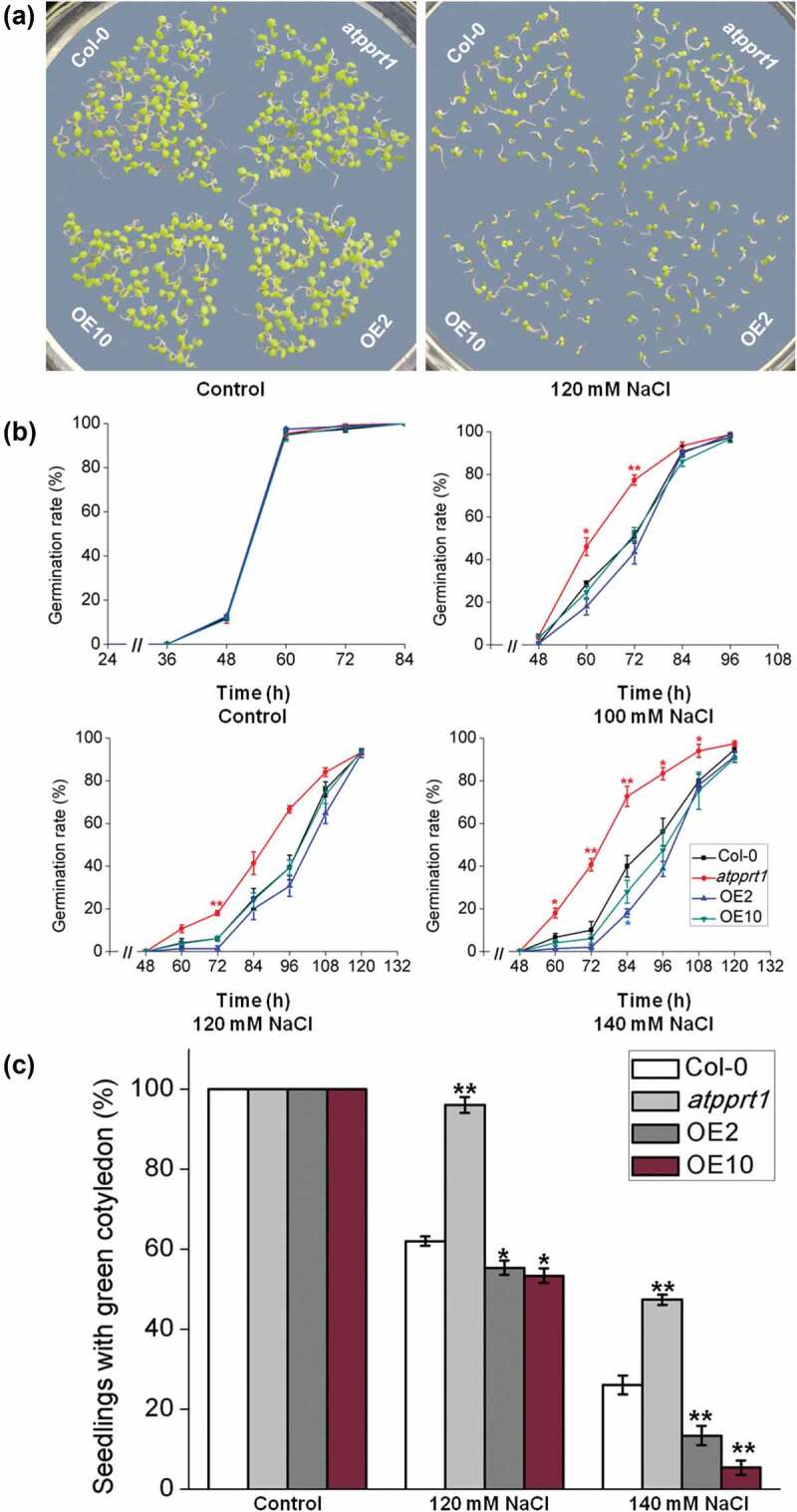
The germination and cotyledon greening rates of each line under different NaCl concentrations. (a) Col-0, atpprt1, OE2 and OE10 were monitored for 7 days on MS medium plates with and without salt; the germination (b) and cotyledon greening (c) rates of Col-0, atpprt1, OE2 and OE10 on MS medium plates supplemented with different concentrations of NaCl. Error bars represent ± SD (n = 50, * p < .05 and ** p < .01, t-test).
Figure 4.
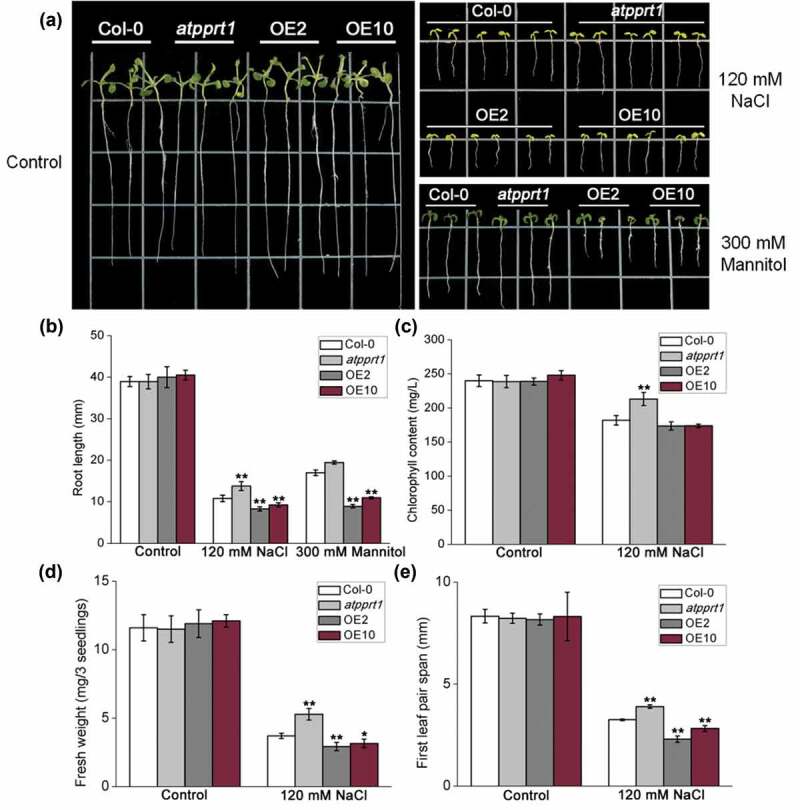
AtPPRT1 negatively affects salt stress tolerance in Arabidopsis seedlings. (a) Col-0, atpprt1, OE2, and OE10 grown vertically on 1/2 MS for 3 days were transferred to 1/2 MS without NaCl or supplemented with 120 mM NaCl or 300 mM mannitol for another 7 days; (b) measurement of root lengths under 120 mM NaCl treatment or 300 mM mannitol treatment; measurement of (c) chlorophyll content, (d) fresh weight and (e)) first leaf pair span under 120 mM NaCl in Col-0, atpprt1, OE2, and OE10. The values are the average of three individual biological replications. Error bars represent ± SD (n = 21, *p < .05 and **p < .01, t-test).
The chlorophyll content of each line was almost equal under untreated conditions. Seedlings of atpprt1 showed increased chlorophyll retention under salinity stress with regard to the salinity treated Col-0 and OE lines (Figure 4(c)). To investigate the role of AtPPRT1 in more detail, the seedlings fresh weight and first leaf pair span of Col-0, atpprt1 and OE lines, were measured. Fresh weight and first leaf pair span of atpprt1 seedlings were significantly increased at 120 mM NaCl compared to Col-0, and OE lines were decreased considerably compared to Col-0 (Figure 4(d,e)). These results indicate that overexpression of AtPPRT1 leads to decreased salt tolerance, whereas loss-of-function in AtPPRT1 results in salt insensitivity, suggesting that AtPPRT1 plays a negative regulatory role in the salt stress response in Arabidopsis seedlings.
3.4. AtPPRT1 may decrease salinity tolerance by reducing osmotic tolerance
When plants suffer from salt stress in their seedling stage, their growth and development may be inhibited because of osmotic resistance. In the previous experiments, the OE lines showed a more sensitive phenotype on 1/2 MS medium plates supplemented with 300 mM mannitol during seed germination.8 To determine whether AtPPRT1 reduces salinity resistance by decreasing osmotic resistance, we observed the root lengths of Col-0, atpprt1, OE2 and OE10 on 1/2 MS medium plates supplemented with 300 mM mannitol. This experiment showed that the overexpression of AtPPRT1 could inhibit post-germination root growth under osmotic stress (Figure 4(a,b)). These results indicate that AtPPRT1 may play a negative regulatory role in the salt stress response by changing the osmotic resistance.
3.5. AtPPRT1 increases the damage caused by salt stress in Arabidopsis seedlings
To further assess the role of AtPPRT1 in salt stress response, all lines were transferred to 1/2 MS medium plates supplemented with 150 mM NaCl and cultured vertically for 7 days. Then, the survival rates of all lines were recorded and the survival seedlings of each line were selected for DAB staining. The results indicate that nearly 66.6% of the atpprt1 seedlings survive from high salinity, while the survival rates of OE2 and OE10 were reduced to 36.6% and 43.3% (Figure 5(a)).
Figure 5.
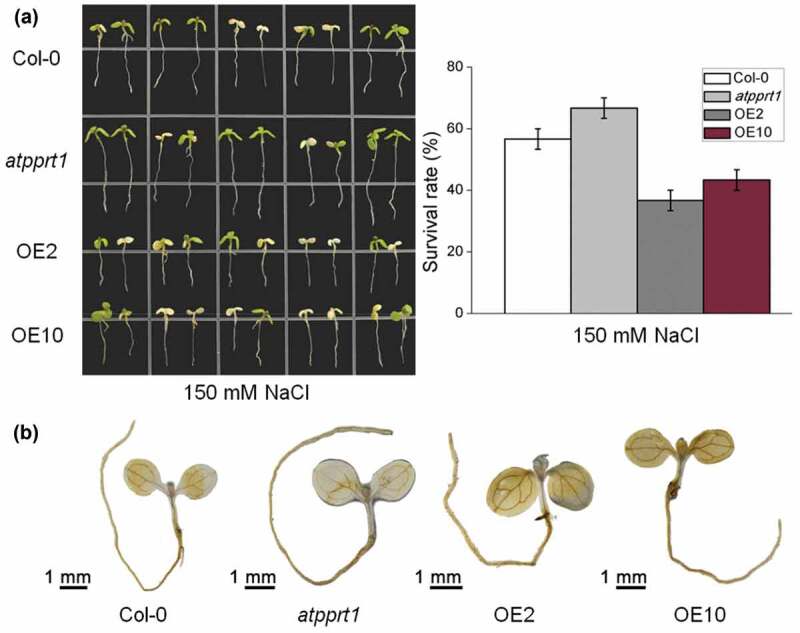
Seedlings survival rate and DAB staining under high salinity. (a) The survival rates of Col-0, atpprt1, OE2 and OE10 under 150 mM NaCl; (b) DAB staining assay of seedlings of Col-0, atpprt1, OE2 and OE10 after 150 mM NaCl treatment.
Under salt stress, the content of hydrogen peroxide (H2O2) directly reflects the degree of plant oxidation and further shows the degree of damage. Additionally, DAB staining showed that a large amount of H2O2 accumulated in the OE2 and OE10 seedlings while less accumulated in the atpprt1 seedlings during salt stress, compared with Col-0 (Figure 5(b)). The above data suggest that AtPPRT1 increases the damage caused by salt stress in Arabidopsis seedlings.
3.6. AtPPRT1 affects the hypocotyl elongation of etiolated seedlings in response to salt stress
Coincidentally, we discovered significant differences in the hypocotyl lengths of etiolated seedlings under salt stress. In the following experiment, the hypocotyl lengths of each line under salt stress were measured, and the results show that there was no significant difference between Col-0, atpprt1 and OE lines on 1/2 MS plates in the dark. Under salt stress, compared with Col-0, the hypocotyl lengths in the atpprt1 were significantly longer, and those in OE lines were shorter (Figure 6(a,b)). These results indicate that loss-of-function in AtPPRT1 leads to increased salt tolerance during cell elongation in the dark. In addition, disruption of AtPPRT1 can increase transcriptional expression levels of AtMAP18 and AtBSK1 under salt stress. Surprisingly, the transcriptional expression levels of AtMAP18 and AtBSK1 in the OE lines were in accordance with those in Col-0 (Figure 6(c)). The detailed molecular mechanism needs to be explored.
Figure 6.
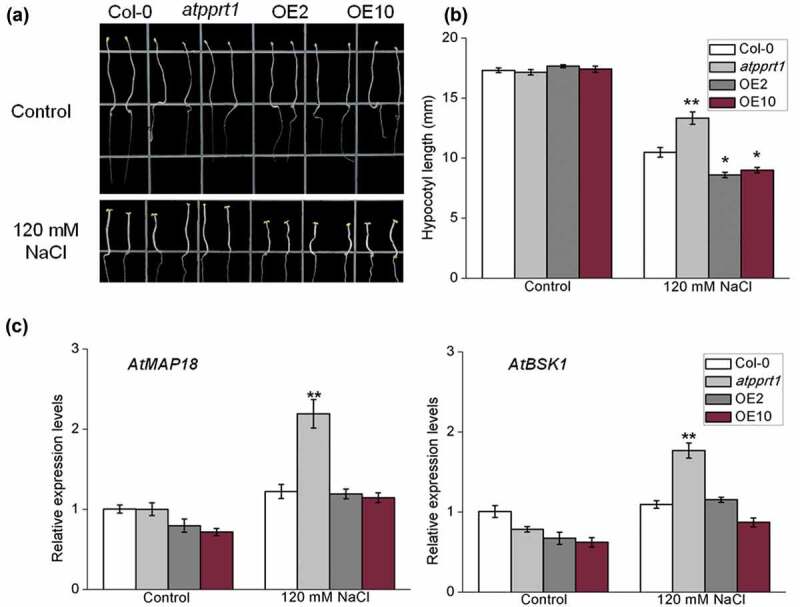
AtPPRT1 affects the elongation of hypocotyl in etiolated seedlings under salt stress. (a) Col-0, atpprt1, OE2, and OE10 were grown for 2 days on 1/2 MS medium plates and then transferred to 1/2 MS supplemented with 120 mM NaCl and grown for 5 days. All the seedlings were grown in the dark; (b) the hypocotyl lengths of samples used in (a) were measured. These experiments were repeated three times with similar results. Error bars represent ± SD (n = 21, * p < .05 and ** p < .01, t-test); (c) qRT-PCR analysis of the transcriptional expression levels of AtMAP18 and AtBSK1 in Col-0, atpprt1, OE2, and OE10 in 120 mM NaCl. These experiments were repeated three times with similar results. Error bars represent ± SD (n = 3, *p < .05, **p < .01, t-test).
4. Discussion
In this work, we analyzed the expression pattern of AtPPRT1 in different growth phases under salt stress. The results of GUS histological staining show that AtPPRT1 is primarily expressed in the vascular tissues of seedlings and rarely expressed in the mesophyll under control conditions. However, in mature leaves, it is primarily expressed in the mesophyll and smaller veins rather than the main veins of leaves. These results indicate that AtPPRT1 is expressed in different areas of seedlings and mature tissues, which is consistent with our previous study.8 Under salt stress, 7-day-old seedlings had stronger GUS activity in petioles and hypocotyls. GUS activity in leaves and petals was also enhanced to some extent during the reproductive stage. The transcriptional expression levels of the GUS gene in three homozygous transformant lines were analyzed by qRT-PCR under non-stress or salt stress conditions.14 After salt stress-treated, the expression of AtPPRT1 in the seedlings was induced, and the transcriptional expression levels of the GUS gene in #5 and #21 were significantly up-regulated. In mature leaves and flower tissues, salt stress seems to have little effect on the GUS activity. Therefore, we speculate that AtPPRT1 mainly participates in the salt tolerance response in the seedling stage and not in the reproductive stage. This inference is supported by the results of subsequent experiments. In samples of the atpprt1 mutant and OE lines, the phenotypic differences in the salt tolerance response were mainly observed in terms of seed germination and root lengths. Samples in the reproductive stage did not show significant phenotypic differences after salt stress-treated (the results are not presented). This further suggests that AtPPRT1 is involved in the salt stress response only in the seedling stage.
When the plant is subjected to salt stress, the external osmotic potential is lower than the osmotic potential in the cell; this results in water stress, which causes the physiological drought of the plant.21,22 When plants suffer from salt stress in their seedling stage, their growth and development may be inhibited because of osmotic resistance.23 The previous study showed that OE lines were more sensitive to osmotic stress during seed germination,8 and the results of the root length experiments verify that the OE lines were also more sensitive to mannitol treatment. This suggests that AtPPRT1 may attenuate the response to salt stress by reducing the tolerance to osmotic stress in Arabidopsis seedlings. Additionally, salt stress would generate a large number of reactive oxygen species (ROS), which may cause oxidative damage to membrane lipids.24 DAB staining showed that OE lines produced more H2O2 than Col-0 under high salinity stress, whereas atpprt1 produced less compared with Col-0 (Figure 5(b)), indicating that AtPPRT1 can increase the damage caused by oxidative stress.
The previous studies confirmed that the ABA signaling pathway plays a vital role in the salt stress response;25 thus, we analyzed the transcriptional expression levels of the ABA-induced salt stress-related genes AtMYB102, AtWRKY28 and AtMKK5 after salt stress-treated. The results show that the increased expression levels in the OE lines were lower than those of Col-0 (Figure S1). This finding suggests that AtPPRT1 may play a negative role in the ABA-mediated salt tolerance pathway.
AtPPRT1 was expressed mainly in cotyledons of etiolated seedlings in the absence of salt, while the activity of GUS was significantly enhanced in the hypocotyl of etiolated seedlings in the presence of 120 mM NaCl. These results suggests that AtPPRT1 is involved in response to salt stress during cell elongation in the dark. AtMAP18, which encodes proteins that bind to cortical microtubules and inhibit tubulin polymerization in Arabidopsis, are primarily expressed in expanded cells. Previous studies have reported many microtubule-regulating genes can alter the stability of cortical microtubules.26–28 For example, loss-of-function in AtMAP18 leads to abnormal growth of the hypocotyl during cell elongation.29 AtBRI1 encodes a receptor kinase involved in brassinosteroid signaling and promotes cell growth.30 AtBSK1 is also involved in cell growth and development by mediating signal transduction from the receptor kinase AtBRI1 to act as a substrate of BRI1.31 After salt stress-treated, the transcriptional expression levels of AtMAP18 and AtBSK1 in atpprt1 were up-regulated and had greater increases compared with the Col-0 and OE lines. This may help to enhance the resistance to salt stress in etiolated Arabidopsis seedlings during cell elongation.
5. Conclusions
We have presented evidence that AtPPRT1, a putative C3HC4 zinc-finger ubiquitin E3 ligase encoded by At1G68820, is induced by salt stress. Compared to Col-0, atpprt1 seedlings showed a significant growth advantage under salt stress, whereas seedlings of OE lines displayed hypersensitive under salt stress. According to the experimental data and the previous study, the overexpression of AtPPRT1 can enhance sensitive responses to osmotic stress at seed germination, and inhibit post-germination root growth. These results show that AtPPRT1 may reduce salinity resistance by relieving osmotic stress tolerance. Additionally, DAB staining shows that AtPPRT1 can increase the damage caused by oxidative stress. Under salt stress and dark conditions, the hypocotyl lengths in the atpprt1 were significantly longer than the Col-0 and OE lines. These results indicate that AtPPRT1 reduces salinity resistance during cell elongation in etiolated Arabidopsis seedlings. Therefore, AtPPRT1 negatively regulates salt stress response in Arabidopsis seedlings.
Funding Statement
This research was supported by grants from the Nation Natural Science Foundation of China [31870240 to Y. Y.] and The National Transgene Project [2016ZX08009003-002 to X. L.]
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:1–10. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuromori T, Seo M, Shinozaki K.. ABA transport and plant water stress responses. Trends Plant Sci. 2018;23:513–522. doi: 10.1016/j.tplants.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Zhu J-K. Abiotic stress signaling and responses in plants. CELL. 2016;167:313–324. doi: 10.1016/j.cell.2016.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 5.Zhu JK. Plant salt tolerance. Trends Plant Sci. 2001;6:66–71. doi: 10.1016/S1360-1385(00)01838-0. [DOI] [PubMed] [Google Scholar]
- 6.Eberhardt HJ, Wegmann K. Effects of abscisic acid and proline on adaptation of tobacco callus cultures to salinity and osmotic shock. Physiol Plantarum. 1989;76:283–288. doi: 10.1111/j.1399-3054.1989.tb06192.x. [DOI] [Google Scholar]
- 7.Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10:385–397. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- 8.Pei L, Peng L, Wan X, Xiong J, Liu Z, Li X, Yang Y, Wang J. Expression pattern and function analysis of AtPPRT1, a novel negative regulator in ABA and drought stress responses in arabidopsis. Int J Mol Sci. 2019;20:2–394. doi:10.3390/ijms20020394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park YC, Lim SD, Moon JC, Jang CS. A rice really interesting new gene H2-type E3 ligase, OsSIRH2-14, enhances salinity tolerance via ubiquitin/26S proteasome-mediated degradation of salt-related proteins. Plant Cell Environ. 2019;42:3061–3076. doi: 10.1111/pce.v42.11. [DOI] [PubMed] [Google Scholar]
- 10.Joo H, Lim CW, Han SW, Lee SC. The pepper RING finger E3 ligase, CaDIR1, regulates the drought stress response via ABA-mediated signaling. Front Plant Sci. 2017;8:690. doi: 10.3389/fpls.2017.00690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi S, Lin Q, Zhu H, Gao F, Zhang W, Hua X. The RING finger E3 ligase SpRing is a positive regulator of salt stress signaling in salt-tolerant wild tomato species. Plant Cell Physiol. 2016;57:528–539. doi: 10.1093/pcp/pcw006. [DOI] [PubMed] [Google Scholar]
- 12.Wang B, Li C, Kong X, Li Y, Liu Z, Wang J, Li X, Yang Y. AtARRE, an E3 ubiquitin ligase, negatively regulates ABA signaling in Arabidopsis thaliana. Plant Cell Rep. 2018;37:1269–1278. doi: 10.1007/s00299-018-2311-8. [DOI] [PubMed] [Google Scholar]
- 13.Arnon DI. COPPER ENZYMES IN ISOLATED CHLOROPLASTS. POLYPHENOLOXIDASE IN BETA VULGARIS. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tada Y. The HKT transporter gene from Arabidopsis, AtHKT1;1, is dominantly expressed in shoot vascular tissue and root tips and is mild salt stress-responsive. Plants (Basel). 2019;8:7–204. doi:10.3390/plants8070204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rombauts S, Déhais P, Van Montagu M, Rouzé P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999;27:295–296. doi: 10.1093/nar/27.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zahur M, Maqbool A, Ifran M, Barozai MY, Rashid B, Riazuddin S, Husnain T. [Isolation and functional analysis of cotton universal stress protein promoter in response to phytohormones and abiotic stresses]. Mol Biol (Mosk). 2009;43:628–635. doi: 10.1134/S0026893309040086. [DOI] [PubMed] [Google Scholar]
- 17.Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K, Yamaguchi-Shinozaki K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003;34:137–148. doi: 10.1046/j.1365-313X.2003.01708.x. [DOI] [PubMed] [Google Scholar]
- 18.Manavella PA, Dezar CA, Ariel FD, Chan RL. Two ABREs, two redundant root-specific and one W-box cis-acting elements are functional in the sunflower HAHB4 promoter. Plant Physiol Biochem. 2008;46:860–867. doi: 10.1016/j.plaphy.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 19.Zou M, Guan Y, Ren H, Zhang F, Chen F. A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol Biol. 2008;66:675–683. doi: 10.1007/s11103-008-9298-4. [DOI] [PubMed] [Google Scholar]
- 20.Iwasaki T, Yamaguchi-Shinozaki K, Shinozaki K. Identification of a cis-regulatory region of a gene in Arabidopsis thaliana whose induction by dehydration is mediated by abscisic acid and requires protein synthesis. Mol Gen Genet. 1995;247:391–398. doi: 10.1007/BF00293139. [DOI] [PubMed] [Google Scholar]
- 21.Chinnusamy V, Zhu J, Zhu JK. Salt stress signaling and mechanisms of plant salt tolerance. Genet Eng (N Y). 2006;27:141–177. [DOI] [PubMed] [Google Scholar]
- 22.Cantabella D, Piqueras A, Acosta-Motos JR, Bernal-Vicente A, Hernandez JA, Diaz-Vivancos P. Salt-tolerance mechanisms induced in Stevia rebaudiana Bertoni: effects on mineral nutrition, antioxidative metabolism and steviol glycoside content. Plant Physiol Biochem. 2017;115:484–496. doi: 10.1016/j.plaphy.2017.04.023. [DOI] [PubMed] [Google Scholar]
- 23.Dai X, Xu Y, Ma Q, Xu W, Wang T, Xue Y, Chong K. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 2007;143:1739–1751. doi: 10.1104/pp.106.094532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Apel K, Hirt H. REACTIVE OXYGEN SPECIES: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi:10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 25.Yang L, Liu Q, Liu Z, Yang H, Wang J, Li X, Yang Y. Arabidopsis C3HC4-RING finger E3 ubiquitin ligase AtAIRP4 positively regulates stress-responsive abscisic acid signaling. J Integr Plant Biol. 2016;58:67–80. doi: 10.1111/jipb.12364. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Wang X, Qin T, Zhang Y, Liu X, Sun J, Zhou Y, Zhu L, Zhang Z, Yuan M, et al. MDP25, a novel calcium regulatory protein, mediates hypocotyl cell elongation by destabilizing cortical microtubules in Arabidopsis. Plant Cell. 2011;23:4411–4427. doi: 10.1105/tpc.111.092684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakajima K, Furutani I, Tachimoto H, Matsubara H, Hashimoto T. SPIRAL1 encodes a plant-specific microtubule-localized protein required for directional control of rapidly expanding Arabidopsis cells. Plant Cell. 2004;16:1178–1190. doi: 10.1105/tpc.017830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Zhang J, Yuan M, Ehrhardt DW, Wang Z, Mao T. Arabidopsis microtubule destabilizing protein40 is involved in brassinosteroid regulation of hypocotyl elongation. Plant Cell. 2012;24:4012–4025. doi: 10.1105/tpc.112.103838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Zhu L, Liu B, Wang C, Jin L, Zhao Q, Yuan M. Arabidopsis Microtubule-Associated Protein18 functions in directional cell growth by destabilizing cortical microtubules. Plant Cell. 2007;19:877–889. doi: 10.1105/tpc.106.048579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salazar-Henao JE, Lehner R, Betegon-Putze I, Vilarrasa-Blasi J, Cano-Delgado AI. BES1 regulates the localization of the brassinosteroid receptor BRL3 within the provascular tissue of the Arabidopsis primary root. J Exp Bot. 2016;67:4951–4961. doi: 10.1093/jxb/erw258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang W, Kim TW, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang ZY. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321:557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


