ABSTRACT
Apoplastic barriers in the endodermis, such as Casparian strips and suberin lamellae, control the passage of water and minerals into the stele. Apoplastic barriers are thus thought to contribute to salt exclusion in salt-excluding plants such as sweet sorghum (Sorghum bicolor). However, little is known about the genes involved in the development of the apoplastic barrier. Here, we identified candidate genes involved in Casparian strip and suberin lamella development in the roots of a sweet sorghum line (M-81E). Three distinct developmental regions (no differentiation, developing, and mature) were identified based on Casparian strip and suberin lamella staining in root cross sections. Sequencing of RNA extracted from these distinct sections identified key genes participating in the differentiation of the apoplastic barrier. The different sections were structurally distinct, presumably due to differences in gene expression. Genes controlling the phenylpropanoid pathway, fatty acid elongation, and fatty acid ω-hydroxylation appeared to be directly responsible for the formation of the apoplastic barrier. Our dataset elucidates the molecular processes underpinning apoplastic barrier development and provides a basis for future research on molecular mechanisms of apoplastic barrier formation and salt exclusion.
Abbreviations: SHR, SHORTROOT; MYB, MYB DOMAIN PROTEIN; CIFs, Casparian strip integrity factors; CASP, Casparian strip domain proteins; PER, peroxidase; ESB1, ENHANCED SUBERIN1; CS, Casparian strip; RPKM, reads per kilobase per million reads; DEGs, differentially expressed genes; FDR, false discovery rate; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; RNA-seq, RNA sequencing; PAL, phenylalanine ammonia-lyase; CYP, cytochrome P450 monooxygenases; 4CL, 4-coumarate-CoA ligase; AAE5, ACYL-ACTIVATING ENZYME5; CCR, cinnamoyl CoA reductase; TKPR, TETRAKETIDE ALPHA-PYRONE REDUCTASE1; CAD, cinnamyl alcohol dehydrogenase; HST, shikimate O-hydroxycinnamoyltransferase; PMAT2, PHENOLIC GLUCOSIDE MALONYLTRANSFERASE2; CCOAOMT, caffeoyl-CoA O-methyltransferase; KCS, β-ketoacyl-CoA synthase; CUT1, CUTICULAR PROTEIN1; DET2, 5-alpha-reductase; TAX, 3ʹ-N-debenzoyl-2ʹ-deoxytaxol N-benzoyltransferase; CER1, ECERIFERUM1; FAR, fatty acyl reductase; AF-CoA, alcohol-forming fatty acyl-CoA reductase; ABCG, ATP-binding cassette, subfamily G; ERF, ethylene-responsive transcription factor; HSF, heat stress transcription factor; NTF, NUCLEAR TRANSCRIPTION FACTOR Y SUBUNIT B-5; GPAT, glycerol 3-phosphate acyltransferase.
KEYWORDS: Apoplastic barrier, Casparian strip, phenylpropanoid pathway, suberin lamellae, sweet sorghum
Introduction
Sweet sorghum [Sorghum bicolor (L.) Moench], a variant of sorghum, has a rapid growth rate, high sugar content, and produces high amounts of biomass even when grown under environmental stresses such as salt and flooding.1 Sweet sorghum is also considered an economically important crop because it can be used to produce biofuels such as ethanol,2 in addition to being useful as feed for livestock.1 The yields of most crops are severely reduced in saline environments; however, sweet sorghum can grow well even on barren saline soil, making this land suitable for the production of fuel and feed.3
Plants rely on transpirational pull to absorb water and mineral ions from the soil,4 a process that is affected by various anatomical structures, growth conditions, and the age of the roots themselves.5 Root absorption relies on three major pathways: the symplastic pathway (via membrane-bound transporters and plasmodesmata), the apoplastic pathway (extracellular transportation), and the coupled transcellular pathway.6 Casparian strips and suberin lamellae, the apoplastic barriers in the endodermal layer,7,8 can effectively block water and inorganic salt transport through both the apoplastic and coupled transcellular pathways.
Biotic9 and abiotic stresses10,11 can enhance the biosynthesis of suberin, which is composed of both polyphenolic and polyaliphatic domains.12 The ability of suberin to block water and solute transportation is particularly dependent on the polyaliphatic domain, which is mainly comprised of oxygenated fatty acids and their derivatives.13,14 By contrast, the Casparian strip is mainly composed of lignin,8 a hydrophobic compound biosynthesized from the polymerization of syringyl, guaiacyl, and p-hydroxyphenol via the phenylpropanoid pathway.15
Several molecules involved in the formation of Casparian strip and suberin lamella in Arabidopsis thaliana.16–18 For example, the transcription factor SHORTROOT (SHR) promotes Casparian strip formation by activating MYB DOMAIN PROTEIN36 (MYB36) and SCARECROW expression. Two small peptides known as Casparian strip integrity factors (CIFs) participate in Casparian strip biosynthesis in Arabidopsis,17 while MYB36 regulates the expression of genes encoding the Casparian strip membrane domain protein 1 (CASP1), PEROXIDASE64 (PER64), and ENHANCED SUBERIN1 (ESB1) and thereby regulates lignin polymerization.19,20 Another MYB, MYB41, regulates the biosynthesis and transport of suberin lamella.21
Expression profiling has been widely used to identify candidate genes involved in important biological processes.3,22–24 A recent study of the root transcriptome of sweet sorghum identified many genes putatively associated with apoplastic barrier development;22 however, the developmental pattern of these barriers has not been fully elucidated. Moreover, all of the previously reported genes in sweet sorghum are annotated members of known gene families22,25 and unknown functional genes may be important drivers of apoplastic barrier development.
To elucidate the genetic regulation of apoplastic barrier development, the exact region of the root in which this process takes place must be identified. In this study, we dyed root sections of the sweet sorghum inbred line M-81E to determine the positions at which Casparian strips and suberin lamellae formed. Three distinct developmental regions were observed and separately enriched in transcriptome analyzes to identify candidate genes involved in the differentiation of the apoplast barrier in sweet sorghum. These results may provide another perspective for further study on molecular mechanism of apoplastic barrier developemnt.
Materials and methods
Plant materials and growth conditions
The sweet sorghum inbred line M-81E was used in this study. The seeds were sterilized in 70% (v/v) ethanol for 1 min and 3% (v/v) NaClO for 10 min, after which they were washed with deionized water and germinated in the dark on filter paper in a Petri dish. After 2 days, seedlings with a 3-cm radicle and a 2-cm shoot were transplanted into a hydroponic culture using Hoagland solution, which was replaced each day. All seedlings were cultured at 28 ± 3°C/23 ± 3°C (day/night) at a light intensity of 600 μmol m–2 s–1 (15-h photoperiod) and 70% relative humidity.
Casparian strip and suberin lamella observations in the root
To identify regions of Casparian strip and suberin lamella development in the seminal root, freehand cross sections of the seminal root were made at 10-mm intervals along the root tip. After identifying the region (from 10 to 50 mm) of Casparian strip and suberin lamella development, sections were cut every 5 mm for a more precise determination of the developmental positions.
The sections were cleared with lactic acid saturated with chloral hydrate for 2 h, 26 after which the sections were stained with 0.1% (w/v) berberine hemisulfate for 1 h and then with 0.5% (w/v) aniline blue for another hour.27 The stained sections were observed with a fluorescence microscope (ECLIPSE 80i; Nikon, Japan) under an excitation wavelength of 450 to 490 nm to view the Casparian strip.
To observe the suberin lamella, the sections were stained with 0.01% (w/v) Fluorol Yellow 088 for 1 h28 and then examined using a fluorescence microscope under ultraviolet (UV, C-SHG1; Nikon, Japan) excitation (330–380 nm). All images were directly recorded with a CCD camera (Nikon).
Casparian strip and suberin lamella development was analyzed using the Loess module in SPSS 22.0 (IBM, USA). Based on the different developmental stages, the number of epidermal cells, cortical cell layers, endodermal cells, and protoxylem and metaxylem layers was quantified, and the distribution of double protoxylem was measured.
RNA extraction, illumina library construction, and sequencing
Root sections displaying the three developmental stages of the Casparian strip and suberin lamella were separately enriched for RNA sequencing (RNA-seq). Each sample contains thirty roots in the corresponding stage and prepare them reference by Yang et al. 22 Total RNA was extracted using a Total Plant RNA Extraction Kit (Karroten, Beijing, China), and contaminating genomic DNA was removed in a digestion with DNaseΙ. Magnetic Oligo dT beads were used to collect the mRNA. A NEB fragmentation buffer was included in the thermomixer to cut the mRNA into short fragments at the right temperature. The broken mRNA was used as a template to generate the first-strand cDNA. The double-stranded cDNA was then synthesized by adding buffer, dNTPs and DNA polymerase I and RNase H, and the double-stranded cDNA was purified by AMPure XP beads. After the purified double-stranded cDNA was end-repaired, the A-tail was added and the sequencing linker was ligated. Fragments of 250–300 bp in length were amplified by PCR and used to build the cDNA library. The library quality was examined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), after which it was sequenced on an Illumina HiSeq 4000 (San Diego, CA, USA) at 1GENE.biotech.co.ltd (Hangzhou, China).
Read mapping and data quality assessment
The raw data in FASTQ format were filtered to remove reads containing adaptors, and low-quality reads with 30% or more of the bases having a quality value of ≤ 20 and reads with more than 5% unknown nucleotides were excluded. The clean reads were aligned to the reference data (ftp://ftp.ensemblgenomes. org/pub/plants/release-43/fasta/sorghum_bicolor/)29,30 using SOAP2 (soap2.21)31 and TopHat2 (tophat-2.0.11.Linux_x86_64).32,33 The sequencing quality was assessed and the random read distribution and gene coverage were evaluated. Gene expression levels were calculated as reads per kilobase per million mapped reads (RPKM).34
Differentially expressed genes and functional annotation
The assembled sequences were annotated in Swiss-Prot.35 Differentially expressed genes (DEGs) were identified using the edgeR function in Bioconductor (http://bioconductor.org), in which the count of the sequencing reads has a negative binomial distribution for each gene, providing a theoretical distribution basis for hypothesis testing.36 The DEGs were identified according to the following criteria: |log2Ratio| ≥ 1 and false discovery rate ≤ 0.05. The Gene Ontology (GO) functional enrichment was performed using Bioconductor to determine the biological function of the DEGs.37 A Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was also performed on the DEGs to identify any metabolic pathways or signal transduction pathways enriched in the DEGs.38
Quantitative real-time PCR analysis
Twelve DEGs possible involved in the formation of the apoplastic barrier were selected for quantitative real-time PCR analysis to verify the RNA-seq results. Primers were designed using Beacon Designer (version 7.0) (Table S1). The cDNA was generated from 1 μg total RNA using the cDNA Synthesis Kit (Vazyme, Nanjing, China). The housekeeping sorghum gene β-ACTIN (GenBank ID: X79378) was used as an internal standard. Real-time PCR was conducted with SYBR Premix Ex Taq (Takara, Beijing, China) using a LightCycler 96 system (Roche), as described by Sui et al. 3 The gene expression level was calculated using the 2–△△Ct method.39
Accession numbers
The RNA-seq data generated in this study were deposited in the National Center for Biotechnology Information (NCBI) database under the accession numbers SRR8997148 (Stage I), SRR8997149 (Stage II), and SRR8997150 (Stage III).
Results
Localization of the Casparian strip and suberin lamella formation
We observed Casparian strip and suberin lamella development in root cross sections stained with berberine hemisulfate-aniline blue and Fluorol Yellow 088, respectively (Figure 1). The location of Casparian strip formation relative to the apex was determined based on the number of radial cell walls between two adjacent stained endodermis cells. Suberin lamella differentiation was characterized based on the number of stained endodermal cells. Based on these observations, the sweet sorghum seminal root apoplast barriers were divided into three developmental stages: Stage I (0–10 mm from the root apex), without any Casparian strip or suberin lamella; Stage II (20–25 mm from the root apex), with a developing Casparian strip and suberin lamella; and Stage III (35–45 mm from the root apex), with a developed Casparian strip and suberin lamella.
Figure 1.
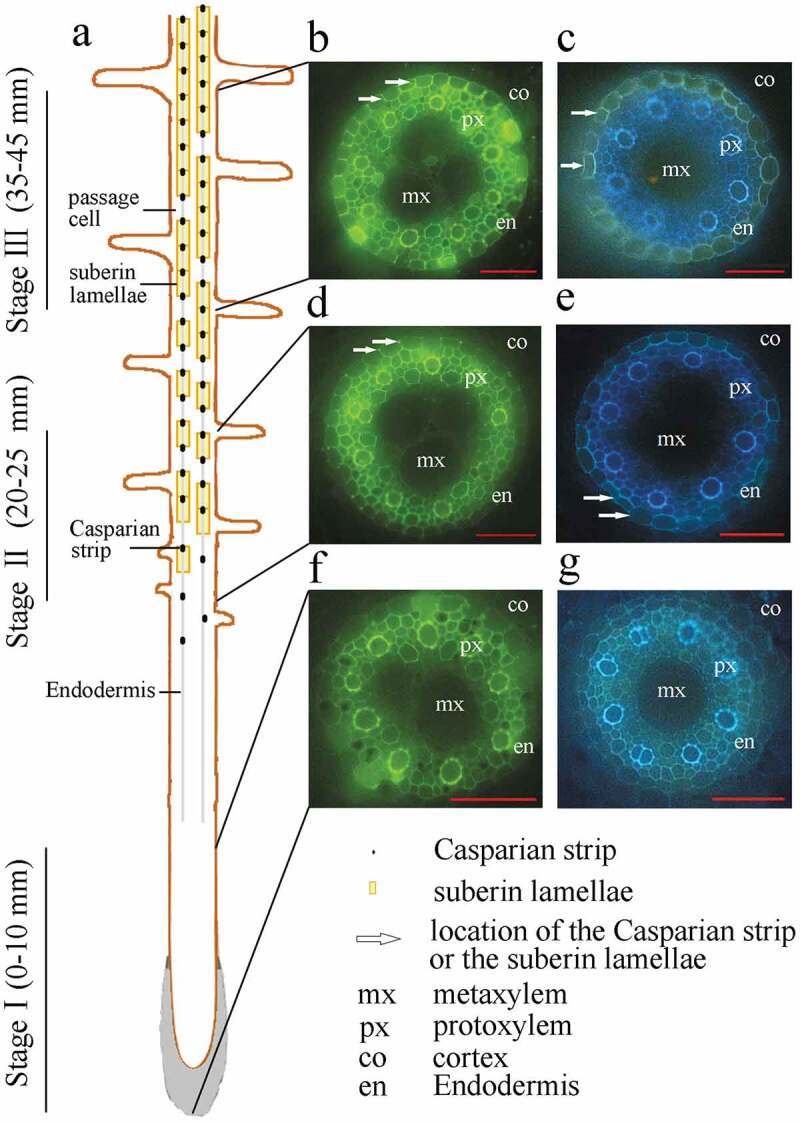
Localization and histochemical staining of the differentiating endodermis and developing Casparian strips and suberin lamellae of sweet sorghum seminal roots. Freehand cross sections were made using 10-day-old seminal roots grown in nutrient solution. (a) Schematic view of endodermal differentiation. Stage I (0–10 mm from the root tip), without any Casparian strips or suberin lamellae; Stage II (20–25 mm from the root tip), with developing Casparian strips and suberin lamellae; Stage III (35–45 mm from the root tip), with developed Casparian strips and suberin lamellae. (b, d, f) Root cross sections stained with berberine aniline blue to show the Casparian bands, viewed under blue light (450 nm). Arrows indicate Casparian strips. (c, e, g) Root cross sections were stained with FY088 to show the suberin lamellae, viewed under UV illumination (365 nm). Arrows indicate suberin lamellae. mx, metaxylem; px, protoxylem; co, cortex; en, endodermis. Bar = 50 μm.
To reduce the overlapping expression patterns of genes involved in each stage of apoplastic barrier formation, a certain segment between the different stages was discarded (21–24 mm from the root apex for Stage II and 26–34 mm from the root apex for Stage III). To further elucidate apoplastic barrier development in the different root sections, we characterized the morphological structures of the seminal root sections at 5-mm intervals from the root apex. No apoplastic barriers were observed in Stage I (Figure 2). The Casparian strip and suberin were first detected in the root at a distance of 20 mm from the apex and were then found to intensively develop at 20 to 30 mm from the apex, particularly in Stage II. The endodermis and Casparian strip had completed their development by the time they were 35 mm from the root apex. The development of the morphological structures of the seminal root sections was further examined by tallying the number of protoxylem layers, metaxylem layers, epidermal cells, cortical cells, and endodermal cells present. There was significant change in the numbers of these morphological features as the distance from the root apex increased (Table S2). The root epidermis was composed of dense cells, with approximately 105 cells comprising the root circumference. Five to six cortical cell layers encircled about 31 endodermal cells (Fig. S1).
Figure 2.
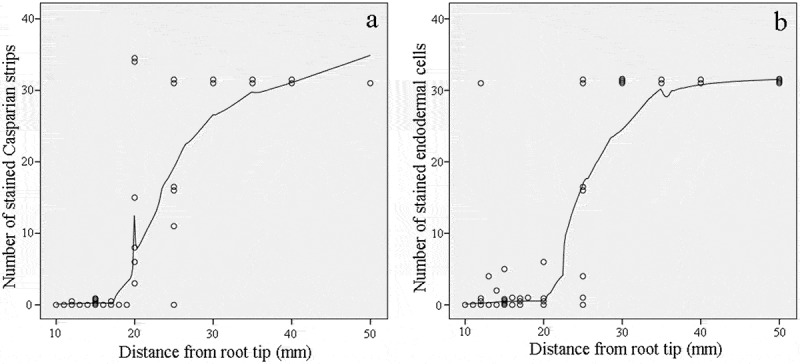
Number of Casparian strips and stained endodermal cells in cross sections along the sweet sorghum seminal root. (a) Number of stained Casparian strips between adjacent endodermis cells. The cells were stained using berberine hemisulfate-aniline blue dye. (b) Number of stained endodermal cells. The cells were stained using Fluorol Yellow 088. Each point represents the corresponding number at this location.
Analysis of DEGs associated with the developmental stages of Casparian strips and suberin lamellae
We divided the sweet sorghum roots into the three developmental sections based on the distances from the root tip, as described above. The number and quality of clean reads from each of the three root sections are presented in Table S3. In total, 24,344 annotated genes were expressed in Stage I, 24,501 in Stage II, and 24,423 in Stage III (Figure 3). A total of 23,246 genes were common among the three sections, while 414 genes were uniquely expressed in Stage I, 418 were unique to Stage II, and 408 were unique to Stage III (Figure 3; Fig. S2). These unique genes may determine the specific differentiation of each section of the root.
Figure 3.
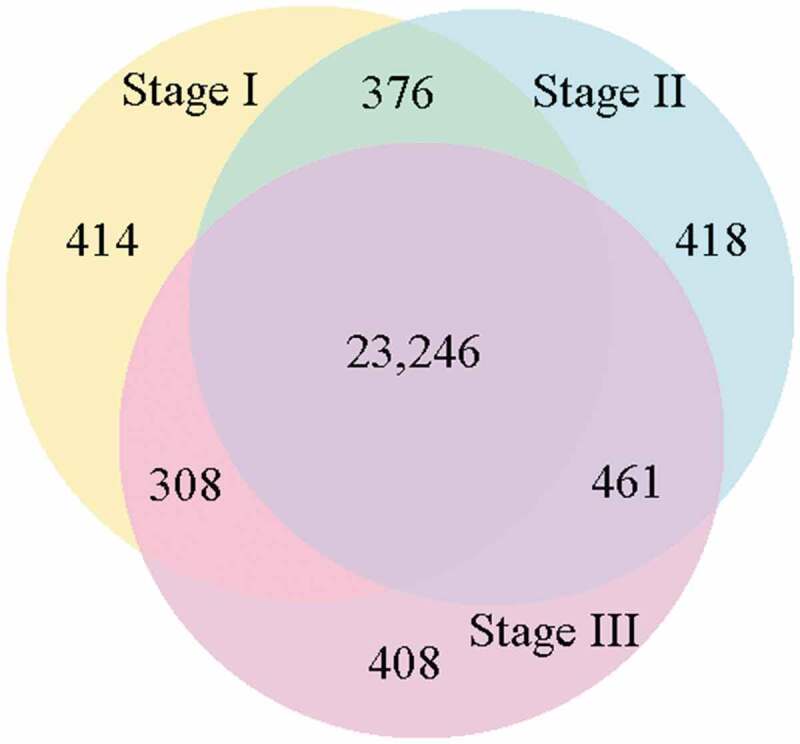
Venn diagram showing the number of shared and unique genes expressed in the three developmental sections of the root (expression threshold, RPKM > 1.0) .
DEGs participating in lignin biosynthesis
Lignin biosynthesis and the deposition of monolignols are essential steps of Casparian strip formation.8 Lignin biosynthesis involves the formation of precursors, monolignol biosynthesis and deposition. Sixty-one genes known to be involved in the phenylpropanoid pathway were differentially expressed between the three root sections (Figures 4 and 5; Tables S4 and S5), including 32 genes that were highly expressed in Stage I, 18 in Stage II, and 11 in Stage III.
Figure 4.
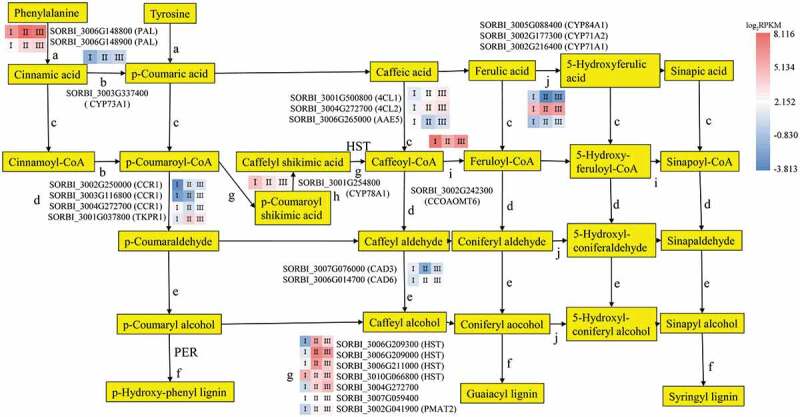
The DEGs mapped to the phenylpropanoid pathway (except peroxidase), with the expression levels in the three developmental sections marked as I, II, and III. The gene SORBI_3004G272700 is annotated as an uncharacterized acetyltransferase. The gene SORBI_3007G059400 is annotated as anthocyanin 5-aromatic acyltransferase. The encoded enzymes of the other genes are presented next to their accession numbers.
Figure 5.
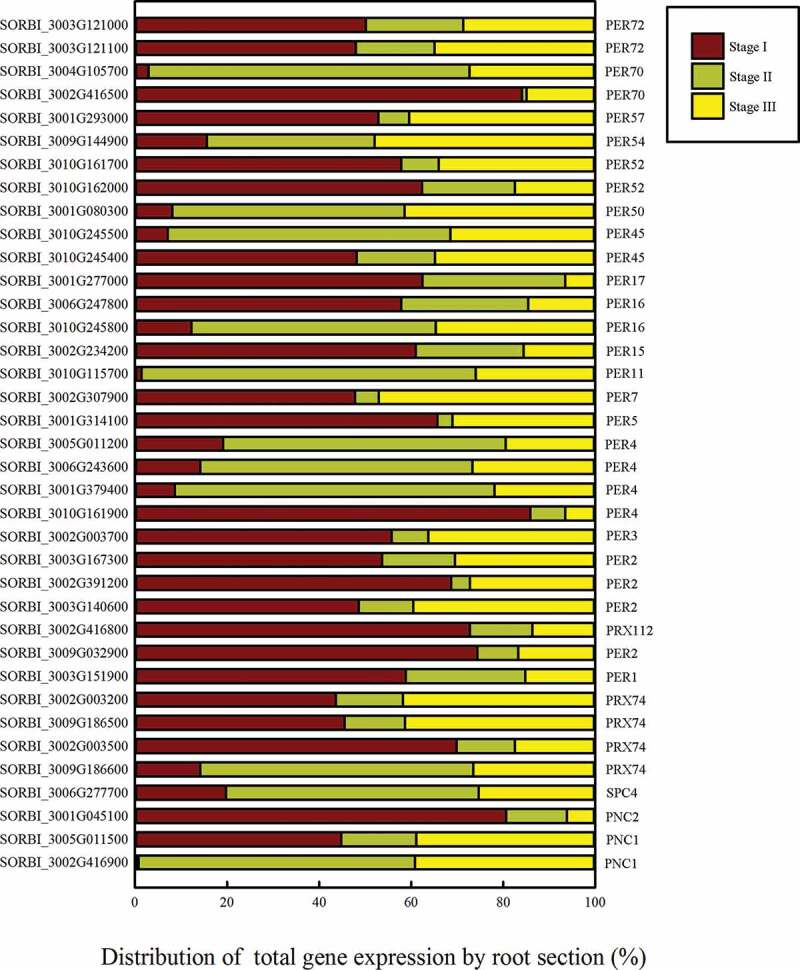
The distribution of the total expression of each gene in the peroxidase portion of the phenylpropanoid pathway. The gene expression level in each section is presented as a proportion of the total RPKM of the three sections.
During precursor biosynthesis, the lignin precursors syringyl, guaiacyl, and p-hydroxyphenol are fed into the phenylpropanoid biosynthesis pathway. Twenty-four of the genes encoding enzymes in this pathway were differentially expressed between the three root sections. Two genes (SORBI_3006G148800 and SORBI_3006G148900) encoding phenylalanine ammonia-lyases (PALs), which promote the conversion of phenylalanine into cinnamic acid and tyrosine into p-cinnamic acid, were highly expressed in the three root sections (Figure 4a); SORBI_3006G148800 was more highly expressed in Stage II, and SORBI_3006G148900 expression gradually increased throughout the three developmental stages but remained lower than the expression level of SORBI_3006G148800. This is the first step of Casparian strip precursor biosynthesis.
The SORBI_3003G337400 gene encoding CYTOCHROME P450 MONOOXYGENASE 73A1 (CYP73A1), which promotes the conversion of cinnamic acid to p-coumaric acid, had decreased expression levels in the three root sections as development progressed (Figure 4b). The 4-coumarate:CoA ligase (4CL) enzymes catalyze the production of coenzyme A esters (Figure 4c). Three genes encoding 4CLs and related enzymes (SORBI_3001G500800, SORBI_3004G272700, and SORBI_3006G265000) showed differential expression levels in the development of the lignin pathway. The expression levels of SORBI_3001G500800 and SORBI_3004G272700, encoding 4CL1 and 4CL2, respectively, increased as development progressed through the three stages; however, the expression of SORBI_3006G265000, which encodes ACYL-ACTIVATING ENZYME5 (AAE5), decreased as root development progressed and had its lowest expression level in Stage II. CINNAMOYL CoA REDUCTASE1 (CCR1) participates in the reduction of three cinnamate CoA esters to produce cinnamaldehyde and is the rate-limiting enzyme for lignin biosynthesis (Figure 4d). Four enzymes participate in the reduction reaction; SORBI_3002G250000 and SORBI_3003G116800 showed increasing levels of expression as root development progressed, SORBI_3004G272700 was most highly expressed in Stage I, and SORBI_3001G037800, encoding TETRAKETIDE ALPHA-PYRONE REDUCTASE1 (TKPR1), was most highly expressed in Stage II. Cinnamyl alcohol dehydrogenases family members (CADs) catalyze the biosynthesis of alcohol in lignin biosynthesis (Figure 4e). SORBI_3007G076000 showed decreased expression in Stage II, while SORBI_3006G014700 expression increased as root development progressed.
The peroxidases catalyze the last step of the formation of the lignin monomers (Figures 4f and 5). After lignin precursor biosynthesis, 37 peroxidase genes in the lignin biosynthesis pathway were found to be differentially expressed between the three root developmental stages outlined above. Eleven such genes were upregulated in Stage II, which suggests that they may participate in the formation of the apoplastic barrier. Seven genes encoding enzymes catalyzing two reactions that convert p-coumaroyl-CoA to caffeoyl-CoA, were differentially expressed (Figure 4g). Four genes encoding shikimate O-hydroxy-cinnamoyl transferase (HST) were differentially expressed in the three root systems, three of which were most highly expressed in Stage II. SORBI_3004G272700, encoding an acetyltransferase, was increasingly upregulated as the root developed. The expression of SORBI_3007G059400, encoding anthocyanin 5-aromatic acyltransferase, was slightly elevated in Stage II. The expression of SORBI_3002G041900, which encodes PHENOLIC GLUCOSIDE MALONYLTRANSFERASE2 (PMAT2), was similarly higher in Stages II and III than in Stage I. SORBI_3001G254800 encodes CYP78A1 and showed a decreasing expression trend across the three Stages (Figure 4h), while SORBI_3002G242300, encoding caffeoyl-CoA O-methyltransferase (CCOAOMT), was most highly expressed in Stage Ι (Figure 4i). The above three enzymatic reactions (Figure 4g–i) promote the production of G-type and S-type lignin. These reactions depend on the CYP family of genes (Figure 4j), which promote the formation of S-type lignin. SORBI_3002G177300 (CYP71A2) was most highly expressed in Stage II, while SORBI_3005G088400 (CYP84A1) shows the contrasting trend. SORBI_3002G216400 (CYP71A1) was increasingly upregulated throughout root development.
DEGs involved in fatty acid elongation and hydroxylation
Fatty acid elongation and hydroxylation are key steps in suberin lamellae biosynthesis.40,41 KCS is the rate-limiting enzyme in the fatty acid elongation pathway.42 Nine DEGs were found to be involved in this process in sorghum (Figure 6; Table S6); the expression levels of six of these genes were elevated in Stage II. SORBI_3005G168700 expression peaked in Stage II, while SORBI_3002G179700 showed the lowest expression level at this stage. SORBI_3009G241000 expression increased as root development progressed. CUTICULAR PROTEIN1 (CUT1; also known as KCS6) was most highly expressed in Stage II. Another DEG in the fatty elongation pathway encodes a steroid 5-alpha-reductase (DET2), which functions downstream of the KCS enzyme. This gene was most highly expressed in Stage I, declined in Stage II, and finally increased again in Stage III.
Figure 6.
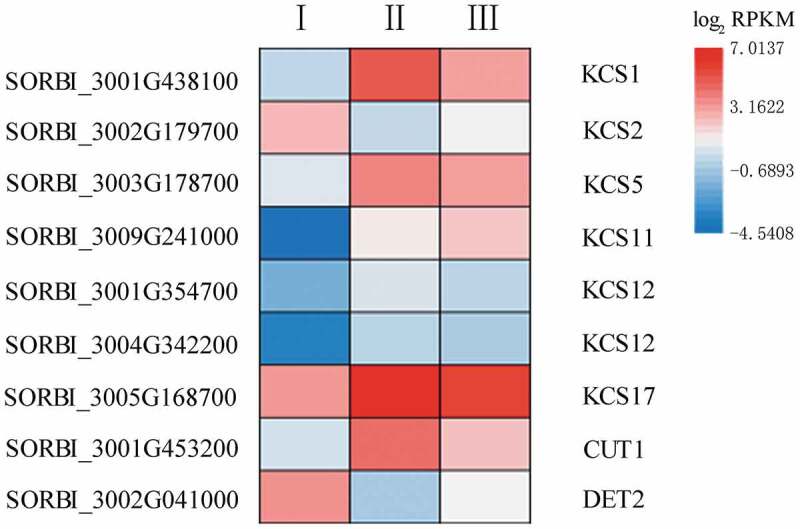
The expression of nine genes involved in fatty acid elongation and possible suberin development. The heatmap shows the log2 RPKM values of the expression levels.
The ω-hydroxylation of fatty acids is an enzymatic step in cuticular suberin and wax biosynthesis. Eleven genes in the cytochrome family are differentially expressed between the different stages of root development (Figure 7; Table S7), five of which were most highly expressed in Stage II. SORBI_3001G213300 (CYP86B1) expression also peaked in Stage II. The expression levels of SORBI_3007G015200 and SORBI_3007G015100 declined in Stage II and then increased in Stage III, while SORBI_3001G137900 and SORBI_3010G197700 expression levels declined throughout root development. By contrast, SORBI_3003G040300 and SORBI_3001G510400 expression increased as root development progressed. Hydroxycinnamoyl-CoA omega-hydroxypalmitic acid O-hydroxycinnamoyltransferase (HHT) catalyzes the conversion of feruloyl-CoA to 16-feruloyloxypalmitic acid in the cutin, suberin, and wax biosynthesis pathway. Seven DEGs (SORBI_3003G408900, SORBI_3003G408900, SORBI_3003G408900, SORBI_3006G211000, SORBI_3010G179900, SORBI_3008G068300, and SORBI_3007G059400) also participate in this enzymatic reaction. Six of these genes (excluding SORBI_3010G179900) were more highly expressed in Stage II, with SORBI_3003G408900 showing the highest level of expression. SORBI_3010G179900 (3ʹ-N-debenzoyl-2ʹ-deoxytaxol N-benzoyltransferase [TAX10]) was more highly expressed as root development progressed. SORBI_3004G342000 encodes ECERIFERUM1 (CER1), which can catalyze the conversion of a long-chain aldehyde to a long-chain alkane, and was most highly expressed in Stage II, while SORBI_3008G076200, also annotated as CER1, had its lowest level of expression in Stage II. This suggests that these two CER1 genes play opposing roles in hydroxylation. Three genes (SORBI_3005G063400, SORBI_3005G063300, and SORBI_3002G134100) were found to encode fatty acyl-CoA reductase (FAR), and all showed an increased level of expression in Stage II, which may be related to the biosynthesis of long-chain alcohols during suberin biosynthesis. The expression of the gene encoding alcohol-forming fatty acyl-CoA reductase (AF-CoA; SORBI_3007G169900) increased as root development progressed.
Figure 7.
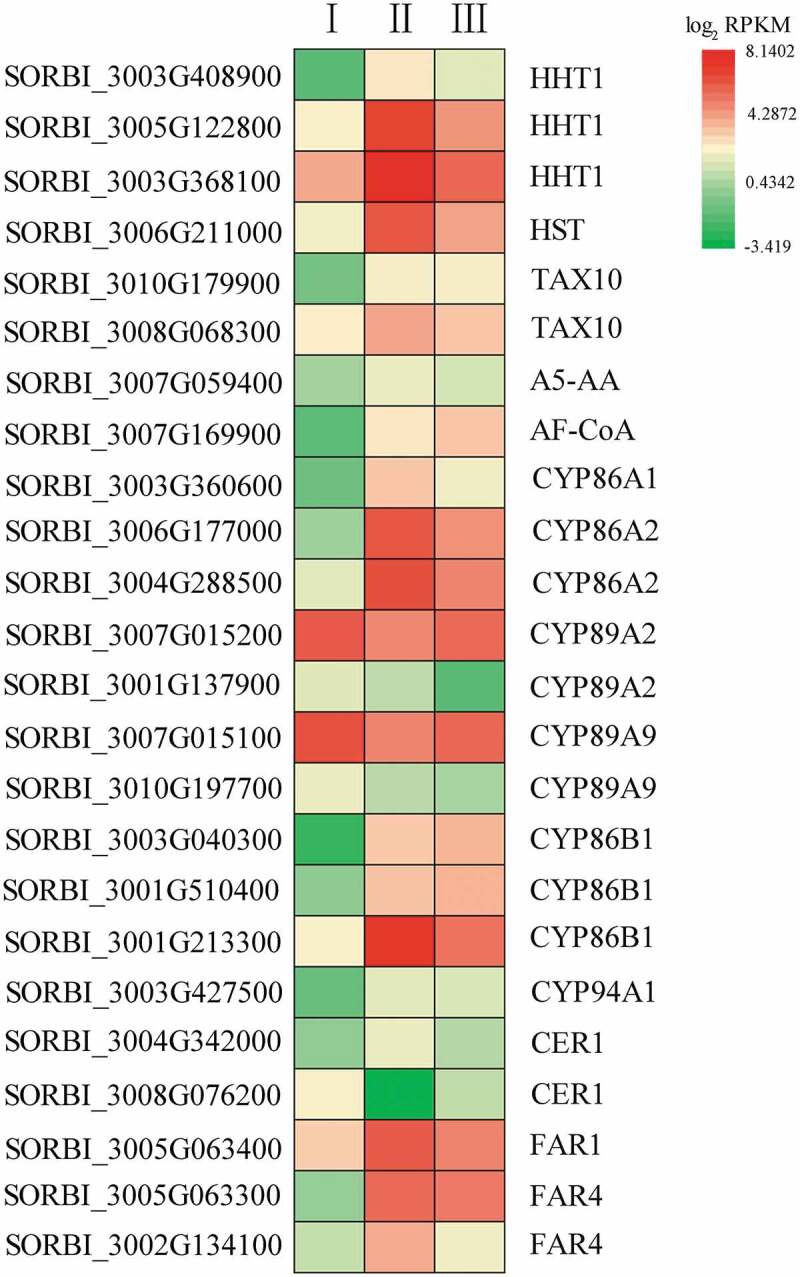
The expression of genes participating in fatty acid ω-hydroxylation and possible suberin development. The heatmap shows the log2 RPKM values of the expression levels.
DEGs involved in the transport of lignin and fatty acid derivatives during the formation of Casparian strips and suberin lamellae
Members of the ATP-binding cassette subfamily G (ABCG) family are thought to mediate the transport of lignin and monolignol fatty acids outside cells to form the Casparian strips and suberin lamellae.43 Five members in this family were differentially expressed throughout root development, with most of them being more highly expressed in Stage II. Two genes encoding ABCG16 showed higher expression than the other genes at each of the three stages. SORBI_3009G022000 showed equally high expression at each of the three stages, while SORBI_3001G413700 and SORBI_3003G348100 were most highly expressed in Stage II (Table S8).
Candidate transcription factors involved in the formation of Casparian strips and suberin lamellae
Forty DEGs encoding transcription factors were upregulated, while 28 DEGs were downregulated (Figure 8 and Table S9), including 12 WRKYs, 12 MYBs, 10 bHLHs, 12 NACs, 5 AP2s, 6 ethylene-responsive transcription factors (ERFs), 6 TFs, 3 heat stress transcription factors (HSFs), 1 GATA, and 1 nuclear transcription factor Y subunit B-5 (NTF). Previous studies have shown that MYBs and NACs are involved in the biosynthesis of the apoplastic barrier;44 however, our data indicated that WRKYs may also participate in the formation of the apoplastic barrier, something that has not been reported previously. Two ERFs were found to have an increased level of expression in Stage II, while four genes encoding other transcription factors (SORBI_3008G172200, SORBI_3003G305000, SORBI_3010G224200, and SORBI_3003G034300) showed increased levels of expression in Stages II and III. Three HSFs (SORBI_3001G496200, SORBI_3010G170600, and SORBI_3002G224200) and one NTF (SORBI_3003G417800) also showed increased expression levels in Stage II.
Figure 8.
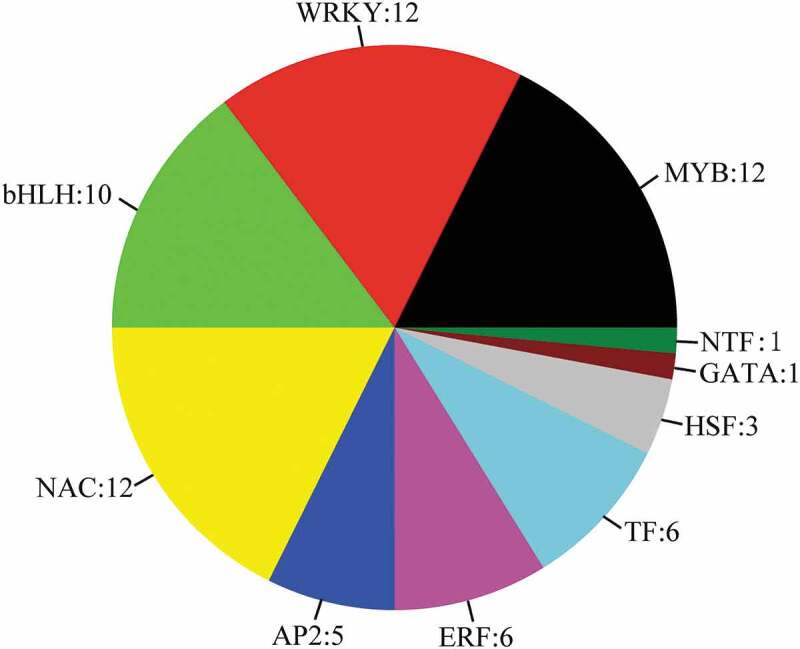
Differential expression of all transcription factors in the alignment of three developmental stages. The number of differentially expressed genes is specified.
Other possible DEGs related to Casparian strip and suberin lamella development
The deposition of monolignols is a key step after the lignin monomers are transported out of the cells, with the CASPs known to be involved in the deposition of lignin on the cell wall.45 Four of the DEGs we identified were annotated as CASP-like proteins. SORBI_3010G214800 and SORBI_3009G088900 were most highly expressed in Stage II, with the latter having a higher expression level than the former. SORBI_3002G113100 and SORBI_3003G162100 expression levels increased during root development (Table S10).
Seven DEGs were annotated as encoding laccases, suggesting that they may contribute to lignin biosynthesis46,47 (Table S11). Laccase-4 and Laccase-25 expression was lowest in Stage II, while Laccase-14, Laccase-8, and Laccase-3 were most highly expressed in Stage II and may participate in the formation of the apoplastic barrier.
Suberin biosynthesis requires the formation and deposition of fatty acid precursors. We identified seven DEGs related to the fatty acid biosynthesis pathway (Table S12), more of which were upregulated than downregulated during root development. No obvious trends were detected in the genes responsible for the fatty acid metabolic pathways (Table S13). The gene encoding GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE5 (GPAT5) was also found to be differentially expressed throughout root development (Table S14). Three genes (SORBI_3004G010300, SORBI_3009G162000, and SORBI_3003G360700) were highly expressed in Stage II, while two genes (SORBI_3001G099300 and SORBI_3001G250200) were highly expressed in Stage III.
qRT-PCR validation of RNA-seq
Based on the transcriptome results and the identified DEGs, 12 genes were randomly selected for qRT-PCR analysis to validate the RNA-seq data. The correlation between the RNA-seq and qRT-PCR data was high (R2 = 0.82, Figure 9; Table S15), indicating that the transcriptomic data were valid.
Figure 9.

Validation of the RNA-seq results using qRT-PCR to analyze the expression of 12 representative candidate genes. R2 represents the correlation between the RNA-seq (RPKM) and qRT-PCR data.
Discussion
The current study is the first to identify the specific developmental sections of the sorghum root in which Casparian strips and suberin lamellae are formed. Using histochemical staining and morphological analyzes, we distinguished three distinct developmental sections, enabling the identification and analysis of putative candidate genes involved in Casparian strip and suberin lamella formation in the endodermis.
Cross sections taken at set distances from the apex of the sorghum seminal root showed no significant differences in morphological structures along the length of the root, which is consistent with the observation that water and inorganic salts are absorbed by the root tip.7 Studies in rice and olive (Olea europaea)11,48 and several research models6,7,49 suggest that Casparian strips form before suberin lamellae. Barley (Hordeum vulgare) roots were previously divided into several sections for the study of suberin lamellae;5 however, in our study, both structures were found to form simultaneously. This rapid formation of both Casparian strips and the suberin lamellae may explain the salt tolerance of sweet sorghum.
We performed an RNA-seq analysis of the three distinct sections of the sorghum seminal root to identify putative candidate genes involved in Casparian strip and suberin lamella development. The formation of the Casparian strip is dependent on the polymerization of lignin,8 which is produced via the phenylpropanoid pathway. CCR and 4CL are the key enzymes in this pathway15 and were highly expressed in Stages II and III of the sorghum roots. These enzymes perform the initial reaction of lignin precursor formation. Members of the HST family, SORBI_3002G242300 (CCOAOMT) and SORBI_3001G254800 (CYP78A1), then further promoted the biosynthesis of the subsequent products based on the lignin precursor. PER64,45 PER02/25/71,50 PER37,51 and PER7252 participate in Casparian strip biosynthesis in Arabidopsis. Here, we discovered many additional peroxidase-encoding genes that were also differentially expressed throughout root development.
The most important reactions in suberin lamella biosynthesis are the elongation and hydroxylation of fatty acids.40,41 Many members of the KCS family are involved in these processes.53–55 Here, KCS2 was found to be highly expressed during Stage I of sorghum root development, consistent with the previous finding that the deletion of KCS2 affects suberin lamella composition in Arabidopsis.53 Recent studies have shown that the hydroxylation of the fatty acid ω sites is mainly catalyzed by the CYP86 and CYP94 families,56–59 many of which were also found to be highly expressed in Stage II. These results indicate that the high expression levels of the above genes may contribute to suberin lamella formation in Stage II. Moreover, FAR was most highly expressed during Stage II and was also previously shown to be involved in suberin lamella formation.60
Five members of the ABCG family were found to be highly expressed in Stage II of sorghum root development. These genes may function in the transport of lignin and fatty acids out of the cells during Casparian strip and suberin lamella formation.61–63
Although a series of genes were mapped to the pathways involved in Casparian strip and suberin lamella formation, most of the identified DEGs were annotated as functioning downstream of these pathways. Some of the upstream signaling genes, such as SHR,17 were not identified as DEGs, possibly due to differences between monocotyledons and dicotyledons. Nevertheless, we identified other candidate genes participating in Casparian strip and suberin lamella development, including transcription factors and those encoding laccases. The 28 DEGs encoding transcription factors that were expressed during Stage I may be involved in initiating the development of the apoplastic barriers, while the 28 that were highly expressed in Stage II may regulate the developmental process.20,44,64–67 The high expression levels of the NAC-type transcription factors in both Stages II and III likely participate in the regulation of the MYB transcription factors in the lignin biosynthesis pathway.67
CASPs play a critical role in the polymerization of the Casparian strip.45,68 We found that most genes encoding CASPs were highly expressed in Stage II, which is consistent with the observation that a CASP-like protein functions in Casparian strip polymerization.47 The laccases were also previously suggested to participate in Casparian strip biosynthesis.47,69 In Arabidopsis, gpat5 mutants had 50% less suberin lamellae than the wild type.70 Here, we found that these genes are highly expressed in Stages II and III, during which Casparian strips form.
Among them, 16 genes (HSTs [SORBI_3006G209300, SORBI_3006G209000, and SORBI_3006G211000], CYP71A2, PER11, PER16, KCS17, HHT1 [SORBI_3005G122800 and SORBI_3003G368100], CYP86B1, MYB39 [SORBI_3007G224800 and SORBI_3004G231700], WRKY70, ABCG16 [SORBI_3001G413700], CASP-like protein [SORBI_3009G088900], and Laccase-3) are highly expressed in Stage II. KCS17 is a homolog of KCS9.71 CYP86B1 can reduce the levels of 16:0, 18:0, and 18:1 ω-hydroxy acids and α,ω-DCAs in the root suberin.58 AtMYB36 is a homolog of AtMYB39.72 Phylogenetic analysis revealed that ABCG16 is closely related to ABCG2/6/20, which have been reported to function in suberin transport.63,73 The CASP-like proteins, laccases, and PERs also participate in the biosynthesis and transport of lignin.47 The physical locations of the candidate genes on the chromosome has been shown in Figure 10.
Figure 10.
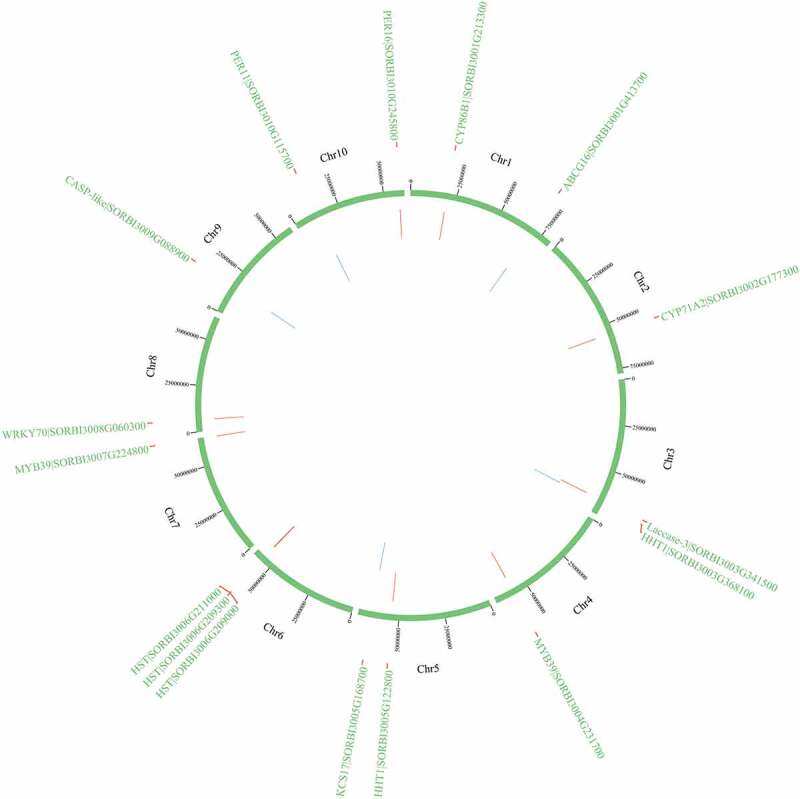
The physical locations of candidate genes involved on chromosomes based on BLAST results against the whole-genome sequencing dataset. Red color indicates the genes on the sense sequence and green color indicates the genes on the antisense strand.
Conclusion
The current research established a method for identifying the key genes involved in apoplastic barrier formation based on different developmental stages of seminal root and the distance from the root apex. In total, 196 genes involved in Casparian strips and suberin lamellae development were identified in the sweet sorghum root (Figure 11). Different genes are expressed in various parts of the roots. Strikingly, 190 unknown genes were highly expressed in the three stages and likely function in apoplastic barrier formation in sweet sorghum (Table S16). Transformation systems22 and CRISPR-Cas974 could be used to verify the functions of these candidate genes in apoplastic barrier formation.
Figure 11.
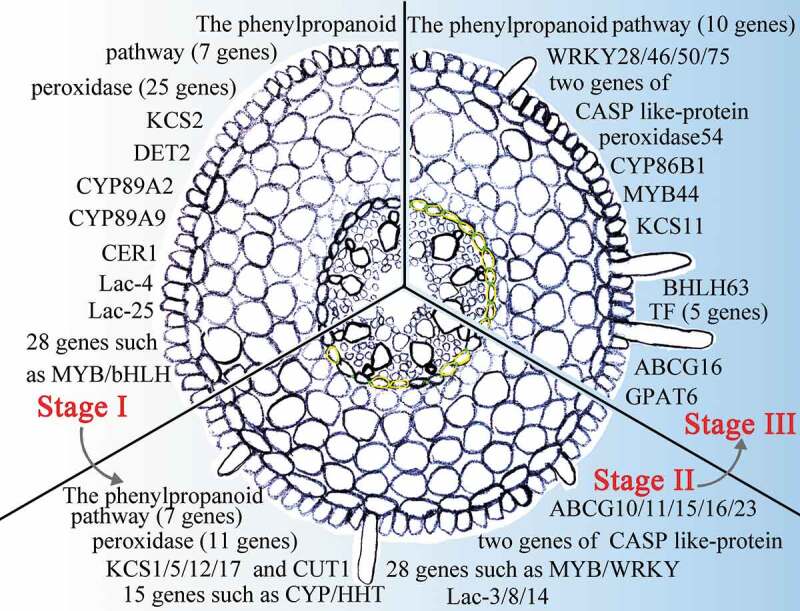
A model of sweet sorghum root Casparian strip and suberin lamella development. The presented genes display high levels of expression in Casparian strip- and suberin lamellae-related pathways in Stage I to Stage III of the root. Green between the endodermal cells indicates that the Casparian strip is stained in this section. The yellow endodermal cells indicate that the suberin lamellae of the cells in this section are stained.
Funding Statement
This work was supported by the NSFC (National Natural Science Research Foundation of China; Project Nos. [31570251, 31600200, and 31770288]), the Shandong Province Key Research and Development Plan [2017CXGC0313], and the Natural Science Research Foundation of Shandong Province [ZR2014CZ002; ZR2017MC003].
Author Contributions
B.W. designed research. X.W. performed and wrote this manuscript. Z.Y., G.H., X.Z. and S.Y. participated in the writing and modification of this manuscript. F.Y. and B.W. revised the paper. All authors read and approved the final manuscript.
Conflict of interest
The authors declare no conflicts of interest.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Basavaraj G, Parthasarathy Rao P, Ravinder Reddy C, Ashok Kumar A, Datta Mazumdar S, Ramana Reddy Y, Srinivasa Rao P, Karuppan Chetty SM, Reddy BV.. Sweet sorghum: A Smart crop to meet the demands for food, fodder, fuel and feed. In: innovative institutions, public policies and private strategies for agro-enterprise development. World Sci. 2015;1–14. doi: 10.1142/9789814596619_0007. [DOI] [Google Scholar]
- 2.Almodares A, Hadi MR.. Production of bioethanol from sweet sorghum: a review. Afr J Agric Res. 2009;4:772–780. doi: 10.5897/AJAR2017.12902. [DOI] [Google Scholar]
- 3.Sui N, Yang Z, Liu M, Wang B.. Identification and transcriptomic profiling of genes involved in increasing sugar content during salt stress in sweet sorghum leaves. BMC Genomics. 2015;16:534. doi: 10.1186/s12864-015-1760-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hasegawa PM. Sodium (Na+) homeostasis and salt tolerance of plants. Environ Exp Bot. 2013;92:19–31. doi: 10.1016/j.envexpbot.2013.03.001. [DOI] [Google Scholar]
- 5.Kreszies T, Shellakkutti N, Osthoff A, Yu P, Baldauf JA, Zeisler‐Diehl VV, Ranathunge K, Hochholdinger F, Schreiber L.. Osmotic stress enhances suberization of apoplastic barriers in barley seminal roots: analysis of chemical, transcriptomic and physiological responses. New Phytol. 2019;221:180–194. doi: 10.1111/nph.15351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barberon M, Geldner N. Radial transport of nutrients: the plant root as a polarized epithelium. Plant Physiol. 2014;166:528–537. doi: 10.1104/pp.114.246124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersen TG, Barberon M, Geldner N. Suberization - the second life of an endodermal cell. Curr Opin Plant Biol. 2015;28:9–15. doi: 10.1016/j.pbi.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Naseer S, Lee Y, Lapierre C, Franke R, Nawrath C, Geldner N (2012) Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc Natl Acad Sci U.S.A. 109:10101–10106. doi: 10.1073/pnas.1205726109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lanoue A, Burlat V, Henkes GJ, Koch I, Schurr U, Röse US. De novo biosynthesis of defense root exudates in response to Fusarium attack in barley. New Phytol. 2010;185:577–588. doi: 10.1111/j.1469-8137.2009.03066.x. [DOI] [PubMed] [Google Scholar]
- 10.Krishnamurthy P, Ranathunge K, Franke R, Prakash HS, Schreiber L, Mathew MK. The role of root apoplastic transport barriers in salt tolerance of rice (Oryza sativa L.). Planta. 2009;230:119–134. doi: 10.1007/s00425-009-0930-6. [DOI] [PubMed] [Google Scholar]
- 11.Krishnamurthy P, Ranathunge K, Nayak S, Schreiber L, Mathew MK. Root apoplastic barriers block Na+ transport to shoots in rice (Oryza sativa L.). J Exp Bot. 2011;62:4215–4228. doi: 10.1093/jxb/err135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernards MA. Demystifying suberin. Can J Bot. 2002;80:227–240. doi: 10.1139/B02-017. [DOI] [Google Scholar]
- 13.Hose E, Clarkson DT, Steudle E, Schreiber L, Hartung W. The exodermis: a variable apoplastic barrier. J Exp Bot. 2001;52:2245–2264. doi: 10.1093/jexbot/52.365.2245. [DOI] [PubMed] [Google Scholar]
- 14.Zimmermann HM, Hartmann K, Schreiber L, Steudle E. Chemical composition of apoplastic transport barriers in relation to radial hydraulic conductivity of corn roots (Zea mays L.). Planta. 2000;210:302–311. doi: 10.1007/PL00008138. [DOI] [PubMed] [Google Scholar]
- 15.Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annu Rev Plant Biol. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 16.Doblas VG, Geldner N, Barberon M. The endodermis, a tightly controlled barrier for nutrients. Curr Opin Plant Biol. 2017;39:136–143. doi: 10.1016/j.pbi.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 17.Li P, Yu Q, Gu X, Xu C, Qi S, Wang H, Zhong F, TI Baskin, Rahman A, Wu S.. Construction of a functional casparian strip in non-endodermal lineages is orchestrated by two parallel signaling systems in Arabidopsis thaliana. Curr Biol. 2018;28(2777–2786):e2772. doi: 10.1016/j.cub.2018.07.028. [DOI] [PubMed] [Google Scholar]
- 18.Nakayama T, Shinohara H, Tanaka M, Baba K, Ogawa-Ohnishi M, Matsubayashi Y. A peptide hormone required for Casparian strip diffusion barrier formation in Arabidopsis roots. Science. 2017;355:284–286. doi: 10.1126/science.aai9057. [DOI] [PubMed] [Google Scholar]
- 19.Franke RB. Caspary’s conductor. Proc Natl Acad Sci U S A. 2015;112:10084–10085. doi: 10.1073/pnas.1512640112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamiya T, Borghi M, Wang P, Danku JM, Kalmbach L, PS Hosmani, Naseer S, Fujiwara T, Geldner N, Salt DE. The MYB36 transcription factor orchestrates Casparian strip formation. Proc Natl Acad Sci U S A. 2015;112:10533–10538. doi: 10.1073/pnas.1507691112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vishwanath SJ, Delude C, Domergue F, Rowland O. Suberin: biosynthesis, regulation, and polymer assembly of a protective extracellular barrier. Plant Cell Rep. 2015;34:573–586. doi: 10.1007/s00299-014-1727-z. [DOI] [PubMed] [Google Scholar]
- 22.Yang Z, Zheng H, Wei X, Song J, Wang B, Sui N. Transcriptome analysis of sweet Sorghum inbred lines differing in salt tolerance provides novel insights into salt exclusion by roots. Plant Soil. 2018;430:423–439. doi: 10.1007/s11104-018-3736-0. [DOI] [Google Scholar]
- 23.Yuan F, Lyu MJ, Leng BY, Zheng GY, Feng ZT, Li PH, Zhu XG, Wang BS. Comparative transcriptome analysis of developmental stages of the Limonium bicolor leaf generates insights into salt gland differentiation. Plant Cell Environ. 2015;38:1637–1657. doi: 10.1111/pce.12514. [DOI] [PubMed] [Google Scholar]
- 24.Yuan H, Yu H, Huang T, Shen X, Xia J, Pang F, Wang J, Zhao M. The complexity of the Fragaria x ananassa (octoploid) transcriptome by single-molecule long-read sequencing. Hortic Res. 2019;6:46. doi: 10.1038/s41438-019-0126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo Y, Song Y, Zheng H, Zhang Y, Guo J, Sui N. NADP-malate dehydrogenase of sweet sorghum improves salt tolerance of Arabidopsis thaliana. J Agric Food Chem. 2018;66:5992–6002. doi: 10.1021/acs.jafc.8b02159. [DOI] [PubMed] [Google Scholar]
- 26.Lux A, Morita S, Abe J, Ito K. An improved method for clearing and staining free-hand sections and whole-mount samples. Ann Bot. 2005;96:989–996. doi: 10.1093/aob/mci266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brundrett MC, Enstone DE, Peterson CA. A berberine-aniline blue fluorescent staining procedure for suberin, lignin, and callose in plant tissue. Protoplasma. 1988;146:133–142. doi: 10.1007/BF01405922. [DOI] [Google Scholar]
- 28.Brundrett MC, Kendrick B, Peterson CA. Efficient lipid staining in plant material with sudan red 7B or fluoral yellow 088 in polyethylene glycol-glycerol. Biotech Histochem. 1991;66:111–116. doi: 10.3109/10520299109110562. [DOI] [PubMed] [Google Scholar]
- 29.Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 30.Zheng LY, Guo XS, He B, Sun LJ, Peng Y, Dong SS, Liu TF, Jiang S, Ramachandran S, Liu CM, et al. Genome-wide patterns of genetic variation in sweet and grain sorghum (Sorghum bicolor). Genome Biol. 2011;12:R114. doi: 10.1186/gb-2011-12-11-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 32.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 35.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. UniProt: the Universal Protein knowledgebase. Nucl Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson MD, Mccarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010:26: 139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alexa A, Rahnenfuhrer J. topGO: enrichment analysis for Gene Ontology. R Package Version. 2010;2:2010. doi: 10.18129/B9.bioc.topGO. [DOI] [Google Scholar]
- 38.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007;36:480–484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.Bonaventure G, Beisson F, Ohlrogge J, Pollard M. Analysis of the aliphatic monomer composition of polyesters associated with Arabidopsis epidermis: occurrence of octadeca-cis-6, cis-9-diene-1,18-dioate as the major component. Plant J. 2004;40:920–930. doi: 10.1111/j.1365-313X.2004.02258.x. [DOI] [PubMed] [Google Scholar]
- 41.Franke R, Briesen I, Wojciechowski T, Faust A, Yephremov A, Nawrath C, Schreiber L. Apoplastic polyesters in Arabidopsis surface tissues - A typical suberin and a particular cutin. Phytochemistry. 2005;66:2643–2658. doi: 10.1016/j.phytochem.2005.09.027. [DOI] [PubMed] [Google Scholar]
- 42.Millar AA, Kunst L. Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 1997;12:121–131. doi: 10.1046/j.1365-313X.1997.12010121.x. [DOI] [PubMed] [Google Scholar]
- 43.Do THT, Martinoia E, Lee Y. Functions of ABC transporters in plant growth and development. Curr Opin Plant Biol. 2018;41:32–38. doi: 10.1016/j.pbi.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 44.McCarthy RL, Zhong R, Ye Z-H. MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 2009;50:1950–1964. doi: 10.1093/pcp/pcp139. [DOI] [PubMed] [Google Scholar]
- 45.Lee Y, Rubio MC, Alassimone J, Geldner N. A mechanism for localized lignin deposition in the endodermis. Cell. 2013;153:402–412. doi: 10.1016/j.cell.2013.02.045. [DOI] [PubMed] [Google Scholar]
- 46.Liu CJ. Deciphering the enigma of lignification: precursortransport, oxidation, and the topochemistry of lignin assembly. Mol Plant. 2012;5:304–317. doi: 10.1093/mp/ssr121. [DOI] [PubMed] [Google Scholar]
- 47.Tobimatsu Y, Schuetz M. Lignin polymerization: how do plants manage the chemistry so well? Curr Opin Biotechnol. 2019;56:75–81. doi: 10.1016/j.copbio.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 48.Rossi L, Francini A, Minnocci A, Sebastiani L. Salt stress modifies apoplastic barriers in olive (Olea europaea L.): A comparison between a salt-tolerant and a salt-sensitive cultivar. Sci Hortic (Amsterdam). 2015;192:38–46. doi: 10.1016/j.scienta.2015.05.023. [DOI] [Google Scholar]
- 49.Robbins NE, Trontin C, Duan L, Dinneny JR. Beyond the barrier: communication in the root through the endodermis. Plant Physiol. 2014;166:551–559. doi: 10.1104/pp.114.244871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shigeto J, Kiyonaga Y, Fujita K, Kondo R, Tsutsumi Y. Putative cationic cell-wall-bound peroxidase homologues in Arabidopsis, AtPrx2, AtPrx25, and AtPrx71, are involved in lignification. J Agric Food Chem. 2013;61:3781–3788. doi: 10.1021/jf400426g. [DOI] [PubMed] [Google Scholar]
- 51.Pedreira J, Herrera MT, Zarra I, Revilla G. The overexpression of AtPrx37, an apoplastic peroxidase, reduces growth in Arabidopsis. Physiol Plant. 2011;141:177–187. doi: 10.1111/j.1399-3054.2010.01427.x. [DOI] [PubMed] [Google Scholar]
- 52.Herrero J, Fernández-Pérez F, Yebra T, Novo-Uzal E, Pomar F, Pedreño MÁ, Cuello J, Guéra A, Esteban-Carrasco A, Zapata JM.. Bioinformatic and functional characterization of the basic peroxidase 72 from Arabidopsis thaliana involved in lignin biosynthesis. Planta. 2013;237:1599–1612. doi: 10.1007/s00425-013-1865-5. [DOI] [PubMed] [Google Scholar]
- 53.Franke R, Hofer RI, Emsermann M, Efremova N, Yephremov A, Schreiber L. The DAISY gene from Arabidopsis encodes a fatty acid elongase condensing enzyme involved in the biosynthesis of aliphatic suberin in roots and the chalaza-micropyle region of seeds. Plant J. 2009;57:80–95. doi: 10.1111/j.1365-313X.2008.03674.x. [DOI] [PubMed] [Google Scholar]
- 54.Lee SB, Jung SJ, Go YS, Kim HU, Kim JK, Cho HJ, Park OK, Suh MC. Two Arabidopsis 3-ketoacylCoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J. 2009;60:462–475. doi: 10.1111/j.1365-313X.2009.03973.x. [DOI] [PubMed] [Google Scholar]
- 55.Serra O, Soler M, Hohn C, Franke R, Schreiber L, Prat S, Molinas M, Figueras M. Silencing of StKCS6 in potato periderm leads to reduced chain lengths of suberin and wax compounds and increased peridermal transpiration. J Exp Bot. 2008;60:697–707. doi: 10.1093/jxb/ern314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benveniste I, Bronner R, Wang Y, Compagnon V, Michler P, Schreiber L, Salaün JP, Durst F, Pinot F. CYP94A1, a plant cytochrome P450-catalyzing fatty acid omega-hydroxylase, is selectively induced by chemical stress in Vicia sativa seedlings. Planta. 2005;221:881–890. doi: 10.1007/s00425-005-1503-y. [DOI] [PubMed] [Google Scholar]
- 57.Benveniste I, Tijet N, Adas F, Philipps G, Salaün JP, Durst F. CYP86A1 from Arabidopsis thaliana encodes a cytochrome P450-dependent fatty acid omega-hydroxylase. Biochem Biophys Res Commun. 1998;243:688–693. doi: 10.1006/bbrc.1998.8156. [DOI] [PubMed] [Google Scholar]
- 58.Compagnon V, Diehl P, Benveniste I, Meyer D, Schaller H, Schreiber L, Franke R, Pinot F. CYP86B1 is required for very long chain ω-hydroxyacid and α,ω-dicarboxylic acid synthesis in root and seed suberin polyester. Plant Physiol. 2009;150:1831–1843. doi: 10.1104/pp.109.141408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Le Bouquin R, Skrabs M, Kahn R, Benveniste I, JP Salaün, Schreiber L, Durst F, Pinot F. CYP94A5, a new cytochrome P450 from Nicotiana tabacum is able to catalyze the oxidation of fatty acids to the ω-alcohol and to the corresponding diacid. Febs J. 2001;268:3083–3090. doi: 10.1046/j.1432-1327.2001.02207.x. [DOI] [PubMed] [Google Scholar]
- 60.Domergue F, Vishwanath SJ, Joubès J, Ono J, JA Lee, Bourdon M, Alhattab R, Lowe C, Pascal S, Lessire R, et al. Three Arabidopsis fatty acyl-coenzyme A reductases, FAR1, FAR4, and FAR5, generate primary fatty alcohols associated with suberin deposition. Plant Physiol. 2010;153:1539–1554. doi: 10.1104/pp.110.158238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miao Y-C, Liu C-J. ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc Natl Acad Sci U S A. 2010;107:22728–22733. doi: 10.1073/pnas.1007747108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shiono K, Ando M, Nishiuchi S, Takahashi H, Watanabe K, Nakamura M, Matsuo Y, Yasuno N, Yamanouchi U, Fujimoto M, et al. RCN1/OsABCG5, an ATP-binding cassette (ABC) transporter, is required for hypodermal suberization of roots in rice (Oryza sativa). Plant J. 2015;80:40–51. doi: 10.1111/tpj.12614. [DOI] [PubMed] [Google Scholar]
- 63.Yadav V, Molina I, Ranathunge K, Castillo IQ, Rothstein SJ, Reed JW. ABCG transporters are required for suberin and pollen wall extracellular barriers in Arabidopsis. Plant Cell. 2014;26:3569–3588. doi: 10.1105/tpc.114.129049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kosma DK, Jhadeswar M, Razeq FM, Patricia S, Richard B, Isabel M, Owen R. AtMYB41 activates ectopic suberin synthesis and assembly in multiple plant species and cell types. Plant J Cell Mol Biol. 2014;80:216–229. doi: 10.1111/tpj.12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lashbrooke J, Cohen H, Levy-Samocha D, Tzfadia O, Panizel I, Zeisler V, Massalha H, Stern A, Trainotti L, Schreiber L, et al. MYB107 and MYB9 homologs regulate suberin deposition in angiosperms. Plant Cell. 2016;28:2097–2116. doi: 10.1105/tpc.16.00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Legay S, Guerriero G, André C, Guignard C, Cocco E, Charton S, Boutry M, Rowland O, Hausman JF. MdMyb93 is a regulator of suberin deposition in russeted apple fruit skins. New Phytol. 2016;212:977–991. doi: 10.1111/nph.14170. [DOI] [PubMed] [Google Scholar]
- 67.Zhong R, Ye Z-H. Transcriptional regulation of lignin biosynthesis. Plant Signal Behav. 2009;4:1028–1034. doi: 10.4161/psb.4.11.9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roppolo D, De Rybel B, Tendon VD, Pfister A, Alassimone J, Vermeer JE, Yamazaki M, Stierhof YD, Beeckman T, Geldner N. A novel protein family mediates Casparian strip formation in the endodermis. Nature. 2011;473:380. doi: 10.1038/nature10070. [DOI] [PubMed] [Google Scholar]
- 69.Sterjiades R, Dean JFD, Eriksson KEL. Laccase from sycamore maple (Acer pseudoplatanus) polymerizes monolignols. Plant Physiol. 1992;99:1162–1168. doi: 10.1104/pp.99.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beisson F, Li Y, Bonaventure G, Pollard M, Ohlrogge JB. The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell. 2007;19:351–368. doi: 10.1105/tpc.106.048033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim J, Jung JH, Lee SB, Go YS, Kim HJ, Cahoon R, Markham JE, Cahoon EB, Suh MC. Arabidopsis 3-ketoacyl-coenzyme a synthase9 is involved in the synthesis of tetracosanoic acids as precursors of cuticular waxes, suberins, sphingolipids, and phospholipids. Plant Physiol. 2013;162:567–580. doi: 10.1104/pp.112.210450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 73.Kang J, Park J, Choi H, Burla B, Kretzschmar T, Lee Y, Martinoia E. Plant ABC transporters. In: The Arabidopsis book/American Society of Plant Biologists. 2011. P. 9. doi: 10.1199/tab.0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41:e188–e188. doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


