ABSTRACT
ProACO4–GUS expression and RT-PCR analysis revealed that ACO4 is predominantly expressed in shoots of Arabidopsis seedlings under light conditions. ACO4-overexpressed mutant 35S–ACO4 produced more ethylene relative to the wild-type, which resulted in reduced growth of Arabidopsis seedlings. The abnormal growth of seedlings recurred after the application of Co2+ ions, suggesting that ACO4 is a functional ACO necessary to regulate the growth and development of Arabidopsis seedlings. Exogenously-applied brassinosteroids (BRs) inhibited the expression of ACO4, and an enhanced ACO4 expression was found in det2, a BR-deficient mutant. Additionally, expression of ACO4 was decreased in bzr1-D (a BZR1-dominant mutant), implying that BR signaling negatively regulates ACO4 expression via BZR1 in Arabidopsis. In the intergenic region of ACO4, four E-boxes and a BR regulatory element (BRRE) are found. Electrophoretic mobility shift and chromatin immunoprecipitation assays showed that BZR1 binds directly to the BRRE in the putative promoter region of ACO4. By binding of BZR1 to BRRE, less ethylene was produced, which seems to regulate the growth and development of Arabidopsis seedlings.
KEYWORDS: ACC oxidase 4, Arabidopsis thaliana, Brassinosteroids, BZR1, ethylene production
Introduction
Ethylene, a gaseous plant hormone, regulates various growth and developmental processes during the plant life cycle.1–3 In plants, ethylene is biosynthesized from S-adenosyl-L-methionine (SAM) through the intermediate 1-aminocyclopropane-1-carboxylic acid (ACC). ACC synthases (ACSs) mediate the conversion of SAM to ACC, which is followed by the conversion of ACC to ethylene by the ACC oxidases (ACOs).4–9 In the ethylene biosynthesis pathway, the conversion of SAM to ACC is considered a rate-limiting step.10–13 The conversion of ACC to ethylene catalyzed by ACOs is the final regulatory step in ethylene biosynthesis. However, compared with ACSs, the biochemical and physiological function of ACOs remain largely unknown.
Exogenously-applied brassinosteroids (BRs) have been shown to increase both ethylene production and endogenous levels of ACC in Arabidopsis thaliana.14,15 The expression of several ACSs, such as ACS4, ACS5, and ACS6, is enhanced by BR application, suggesting that BRs regulate ethylene production via ACSs in A. thaliana.16–19 Transcriptome analysis has revealed that BRs alter the transcript levels of ACOs, as well as ACSs in A. thaliana, indicating that the biosynthetic conversion of ACC to ethylene by ACOs might also be regulated by BRs in this species.15,20 However, the mechanism of ethylene production by BRs via ACSs and ACOs is still largely unknown.
The Arabidopsis genome contains 13 ACOs as multigene family members. Among them, ACO1, ACO2, and ACO4 (formerly ethylene-forming enzyme, EFE) are considered as functional ACOs in Arabidopsis, implying that BR-induced ethylene production via ACOs is likely to be mediated by these functional ACOs in the plant.20 We recently demonstrated that ACO1, which is dominantly expressed in Arabidopsis root tips, is up-regulated by directly binding of a BR transcription factor BRASSINOSTEROID INSENSITIVE 1-EMS-SUPRESSOR1 (BES1), to E-box sequences in the ACO1 promoter region.10,21 Binding of BES1 to the ACO1 promoter increased ethylene production, which, in turn, activates the gravitropic response in Arabidopsis roots. These findings infer that ACO2 and ACO4 might also be regulated by BR signaling to exert physiologies associated with ethylene in Arabidopsis. To expand the knowledge on BR-induced ethylene production via ACOs, molecular regulation of ACO4 by BR signaling was investigated in this study.
Materials and methods
Plant materials and growth conditions
The seeds of A. thaliana were surface-sterilized with EtOH–H2O (70:30, v/v) for 5 min, washed with distilled H2O and stratified at 4°C for 3 days. The surface-sterilized seeds were planted on 0.8% agar medium (Phytagel, Sigma, Saint Louis, MO, USA) containing 0.5X Murashige and Skoog (MS) salt medium and 1% (w/v) sucrose, and grown under light (120 μmol m−2 s−1 at 22 ± 1°C for 16 h) and dark conditions (at 20 ± 1°C for 8 h), respectively, in an environmental growth chamber (Sanyo, Osaka, Japan). Seedlings were incubated for 2 days in the presence of brassinolide (BL; 10−8–10−10 M), 1 μM ACC, and 1 μM Co2+ ions. The seedlings were grown for 7 days on MS plates and photographed for measurement of the length of hypocotyls and roots.
Total RNA isolation and quantitative RT-PCR analysis
Total RNA was extracted from the wild-type, mutant, and BL-treated seedlings using TRI reagent (Sigma) according to the manufacturer’s instructions. For BL treatment, 7-day-old seedlings were transferred to 1X liquid MS medium with or without 0.01 μM BL and incubated for 6 h. For quantitative RT-PCR, first-strand cDNA was synthesized using 5 μg of total RNA and M-MLV reverse transcriptase (Promega, Madison, WI, USA). PCR was performed in a reaction containing 1 μL of cDNA, 0.25 μL of Taq DNA polymerase (Real Biotech Corporation, Taiwan), 2.5 μL of 2.5 mM dNTP mixture, 2.5 μL of 10X buffer (Takara Bio, Shiga, Japan), and 1 μL of 10 pmol of each primer, in a 25-μL reaction. The following gene-specific primers were used for the ACO4 genes: ACO4 forward (5′-GAATTCCGTCTGACTGAGTAATAATATAC-3′) and reverse (5′-CCATGGCTCTCTCTCTTTTTTTTTAAATG-3′. As a loading control, UBQ5 was amplified as well, with the following primers: UBQ5 forward (5′-GACCATAACCCTTGAGGTTGAATC-3′) and reverse (5′-AGAGAGAAAGAGAAGGATCGATC-3′). From a total of 25 μL of PCR products, 10 μL was loaded onto 0.8 or 1% agarose gel and stained with ethidium bromide (EtBr). Stained-bands in the gel were scanned and analyzed for densitometry using the Quantity One program (Bio-Rad, Hercules, CA, USA).
Histochemical β-glucuronidase (GUS) staining
Histochemical and quantitative analyzes of the transgenic lines expressing the GUS gene were performed, as described previously.22 For histochemical GUS staining, samples were incubated in a staining solution containing 2 mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid (Duchefa, Haarlem, The Netherlands) in a 50 mM Na2HPO4 buffer (pH 7.2), 2 mM potassium ferrocyanide, 2 mM potassium ferricyanide, and 0.2% Triton X-100 at 37°C overnight after vacuum infiltration on ice for 20 min. The samples were cleared using 70% EtOH and observed under a dissecting microscope (Olympus SZ-PT) and a light microscope (Olympus CX21).
Electrophoretic mobility shift assays (EMSAs)
To prepare the samples for EMSAs, first, the N-terminal of full-length BES1 was fused to maltose-binding protein (MBP) using a pMALc2X vector (New England Biolabs, Ipswich, MA, USA). The recombinant protein was then expressed and affinity-purified from Escherichia coli (BL21-CodonPlus [DE3]-RIL) using amylose resin. MBP, which was used as a negative control in the assays, was purchased from NEB. The probes of pACO4 used for the EMSA are described in Supplementary Table S1. These probes were labeled with γ-32P-dATP (10 μCi μL−1) using the DNA 5ʹ-end labeling system (Promega), according to the manufacturer’s instructions. The protein was mixed with 5X binding buffer (50 mM Tris–HCl [pH 7.5], 250 mM NaCl, 2.5 mM EDTA, 2.5 mM DTT, 5 mM MgCl2, and 10% glycerol), and incubated at room temperature (RT) for 10 min. Radioisotope-labeled probes were added to this mixture and incubated at RT for 10 min to allow the binding reaction. The reactions were resolved using 4% native polyacrylamide gels with a 0.5X TBE buffer (5.4 g L−1 Tris base, 2.75 g L−1 boric acid, 1 mM EDTA [pH 8.0]) and exposed to a phosphoimager.
Chromatin immunoprecipitation (ChIP)
The wild-type Col-0 plant and transgenic plants expressing the BES1–YFP, BRASSINAZOLE-RESISTANT1 (BZR1)–YFP were used for the ChIP assay procedure, described by Saleh et al.23 A chromatin–protein complex was isolated from the plant tissue by incubation in cross-linking buffer under vacuum. The buffer contents used in this assay are described in Supplementary Table S2. Tissues were ground to a fine powder with liquid nitrogen and resuspended in cold nuclei isolation buffer. The homogenates were filtered and centrifuged at 11,000 g, at 4°C, for 20 min. The supernatants were discarded, and the pellet was resuspended in cold nuclei lysis buffer. The resuspended samples (DNA) were sheared into ~500-bp fragments by ultrasonication. The samples were mixed with ChIP-grade YFP antibody (Abcam, Cambridge, UK) and incubated at 4°C for at least 5 h under gentle rotation. The immunocomplexes were precipitated with pre-equilibrated protein A agarose beads (Santa Cruz Biotechnology, Dallas, TX, USA). The precipitated DNA was purified using a PCR purification kit (LaboPass, Seoul, Korea) and amplified by PCR for 40 cycles. The primers used for amplifying the pACO4 regions in this assay are described in Supplementary Table S3. Before immunoprecipitation, aliquots of the samples were used as inputs and amplified using primers for the ACO4 promoter.
Measurement of ethylene production
Seven days after germination (DAG), the seedlings of plants were harvested in 25-mL silicon-capped vials, and 200 μL of buffer (100 mM MES, pH 6.8, 1.5 mM chloramphenicol) was added. The vials were incubated in the dark at 27 ± 1 °C with gentle shaking (170 rpm). Ethylene was measured as described elsewhere.24 The air was withdrawn from the sample (1 mL) vials using a syringe and injected into a gas chromatograph (HP5890 Series II; Hewlett Packard, USA) equipped with an alumina column and an 80/100 Porapak-Q column. The operating conditions were set as follows: oven temperature, 120 °C; injector temperature, 150 °C; detector temperature, 280 °C).
Results and discussion
ACO4 was the first enzyme predicted to have ACO catalytic activity, but its biochemical and physiological function in ethylene biology remains to be elucidated. To investigate the functional and physiological relevance of ACO4, we prepared transgenic Arabidopsis plants expressing the GUS gene driven by the 708-bp ACO4 promoter. Using ProACO4–G708 USD transgenic plants, the spatiotemporal expression of ACO4 was analyzed. In 5 DAG seedlings grown under dark, strong GUS activities were detected in the cotyledons (Figure 1a and 1b). Very weak GUS activities were detected in the vascular system of the hypocotyl, hypocotyl–root junction, and mature zone of roots (Figure 1c). In the elongation zone and root tip, little GUS activities were detected (Figure 1d). In 5 DAG seedlings grown under light, strong GUS activities were expressed in the vascular streams from the cotyledon to the elongation zone in root via the hypocotyl, hypocotyl–root junction, and root maturation zone (Figure 1e–i). No GUS activities were detected in the root tip (Figure 1i). These results suggest that light induces diffusion of ACO4 from the cotyledon to root apical meristematic zone via the vascular system to exert its physiological activity in A. thaliana.
Figure 1.
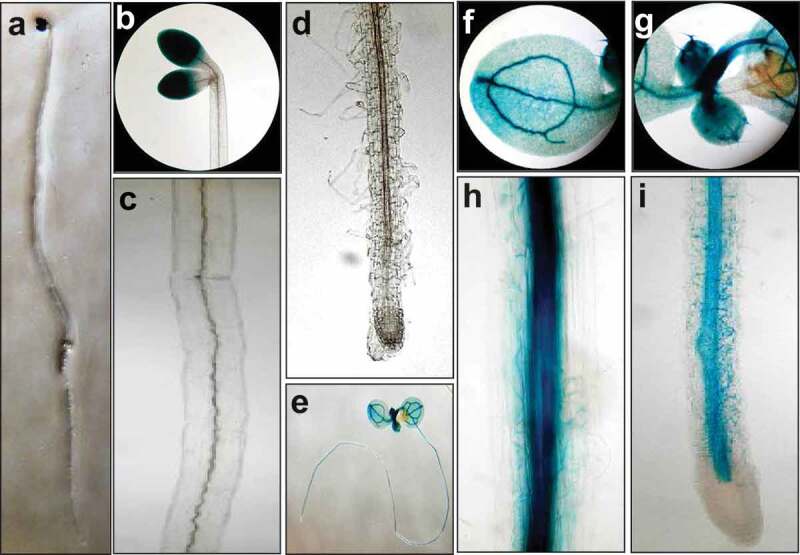
Spatiotemporal expression pattern for ACO4. (a-d) ACO4 promoter tissue-specific activity in 5 DAG ProACO4-G708 USD plants under dark conditions. Images show whole seedling (a), cotyledon (b), hypocotyl (c) and primary root (d). (e-i) Tissue-specific activity for ACO4 promoter in the transgenic plants under light conditions. Whole seedling (e), cotyledon (f), developing leaf (g), hypocotyl (h) and primary root (i).
Consistent with the ProACO4–GUS expression, RT-PCR analysis indicated that ACO4 was more highly expressed in shoot than the root of Arabidopsis seedlings grown under light (Figure 2a and 2b). Exogenously-applied ACC activated expression of ACO4, while Co2+ ions inhibited the gene expression in Arabidopsis seedlings (Figure 2c). In addition, overexpressed ACO4 (35 S–ACO4) increased ethylene production relative to wild-type seedlings (Figure 2d and e), indicating that ACO4 plays a role in controlling ethylene production in Arabidopsis seedlings. Under the dark condition, seedlings of 35 S–ACO4 showed reduced hypocotyls when compared with their wild-type counterparts, which was diminished by the application of Co2+ ions (figure 2f). Similarly, under light condition, 35 S–ACO4 seedlings exhibited shorter hypocotyls with inhibited primary roots compared with those in wild-type seedlings (Figure 2g). The retarded growth of 35 S–ACO4 was recovered to that of wild-type seedlings by application of Co2+ ion in seedlings grown in both dark and the light (figure 2f and g). These observations suggest that ethylene production by ACO4 is physiologically important to regulate the growth and development of Arabidopsis seedlings.
Figure 2.
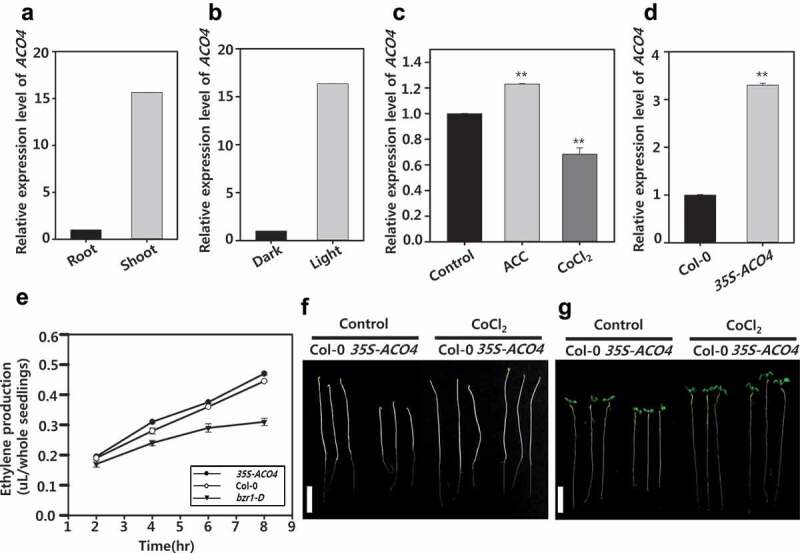
Expression and physiological functions of ACO4 under various conditions. (a) Comparison of ACO4 expression in the shoot and root of 5 DAG seedlings. (b) ACO4 expression in the seedlings grown under different light conditions. (c) Effect of ACC (1 µM) and CoCl2 (1 µM) on expression of ACO4 in 5 DAG seedlings under light condition. (d) Comparison of ACO4 expression in wild type (Col-0) and 35 S-ACO4 transgenic plants. Semi-quantitative RT-PCR was performed with total RNA. UBQ5 was used to normalize the expression level. Two biological repeats along with three technical repeats were performed for quantification of ACO4 expression level. The asterisks indicate significant difference between control and chemical-treated at P < .001 using t-test. (e) Measurement of ethylene production in the seedlings of wild type, 35 S-ACO4 and bzr1-D. Values are expressed as the mean of three biological replication ± S.E. (f and g) Seedling growth of wild type and 35 S-ACO4 in the absence (Control) and presence of 1 µM CoCl2 under dark (f) and light condition (g). The scale bar in F and G represents 1 cm.
Exogenously-applied BL, the most active BR, reduced expression of ACO4 in a concentration-dependent manner in seedlings of A. thaliana (Figure 3a). In det2, a BR-deficient mutant, the expression of ACO4 was increased more than that in wild-type seedlings (Figure 3b). The enhanced ACO4 expression in det2 was decreased significantly by the application of BL, suggesting that BR negatively regulates the expression of ACO4 in Arabidopsis. In bes1-D (a BES1-dominant mutant), no change in ACO4 expression was detected compared with that in wild-type seedlings. In contrast, significantly reduced ACO4 expression was found in bzr1-D (a BZR1-dominant mutant), indicating that BR-regulated ACO4 is likely to be mediated by the BZR1 transcription factor (Figure 3c).
Figure 3.
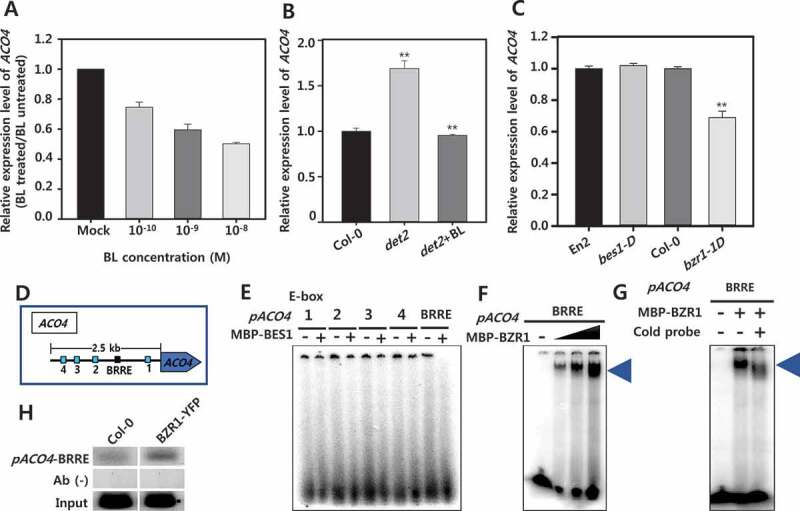
Regulation of ACO4 expression by BR signaling via BZR1 in Arabidopsis seedlings. (a) Effect of exogenously applied BL on expression of ACO4. (b) Comparison of ACO4 expression in wild type and det2. (c) Relative expression of ACO4 in bes1-D and bzr1-D compared to its wild type (En2 and Col-0 for bes1-D and bzr1-1D, respectively). Semi-quantitative RT-PCR was performed with total RNA. UBQ5 was used to normalize the expression level. Two biological repeats along with three technical repeats were performed for quantification of ACO4 expression level. The asterisks indicate significant difference between control and chemical-treated at P < .001 using t-test. (d) Cis-elements located on putative promoter region for ACO4. The blue and black box indicates E-box and BRRE element, respectively. (e-g) Electrophoretic mobility shift assays (EMSAs) with BES1 and BZR1 protein and the ACO4 promoter. (e) Probes containing the E-boxes indicated in (D) were incubated with (+) or without (-) 0.5 μg of recombinant BES1 and BZR1 protein. (f) 0.5, 1, and 2 μg of recombinant BZR1 protein incubated with a probe containing BRRE. The triangle represents increasing protein concentration. (g) 1 µg of BZR1 protein incubated with a cold-probe for competition assay. (h) Chromatin immunoprecipitation (ChIP) assays with wild-type and BZR1-YFP transgenic plants using an anti-GFP antibody (AB). Input: positive control (genomic DNA), AB (-): negative control (without antibody).
To determine the regulatory region required for BR-regulated ACO4 expression in Arabidopsis, cis-regulatory elements in the putative promoter region (2558-bp 5′-intergenic region from the start codon of ACO4 to the 3′-untranslated region of the adjacent gene) for ACO4 expression was analyzed. This putative promoter contains four E-box motifs (CANNTG, E1–E4) and a BR regulatory element (BRRE; CGTGTG) motif, both regulated by BR (Figure 3d). To investigate the interaction between the ACO4 promoter and BES1 and BZR1 protein, radioisotope-labeled probes (20-bp DNA probes containing E1–E4 and BRRE) for the DNA fragments were prepared. MBP–BES1 and MBP–BZR1 were obtained from E. coli using a construct in which MBP was fused to the N-terminal of full-length BES1 and BZR1, and EMSAs were conducted. As shown in Figure 3e, MBP–BES1 protein was not bound to any E-boxes and BRRE probes (Figure 3e). When MBP–BZR1 was used, binding of BZR1 to the BRRE probe occurred in a concentration-dependent manner (figure 3f). To clarify the binding of BZR1 to the BRRE, the BRRE sequence was then mutated to TTTTTT, and EMSAs were carried out again. As shown in Figure 3g, the binding of BZR1 to BRRE was greatly reduced by the mutation, which confirms that BZR1 directly binds to BRRE in the ACO4 promoter. Next, to confirm that BZR1 binds to the ACO4 promoter in vivo, ChIP assays were performed using transgenic plant specimens in which the BZR1–YFP fusion protein was expressed. As shown in Figure 3h, the in vivo binding of BZR1 to the ACO4 promoter was identified. Taken together, the results indicate that BZR1 is a BR transcription factor that directly binds to the ACO4 promoter to regulate the expression of ACO4 in A. thaliana. In bzr1-D, ethylene production was noticeably reduced relative to wild-type seedlings (Figure 2e). Hence, BZR1-mediated down-regulation of ACO4 seems to function in ethylene biology during the growth and development of Arabidopsis seedlings.
Tissue-specific and regulatory expression by light and BR of ACO4 was different from those of ACO1 in Arabidopsis. Stronger ACO4 expression was detected in shoots than roots. In contrast, ACO1 expression was much lower in shoots than roots.10 No ACO4 expression was detected in root tips, while ACO1 expression was concentrated in the tips, which promotes the gravitropic response of Arabidopsis roots. The expression of ACO4 was activated by light, whereas ACO1 expression was inhibited. By BR signaling, the expression of ACO4 was reduced because of the direct binding of BZR1 to BRRE in the promoter region of ACO4. ACO1 expression was enhanced by the binding of BES1 to E-boxes in the ACO1 promoter region. Recently, we found that expression of another functional ACO2 is strongly expressed in aerial parts under light condition, which is down-regulated by BR (data published elsewhere). These findings imply that ACO family members, which include at least three functional ACOs, may be optimized to perform different functions in various tissues and organs under the control of external (light) and internal stimuli (plant hormones) in A. thaliana.
Funding Statement
This work was supported by a grant from the Next-Generation BioGreen 21 Program (SSAC, grant#: PJ01320801), Rural Development Administration, Republic of Korea and by the Chung-Ang University Graduate Research Scholarship in 2018;Chung-Ang University [2018].
Disclosure Statement
No potential conflicts of interest were disclosed.
Supplemental material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Bleecker AB, Kende H.. Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol. 2000;16:1–6. doi: 10.1146/annurev.cellbio.16.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Yin C-C, Zhao H, Ma B, Chen S-Y, Zhang J-S.. Diverse roles of ethylene in regulating agronomic traits in rice. Front Plant Sci. 2017;8:1676. doi: 10.3389/fpls.2017.01676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubois M, Van den Broeck L, Inzé D. The pivotal role of ethylene in plant growth. Trends Plant Sci. 2018;23:311–323. doi: 10.1016/j.tplants.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol. 1984;35:155–189. doi: 10.1146/annurev.pp.35.060184.001103. [DOI] [Google Scholar]
- 5.Kende H. Ethylene biosynthesis. Annu Rev Plant Biol. 1993;44:283–307. doi: 10.1146/annurev.pp.44.060193.001435. [DOI] [Google Scholar]
- 6.Sato T, Theologis A. Cloning the mRNA encoding 1-aminocyclopropane-1-carboxylate synthase, the key enzyme for ethylene biosynthesis in plants. Proc Natl Acad Sci USA. 1989;86:6621–6625. doi: 10.1073/pnas.86.17.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamilton A, Bouzayen M, Grierson D. Identification of a tomato gene for the ethylene-forming enzyme by expression in yeast. Proc Natl Acad Sci USA. 1991;88:7434–7437. doi: 10.1073/pnas.88.16.7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spanu P, Reinhardt D, Boller T. Analysis and cloning of the ethylene‐forming enzyme from tomato by functional expression of its mRNA in Xenopus laevis oocytes. Embo J. 1991;10:2007–2013. doi: 10.1002/embj.1991.10.issue-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson PR, Ecker JR. The ethylene gas signal transduction pathway: a molecular perspective. Annu Rev Genet. 1998;32:227–254. doi: 10.1146/annurev.genet.32.1.227. [DOI] [PubMed] [Google Scholar]
- 10.Park CH, Roh J, Youn J-H, Son S-H, Park JH, Kim SY, Kim TW, Kim S-K. Arabidopsis ACC oxidase 1 coordinated by multiple signals mediates ethylene biosynthesis and is involved in root development. Mol Cells. 2018;41:923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van de Poel B, Van Der Straeten D. 1-aminocyclopropane-1-carboxylic acid (ACC) in plants: more than just the precursor of ethylene! Front Plant Sci. 2014;5:640. doi: 10.3389/fpls.2014.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang KL-C, Li H, Ecker JR. Ethylene biosynthesis and signaling networks. Plant Cell. 2002;14:S131–S51. doi: 10.1105/tpc.001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams D, Yang S. Ethylene biosynthesis: identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc Natl Acad Sci USA. 1979;76:170–174. doi: 10.1073/pnas.76.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joo S, Seo YS, Kim SM, Hong DK, Park KY, Kim WT. Brassinosteroid induction of AtACS4 encoding an auxin‐responsive 1‐aminocyclopropane‐1‐carboxylate synthase 4 in Arabidopsis seedlings. Physiol Plant. 2006;126:592–604. [Google Scholar]
- 15.Müssig C, Shin G-H, Altmann T. Brassinosteroids promote root growth in Arabidopsis. Plant Physiol. 2003;133:1261–1271. doi: 10.1104/pp.103.028662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen M, Chae HS, Kieber JJ. Regulation of ACS protein stability by cytokinin and brassinosteroid. Plant J. 2009;57:606–614. doi: 10.1111/tpj.2009.57.issue-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arteca RN, Bachman JM. Light inhibition of brassinosteroid-induced ethylene production. J Plant Physiol. 1987;129:13–18. doi: 10.1016/S0176-1617(87)80097-4. [DOI] [Google Scholar]
- 18.Lv B, Tian H, Zhang F, Liu J, Lu S, Bai M, Li C, Ding Z. Brassinosteroids regulate root growth by controlling reactive oxygen species homeostasis and dual effect on ethylene synthesis in Arabidopsis. PLoS Genet. 2018;14:e1007144. doi: 10.1371/journal.pgen.1007144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 20.Jiroutova P, Oklestkova J, Strnad M. Crosstalk between brassinosteroids and ethylene during plant growth and under abiotic stress conditions. Int J Mol Sci. 2018;19:3283. doi: 10.3390/ijms19103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park CH, Seo C, Park YJ, Youn J-H, Roh J, Moon J, Kim S-K. BES1 directly binds to the promoter of the ACC oxidase 1 gene to regulate gravitropic response in the roots of Arabidopsis thaliana. Plant Signal Behav. 15:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glazebrook J, Weigel D. Arabidopsis: A laboratory manual. New York, NY: Cold Spring Harbo r Laboratory Press; 2002. [Google Scholar]
- 23.Saleh A, Alvarez-Venegas R, Avramova Z. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc. 2008;3:1018. doi: 10.1038/nprot.2008.66. [DOI] [PubMed] [Google Scholar]
- 24.Yun HR, Joo S-H, Park CH, Kim S-K, Chang SC, Kim SY. Effects of brassinolide and IAA on ethylene production and elongation in maize primary roots. J Plant Biol. 2009;52:268–274. doi: 10.1007/s12374-009-9032-z. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


