ABSTRACT
Phytochrome A (phyA) is the primary photoreceptor mediating various plant responses to far-red (FR) light. The defense-related phytohorme jasmonic acid (JA) has been shown recently to play a role in regulating phyA-mediated FR signaling. However, the detailed molecular mechanisms governing phyA- and JA-mediated signaling cross talks are still not well understood. Here, we uncover a molecular cascade in which JAZ1 inactivates phyA signaling through repressing the transcriptional activity of FHY3 on FHY1 and FHL. Furthermore, we demonstrate that the expression levels of FHY1 and FHL, and some FR response genes are reduced in the coi1 mutant. These findings unveil a previously unrecognized mechanism whereby JA modulates phyA signaling through repressing the activities of FHY3 by JAZs.
KEYWORDS: FHY3, JAZ1, FHY1/FHL, phyA
Plant growth and development follows a highly plastic program, which is regulated by both internal cues (such as hormones) and external signals (such as light). Light and hormone coordinately control a set of plant growth and development processes, including seed germination, seedling de-etiolation, stomata and chloroplast movement, stem elongation, circadian rhythms and flowering.1 Plants have evolved an array of photoreceptors to perceive the light information. Among them, phytochromes are responsible for detecting red and far-red light. Phytochromes exist in two photoreversible forms: the inactive red light-absorbing form (Pr) and the active far-red light-absorbing form (Pfr). Upon light irradiation, inactive Pr form is converted to the Pfr form which is translocated from the cytosol into the nucleus, triggering downstream signaling cascade.2 There are five phytochromes exist in Arabidopsis, which are designated phyA–E. PhyA is the primary photoreceptor for perceiving FR light. Two small plant-specific proteins, FAR-RED ELONGATED HYPOCOTYL1 (FHY1) and its homolog FHY1-LIKE (FHL), are essential for the nuclear accumulation of light-activated phyA and subsequent FR light responses.3–5 The activation and repression of FHY1/FHL-phyA signaling is energetically demanding. FHY3, a transposase-derived transcription factor, activate FHY1/FHL gene expression directly, which in turn facilitating phyA nuclear accumulation upon FR light irradiation.6 Our prior work revealed that a group of JA signaling repressors, JAZ proteins can physically interact with FHY3 and repress its function in shade avoidance response.7 As FHY3 is essential for FHY1 and FHL genes expression, thus, we speculated that JAZ proteins might also affect FHY3-mediated FHY1 and FHL transcription. To test this, we performed a transient gene expression assay in N. benthamiana leaf to examine the effect of JAZ1 on the ability of FHY3 to promote FHY1 and FHL expression. As expected, our results showed that FHY3 could effectively induce the expression of the FHY1p:LUC and FHLp:LUC reporter genes, whereas co-expression of JAZ1 with FHY3 significantly repressed the expression of the FHY1p:LUC and FHLp:LUC reporter genes (Figure 1(a–d), indicating that JAZ1 can suppress the transcriptional activation activity of FHY3 on FHY1 and FHL in planta. Consistent with the observation, JAZ1 overexpression and coi1-2 mutant line (in which the JAZs protein are not degraded) exhibited an impaired phyA signaling phenotype: longer hypocotyl than wild type when seedlings were grown in continuous FR (FRc) conditions (Figure 2(a,b)). Furthermore, the JAZ1D3A transgenic plant, in which the overexpressed JAZ1 does not contain the JAS domain and dominantly represses JA responses probably by forming stable dimers with the native JAZ proteins,8,9 also displayed a more pronounced long hypocotyl phenotype than wild-type seedlings when grew under FRc conditions (Figure 2(a,b)). These observations suggest that inactivation of JA signaling or over accumulation of JAZ proteins can attenuate phyA signaling, at least partially through inhibition of FHY3 activity.
Figure 1.
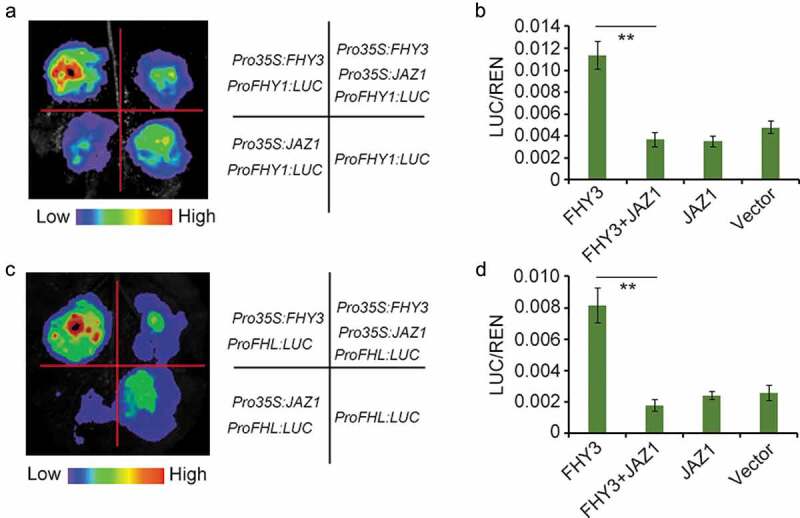
JAZ1 represses the transcriptional activation activity of FHY3 on FHY1 and FHL expression.
(a) and (b) JAZ1 suppresses the activation activity of FHY3 on FHY1 expression in N. Benthamiana leaves. (c) and (d) JAZ1 suppresses the activation activity of FHY3 on FHL expression in N. Benthamiana leaves. The relative LUC activities were normalized to the REN activity (LUC/REN). Significant differences are indicated: **, P < .01, Student’s t-test. Values are mean ± SD; n = 3.
Figure 2.
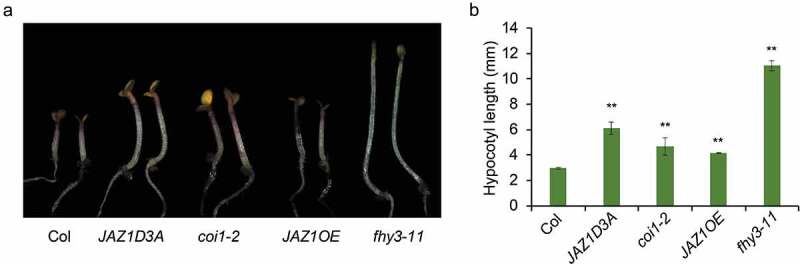
The JAZ1D3A, coi1-2, JAZ1OE and fhy3-11 mutants exhibit exaggerated hypocotyl elongation under FRc conditions.
(a) Hypocotyl length of the wild type (Col) and JAZ1D3A, coi1-2, JAZ1OE, fhy3-11 mutants grown under FRc conditions. (b) Quantification of hypocotyl length. Asterisks indicate significant differences compared with the wild type by Student’s t-test (p < .01). Data are presented as means ±SD, n > 15.
Next, we investigated the effect of JAZ overexpression in regulating FHY1 and FHL expression in vivo. Previous studies showed that the transcript levels of FHY1 reduced when dark-grown seedling were transferred to FRc conditions.6,10 Quantitative reverse transcriptase-PCR (qRT-PCR) analysis revealed that FHY1 and FHL transcript levels were significantly reduced in the coi1-2 plants (Figure 3(a,b)). Given that FHY1 and FHL are essential for phyA nuclear accumulation and subsequent FR signaling, we deduced that the expression of FR-responsive genes should be compromised in the coi1-2 mutant. To test this, we examined the expression of several representative FR-responsive genes in wild-type and coi1-2 seedlings grew under darkness and then transferred to FRc conditions. As shown in Figure 3(c–f), four FR-responsive genes CAB2, CO, HY5 and PIL1, showed reduced expression levels in the coi1-2 mutant relative to the wild-type. Together, these data suggest that JAZ proteins can antagonize FHY3-mediated activation of FHY1/FHL expression, thereby modulating the phyA signaling pathway. Thus, this work uncovers a previously unrecognized mechanism whereby JA modulates phyA signaling.
Figure 3.
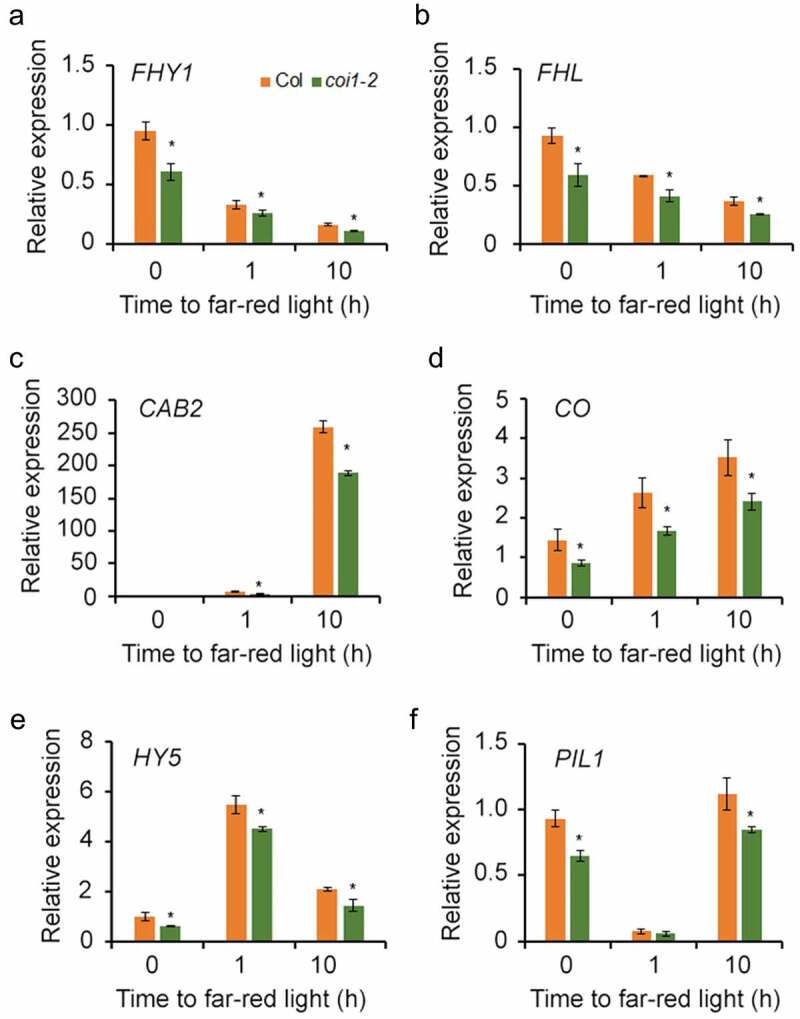
Expression of FHY1/FHL and FR-responsive genes is reduced in the coi1-2 mutant.
(a) and (b) FHY1 and FHL expression is significantly reduced in the coi1-2 mutant compared with wild type. (C)–(F) qRT-PCR analysis of far-red responsive gene expression (CAB2, CO, HY5 and PIL1) in wild-type and the coi1-2 mutant. All the seedlings were grown in darkness for 4 d and then transferred to FRc for various time periods. The comparative CT method was used to determine the relative gene expression, with the expression of PP2A (At1g13320) used as the internal control. Asterisks indicate significant differences from the wild-type plants (p < .05, Student’s t-test). Values are means ± SD; n = 3.
Funding Statement
This work was supported by the National Natural Science Foundation of China [31430008].
References
- 1.Lau OS, Deng XW.. Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol. 2010;13(5):1–4. doi: 10.1016/j.pbi.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Whitelam GC, Patel S, Devlin PF.. Phytochromes and photomorphogenesis in Arabidopsis. Phil Trans Roy Soc Lond B. 1998;353(1374):1445–1453. doi: 10.1098/rstb.1998.0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hiltbrunner A, Tscheuschler A, Viczián A, Kunkel T, Kircher S, Schäfer E. FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol. 2006;47(8):1023–1034. doi: 10.1093/pcp/pcj087. [DOI] [PubMed] [Google Scholar]
- 4.Hiltbrunner A, Viczián A, Bury E, Tscheuschler A, Kircher S, Tóth R, Honsberger A, Nagy F, Fankhauser C, Schäfer E. Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr Biol. 2005;15(23):2125–2130. doi: 10.1016/j.cub.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 5.Rösler J, Klein I, Zeidler M. Arabidopsis fhl/fhy1 double mutant reveals a distinct cytoplasmic action of phytochrome A. Proc Natl Acad Sci USA. 2007;104(25):10737–10742. doi: 10.1073/pnas.0703855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H. Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science. 2007;318(5854):1302–1305. doi: 10.1126/science.1146281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, Wei H, Ma M, Li Q, Kong D, Sun J, Ma X, Wang B, Chen C, Xie Y, et al. Arabidopsis FHY3 and FAR1 regulate the balance between growth and defense responses under shade conditions. Plant Cell. 2019;31(9):2089–2106. doi: 10.1105/tpc.18.00991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature. 2007;448(7154):661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 9.Chung HS, Howe GA. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM domain protein JAZ10 in Arabidopsis. Plant Cell. 2009;21(1):131–145. doi: 10.1105/tpc.108.064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Li G, Gao S, Martinez C, He G, Zhou Z, Huang X, Lee JH, Zhang H, Shen Y, et al. Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell. 2010;22(11):3634–3649. doi: 10.1105/tpc.110.075788. [DOI] [PMC free article] [PubMed] [Google Scholar]


