The substantial discrepancy between the strong effects of functional foods and various drugs, especially traditional Chinese medicines (TCMs), and the poor bioavailability of these substances remains a perplexing problem. Understanding the gut microbiota, which acts as an effective bioreactor in the human intestinal tract, provides an opportunity for the redefinition of bioavailability. Here, we discuss four different pathways associated with the role of the gut microbiota in the transformation of parent compounds to beneficial or detrimental small molecules, which can enter the body’s circulatory system and be available to target cells, tissues, and organs.
KEYWORDS: bioavailability, gut microbiota, host metabolism, nutrient and drug outcomes, short-chain fatty acid
SUMMARY
The substantial discrepancy between the strong effects of functional foods and various drugs, especially traditional Chinese medicines (TCMs), and the poor bioavailability of these substances remains a perplexing problem. Understanding the gut microbiota, which acts as an effective bioreactor in the human intestinal tract, provides an opportunity for the redefinition of bioavailability. Here, we discuss four different pathways associated with the role of the gut microbiota in the transformation of parent compounds to beneficial or detrimental small molecules, which can enter the body’s circulatory system and be available to target cells, tissues, and organs. We further describe and propose effective strategies for improving bioavailability and alleviating side effects with the help of the gut microbiota. This review also broadens our perspectives for the discovery of new medicinal components.
INTRODUCTION
Considerable attention has been paid to the substantial discrepancy between the strong biological effects of some drugs and functional foods and the poor bioavailability of these substances. Recent studies by Zimmermann et al. have opened the door to a revolution in understanding the important roles of the gut microbiota in the metabolism of many pharmaceuticals, which could lead to a possible redefinition of bioavailability (1, 2).
For orally administered drugs and functional foods, bioavailability generally summarizes the quantity or proportion of the ingested dose that is directly absorbed in the small intestine to enter the living system (into circulation) (Fig. 1) (3, 4). However, the parent ingredients of compounds, especially many Chinese herbs and medicinal foods, are difficult to detect in the circulatory system after oral administration, while many Chinese herbs and medicinal foods have been proven to be efficient for thousands of years. Troubleshooting and other strategies, such as modification of structures and delivery systems, have been carried out to improve bioavailability in the small intestine, but most of these attempts have been in vain. Recent great progress in studies on the gut microbiota has shed light on understanding the discrepancy between the explicit effects of parent compounds and low plasma concentrations. It has been reported that the influence of intestinal commensal bacteria on the utilization of these compounds might explain such a discrepancy (5–7).
FIG 1.
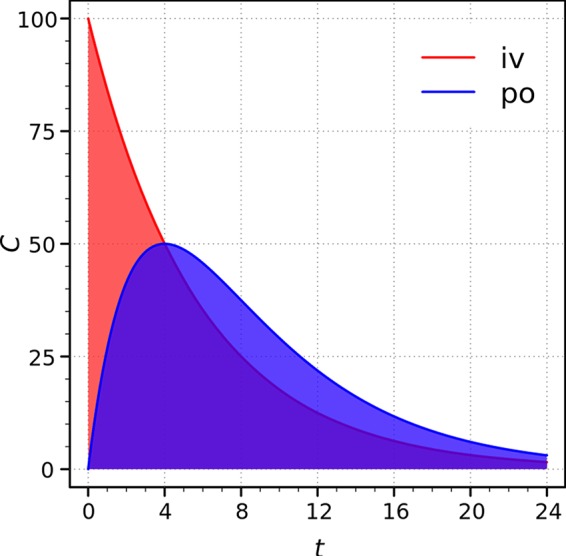
General understanding of bioavailability: the ratio of the areas under the curves. iv, intravenous administration; po, oral route; C, plasma concentration (in arbitrary units) (3, 4).
Zimmermann et al. utilized a combination of germfree animals and Bacteroides thetaiotaomicron transposon mutants deficient in enzymes that could convert brivudine (BRV) to bromovinyluracil (BVU) as a pharmacokinetic paradigm to quantitatively predict host and microbiome contributions to drug metabolic transformation. Astonishingly, the contributions of the microbiome to the metabolism of some drugs are much more than 50% (1). Chemical modifications of 271 oral drugs under microbiome-encoded enzymes of 76 diverse gut bacteria were further identified as being the results of oxidation, reduction, deacetylation, hydrogenation, hydroxylation, acetylation, and propionylation, which changed the masses of metabolites with respect to the corresponding parent drugs. This revealed causal links between the microbiota gene content and interpersonal differences in drug metabolism and drug responses (2). In addition, another model of a special population can demonstrate the roles of the intestinal microbiota in the human body in the regulation of bioavailability. Ileostomists, first reported by Hollman et al. in 1995 (6), are patients who have their colons surgically isolated from their bodies. Ileostomists are ideal model systems for examination of the absorption ratios of dietary substances in the human small and large intestines. During the 24 h after ingestion of coffee, the ratio of the metabolites presented in the urine of ileostomy patients to chlorogenic acid intake was 8% ± 1%, while this ratio was 29% ± 4% for the group consisting of healthy subjects (8). With the help of such a model for humans, Stalmach et al. indicated that the absorption of chlorogenic acid in instant coffee occurred in both the small and large intestines and that absorption occurred primarily in the large intestine (9). Thus, the microbiota in both the small intestine and colon should be considered to influence the bioavailability of foods or drugs. Therefore, the classic conceptual framework of bioavailability needs to be reexamined. The important roles of the gut microbiota in the metabolism of many pharmaceuticals and foods have led to a possible redefinition of bioavailability. Here, we summarize four pathways in the regulation of bioavailability by the gut microbiota and present constructive proposals for the improvement of bioavailability based on modulation of the gut microbiota.
THE GUT MICROBIOTA: A BIOREACTOR OF VARIOUS BIOACTIVE METABOLITES FROM PARENT COMPOUNDS
A vast number of gut bacteria have colonized the human intestinal tract throughout human history. These microbes are referred to as our second genome due to their high abundance and vital role. The gut microbiota was found to be a strong regulator of energy harvesting in 2004 (10); accordingly, these microbes have shown strong correlations with processes such as lipometabolism (11–14), glycometabolism (15–18), intestinal infection (19–21), brain function (22–26), immunity (27–29), and tumorigenesis (30–32). The gut microbiota is influenced by both genetic (33) and environmental (34, 35) factors, although the latter are more important than the former.
The gut microbiota can help hosts to digest ingested foods or drugs via enzymes that are secreted only by bacteria and via the excretion of fecal energy. The main metabolic reactions have been reported to be conducted by intestinal microbial enzymes, including β-glucosidase, β-glucuronidase, azoreductase, sulfatase, nitroreductase, and nitrate reductase (36, 37). Important biotransformations, including reductive metabolism, hydrolytic reactions, demethylation, deamination, dehydroxylation, deacylation, decarboxylation, and oxidation, are considered to be conducted by specific gut microbes (36, 38–40). Although the metabolites and related enzymes that participate in these biotransformations have been described for some reactions, an understanding of the roles of the enteric community of bacteria in these biochemical reactions remains largely uncharacterized (41).
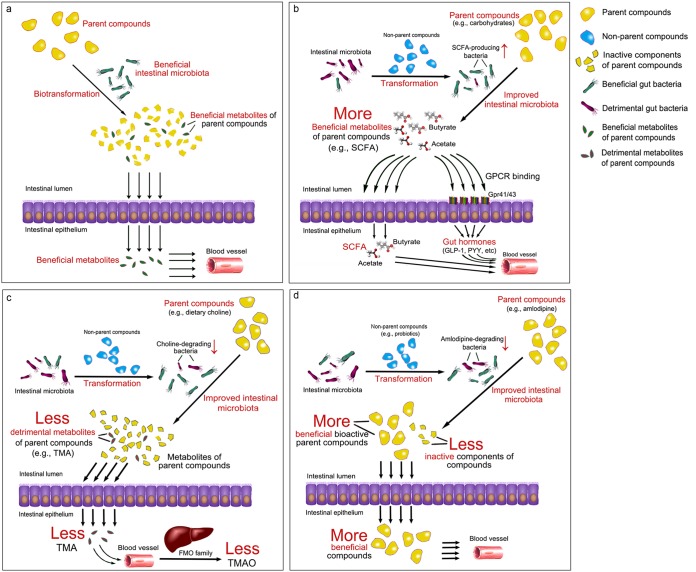
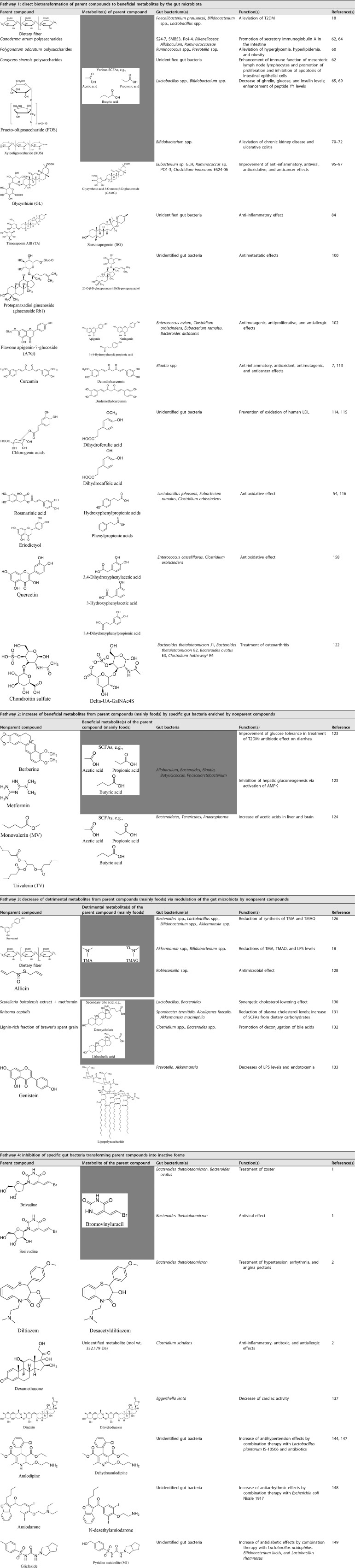
The intestinal microbiota has significant effects on the digestion of foods and the synthesis of beneficial bioactive substances, such as short-chain fatty acids (SCFAs) (18, 42–44) and vitamins (45, 46), and detrimental molecules, such as lipopolysaccharides (LPSs) (47, 48), branched-chain amino acids (BCAAs) (49), bile acids (50, 51), and trimethylamine (TMA), the precursor of TMA N-oxide (TMAO) (52, 53). Therefore, despite the direct effects of some substances on human health, many biological functions are performed by small bioactive molecules transformed from either foods or drugs by the gut microbiota. We propose that the intestinal microbiota has significant effects on the formation of bioactive small molecules via four pathways (Fig. 2): the gut microbiota biotransforms the parent functional foods and drugs directly into bioactive compounds (pathway 1), nonparent components trigger the metabolism of the parent nutrients by beneficial gut bacteria to produce additional beneficial molecules (pathway 2), the gut microbiota is modulated by nonparent molecules to decrease the entry of detrimental metabolites from the parent drugs or foods into the bloodstream (pathway 3), and specific gut bacteria that can transform the parent drugs into inactive compounds are inhibited by nonparent molecules to increase the entry of drugs into the circulatory system (pathway 4). Notably, in pathway 1, the investigated components are the parent molecules, the metabolites of which are the bioactive molecules, and in pathway 4, the investigated compounds are the bioactive components themselves. In contrast, in pathways 2 and 3, the investigated compounds are the nonparent compounds that affect the bioavailability of the parent compounds (frequently from the daily diet) via modulation of the gut microbiota. Therefore, when discussing bioavailability, we should distinguish parent compounds from nonparent compounds. Furthermore, the four pathways (especially pathways 1 and 2) are seldom mutually exclusive and occur separately.
FIG 2.
Four pathways by which the gut microbiota alters the bioavailability of food and medicine. (a) Pathway 1: direct biotransformation of parent compounds to beneficial metabolites by the gut microbiota. (b) Pathway 2: increase of beneficial metabolites from parent compounds by specific gut bacteria enriched by nonparent compounds. (c) Pathway 3: decrease of detrimental metabolites from parent compounds via modulation of the gut microbiota by nonparent compounds. (d) Pathway 4: inhibition of specific gut bacteria transforming parent compounds into inactive forms by nonparent compounds.
Most of the interactions between parent foods or drugs and the intestinal microbiota can potentially be classified into one of the above-described four pathways. Therefore, it is crucial to reasonably improve the bioavailability of parent foods or drugs effectively based on one of these four pathways to improve the therapeutic effects of these substances. Unquestionably, such a development would rapidly expand the market for researchers and producers of functional foods and pharmaceuticals.
THE FOUR PATHWAYS: A BREAKTHROUGH IN THE SEARCH FOR THE EXACT BIOACTIVE MOLECULES
The gut microbiota improves the bioavailability of the parent ingested substances via four pathways to promote the therapeutic effects of these substances. These four pathways also lay the foundation for subsequent breakthroughs in the search for functional foods and new drugs. All the parent compounds, metabolites, and key gut bacteria involved in the improvement of bioavailability mentioned in this review are summarized in Table 1.
TABLE 1.
Participants involved in the four pathways to improve bioavailability
Pathway 1: Direct Biotransformation of Parent Compounds into Beneficial Metabolites by the Gut Microbiota
An abundance of evidence has demonstrated that the gut microbiota can interact with indigestible dietary compounds and herbal medicines, such as polysaccharides, oligosaccharides, saponins, and phenolic compounds, directly transforming these molecules into more active compounds and improving their oral bioavailability (54–58).
Indigestible carbohydrates.
Indigestible carbohydrates, including dietary fiber, polysaccharides, and oligosaccharides, have been proven to be the active components of a large number of pharmaceuticals and functional foods. However, these compounds cannot be easily digested in the small intestine. Many studies have shown that the important metabolites, SCFAs, are the primary final products of the fermentation of polysaccharides or oligosaccharides by potential beneficial bacteria (59).
Dietary fiber has been reported to greatly enrich acetic acid- and butyric acid-producing bacteria in a randomized controlled clinical study. Accordingly, the concentrations of these two SCFAs also increased during the intervention period. Alleviation of type 2 diabetes mellitus (T2DM) was correlated with an improvement in the gut microbiota, an upregulation of SCFA production, and increased serum glucagon-like peptide 1 (GLP-1) and peptide YY (PYY) levels. Notably, an elevation of an index based on mathematical modeling of the 15 SCFA producers could predict a reduction in glycosylated hemoglobin (HbA1c, an important glycometabolism parameter), indicating significant roles of the SCFAs derived from beneficial gut bacteria during the alleviation of T2DM (18).
Polysaccharides in medicinal and edible plants and in marine organisms not only are metabolized to produce high levels of SCFAs but also reduce metabolic syndrome or play immunoregulatory functions; in addition, these compounds exhibit improved therapeutic effects in clinical applications via the enrichment of SCFA-producing gut bacteria (60–63). For instance, polysaccharides from Ganoderma atrum might regulate the gut microbiota, play a core role in the bioactive effects, and enhance the concentrations of SCFAs and secretory immunoglobulin A in the intestinal tracts of rats (62, 64). Polygonatum odoratum polysaccharides can restore the composition of the impaired intestinal microbiota of high-fat diet (HFD)-fed rats to that of the control group by increasing the abundance of SCFA-producing bacteria, which is correlated with an improvement in obesity parameters (60). Cordyceps sinensis polysaccharides significantly enhanced butyrate levels produced by the gut microbiota and not only improved histone H3 acetylation, mediating modulatory T (Treg) cell-specific Foxp3, but also markedly reversed the increases in interleukin-17 (IL-17) and IL-21 levels induced by cyclophosphamide in mice (62).
Oligosaccharides exhibit considerable modulatory effects on the gut microbiota, leading to the production of various SCFAs and a favorable influence on metabolic disorders and colonic motility. For example, fructo-oligosaccharide (FOS), galactose-oligosaccharide (GOS), and isomaltose-oligosaccharide (IMO) treatments increased the levels of SCFA-producing bacteria, including Lactobacillus and Bifidobacterium (65). SCFAs such as butyric acid could rectify motility in germfree mice (66), possibly by increasing histone H3 acetylation in enteric neurons (67), leading to the alleviation of constipation by the enhancement of vagal activity (68).
Furthermore, FOS intake facilitated decreases in the levels of ghrelin, glucose, and insulin and an increase in the level of PYY (69). Xylo-oligosaccharide (XOS) and inulin-type fructans can also increase the abundance of Bifidobacterium and the levels of butyric acid and acetic acid for significant alleviation of chronic kidney disease and ulcerative colitis (70–72). Therefore, although the bioavailability of herbs and functional foods containing polysaccharides or oligosaccharides is low, SCFAs metabolized from these two complex-carbohydrate components by beneficial bacteria might be key bioactive molecules. Detecting SCFA levels might be much more important than focusing on the bioavailability of herbs and functional foods containing these two complex carbohydrates.
SCFAs such as propionic acid are biotransformed from carbohydrates by the gut microbiota primarily via the succinate pathway, the acrylate pathway, and the propanediol pathway. The succinate pathway is the primary pathway adopted by a number of Bacteroidetes species to synthesize propionate (73). The succinate pathway also exists in Ruminococcus flavefaciens, which generates succinate, but not propionate, and succinate is a precursor of propionate (74). The conversion of lactate to propionate involves the succinate pathway and the acrylate pathway, as observed in Veillonella spp. (via the succinate pathway) and Megasphaera spp. (via the acrylate pathway) (75). Escherichia coli, Anaerostipes rhamnosivorans, and Bacteroides species can all degrade deoxy sugars via the propanediol pathway (76, 77).
SCFAs can bind to G-protein-coupled receptors (GPCRs), mainly GPR41 (referred to as FFAR3) and GPR43 (referred to as FFAR2), on intestinal epithelial cells; regulate inflammatory responses (78, 79); and trigger the secretion of gut hormones, mainly GLP-1 and PYY (80, 81). GPR41 and GPR43 have been discovered on several cells other than intestinal cells, such as adipocytes, endocrine cells, and immune cells. SCFAs in the bloodstream could reach these cells and bind to these SCFA-sensing receptors.
Saponins.
Many poor-circulatory bioavailable herbs have been found to exert pharmacological effects without SCFA production. Recently, increasing numbers of bioactive molecules have been gradually discovered to be metabolized from these functional herbs by the gut microbiota. Saponins are valuable bioactive components that exhibit anti-inflammatory and anticancer activities. However, these molecules are poorly absorbed into human blood, resulting in very low efficacy in human tissues (82, 83).
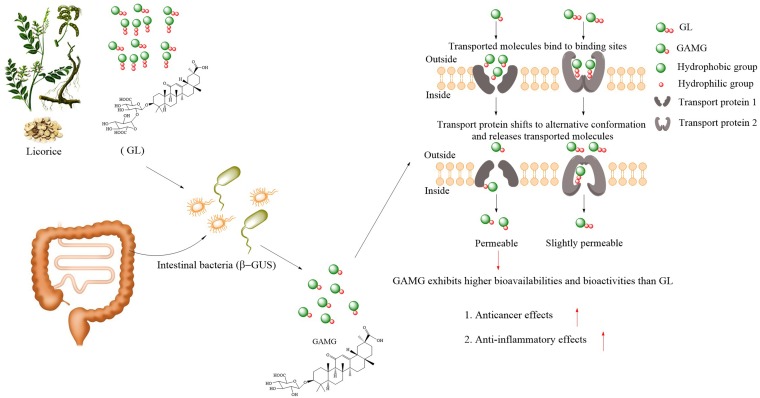
The gut microbiota provides new opportunities to overcome the poor bioavailability of saponins by transforming these compounds into beneficial metabolites (84, 85). Among these saponin compounds, glycyrrhizin (GL), a typical and well-known triterpenoid saponin extracted from licorice, has many valuable pharmacological effects, such as antiviral (86), antioxidative (87), anticancer (88), and anti-inflammatory (89) effects, and has been attracting our attention. It is also one of the few ingredients whose fate is relatively clear after metabolism by specific gut microbiota. Based on this discovery, glycyrrhetic acid 3-O-mono-β-d-glucuronide (GAMG), a metabolite with higher bioavailability than the parent compound GL, has been commercially developed through direct biotransformation in the food and pharmaceutical industries in Japan (90–93). GL is not easily absorbed from the intestine into the blood when administered orally to humans, as the structure, consisting of two glucuronide molecules, is strongly polar (94). However, one distal glucuronic acid of GL can be hydrolyzed by β-d-glucuronidase (β-GUS) derived from intestinal bacteria and then converted to GAMG, which exhibits moderate membrane permeability with suitable molecular polarity, possibly because of the sugar conjugation in this molecule, leading to an elevation in bioavailability (95, 96). Gut bacteria, including Eubacterium sp. strain GLH, Ruminococcus sp. strain PO1-3, and Clostridium innocuum ES24-06, play significant roles in the modification of the chemical structure of GL to produce other relevant intestinally absorbed metabolites (such as GAMG, 3α-hydroxyglycyrrhetic acid [3α-hydroxyGA], and 3-oxo-glycyrrhetic acid [3-oxoGA]) with relatively strong pharmacological activities (97) (Fig. 3). Timosaponin AIII (TA), another saponin compound, is metabolized to its active metabolite sarsasapogenin (SG) in the intestine via cleavage of the glycosyl moieties of TA by the gut microbiota to exert diverse pharmacological effects. Interestingly, SG exhibits a higher anti-inflammatory effect than the parent compound (TA), mainly via inhibition of NF-κB activation and proinflammatory cytokine (tumor necrosis factor alpha [TNF-α], IL-1β, and IL-6) expression in LPS-stimulated macrophages (84). Ginseng, whose pharmacological activities are primarily ascribed to ginseng saponins (98), generally exhibits restorative effects, tonicity, and revitalization effects (99). An unexpected phenomenon occurred in which ginseng and a ginseng-derived triterpenoid saponin, ginsenoside Rb1 (Rb1), exhibited different efficacies among individual patients based on the different Rb1 hydrolysis potentials of the intestinal bacteria. The results also indicated that Rb1 cannot be highly absorbed in its native forms to exert health benefits after oral administration as a natural prodrug. Rb1 can be hydrolyzed to its active form, 20-O-β-d-glucopyranosyl-20(S)-protopanaxadiol, by Rb1-hydrolyzing intestinal bacteria via cleavage of the glycosyl moieties, allowing Rb1 to reach the plasma and thereby exert its strong antimetastatic activity (100). These studies reveal that parent saponins are deglycosylated into their relevant metabolites to exert biological activities, which are mediated mainly by digestive enzyme secretion by the gut microbiota (55).
FIG 3.
Representative example with a promising application of pathway 1: metabolism of a parent compound (glycyrrhizin, for example) into value-added compounds by regulation of the gut microbiota.
Phenolic compounds.
Phenolic compounds, classified as flavonoids and nonflavonoids, are commonly detected in food plants. These compounds exhibit potent antioxidative effects and affect human health. However, the bioavailability of these compounds varies due to their vast structural diversity. A portion of phenolic compounds is transformed by host-derived enzymes into bioactive metabolites, while the others are not absorbed in the small intestine and may be fermented and transformed by the gut microbiota (101).
Flavones are the major flavonoids, and apigenin is a flavone aglycone. Apigenin-7-glucoside (A7G), which is the parent compound of apigenin, exhibits antimutagenic, antiproliferative, and antiallergic effects (102). By applying A7G to germfree and human-microbiota-associated rats, Hanske et al. (102) showed that relatively few A7G metabolites were present in the urine and feces of germfree rats; the main A7G metabolites in germfree and human-microbiota-associated rats were apigenin and 3-(4-hydroxyphenyl)propionic acid, respectively, and the compounds detected in the blood samples of germfree rats were apigenin conjugates, while the compound detected in blood samples of human-microbiota-associated rats was phloretin. These results indicate that the human intestinal microbiota influenced A7G metabolism and impacted the bioavailability of flavones. Anthocyanins, another type of flavonoid, have been mistaken to be considerably less bioavailable than other flavonoid subclasses in previous studies. Recent studies have suggested that anthocyanins are extremely potent and that the bioavailability of these compounds has been underestimated (103). Most of the ingested anthocyanins possibly arrive at the colon and are metabolized by the colonic microbiota (104). By feeding ileostomy patients raspberries, blueberries, and grapes, approximately 40% of the ingested anthocyanins were found to remain in the ileal effluent (105–107). In healthy humans with intact colons, anthocyanins can enter the colon and be deglycosylated. The dissociation of the C ring leads to the decomposition of aglycones and the conversion of these molecules into several phenolic constituents with additional effects (108).
As another example of a flavonoid compound, curcumin, which exhibits antioxidant, anti-inflammatory, antiviral, antibacterial, and beneficial effects in the treatment of some diseases, including cancers, cardiovascular diseases, diabetes, liver diseases, and neurodegenerative diseases, has been receiving considerable attention (109–111). Due to the β-diketones present in the structure of curcumin, this compound exhibits high hydrophobicity and poor solubility and “bioavailability.” Therefore, a high daily intake of curcumin is necessary to observe strong health-promoting effects. Unfortunately, a high intake of curcumin may have harmful effects and reduce effectiveness, which has limited the utilization of curcumin for illness prevention (112). The metabolites of curcumin generated by gut bacteria, rather than the original forms of curcumin, exhibit the biological effects. Curcumin is reported to be converted to demethylcurcumin and bisdemethylcurcumin via methyl aryl ether cleavage caused by the human intestinal bacterium Blautia sp. strain MRG-PMF1 (7). The evidence indicates that unabsorbed curcumin can modulate the colonic microbiota indirectly, with beneficial effects on multifarious diseases by producing additional bioavailable and bioactive molecules (such as di-O-demethylcurcumin and dimethoxycurcumin) (113).
As a familiar example of nonflavonoid compounds, chlorogenic acids, which are richly contained in coffee beans, can prevent the oxidation of human low-density lipoprotein (LDL), which plays a pivotal role in the formation of atherosclerotic plaques (101). Stalmach et al. indicated that chlorogenic acid absorption occurred in both the small and large intestines but primarily in the large intestine (9). Most chlorogenic acids arrived in the colon intact, and several bacteria produce a number of esterases for the hydrolysis of phenol-quinic acid linkages (114). Released chlorogenic acids are easily transformed by bacteria to their dihydro forms, such as dihydroferulic acid and dihydrocaffeic acid, and are then absorbed by the colonic epithelium. Next, dihydroferulic acid, dihydroferulic acid-4′-O-sulfate, and dihydrocaffeic acid-3′-O-sulfate can enter the circulation at high concentrations (115). Moreover, some other phenolic compounds with low bioavailability, such as rosmarinic acid, eriodictyol, and some quercetin derivatives, are fermented into absorbable and bioactive phenolic acids by the colon microbiota, e.g., hydroxyphenylpropionic acids, phenylpropionic acids, and 3,4-dihydroxyphenylacetic acid (54, 116). These bioactive microbial metabolites may be absorbed and transported by the circulatory system to organs and tissues or exert their effects in the intestinal lumen (54, 102, 116, 117).
All of the above-mentioned examples indicate that improvement of the gut microbiota is potentially a good strategy for the evaluation of the bioavailability of parent substances. However, the metabolites of a large number of functional foods and drugs with low bioavailability remain unknown. Furthermore, the characteristics of individual gut microbes should be considered to identify methods to increase the bioavailability of foods and drugs.
Specifically, oral formulations of chondroitin sulfate (CS) (a high-molecular-weight glycosaminoglycan) have been used as drugs for a long time to treat osteoarthritis (118). Regrettably, 1,200 mg/day has been required to observe the expected curative properties in clinical trials (119). The absorption of CS in the small intestine is low, with the bioavailability of CS estimated to be only 0 to 13% after oral administration (118–120). CS is nondegradable in the stomach and small intestine and is mostly removed or degraded by the colonic microbiota after oral administration (121). By analyzing the degradation of CS by the intestinal microbiota in the distal gastrointestinal (GI) tracts of six healthy subjects, Shang et al. found that each individual’s CS-degrading bacteria (Bacteroides thetaiotaomicron J1, Bacteroides thetaiotaomicron 82, Bacteroides ovatus E3, and Clostridium hathewayi R4) had different degradation effects, but 2-acetamido-2-deoxy-3-O-(β-d-gluco-4-Δ enepyranosyluronic acid)-4-O-sulpho-d-galactose (Δ-UA-GalNAc4S) was the product in all cases (122). This study indicated that although different individuals carried bacteria with the same function, the species might determine the degradation rates of the drugs, leading to differences in bioavailability. Therefore, we believe that the bioavailability of a food or drug is determined not simply by the substance itself but also by an individual’s functional gut bacteria.
Pathway 2: Increase of Beneficial Metabolites from Parent Compounds by Specific Gut Bacteria Enriched by Nonparent Compounds
Some medicinal components, such as alkaloids (berberine, etc.) and metformin, cannot be transformed by the gut microbiota into SCFAs, but these compounds can trigger an increase in the production of SCFAs from dietary carbohydrates by the gut microbiota. Berberine, an extract from traditional Chinese herbal drugs such as Coptis chinensis, previously regarded as an antibiotic drug for diarrhea in China, was recently found to improve glucose tolerance in the treatment of T2DM. However, the clinical efficacy of berberine could not be easily explained because of the low oral bioavailability of this compound. Compared to berberine, metformin has high oral bioavailability, and the mechanisms of action of metformin in the treatment of metabolic diseases are well understood, i.e., the inhibition of hepatic gluconeogenesis via the activation of AMP-activated protein kinase (AMPK). Recent studies have indicated that the intestinal microbiota has an important effect on the efficacy of metformin. Metformin and berberine exhibit contrasting effects on HFD-induced alterations of the composition of the intestinal microbiota. Putative SCFA-producing bacteria, including Allobaculum, Bacteroides, Blautia, Butyricicoccus, and Phascolarctobacterium species, were enriched by berberine and metformin (123). Interestingly, some types of SCFAs could even trigger the production of other types of SCFAs or organic acids by the gut microbiota. The addition of monovalerin (MV) and trivalerin (TV) to an HFD improved not only valeric acid levels but also acetic acid levels in the brain. Although the increment of valeric acid was considered to have been released from the delivered esters, increased amounts of acetic acid, which did not originate from MV or TV, were also observed in the serum and liver in both the MV and TV groups. However, MV and TV tended to decrease the concentrations of acetic acid in the cecum, which indicated that cecal acetate could be transferred to the brain. Furthermore, acetic acid levels in the brain were notably negatively correlated with the abundances of TM7, the S24-7 family, and rc4-4 and positively correlated with the abundances of Anaeroplasma and Tenericutes in the cecal microbiota. These results implied that the promotion of acetate levels in the brain might be traced back to the microbial alteration in the cecum induced by MV and TV (124).
In summary, several studies have shown that the extracts of some drugs and functional foods exhibit poor bioavailability but strong effects on host health. Nonparent compounds can enhance biological effects by regulating the gut microbiota to metabolize parent compounds rather than having a direct impact on the host. Furthermore, the functions of some bioactive molecules have sometimes originated from the metabolites of the parent compounds produced by beneficial bacteria, such as polysaccharides and oligosaccharides, rather than from the parent compounds themselves. However, bioactive effects can frequently be detected only after the intake of beneficial foods or drugs. To date, we have seldom been able to determine whether beneficial molecules exert their effects directly or indirectly via metabolism by the gut microbiota. Therefore, it is not easy to distinguish between pathways 1 and 2. In our opinion, it is necessary to identify therapeutic molecules via metabolomics studies and correlation analysis of each separate metabolite with the host’s phenotype and try to increase the distribution of these molecules in the bloodstream.
Pathway 3: Decrease of Detrimental Metabolites from Parent Compounds via Modulation of the Gut Microbiota by Nonparent Compounds
In addition to the upregulation of beneficial molecules by modulating the gut microbiota, downregulation of the absorption of detrimental molecules by the microbiota could also be beneficial for human health. Detrimental microbes also interact with the host via a number of predictable pathways.
Some foods, such as a red-meat-rich diet, can induce the generation of harmful molecules by the gut microbiota, such as TMA/TMAO and bile acid/cholesterol. Increasing evidence indicates that phenolic phytochemicals with low bioavailability may reduce the levels of harmful compounds, primarily via remodeling of the gut microbiota. TMA, derived from the degradation of choline and l-carnitine by the gut microbiota, is absorbed into the bloodstream and then rapidly oxidized to TMAO by flavin-containing monooxygenase 3 (FMO3), which is a hepatic enzyme (52, 125). Resveratrol (a natural phenolic compound) reduces the synthesis of TMA and TMAO by reshaping the gut microbiota. A genus-level analysis indicated that resveratrol increased the relative abundances of Bacteroides, Lactobacillus, Bifidobacterium, and Akkermansia in mice. Simultaneously, resveratrol administration caused declines in the relative abundances of Prevotella and Ruminococcaceae and in the synthesis of TMA and TMAO (126). In addition to natural drugs, dietary fiber also reduces TMAO levels by altering the gut microbiome. A study demonstrated that dietary fiber feeding reshaped the intestinal microbial ecology; enhanced the growth of Akkermansia and Bifidobacterium, etc.; and restrained the growth of harmful species to reduce TMA and TMAO metabolism via remodeling of the gut microbiota structure in mice (127). Furthermore, some bioactive molecules from foods, such as allicin, also influence TMA and TMAO production (128).
Some intestinal bacteria metabolize primary bile acids and produce secondary bile acids, which can lead to dysbiosis of glycometabolism and lipid metabolism after excessive accumulation in human serum (129). K. Han et al. discovered that the coadministration of a Scutellaria baicalensis extract and metformin has a synergetic cholesterol-lowering effect in rats by the excretion of bile acids through feces (130). Excess cholesterol is discharged into the intestinal lumen by primary bile acid secretion. The abundances of some beneficial bacteria in the intestinal tract were increased due to this combination treatment, including Lactobacillus and Bacteroides, which may promote the deconjugation of bile acids that eventually failed to be reabsorbed in the blood. In contrast, Clostridium and Enterobacter showed the opposite effect (130). Rhizoma coptidis alkaloids, which contain berberine, coptisine, palmatine, and epiberberine, clearly enhanced the abundances of Sporobacter termitidis, Alcaligenes faecalis, and Akkermansia muciniphila in the intestinal tracts of fed mice. However, the growth of Escherichia coli, Desulfovibrio sp. strain C21_c20, and Parabacteroides distasonis was inhibited, which promoted the deconjugation of bile acids to reduce cholesterol levels in the blood (131). Therefore, R. coptidis alkaloids and metformin can not only trigger an increase in the production of SCFAs by enriching a group of SCFA-producing gut bacteria but also reduce the absorption of harmful substances by the body via regulation of the gut microbiota. Furthermore, a lignin-rich fraction of brewer’s spent grain (BSG), as a special dietary fiber containing β-glucan and arabinoxylan, could also promote the deconjugation of bile acids (132).
LPSs, which are membrane components of Gram-negative bacteria, are another class of detrimental substances that are not intestinal metabolites. In 2018, Zhao et al. revealed that dietary fiber intake inhibited the growth of potentially detrimental bacteria and reduced LPS levels but enriched SCFA-producing bacteria in T2DM patients (18). Lopez et al. indicated that genistein, as a dietary bioactive compound in soy, can regulate the intestinal microbiota in HFD-fed mice by increasing the abundances of the genera Prevotella and Akkermansia. These increases resulted in decreases in circulating levels of LPS and reduced metabolic endotoxemia (133). In addition, many other harmful metabolites in the bloodstream, such as microbiome-generated indoxyl sulfate and p-cresol sulfate, can be reduced by remodeling the gut microbiota in patients with chronic renal disease (134, 135).
In summary, functional components can reduce the production of harmful intestinal metabolites by remodeling the intestinal microbiota. To fully understand bioavailability, it is essential to take into account the declining levels of harmful bioavailable molecules. Therefore, a reduction in the bioavailability of detrimental molecules can also alleviate metabolic diseases. In many cases, some functional foods or drugs can not only increase the abundance of beneficial bacteria (frequently SCFA producers) but also decrease the abundance of detrimental bacteria to improve various diseases.
Pathway 4: Inhibition of Specific Gut Bacteria Transforming Parent Compounds into Inactive Forms by Nonparent Compounds
In pathway 1, we describe a group of intestinal bacteria that can transform the parent inactive components into highly beneficial bioactive and absorbable compounds. Conversely, the degradation of some functional components in drugs or foods by specific intestinal bacteria can decrease the bioavailability of these compounds. In contrast to the strategy for the enrichment of beneficial bacteria in pathway 1, effective methods to inhibit intestinal bacteria that degrade bioactive parent drugs might increase the bioavailability of these drugs.
After oral administration, drugs experience first-pass metabolism in the gut and liver, which mostly affects the outcomes and adverse effects of drugs. Many drugs enter the intestine and are metabolized by the gut microbiota, leading to reduced efficacy. For example, Zimmermann et al. found that among eight bacterial species representing five dominant phyla in the mammalian gut microbiota, Bacteroides thetaiotaomicron and B. ovatus show the highest metabolic activity to convert BRV to BVU (1), while the latter can interact with some other drugs and trigger fatal effects. Sorivudine (SRV), which is structurally similar to BRV, can be slowly converted to BVU by B. thetaiotaomicron. In addition, B. thetaiotaomicron can also participate in a diltiazem deacetylation reaction generating desacetyldiltiazem (2). Besides, sulindac is reduced by the intestinal microbiota to sulindac sulfide (136). Digoxin can be converted to dihydrodigoxin and dihydrodigoxigenin in vitro. These derivatives result in decreased cardiac activity (137). Some studies have illustrated that some bonds can be transformed by enzymes carried by specific gut bacteria. For example, the azo bond of sulfasalazine is reduced by colonic bacterial azoreductases to form mesalazine and sulfapyridine (138, 139). The absorption of sulfapyridine in the colon can result in some adverse effects, such as nausea, skin rash, headache, dizziness, and decreased appetite (140). Nitrazepam experiences a nitroreduction catalyzed by gut bacterial enzymes, but the products of this reaction have teratogenic effects (141). The intestinal microbiota is mainly involved in a modification in which zonisamide is primarily converted to 2-sulfamoylacetyl-phenol by the reduction of the benzisoxazole ring (142).
Another example tactfully demonstrated the effects of the gut microbiota on the degradation of active parent drugs into inactive components. Amlodipine, one of the most frequently prescribed drugs for the treatment of hypertension, is absorbed in the GI tract, with a bioavailability of approximately 60% after oral administration (143).
Some gut microbiota-targeted methods have been developed to improve bioavailability. It has been found that coadministration of amlodipine with antibiotics resulted in increased amlodipine absorption in the GI tract. Researchers found that amlodipine could be metabolized by intestinal microbial enzymes and yield the major pyridine metabolite, which suggested that the intestinal microbiota is associated with the metabolism of amlodipine. Furthermore, coadministration of amlodipine and antibiotics increased the human plasma concentration of amlodipine to almost twice that of the amlodipine monotherapy group (144). The use of antibiotics could change the intestinal microbiota, leading to metabolic changes in the coadministered antihypertension drugs. This finding strongly indicated that the gut microbiota might affect the pharmacokinetics of antihypertensive drugs. However, antibiotics have an associated risk of dysregulation of the gut microbiota (145, 146), which limits the use of these compounds to increase the bioavailability of amlodipine. Compared with amlodipine monotherapy, coadministration with Lactobacillus plantarum IS-10506 in rabbits significantly enhanced the amlodipine concentration. The authors speculated that L. plantarum IS-10506 could enhance red blood cells and hematocrit, leading to a lower sedimentation rate and increased levels of plasma proteins, which bind amlodipine (147). In addition, a live bacterial suspension of the probiotic Escherichia coli Nissle 1917 enhanced the bioavailability of amiodarone, which is an antiarrhythmic drug, in rats (148), and mixed cultures of Lactobacillus acidophilus, Bifidobacterium lactis, and Lactobacillus rhamnosus increased the bioavailability of gliclazide, an antidiabetic drug, in diabetic rats (149). Beneficial bacteria can limit the expansion of competing opportunistic pathogens (150, 151). Thus, in our opinion, this improvement in bioavailability might be associated with a decreased abundance of drug-degrading bacteria due to competition pressure from beneficial bacteria.
Prebiotics are also able to modulate the gut microbiota. The use of prebiotics has also been reported to enhance the absorption of some parent compounds. For example, the coadministration of genistin and FOS improved the absorption of genistin, leading to this compound reaching its target tissues, such as bone, with high effectiveness (152). However, the underlying mechanism has not been explained.
In addition, gut microbes can interact with receptors in the liver and other organs via the corresponding metabolites. Indoles, which are microbial tryptophan metabolites, have been shown to induce some cytochrome P450s, which are responsible for the metabolism of most therapeutic drugs and play vital roles in bioavailability and drug-drug interactions via an aryl hydrocarbon receptor-mediated mechanism in the liver. Furthermore, antibiotics decreased the hepatic expression and enzymatic activity of cytochrome P450 3a (153). Regardless of the direct (in the intestine) or indirect (in the liver) influence on first-pass metabolism, the gut microbiota might play significant roles in these processes.
Taken together, these results show that antibiotics, beneficial bacteria, and prebiotics are able to increase the absorption of some oral drugs by inhibiting the abundance of drug-degrading bacteria.
REDEFINITION AND IMPROVEMENT OF BIOAVAILABILITY
To address the perplexingly low bioavailability of pharmaceuticals and functional foods, various strategies have been carried out to promote bioavailability. However, high bioavailability frequently does not lead to health improvements. On the other hand, despite poor bioavailability, some drugs or functional foods continue to exhibit strong therapeutic effects. Thus, the mechanisms by which these substances function in vivo are puzzling to researchers. Recently, the correlation of gut microbiota dysbiosis and the occurrence and progression of a series of diseases was gradually verified. Poor bioavailability of drugs or foods might provide an opportunity for cross talk among these nonabsorbable components and bacteria in both the small and large intestines. Thus, determining whether such cross talk has a significant effect on the biotransformation of these substances with low bioavailability is of great interest. This review demonstrates that four pathways associated with these medicines or foods with low bioavailability determine the effects of these substances aided by the gut microbiota. First, the functional components derived from these drugs or foods by the gut microbiota exhibit key effects on the improvement of health. In other words, the high availability of these functional metabolites successfully overcomes the low availability of the parent substances. Second, many drugs or functional foods can modulate the structure and function of the gut microbiota and facilitate the production of beneficial metabolites from daily nutrient substances via the gut microbiota. Thus, highly bioavailable beneficial components (SCFAs, etc.) metabolized from healthy foods consumed daily (dietary fiber, etc.), which are different from the investigated drugs or functional foods, by the improved intestinal microbiota play key roles in health promotion. Third, the improved gut microbiota modulated by drugs or functional foods can downregulate the production of some harmful molecules metabolized from unhealthy foods (dietary choline, etc.) by the gut microbiota. We further propose that the understanding of bioavailability can be expanded to include the downregulation of the availability of detrimental metabolites. Thus, in addition to the enhancement of the bioavailability of beneficial components, inhibition of the bioavailability of detrimental components might be an effective method for the promotion of human health. Fourth, the “binding partner” approach for food or drugs and targeted inhibitors can be considered to improve bioavailability and therapeutic effects. Targeted inhibitory substances in the intestine specifically inhibit bacterial degradation of functional components, allowing functional components to “escape” degradation by the gut microbiota and enter the circulatory system with higher absorption. This approach will facilitate developmental breakthroughs to improve the bioavailability of drugs and foods that could be degraded by the intestinal microbiota. Of course, we also need to consider the possible interactions between the two substances and the side effects on the organisms.
Regardless of the pathways mentioned above, gut microbes are effective modulators of the bioavailability of these pharmaceuticals or foods. For example, dietary fiber can be directly metabolized into biologically active substances, i.e., substances that have biological effects on the enhancement of the levels of SCFAs, which might be derived from dietary carbohydrates via the enrichment of SCFA producers. On the other hand, based on the functions of the gut microbiota, dietary fiber can also reduce TMA, TMAO, and LPS metabolism by remodeling the gut microbiota structure, which suggests that pathways 1 and 3 play important roles in the bioavailability-promoting effects of dietary fiber (18, 127). Metformin, the exact pharmacological mechanism of which is unknown, can also increase the abundance of SCFA producers. Additionally, metformin can reduce the levels of secondary bile acids, which implies that metformin can have beneficial effects via both pathways 2 and 3 (123, 130). Therefore, when discussing the bioavailability of components, we should pay attention to various bioavailability-promoting pathways based on the multidimensional effects of the gut microbiota. Importantly, even if we find that some water extracts or crude extracts of functional foods or drugs have biological effects, there is a strong possibility that these extracts themselves do not exhibit the effects. Instead, it is likely that small bioactive substances produced by the gut microbiota play roles in these biological processes. Simultaneously, these phenomena also remind us that we cannot be satisfied with the biological effects of crude extracts, and we should study the small-molecule bioactive substances that enter the bloodstream to improve the bioavailability and, ultimately, the therapeutic effects of these molecules. These substances can be extracted and used in the pharmaceutical industry.
In addition, almost all traditional Chinese medicines (TCMs) have poor bioavailability, so the medical effects of these medicines cannot be easily explained based on Western medicine. However, TCMs have helped Asian doctors cure patients for thousands of years. Some unknown mechanisms might play important roles in the therapeutic processes. The effects of the human commensal microbiota may explain the mechanisms underlying these processes and help us understand the effects of TCMs.
Notably, in pathway 4, some modulators of the gut microbiota may be used for the downregulation of drug-degrading bacteria to both elevate the bioavailability and reduce the side effects of parent drugs. Furthermore, some microbial metabolites might induce cytochrome P450s and decrease first-pass metabolism in the liver.
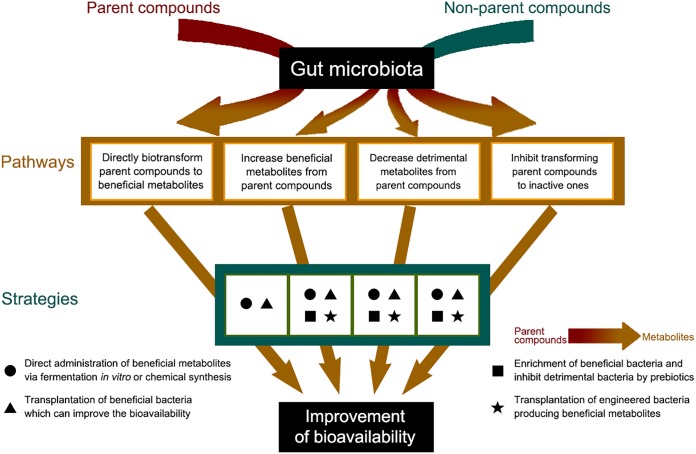
In addition to the in vivo (1), in vitro (2), and human (8) models mentioned above, some other models, such as a simulator of the human intestinal tract (154, 155), continuous-culture systems (156), a GI-targeted release model (extended or immediate release) (157), and an in vitro culture combination of key functional strains, might be potential tools to illustrate the roles of the gut microbiota in bioavailability. In this review, bioavailability can be improved by either increasing the abundance of beneficial gut bacteria or decreasing the concentration of detrimental bacteria. Some potential methods, in our opinion, may be launched. First, beneficial metabolites from parent or nonparent compounds by the gut bacteria in pathways 1 and 2 can be produced via in vitro fermentation or by chemical synthesis for direct administration. Second, targeted beneficial bacteria for directly improving bioavailability need to be identified according to studies on the cross talk of these bacteria and the parent compounds in pathways 1 and 2. The isolation, culture, and transplantation of these beneficial bacteria might be a promising method. On the other hand, although targeted inhibition of the detrimental bacteria in pathways 3 and 4 is difficult, the utilization of some competitive beneficial bacteria can be expected. Third, prebiotics can also be used to enrich beneficial bacteria in pathways 1 and 2 and thereafter inhibit detrimental bacteria in pathways 3 and 4. Last, there are broad prospects in using engineered bacteria harboring specific genes to produce beneficial metabolites to be used in pathways 1 and 2.
In summary, based on the interactions between drugs or functional foods and the gut microbiota, a redefinition of bioavailability will provide novel insights into the roles of many medicines or functional foods in human health. The modulation of the structure of the gut microbiota will be a novel strategy for the promotion of bioavailability, the alleviation of side effects, and the discovery and design of novel pharmaceuticals (Fig. 4).
FIG 4.
Strategies for improving the bioavailability of food and medicine via modulation of the gut microbiota toward each pathway.
ACKNOWLEDGMENTS
We gratefully acknowledge the financial support provided by the National Key Research and Development Program of China (2017YFC1601704); projects 31522044, 31671909, 31772034, and 81870544 of the National Natural Science Foundation of China; the Natural Science Foundation of Jiangsu Province (BK20181132); the Research Project of Maternal and Child Health of Jiangsu Province (F201741); the Program of the Jiangsu Key Laboratory of Advanced Food Manufacturing Equipment and Technology (FMZ201904); and the National First-Class Discipline Program of Food Science and Technology (JUFSTR20180205).
We declare no competing financial interests.
Biographies

Feng Zhang studies the roles of gut microbiota in metabolic diseases, including obesity, type 2 diabetes, prediabetes, gestational diabetes mellitus, and polycystic ovarian syndrome. As the coauthor of research published in Science entitled “Gut Bacteria Selectively Promoted by Dietary Fibers Alleviate Type 2 Diabetes,” he and his collaborators focused on the effects of dietary fiber on the enrichment of SCFA-producing bacteria, which could enhance GLP-1 levels and thereafter alleviate type 2 diabetes. Recently, he has been interested in the roles of the gut microbiota in the alleviation of metabolic diseases by functional foods.

Fang He studies the roles of functional foods and nutrients in human health and wellness, especially the regulation of the gut microbiota by these compounds. She has been active in the interaction between the gut microbiota and the host for several years. Currently, she is mainly studying the effects and mechanisms of prebiotics such as xylo-oligosaccharides in the alleviation of type 2 diabetes by modulating the gut microbiota. She has screened several gut bacteria that have the potential to treat obesity and diabetes.

Li Li studies learning and memory, individual variations in learning ability, and their underlying mechanisms. She has a particular interest in the interactions among the gut microbiota, the brain, and cognitive performance. She obtained her Ph.D. in Biological Sciences in 2017 at the Queen Mary University in London, United Kingdom, with a Ph.D. dissertation entitled The Neural Mechanisms Underlying Bumblebee Visual Learning and Memory. After her Ph.D., she has worked as a postdoctoral researcher in the School of Food Science and Technology at Jiangnan University, China, focusing on the brain-gut-microbiome axis. Two research grants sponsor her current work, and she has accumulated abundant knowledge about the effects of the gut microbiota on brain function and the potential pathways.

Lichun Guo studies the roles of the gut microbiota and enzymatic modifications to overcome the poor bioavailability of saponins such as glycyrrhizin (GL). She focuses on research on the enzymes derived from microbes that display great potential to transform these compounds into beneficial metabolites, especially glycyrrhetic acid 3-O-mono-β-d-glucuronide (GAMG), which has higher bioactivity than the parent compound GL. Currently, she is studying the effects and mechanisms of hypoglycemic drugs in the treatment of obesity and diabetes, especially the regulation of the gut microbiota by these compounds. She has found a beneficial metabolite modified by the gut microbiota that has potential in the prevention and treatment of diabetes and related symptoms.

Bin Zhang studies the gut microbiota and their metabolites. She has been pursuing a Ph.D. in Food Science and Technology at Jiangnan University since 2018. Her research focuses on the regulatory mechanism of hydrogen sulfide (a kind of metabolite produced by the gut microbiota) with glycometabolism in type 2 diabetes.

Shuhuai Yu studies the structure, function, catalytic mechanism, and evolutionary relationship of enzymes involved in the metabolic process of carbohydrates. In this field, he revealed the mechanism and evolutionary relationship of difructose anhydride III hydrolase and inulin fructotransferase using crystallography, the result of which was published in ACS Catalysis (2018) entitled “Structural and Functional Basis of Difructose Anhydride III Hydrolase, Which Sequentially Converts Inulin Using the Same Catalytic Residue.” At Harvard University, he studied molecular evolution with experimental and computational methodologies in the Department of Chemistry and Chemical Biology. After his Ph.D., he has worked as an associate professor in the School of Food Science and Technology at Jiangnan University, China, continuing his investigation of enzymes, including their exploitation and modification. Recently, he has been interested in the ecology of bacteriophage interactions with bacterial populations, finally applying this to phage therapy.

Wei Zhao studies the relationship between nutrition and health and the efficacy and mechanisms of bioactive compounds in the body. He has been active in these studies for almost 15 years and has published more than 150 peer-reviewed papers. Dr. Zhao is currently a full professor in Food Science at Jiangnan University. He has commercially developed several prebiotics and functional food ingredients in industry and has successfully applied them to alleviate metabolic diseases. Recently, he has become interested in the roles of the gut microbiota in metabolic diseases, including obesity and type 2 diabetes.
REFERENCES
- 1.Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL. 2019. Separating host and microbiome contributions to drug pharmacokinetics and toxicity. Science 363:eaat9931. doi: 10.1126/science.aat9931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL. 2019. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 570:462–467. doi: 10.1038/s41586-019-1291-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heaney R. 2001. Factors influencing the measurement of bioavailability, taking calcium as a model. J Nutr 131:1344S–1348S. doi: 10.1093/jn/131.4.1344S. [DOI] [PubMed] [Google Scholar]
- 4.Srinivasan V. 2001. Bioavailability of nutrients: a practical approach to in vitro demonstration of the availability of nutrients in multivitamin-mineral combination products. J Nutr 131:1349S–1350S. doi: 10.1093/jn/131.4.1349S. [DOI] [PubMed] [Google Scholar]
- 5.Wu TR, Lin CS, Chang CJ, Lin TL, Martel J, Ko YF, Ojcius DM, Lu CC, Young JD, Lai HC. 2019. Gut commensal Parabacteroides goldsteinii plays a predominant role in the anti-obesity effects of polysaccharides isolated from Hirsutella sinensis. Gut 68:248–262. doi: 10.1136/gutjnl-2017-315458. [DOI] [PubMed] [Google Scholar]
- 6.Hollman PC, de Vries JH, van Leeuwen SD, Mengelers MJ, Katan MB. 1995. Absorption of dietary quercetin glycosides and quercetin in healthy ileostomy volunteers. Am J Clin Nutr 62:1276–1282. doi: 10.1093/ajcn/62.6.1276. [DOI] [PubMed] [Google Scholar]
- 7.Burapan S, Kim M, Han J. 2017. Curcuminoid demethylation as an alternative metabolism by human intestinal microbiota. J Agric Food Chem 65:3305–3310. doi: 10.1021/acs.jafc.7b00943. [DOI] [PubMed] [Google Scholar]
- 8.Veeriah S, Balavenkatraman KK, Bohmer FD, Kahle K, Glei M, Richling E, Scheppach W, Pool-Zobel BL. 2008. Intervention with cloudy apple juice results in altered biological activities of ileostomy samples collected from individual volunteers. Eur J Nutr 47:226–234. doi: 10.1007/s00394-008-0726-7. [DOI] [PubMed] [Google Scholar]
- 9.Stalmach A, Steiling H, Williamson G, Crozier A. 2010. Bioavailability of chlorogenic acids following acute ingestion of coffee by humans with an ileostomy. Arch Biochem Biophys 501:98–105. doi: 10.1016/j.abb.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 10.Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. 2004. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A 101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanton LV, Charbonneau MR, Salih T, Barratt MJ, Venkatesh S, Ilkaveya O, Subramanian S, Manary MJ, Trehan I, Jorgensen JM, Fan Y, Henrissat B, Leyn SA, Rodionov DA, Osterman AL, Maleta KM, Newgard CB, Ashorn P, Dewey KG, Gordon JI. 2016. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 351:aad3311. doi: 10.1126/science.aad3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. 2005. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A 102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 14.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carvalho BM, Guadagnini D, Tsukumo DML, Schenka AA, Latuf-Filho P, Vassallo J, Dias JC, Kubota LT, Carvalheira JBC, Saad MJA. 2012. Modulation of gut microbiota by antibiotics improves insulin signalling in high-fat fed mice. Diabetologia 55:2823–2834. doi: 10.1007/s00125-012-2648-4. [DOI] [PubMed] [Google Scholar]
- 16.Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, Prifti E, Vieira-Silva S, Gudmundsdottir V, Pedersen HK, Arumugam M, Kristiansen K, Voigt AY, Vestergaard H, Hercog R, Costea PI, Kultima JR, Li J, Jørgensen T, Levenez F, Dore J, MetaHIT Consortium, Nielsen HB, Brunak S, Raes J, Hansen T, Wang J, Ehrlich SD, Bork P, Pedersen O. 2015. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528:262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto J-M, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, et al. 2012. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 18.Zhao LP, Zhang F, Ding XY, Wu GJ, Lam YY, Wang XJ, Fu HQ, Xue XH, Lu CH, Ma JL, Yu LH, Xu CM, Ren ZY, Xu Y, Xu SM, Shen HL, Zhu XL, Shi Y, Shen QY, Dong WP, Liu R, Ling YX, Zeng Y, Wang XP, Zhang QP, Wang J, Wang LH, Wu YQ, Zeng BH, Wei H, Zhang MH, Peng YD, Zhang CH. 2018. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 359:1151–1156. doi: 10.1126/science.aao5774. [DOI] [PubMed] [Google Scholar]
- 19.Gao X, Cao Q, Cheng Y, Zhao D, Wang Z, Yang H, Wu Q, You L, Wang Y, Lin Y, Li X, Wang Y, Bian JS, Sun D, Kong L, Birnbaumer L, Yang Y. 2018. Chronic stress promotes colitis by disturbing the gut microbiota and triggering immune system response. Proc Natl Acad Sci U S A 115:E2960–E2969. doi: 10.1073/pnas.1720696115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsiao A, Ahmed AMS, Subramanian S, Griffin NW, Drewry LL, Petri WA Jr, Haque R, Ahmed T, Gordon JI. 2014. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 515:423–426. doi: 10.1038/nature13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vich Vila A, Imhann F, Collij V, Jankipersadsing SA, Gurry T, Mujagic Z, Kurilshikov A, Bonder MJ, Jiang X, Tigchelaar EF, Dekens J, Peters V, Voskuil MD, Visschedijk MC, van Dullemen HM, Keszthelyi D, Swertz MA, Franke L, Alberts R, Festen EAM, Dijkstra G, Masclee AAM, Hofker MH, Xavier RJ, Alm EJ, Fu J, Wijmenga C, Jonkers DMAE, Zhernakova A, Weersma RK. 2018. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci Transl Med 10:eaap8914. doi: 10.1126/scitranslmed.aap8914. [DOI] [PubMed] [Google Scholar]
- 22.Yang XD, Wang LK, Wu HY, Jiao L. 2018. Effects of prebiotic galacto-oligosaccharide on postoperative cognitive dysfunction and neuroinflammation through targeting of the gut-brain axis. BMC Anesthesiol 18:177. doi: 10.1186/s12871-018-0642-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mailing LJ, Allen JM, Pence BD, Rytych J, Sun Y, Bhattacharya TK, Park P, Cross T-WL, McCusker RH, Swanson KS, Fahey GC, Rhodes JS, Kelley KW, Johnson RW, Woods JA. 2019. Behavioral response to fiber feeding is cohort-dependent and associated with gut microbiota composition in mice. Behav Brain Res 359:731–736. doi: 10.1016/j.bbr.2018.09.012. [DOI] [PubMed] [Google Scholar]
- 24.Cryan JF, Dinan TG. 2012. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci 13:701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 25.Coello K, Hansen TH, Sorensen N, Munkholm K, Kessing LV, Pedersen O, Vinberg M. 2019. Gut microbiota composition in patients with newly diagnosed bipolar disorder and their unaffected first-degree relatives. Brain Behav Immun 75:112–118. doi: 10.1016/j.bbi.2018.09.026. [DOI] [PubMed] [Google Scholar]
- 26.Perry RJ, Peng L, Barry NA, Cline GW, Zhang DY, Cardone RL, Petersen KF, Kibbey RG, Goodman AL, Shulman GI. 2016. Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature 534:213–217. doi: 10.1038/nature18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Markle JGM, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, von Bergen M, McCoy KD, Macpherson AJ, Danska JS. 2013. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 28.Thaiss CA, Zmora N, Levy M, Elinav E. 2016. The microbiome and innate immunity. Nature 535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 29.Honda K, Littman DR. 2016. The microbiota in adaptive immune homeostasis and disease. Nature 535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 30.Peters BA, Dominianni C, Shapiro JA, Church TR, Wu J, Miller G, Yuen E, Freiman H, Lustbader I, Salik J, Friedlander C, Hayes RB, Ahn J. 2016. The gut microbiota in conventional and serrated precursors of colorectal cancer. Microbiome 4:69. doi: 10.1186/s40168-016-0218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomkovich S, Yang Y, Winglee K, Gauthier J, Muhlbauer M, Sun XL, Mohamadzadeh M, Liu XL, Martin P, Wang GP, Oswald E, Fodor AA, Jobin C. 2017. Locoregional effects of microbiota in a preclinical model of colon carcinogenesis. Cancer Res 77:2620–2632. doi: 10.1158/0008-5472.CAN-16-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R, Hannani D, Enot DP, Pfirschke C, Engblom C, Pittet MJ, Schlitzer A, Ginhoux F, Apetoh L, Chachaty E, Woerther PL, Eberl G, Berard M, Ecobichon C, Clermont D, Bizet C, Gaboriau-Routhiau V, Cerf-Bensussan N, Opolon P, Yessaad N, Vivier E, Ryffel B, Elson CO, Dore J, Kroemer G, Lepage P, Boneca IG, Ghiringhelli F, Zitvogel L. 2013. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Thingholm LB, Skiecevičienė J, Rausch P, Kummen M, Hov JR, Degenhardt F, Heinsen F-A, Rühlemann MC, Szymczak S, Holm K, Esko T, Sun J, Pricop-Jeckstadt M, Al-Dury S, Bohov P, Bethune J, Sommer F, Ellinghaus D, Berge RK, Hübenthal M, Koch M, Schwarz K, Rimbach G, Hübbe P, Pan W-H, Sheibani-Tezerji R, Häsler R, Rosenstiel P, D’Amato M, Cloppenborg-Schmidt K, Künzel S, Laudes M, Marschall H-U, Lieb W, Nöthlings U, Karlsen TH, Baines JF, Franke A. 2016. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet 48:1396–1406. doi: 10.1038/ng.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie HL, Guo RJ, Zhong HZ, Feng Q, Lan Z, Qin BC, Ward KJ, Jackson MA, Xia Y, Chen X, Chen B, Xia HH, Xu CL, Li F, Xu X, Al-Aama JY, Yang HM, Wang J, Kristiansen K, Wang J, Steves CJ, Bell JT, Li JH, Spector TD, Jia HJ. 2016. Shotgun metagenomics of 250 adult twins reveals genetic and environmental impacts on the gut microbiome. Cell Syst 3:572–584. doi: 10.1016/j.cels.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, Shilo S, Lador D, Vila AV, Zmora N, Pevsner-Fischer M, Israeli D, Kosower N, Malka G, Wolf BC, Avnit-Sagi T, Lotan-Pompan M, Weinberger A, Halpern Z, Carmi S, Fu JY, Wijmenga C, Zhernakova A, Elinav E, Segal E. 2018. Environment dominates over host genetics in shaping human gut microbiota. Nature 555:210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 36.Jourova L, Anzenbacher P, Anzenbacherova E. 2016. Human gut microbiota plays a role in the metabolism of drugs. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 160:317–326. doi: 10.5507/bp.2016.039. [DOI] [PubMed] [Google Scholar]
- 37.Sousa T, Paterson R, Moore V, Carlsson A, Abrahamsson B, Basit AW. 2008. The gastrointestinal microbiota as a site for the biotransformation of drugs. Int J Pharm 363:1–25. doi: 10.1016/j.ijpharm.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 38.Saad R, Rizkallah MR, Aziz RK. 2012. Gut pharmacomicrobiomics: the tip of an iceberg of complex interactions between drugs and gut-associated microbes. Gut Pathog 4:16. doi: 10.1186/1757-4749-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson ID, Nicholson JK. 2017. Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl Res 179:204–222. doi: 10.1016/j.trsl.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dorrestein PC, Mazmanian SK, Knight R. 2014. Finding the missing links among metabolites, microbes, and the host. Immunity 40:824–832. doi: 10.1016/j.immuni.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. 2009. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A 106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. 2013. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 43.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zarrinpar A, Chaix A, Xu ZZ, Chang MW, Marotz CA, Saghatelian A, Knight R, Panda S. 2018. Antibiotic-induced microbiome depletion alters metabolic homeostasis by affecting gut signaling and colonic metabolism. Nat Commun 9:2872. doi: 10.1038/s41467-018-05336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang Y-L, Rossetti M, Vlamakis H, Casero D, Sunga G, Harre N, Miller S, Humphries R, Stappenbeck T, Simpson KW, Sartor RB, Wu G, Lewis J, Bushman F, McGovern DPB, Salzman N, Borneman J, Xavier R, Huttenhower C, Braun J. 2019. A screen of Crohn’s disease-associated microbial metabolites identifies ascorbate as a novel metabolic inhibitor of activated human T cells. Mucosal Immunol 12:457–467. doi: 10.1038/s41385-018-0022-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karl JP, Meydani M, Barnett JB, Vanegas SM, Barger K, Fu XY, Goldin B, Kane A, Rasmussen H, Vangay P, Knights D, Jonnalagadda SS, Saltzman E, Roberts SB, Meydani SN, Booth SL. 2017. Fecal concentrations of bacterially derived vitamin K forms are associated with gut microbiota composition but not plasma or fecal cytokine concentrations in healthy adults. Am J Clin Nutr 106:1052–1061. doi: 10.3945/ajcn.117.155424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fabbiano S, Suárez-Zamorano N, Chevalier C, Lazarević V, Kieser S, Rigo D, Leo S, Veyrat-Durebex C, Gaïa N, Maresca M, Merkler D, Gomez de Agüero M, Macpherson A, Schrenzel J, Trajkovski M. 2018. Functional gut microbiota remodeling contributes to the caloric restriction-induced metabolic improvements. Cell Metab 28:907–921. doi: 10.1016/j.cmet.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cani PD, Bibiloni R, Knauf C, Neyrinck AM, Neyrinck AM, Delzenne NM, Burcelin R. 2008. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 49.Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BAH, Forslund K, Hildebrand F, Prifti E, Falony G, Le Chatelier E, Levenez F, Dore J, Mattila I, Plichta DR, Poho P, Hellgren LI, Arumugam M, Sunagawa S, Vieira-Silva S, Jorgensen T, Holm JB, Trost K, MetaHIT Consortium, Kristiansen K, Brix S, Raes J, Wang J, Hansen T, Bork P, Brunak S, Oresic M, Ehrlich SD, Pedersen O. 2016. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 535:376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 50.Gu YY, Wang XK, Li JH, Zhang YF, Zhong HZ, Liu RX, Zhang DY, Feng Q, Xie XY, Hong J, Ren HH, Liu W, Ma J, Su Q, Zhang HM, Yang JL, Wang XL, Zhao XJ, Gu WQ, Bi YF, Peng YD, Xu XQ, Xia HH, Li F, Xu X, Yang HM, Xu GW, Madsen L, Kristiansen K, Ning G, Wang WQ. 2017. Analyses of gut microbiota and plasma bile acids enable stratification of patients for antidiabetic treatment. Nat Commun 8:1785. doi: 10.1038/s41467-017-01682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sayin SI, Wahlstrom A, Felin J, Jantti S, Marschall HU, Bamberg K, Angelin B, Hyotylainen T, Oresic M, Backhed F. 2013. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab 17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 52.Wang ZN, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu XM, Chung YM, Wu YP, Schauer P, Smith JD, Allayee H, Tang WHW, DiDonato JA, Lusis AJ, Hazen SL. 2011. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu WF, Gregory JC, Org E, Buffa JA, Gupta N, Wang ZN, Li L, Fu XM, Wu YP, Mehrabian M, Sartor RB, McIntyre TM, Silverstein RL, Tang WHW, DiDonato JA, Brown JM, Lusis AJ, Hazen SL. 2016. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell 165:111–124. doi: 10.1016/j.cell.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aura A-M. 2008. Microbial metabolism of dietary phenolic compounds in the colon. Phytochem Rev 7:407–429. doi: 10.1007/s11101-008-9095-3. [DOI] [Google Scholar]
- 55.Chen M-Y, Shao L, Zhang W, Wang C-Z, Zhou H-H, Huang W-H, Yuan C-S. 2018. Metabolic analysis of Panax notoginseng saponins with gut microbiota-mediated biotransformation by HPLC-DAD-Q-TOF-MS/MS. J Pharm Biomed Anal 150:199–207. doi: 10.1016/j.jpba.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 56.De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, Bäckhed F, Mithieux G. 2014. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 156:84–96. doi: 10.1016/j.cell.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 57.Koppel N, Maini Rekdal V, Balskus EP. 2017. Chemical transformation of xenobiotics by the human gut microbiota. Science 356:eaag2770. doi: 10.1126/science.aag2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martel J, Ojcius DM, Chang CJ, Lin CS, Lu CC, Ko YF, Tseng SF, Lai HC, Young JD. 2017. Anti-obesogenic and antidiabetic effects of plants and mushrooms. Nat Rev Endocrinol 13:149–160. doi: 10.1038/nrendo.2016.142. [DOI] [PubMed] [Google Scholar]
- 59.Hold GL. 2014. The gut microbiota, dietary extremes and exercise. Gut 63:1838–1839. doi: 10.1136/gutjnl-2014-307305. [DOI] [PubMed] [Google Scholar]
- 60.Wang Y, Fei Y, Liu L, Xiao Y, Pang Y, Kang J, Wang Z. 2018. Polygonatum odoratum polysaccharides modulate gut microbiota and mitigate experimentally induced obesity in rats. Int J Mol Sci 19:3587. doi: 10.3390/ijms19113587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang X, Wang X, Jiang H, Cai C, Li G, Hao J, Yu G. 2018. Marine polysaccharides attenuate metabolic syndrome by fermentation products and altering gut microbiota: an overview. Carbohydr Polym 195:601–612. doi: 10.1016/j.carbpol.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 62.Fan ST, Nie SP, Huang XJ, Wang S, Hu JL, Xie JH, Nie QX, Xie MY. 2018. Protective properties of combined fungal polysaccharides from Cordyceps sinensis and Ganoderma atrum on colon immune dysfunction. Int J Biol Macromol 114:1049–1055. doi: 10.1016/j.ijbiomac.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 63.Liu J, Kandasamy S, Zhang J, Kirby CW, Karakach T, Hafting J, Critchley AT, Evans F, Prithiviraj B. 2015. Prebiotic effects of diet supplemented with the cultivated red seaweed Chondrus crispus or with fructo-oligo-saccharide on host immunity, colonic microbiota and gut microbial metabolites. BMC Complement Altern Med 15:279. doi: 10.1186/s12906-015-0802-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin M, Zhu Y, Shao D, Zhao K, Xu C, Li Q, Yang H, Huang Q, Shi J. 2017. Effects of polysaccharide from mycelia of Ganoderma lucidum on intestinal barrier functions of rats. Int J Biol Macromol 94:1–9. doi: 10.1016/j.ijbiomac.2016.09.099. [DOI] [PubMed] [Google Scholar]
- 65.Wang LL, Hu LJ, Yan S, Jiang T, Fang SG, Wang G, Zhao JX, Zhang H, Chen W. 2017. Effects of different oligosaccharides at various dosages on the composition of gut microbiota and short-chain fatty acids in mice with constipation. Food Funct 8:1966–1978. doi: 10.1039/c7fo00031f. [DOI] [PubMed] [Google Scholar]
- 66.Vincent AD, Wang XY, Parsons SP, Khan WI, Huizinga JD. 2018. Abnormal absorptive colonic motor activity in germ-free mice is rectified by butyrate, an effect possibly mediated by mucosal serotonin. Am J Physiol Gastrointest Liver Physiol 315:G896–G907. doi: 10.1152/ajpgi.00237.2017. [DOI] [PubMed] [Google Scholar]
- 67.Soret R, Chevalier J, De Coppet P, Poupeau G, Derkinderen P, Segain JP, Neunlist M. 2010. Short-chain fatty acids regulate the enteric neurons and control gastrointestinal motility in rats. Gastroenterology 138:1772–1782. doi: 10.1053/j.gastro.2010.01.053. [DOI] [PubMed] [Google Scholar]
- 68.Wang X, Yang B, Yin J, Wei W, Chen JDZ. 2019. Electroacupuncture via chronically implanted electrodes improves gastrointestinal motility by balancing sympathovagal activities in a rat model of constipation. Am J Physiol Gastrointest Liver Physiol 316:G797–G805. doi: 10.1152/ajpgi.00018.2018. [DOI] [PubMed] [Google Scholar]
- 69.Parnell JA, Reimer RA. 2009. Weight loss during oligofructose supplementation is associated with decreased ghrelin and increased peptide YY in overweight and obese adults. Am J Clin Nutr 89:1751–1759. doi: 10.3945/ajcn.2009.27465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miguez B, Gomez B, Parajo JC, Alonso JL. 2018. Potential of fructooligosaccharides and xylooligosaccharides as substrates to counteract the undesirable effects of several antibiotics on elder fecal microbiota: a first in vitro approach. J Agric Food Chem 66:9426–9437. doi: 10.1021/acs.jafc.8b02940. [DOI] [PubMed] [Google Scholar]
- 71.Valcheva R, Koleva P, Martinez I, Walter J, Ganzle MG, Dieleman LA. 2019. Inulin-type fructans improve active ulcerative colitis associated with microbiota changes and increased short-chain fatty acids levels. Gut Microbes 10:334–357. doi: 10.1080/19490976.2018.1526583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang J, Li Q, Henning SM, Zhong J, Hsu M, Lee R, Long J, Chan B, Nagami GT, Heber D, Li Z. 2018. Effects of prebiotic fiber xylooligosaccharide in adenine-induced nephropathy in mice. Mol Nutr Food Res 62:1800014. doi: 10.1002/mnfr.201800014. [DOI] [PubMed] [Google Scholar]
- 73.Salonen A, Lahti L, Salojarvi J, Holtrop G, Korpela K, Duncan SH, Date P, Farquharson F, Johnstone AM, Lobley GE, Louis P, Flint HJ, de Vos WM. 2014. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J 8:2218–2230. doi: 10.1038/ismej.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Macfarlane GT, Gibson GR. 1997. Carbohydrate fermentation, energy transduction and gas metabolism in the human large intestine. Gastrointest Microbiol doi: 10.1007/978-1-4615-4111-0_9. [DOI] [Google Scholar]
- 75.Reichardt N, Duncan SH, Young P, Belenguer A, Leitch CM, Scott KP, Flint HJ, Louis P. 2014. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J 8:1323–1335. doi: 10.1038/ismej.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rodionova IA, Li XQ, Thiel V, Stolyar S, Stanton K, Fredrickson JK, Bryant DA, Osterman AL, Best AA, Rodionov DA. 2013. Comparative genomics and functional analysis of rhamnose catabolic pathways and regulons in bacteria. Front Microbiol 4:407. doi: 10.3389/fmicb.2013.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saxena RK, Anand P, Saran S, Isar J, Agarwal L. 2010. Microbial production and applications of 1,2-propanediol. Indian J Microbiol 50:2–11. doi: 10.1007/s12088-010-0017-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S, Maruya M, McKenzie CI, Hijikata A, Wong C, Binge L, Thorburn AN, Chevalier N, Ang C, Marino E, Robert R, Offermanns S, Teixeira MM, Moore RJ, Flavell RA, Fagarasan S, Mackay CR. 2015. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat Commun 6:6734. doi: 10.1038/ncomms7734. [DOI] [PubMed] [Google Scholar]
- 79.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, Xavier RJ, Teixeira MM, Mackay CR. 2009. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nohr MK, Pedersen MH, Gille A, Egerod KL, Engelstoft MS, Husted AS, Sichlau RM, Grunddal KV, Poulsen SS, Han S, Jones RM, Offermanns S, Schwartz TW. 2013. GPR41/FFAR3 and GPR43/FFAR2 as cosensors for short-chain fatty acids in enteroendocrine cells vs FFAR3 in enteric neurons and FFAR2 in enteric leukocytes. Endocrinology 154:3552–3564. doi: 10.1210/en.2013-1142. [DOI] [PubMed] [Google Scholar]
- 81.Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, Hammer RE, Williams SC, Crowley J, Yanagisawa M, Gordon JI. 2008. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci U S A 105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Singh D, Chaudhuri PK. 2018. Structural characteristics, bioavailability and cardioprotective potential of saponins. Integr Med Res 7:33–43. doi: 10.1016/j.imr.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao S, Basu S, Yang Z, Deb A, Hu M. 2012. Bioavailability challenges associated with development of saponins as therapeutic and chemopreventive agents. Curr Drug Targets 13:1885–1899. doi: 10.2174/138945012804545498. [DOI] [PubMed] [Google Scholar]
- 84.Lim SM, Jeong JJ, Kang GD, Kim KA, Choi HS, Kim DH. 2015. Timosaponin AIII and its metabolite sarsasapogenin ameliorate colitis in mice by inhibiting NF-κB and MAPK activation and restoring Th17/Treg cell balance. Int Immunopharmacol 25:493–503. doi: 10.1016/j.intimp.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 85.Wang Q, Xiao BX, Pan RL, Liu XM, Liao YH, Feng L, Cao FR, Chang Q. 2015. An LC-MS/MS method for simultaneous determination of three Polygala saponin hydrolysates in rat plasma and its application to a pharmacokinetic study. J Ethnopharmacol 169:401–406. doi: 10.1016/j.jep.2015.04.033. [DOI] [PubMed] [Google Scholar]
- 86.Dastagir G, Rizvi MA. 2016. Review—Glycyrrhiza glabra L. (liquorice). Pak J Pharm Sci 29:1727–1733. [PubMed] [Google Scholar]
- 87.Michaelis M, Geiler J, Naczk P, Sithisarn P, Leutz A, Doerr HW, Cinatl J Jr.. 2011. Glycyrrhizin exerts antioxidative effects in H5N1 influenza A virus-infected cells and inhibits virus replication and pro-inflammatory gene expression. PLoS One 6:e19705. doi: 10.1371/journal.pone.0019705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roohbakhsh A, Iranshahy M, Iranshahi M. 2016. Glycyrrhetinic acid and its derivatives: anti-cancer and cancer chemopreventive properties, mechanisms of action and structure-cytotoxic activity relationship. Curr Med Chem 23:498–517. doi: 10.2174/0929867323666160112122256. [DOI] [PubMed] [Google Scholar]
- 89.Kohanpour M-AP, Peeri M, Azarbayjani M-A. 2017. The effects of Glycyrrhiza glabra L. extract use with aerobic training on inflammatory factors and cognitive state in elderly with mild cognitive impairment. J Herbmed Pharmacol 6:179–184. [Google Scholar]
- 90.Kuramoto T, Yamamoto M. 1990. Monoglucuronyl glycyrretinic acid (MGGR) from licorice and its use as a sweetener in foods. Shokuhin Kogyo 33:46–50. [Google Scholar]
- 91.Liu H-M, Akiyama T, Sugimoto N, Maitani T. 2001. Isolation and identification of main constituents in an enzymatically hydrolysed licorice extract sweetener. Food Addit Contam 18:281–284. doi: 10.1080/02652030116955. [DOI] [PubMed] [Google Scholar]
- 92.Liu HM, Sugimoto N, Akiyama T, Maitani T. 2000. Constituents and their sweetness of food additive enzymatically modified licorice extract. J Agric Food Chem 48:6044–6047. doi: 10.1021/jf000772e. [DOI] [PubMed] [Google Scholar]
- 93.Yang YA, Tang WJ, Zhang X, Yuan JW, Liu XH, Zhu HL. 2014. Synthesis, molecular docking and biological evaluation of glycyrrhizin analogs as anticancer agents targeting EGFR. Molecules 19:6368–6381. doi: 10.3390/molecules19056368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang C, Guo XX, Wang XY, Qi F, Feng SJ, Li C, Zhou XH. 2013. Isolation and characterization of three fungi with the potential of transforming glycyrrhizin. World J Microbiol Biotechnol 29:781–788. doi: 10.1007/s11274-012-1233-9. [DOI] [PubMed] [Google Scholar]
- 95.Akao T. 2000. Differences in the metabolism of glycyrrhizin, glycyrrhetic acid and glycyrrhetic acid monoglucuronide by human intestinal flora. Biol Pharm Bull 23:1418–1423. doi: 10.1248/bpb.23.1418. [DOI] [PubMed] [Google Scholar]
- 96.Guo L, Katiyo W, Lu L, Zhang X, Wang M, Yan J, Ma X, Yang R, Zou L, Zhao W. 2018. Glycyrrhetic acid 3-O-mono-β-d-glucuronide (GAMG): an innovative high-potency sweetener with improved biological activities. Compr Rev Food Sci Food Saf 17:905–919. doi: 10.1111/1541-4337.12353. [DOI] [PubMed] [Google Scholar]
- 97.Akao T. 2001. Effect of pH on metabolism of glycyrrhizin, glycyrrhetic acid and glycyrrhetic acid monoglucuronide by collected human intestinal flora. Biol Pharm Bull 24:1108–1112. doi: 10.1248/bpb.24.1108. [DOI] [PubMed] [Google Scholar]
- 98.Shin BK, Kwon SW, Park JH. 2015. Chemical diversity of ginseng saponins from Panax ginseng. J Ginseng Res 39:287–298. doi: 10.1016/j.jgr.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hu SY. 1977. A contribution to our knowledge of ginseng. Am J Chin Med (Gard City N Y) 5:1–23. doi: 10.1142/s0192415x77000026. [DOI] [PubMed] [Google Scholar]
- 100.Hasegawa H, Uchiyama M. 1998. Antimetastatic efficacy of orally administered ginsenoside Rb1 in dependence on intestinal bacterial hydrolyzing potential and significance of treatment with an active bacterial metabolite. Planta Med 64:696–700. doi: 10.1055/s-2006-957560. [DOI] [PubMed] [Google Scholar]
- 101.Del Rio D, Rodriguez-Mateos A, Spencer JP, Tognolini M, Borges G, Crozier A. 2013. Dietary (poly)phenolics in human health: structures, bioavailability, and evidence of protective effects against chronic diseases. Antioxid Redox Signal 18:1818–1892. doi: 10.1089/ars.2012.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hanske L, Loh G, Sczesny S, Blaut M, Braune A. 2009. The bioavailability of apigenin-7-glucoside is influenced by human intestinal microbiota in rats. J Nutr 139:1095–1102. doi: 10.3945/jn.108.102814. [DOI] [PubMed] [Google Scholar]
- 103.Czank C, Cassidy A, Zhang QZ, Morrison DJ, Preston T, Kroon PA, Botting NP, Kay CD. 2013. Human metabolism and elimination of the anthocyanin, cyanidin-3-glucoside: a C-13-tracer study. Am J Clin Nutr 97:995–1003. doi: 10.3945/ajcn.112.049247. [DOI] [PubMed] [Google Scholar]
- 104.Gonzalez-Barrio R, Edwards CA, Crozier A. 2011. Colonic catabolism of ellagitannins, ellagic acid, and raspberry anthocyanins: in vivo and in vitro studies. Drug Metab Dispos 39:1680–1688. doi: 10.1124/dmd.111.039651. [DOI] [PubMed] [Google Scholar]
- 105.Gonzalez-Barrio R, Borges G, Mullen W, Crozier A. 2010. Bioavailability of anthocyanins and ellagitannins following consumption of raspberries by healthy humans and subjects with an ileostomy. J Agric Food Chem 58:3933–3939. doi: 10.1021/jf100315d. [DOI] [PubMed] [Google Scholar]
- 106.Kahle K, Kraus M, Scheppach W, Ackermann M, Ridder F, Richling E. 2006. Studies on apple and blueberry fruit constituents: do the polyphenols reach the colon after ingestion? Mol Nutr Food Res 50:418–423. doi: 10.1002/mnfr.200500211. [DOI] [PubMed] [Google Scholar]
- 107.Stalmach A, Edwards CA, Wightman JD, Crozier A. 2013. Colonic catabolism of dietary phenolic and polyphenolic compounds from Concord grape juice. Food Funct 4:52–62. doi: 10.1039/c2fo30151b. [DOI] [PubMed] [Google Scholar]
- 108.Feliciano RP, Boeres A, Massacessi L, Istas G, Ventura MR, dos Santos CN, Heiss C, Rodriguez-Mateos A. 2016. Identification and quantification of novel cranberry-derived plasma and urinary (poly)phenols. Arch Biochem Biophys 599:31–41. doi: 10.1016/j.abb.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 109.Anand P, Sundaram C, Jhurani S, Kunnumakkara AB, Aggarwal BB. 2008. Curcumin and cancer: an “old-age” disease with an “age-old” solution. Cancer Lett 267:133–164. doi: 10.1016/j.canlet.2008.03.025. [DOI] [PubMed] [Google Scholar]
- 110.Hatcher H, Planalp R, Cho J, Torti FM, Torti SV. 2008. Curcumin: from ancient medicine to current clinical trials. Cell Mol Life Sci 65:1631–1652. doi: 10.1007/s00018-008-7452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kocaadam B, Şanlier N. 2017. Curcumin, an active component of turmeric (Curcuma longa), and its effects on health. Crit Rev Food Sci Nutr 57:2889–2895. doi: 10.1080/10408398.2015.1077195. [DOI] [PubMed] [Google Scholar]
- 112.Vareed SK, Kakarala M, Ruffin MT, Crowell JA, Normolle DP, Djuric Z, Brenner DE. 2008. Pharmacokinetics of curcumin conjugate metabolites in healthy human subjects. Cancer Epidemiol Biomarkers Prev 17:1411–1417. doi: 10.1158/1055-9965.EPI-07-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shen L, Ji HF. 2019. Bidirectional interactions between dietary curcumin and gut microbiota. Crit Rev Food Sci Nutr 59:2896–2902. doi: 10.1080/10408398.2018.1478388. [DOI] [PubMed] [Google Scholar]
- 114.Plumb GW, Garcia-Conesa MT, Kroon PA, Rhodes M, Ridley S, Williamson G. 1999. Metabolism of chlorogenic acid by human plasma, liver, intestine and gut microflora. J Sci Food Agric 79:390–392. doi:. [DOI] [Google Scholar]
- 115.Stalmach A, Mullen W, Barron D, Uchida K, Yokota T, Cavin C, Steiling H, Williamson G, Crozier A. 2009. Metabolite profiling of hydroxycinnamate derivatives in plasma and urine after the ingestion of coffee by humans: identification of biomarkers of coffee consumption. Drug Metab Dispos 37:1749–1758. doi: 10.1124/dmd.109.028019. [DOI] [PubMed] [Google Scholar]
- 116.Mosele JI, Martín-Peláez S, Macià A, Farràs M, Valls RM, Catalán Ú, Motilva M-J. 2014. Study of the catabolism of thyme phenols combining in vitro fermentation and human intervention. J Agric Food Chem 62:10954–10961. doi: 10.1021/jf503748y. [DOI] [PubMed] [Google Scholar]
- 117.Serra A, Rubió L, Borràs X, Macià A, Romero MP, Motilva MJ. 2012. Distribution of olive oil phenolic compounds in rat tissues after administration of a phenolic extract from olive cake. Mol Nutr Food Res 56:486–496. doi: 10.1002/mnfr.201100436. [DOI] [PubMed] [Google Scholar]
- 118.Ronca F, Palmieri L, Panicucci P, Ronca G. 1998. Anti-inflammatory activity of chondroitin sulfate. Osteoarthritis Cartilage 6:14–21. doi: 10.1016/S1063-4584(98)80006-X. [DOI] [PubMed] [Google Scholar]
- 119.Messier SP, Mihalko S, Loeser RF, Legault C, Jolla J, Pfruender J, Prosser B, Adrian A, Williamson JD. 2007. Glucosamine/chondroitin combined with exercise for the treatment of knee osteoarthritis: a preliminary study. Osteoarthritis Cartilage 15:1256–1266. doi: 10.1016/j.joca.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 120.Baici A, Horler D, Moser B, Hofer HO, Fehr K, Wagenhauser FJ. 1992. Analysis of glycosaminoglycans in human serum after oral administration of chondroitin sulfate. Rheumatol Int 12:81–88. doi: 10.1007/bf00290259. [DOI] [PubMed] [Google Scholar]
- 121.Barthe L, Woodley J, Lavit M, Przybylski C, Philibert C, Houin G. 2004. In vitro intestinal degradation and absorption of chondroitin sulfate, a glycosaminoglycan drug. Arzneimittelforschung 54:286–292. doi: 10.1055/s-0031-1296972. [DOI] [PubMed] [Google Scholar]
- 122.Shang Q, Yin Y, Zhu L, Li G, Yu G, Wang X. 2016. Degradation of chondroitin sulfate by the gut microbiota of Chinese individuals. Int J Biol Macromol 86:112–118. doi: 10.1016/j.ijbiomac.2016.01.055. [DOI] [PubMed] [Google Scholar]
- 123.Zhang X, Zhao YF, Xu J, Xue ZS, Zhang MH, Pang XY, Zhang XJ, Zhao LP. 2015. Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci Rep 5:14405. doi: 10.1038/srep14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nguyen TD, Prykhodko O, Fak Hallenius F, Nyman M. 2019. Monovalerin and trivalerin increase brain acetic acid, decrease liver succinic acid, and alter gut microbiota in rats fed high-fat diets. Eur J Nutr 58:1545–1560. doi: 10.1007/s00394-018-1688-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zeisel SH, Wishnok JS, Blusztajn JK. 1983. Formation of methylamines from ingested choline and lecithin. J Pharmacol Exp Ther 225:320–324. [PubMed] [Google Scholar]
- 126.Chen M, Yi L, Zhang Y, Zhou X, Ran L, Yang J, Zhu J, Zhang Q, Mi M. 2016. Resveratrol attenuates trimethylamine-N-oxide (TMAO)-induced atherosclerosis by regulating TMAO synthesis and bile acid metabolism via remodeling of the gut microbiota. mBio 7:e02210-15. doi: 10.1128/mBio.02210-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li Q, Wu T, Liu R, Zhang M, Wang R. 2017. Soluble dietary fiber reduces trimethylamine metabolism via gut microbiota and co-regulates host AMPK pathways. Mol Nutr Food Res 61:1700473. doi: 10.1002/mnfr.201700473. [DOI] [PubMed] [Google Scholar]
- 128.Wu W-K, Panyod S, Ho C-T, Kuo C-H, Wu M-S, Sheen L-Y. 2015. Dietary allicin reduces transformation of L-carnitine to TMAO through impact on gut microbiota. J Funct Foods 15:408–417. doi: 10.1016/j.jff.2015.04.001. [DOI] [Google Scholar]
- 129.Tang WH, Kitai T, Hazen SL. 2017. Gut microbiota in cardiovascular health and disease. Circ Res 120:1183–1196. doi: 10.1161/CIRCRESAHA.117.309715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Han K, Bose S, Wang JH, Lim SK, Chin YW, Kim YM, Choi HS, Kim H. 2017. In vivo therapeutic effect of combination treatment with metformin and Scutellaria baicalensis on maintaining bile acid homeostasis. PLoS One 12:e0182467. doi: 10.1371/journal.pone.0182467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.He K, Hu Y, Ma H, Zou Z, Xiao Y, Yang Y, Feng M, Li X, Ye X. 2016. Rhizoma coptidis alkaloids alleviate hyperlipidemia in B6 mice by modulating gut microbiota and bile acid pathways. Biochim Biophys Acta 1862:1696–1709. doi: 10.1016/j.bbadis.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 132.Raza GS, Maukonen J, Makinen M, Niemi P, Niiranen L, Hibberd AA, Poutanen K, Buchert J, Herzig KH. 2019. Hypocholesterolemic effect of the lignin-rich insoluble residue of brewer’s spent grain in mice fed a high-fat diet. J Agric Food Chem 67:1104–1114. doi: 10.1021/acs.jafc.8b05770. [DOI] [PubMed] [Google Scholar]
- 133.Lopez P, Sanchez M, Perez-Cruz C, Velazquez-Villegas LA, Syeda T, Aguilar-Lopez M, Rocha-Viggiano AK, Del Carmen Silva-Lucero M, Torre-Villalvazo I, Noriega LG, Torres N, Tovar AR. 2018. Long-term genistein consumption modifies gut microbiota, improving glucose metabolism, metabolic endotoxemia, and cognitive function in mice fed a high-fat diet. Mol Nutr Food Res 62:e1800313. doi: 10.1002/mnfr.201800313. [DOI] [PubMed] [Google Scholar]
- 134.Nazzal L, Roberts J, Singh P, Jhawar S, Matalon A, Gao Z, Holzman R, Liebes L, Blaser MJ, Lowenstein J. 2017. Microbiome perturbation by oral vancomycin reduces plasma concentration of two gut-derived uremic solutes, indoxyl sulfate and p-cresyl sulfate, in end-stage renal disease. Nephrol Dial Transplant 32:1809–1817. doi: 10.1093/ndt/gfx029. [DOI] [PubMed] [Google Scholar]
- 135.Rossi M, Johnson DW, Morrison M, Pascoe EM, Coombes JS, Forbes JM, Szeto CC, McWhinney BC, Ungerer JP, Campbell KL. 2016. Synbiotics easing renal failure by improving gut microbiology (SYNERGY): a randomized trial. Clin J Am Soc Nephrol 11:223–231. doi: 10.2215/CJN.05240515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Strong HA, Renwick AG, George CF, Liu YF, Hill MJ. 1987. The reduction of sulphinpyrazone and sulindac by intestinal bacteria. Xenobiotica 17:685–696. doi: 10.3109/00498258709043976. [DOI] [PubMed] [Google Scholar]
- 137.Magnusson JO, Bergdahl B, Bogentoft C, Jonsson UE. 1982. Metabolism of digoxin and absorption site. Br J Clin Pharmacol 14:284–285. doi: 10.1111/j.1365-2125.1982.tb01976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Das KM, Eastwood MA, McManus JP, Sircus W. 1973. The metabolism of salicylazosulphapyridine in ulcerative colitis. I. The relationship between metabolites and the response to treatment in inpatients. Gut 14:631–641. doi: 10.1136/gut.14.8.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Peppercorn MA, Goldman P. 1972. The role of intestinal bacteria in the metabolism of salicylazosulfapyridine. J Pharmacol Exp Ther 181:555–562. [PubMed] [Google Scholar]
- 140.Peppercorn MA, Goldman P. 1973. Distribution studies of salicylazosulfapyridine and its metabolites. Gastroenterology 64:240–245. [PubMed] [Google Scholar]
- 141.Takeno S, Sakai T. 1991. Involvement of the intestinal microflora in nitrazepam-induced teratogenicity in rats and its relationship to nitroreduction. Teratology 44:209–214. doi: 10.1002/tera.1420440209. [DOI] [PubMed] [Google Scholar]
- 142.Kitamura S, Sugihara K, Kuwasako M, Tatsumi K. 1997. The role of mammalian intestinal bacteria in the reductive metabolism of zonisamide. J Pharm Pharmacol 49:253–256. doi: 10.1111/j.2042-7158.1997.tb06790.x. [DOI] [PubMed] [Google Scholar]
- 143.Meredith PA, Elliott HL. 1992. Clinical pharmacokinetics of amlodipine. Clin Pharmacokinet 22:22–31. doi: 10.2165/00003088-199222010-00003. [DOI] [PubMed] [Google Scholar]
- 144.Yoo HH, Kim IS, Yoo DH, Kim DH. 2016. Effects of orally administered antibiotics on the bioavailability of amlodipine: gut microbiota-mediated drug interaction. J Hypertens 34:156–162. doi: 10.1097/HJH.0000000000000773. [DOI] [PubMed] [Google Scholar]
- 145.Bartlett JG. 2002. Clinical practice. Antibiotic-associated diarrhea. N Engl J Med 346:334–339. doi: 10.1056/NEJMcp011603. [DOI] [PubMed] [Google Scholar]
- 146.Wistrom J, Norrby SR, Myhre EB, Eriksson S, Granstrom G, Lagergren L, Englund G, Nord CE, Svenungsson B. 2001. Frequency of antibiotic-associated diarrhoea in 2462 antibiotic-treated hospitalized patients: a prospective study. J Antimicrob Chemother 47:43–50. doi: 10.1093/jac/47.1.43. [DOI] [PubMed] [Google Scholar]
- 147.Saputri FA, Kang D, Kusuma ASW, Rusdiana T, Hasanah AN, Mutakin, Surono IS, Koyama H, Abdulah R. 2018. Lactobacillus plantarum IS-10506 probiotic administration increases amlodipine absorption in a rabbit model. J Int Med Res 46:5004–5010. doi: 10.1177/0300060518788994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Matuskova Z, Anzenbacherova E, Vecera R, Tlaskalova-Hogenova H, Kolar M, Anzenbacher P. 2014. Administration of a probiotic can change drug pharmacokinetics: effect of E. coli Nissle 1917 on amidarone absorption in rats. PLoS One 9:e87150. doi: 10.1371/journal.pone.0087150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Al-Salami H, Butt G, Fawcett JP, Tucker IG, Golocorbin-Kon S, Mikov M. 2008. Probiotic treatment reduces blood glucose levels and increases systemic absorption of gliclazide in diabetic rats. Eur J Drug Metab Pharmacokinet 33:101–106. doi: 10.1007/BF03191026. [DOI] [PubMed] [Google Scholar]
- 150.Sassone-Corsi M, Nuccio SP, Liu H, Hernandez D, Vu CT, Takahashi AA, Edwards RA, Raffatellu M. 2016. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 540:280–283. doi: 10.1038/nature20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Litvak Y, Mon KKZ, Nguyen H, Chanthavixay G, Liou M, Velazquez EM, Kutter L, Alcantara MA, Byndloss MX, Tiffany CR, Walker GT, Faber F, Zhu Y, Bronner DN, Byndloss AJ, Tsolis RM, Zhou H, Bäumler AJ. 2019. Commensal Enterobacteriaceae protect against Salmonella colonization through oxygen competition. Cell Host Microbe 25:128–139. doi: 10.1016/j.chom.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hooshmand S, Juma S, Arjmandi BH. 2010. Combination of genistin and fructooligosaccharides prevents bone loss in ovarian hormone deficiency. J Med Food 13:320–325. doi: 10.1089/jmf.2009.0059. [DOI] [PubMed] [Google Scholar]
- 153.Fu ZD, Cui JY. 2017. Remote sensing between liver and intestine: importance of microbial metabolites. Curr Pharmacol Rep 3:101–113. doi: 10.1007/s40495-017-0087-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sun GX, Van de Wiele T, Alava P, Tack FMG, Du Laing G. 2017. Bioaccessibility of selenium from cooked rice as determined in a simulator of the human intestinal tract (SHIME). J Sci Food Agric 97:3540–3545. doi: 10.1002/jsfa.8208. [DOI] [PubMed] [Google Scholar]
- 155.Van de Wiele T, Van den Abbeele P, Ossieur W, Possemiers S, Marzorati M. 2015. The simulator of the human intestinal microbial ecosystem (SHIME), p 305–317. In Verhoeckx K, Cotter P, Lopez-Exposito I, Kleiveland C, Lea T, Mackie A, Requena T, Swiatecka D, Wichers H (ed), The impact of food bioactives on health: in vitro and ex vivo models. Springer, Cham, Switzerland: 10.1007/978-3-319-16104-4_27. [DOI] [PubMed] [Google Scholar]
- 156.Cha KH, Lee EH, Yoon HS, Lee JH, Kim JY, Kang K, Park JS, Jin JB, Ko G, Pan CH. 2018. Effects of fermented milk treatment on microbial population and metabolomic outcomes in a three-stage semi-continuous culture system. Food Chem 263:216–224. doi: 10.1016/j.foodchem.2018.04.095. [DOI] [PubMed] [Google Scholar]
- 157.Bonnet F, Scheen A. 2017. Understanding and overcoming metformin gastrointestinal intolerance. Diabetes Obes Metab 19:473–481. doi: 10.1111/dom.12854. [DOI] [PubMed] [Google Scholar]
- 158.Aura A-M, O’Leary KA, Williamson G, Ojala M, Bailey M, Puupponen-Pimiä R, Nuutila AM, Oksman-Caldentey K-M, Poutanen K. 2002. Quercetin derivatives are deconjugated and converted to hydroxyphenylacetic acids but not methylated by human fecal flora in vitro. J Agric Food Chem 50:1725–1730. doi: 10.1021/jf0108056. [DOI] [PubMed] [Google Scholar]