Abstract
Background and Purpose:
Brain MRI-derived lesions and atrophy are related to multiple sclerosis (MS) disability. In the Serially Unified Multicenter MS Investigation (SUMMIT), from Brigham and Women’s Hospital (BWH) and University of California, San Francisco (UCSF), we assessed whether MRI methodologic heterogeneity may limit the ability to pool multisite data sets to assess 5-year clinical-MRI associations.
Methods:
Patients with relapsing-remitting (RR) MS (n=100 from each site) underwent baseline brain MRI and baseline and 5-year clinical evaluations. Patients were matched on sex (74 women each), age, disease duration, and Expanded Disability Status Scale (EDSS) score. MRI was performed with differences between sites in both acquisition (field strength, voxel size, pulse sequences), and post-processing pipeline to assess brain parenchymal fraction (BPF) and T2 lesion volume (T2LV).
Results:
The UCSF cohort showed higher correlation than the BWH cohort between T2LV and disease duration. UCSF showed a higher inverse correlation between BPF and age than BWH. UCSF showed a higher inverse correlation than BWH between BPF and 5-year EDSS score. Both cohorts showed inverse correlations between BPF and T2LV, with no between-site difference. The pooled but not individual cohort data showed a link between a lower baseline BPF and the subsequent 5-year worsening in disability in addition to other stronger relationships in the data.
Conclusions:
MRI acquisition and processing differences may result in some degree of heterogeneity in assessing brain lesion and atrophy measures in patients with MS. Pooling of data across sites is beneficial to correct for potential biases in individual data sets.
Keywords: Multiple sclerosis, MRI, Brain, Neuroimaging, Disability, Multicenter study
Introduction
Brain MRI is commonly used as a surrogate to monitor the underlying multiple sclerosis (MS) disease process.1 This includes the detection of lesions2 and atrophy,3 both of which are associated with physical disability,4 cognitive impairment,5 risk of disease progression,6 and response to disease-modifying therapy.7 In prospectively designed multicenter MS therapeutic trials, MRI is commonly employed as a supportive outcome measure.8 This involves advanced planning and harmonization of MRI methodology to assure consistent data acquisition across sites, typically followed by centralized image processing using a single processing pipeline at one site.
However, given the valuable information provided by brain MRI, it would be desirable to be able to retrospectively combine heterogeneously acquired and processed MRI data sets across sites to provide deep characterization of the MS disease process in large numbers of patients. In most instances, such data were not consistently obtained, which poses a challenge on several fronts given the potential for divergence of MRI results based on numerous inconsistencies in acquisition and processing between or within sites. For example, studies have shown that field strength,9–11 type of pulse sequence,9,12–14 scanner upgrades,9 scanner vendor,9,15 and voxel size13 may each influence MS-related cerebral lesion and atrophy volumetric measures. Furthermore, the type of post-processing image segmentation pipeline,12,13,16–20 pre-processing image preparation and software version within the same pipeline,9,21 or workstation operating system21 may also add variation to the results. Even when using a consistent field strength, scanner vendor, and high-resolution harmonized pulse sequences, there is considerable variability in manual and automated segmentation results22 partly based on hardware factors such as gradient non-linearity.23 Such challenges have led to intense interest in developing post-hoc statistical and technical approaches to harmonize results from heterogeneously-acquired data sets.23–27
The purpose of this study was to evaluate “real-world” effects on clinical-MRI relationships between two comprehensive care MS centers when the brain MRI methodology used to quantify lesions and atrophy had major differences between sites in both acquisition (field strength, voxel size, pulse sequences), and post-processing pipeline.
Methods
Patients
This study was part of the Serially Unified Multicenter Multiple Sclerosis (SUMMIT) investigation, which includes the Brigham and Women’s Hospital (BWH) and the University of California, San Francisco (UCSF) MS programs.28 We retrospectively identified 100 patients at each site with a diagnosis of relapsing-remitting (RR) MS29 who underwent a neurological examination and brain MRI scan at baseline followed by a repeat clinical examination five years later, as part of prospective studies at each site.28 Clinical examination included determination of the Expanded Disability Status Scale (EDSS) score30 by an MS-specialist neurologist. At the follow-up visit, patients were also evaluated for conversion of their MS subtype from RR to secondary progressive (SP).31 Subject characteristics are summarized in Table 1. Based on study design, the patients were matched as closely as possible at baseline by age, sex, disease duration and EDSS score (Table 1). Because the source data were prospectively collected, all patients signed informed consent to be included in this study, which was approved by each hospital’s research ethics committee.
Table 1.
Patient characteristics: Baseline and 5-year follow-up
| BWH (n=100) | UCSF (n=100) | p value | |
|---|---|---|---|
| Age (years) – baseline | 39.5±8.4 (21-55) | 39.5±8.0 (23-56) | 1.0** |
| Sex (% women) | 74 | 74 | 1.0** |
| Clinical type – baseline | RR (100%) | RR (100%) | 1.0** |
| Clinical type – follow-up | RR (96%)/SP (4%) | RR (93%)/SP (7%) | 0.54 |
| Disease duration* | 7.7±6.8 (0.4-33.7) | 7.2±6.5 (0-34.0) | 0.56** |
| BPF– baseline | 0.86±0.04 (0.77-0.94) | 0.74±0.04 (0.65-0.83) | <0.001 |
| T2LV (ml) – baseline | 5.3±5.7 (0.7-30.7) | 7.9±15.7 (0-132) | 0.12 |
| MRI-clinical follow-up interval (months) | 60.8±2.2 (55-67) | 61.1±2.4 (55-67) | 0.34 |
| EDSS – baseline | 1.2±1.1 (0-5.5) | 1.4±1.2 (0-5) | 0.13** |
| EDSS – follow-up | 1.6±1.5 (0-6.5) | 2.3±1.4 (0-6.5) | 0.001 |
Key: data are shown as mean ± standard deviation (range), except as otherwise indicated;
years from first symptoms; n = number of subjects; BWH = Brigham and Women’s Hospital; UCSF = University of California, San Francisco; BPF = brain parenchymal fraction; T2LV = global cerebral T2 hyperintense lesion volume; EDSS = Expanded Disability Status Scale score;
as part of study design, the patients were matched as closely as possible on these baseline characteristics
Brain MRI acquisition
For the 100 patients from BWH, MRI was performed at 1.5T on a General Electric (GE) Signa scanner (Milwaukee, WI) using a consistently acquired T2-weighted 2D dual-echo series with the following parameters: TR/TE2/TE1: 3000/80/30 ms, voxel size 0.94 × 0.94 × 3 mm, no inter-slice gaps, 54 axial slices. For the 100 patients from UCSF, MRI was performed on a 3T General Electric Signa scanner to acquire T1-weighted 3D images (Inversion Recovery Spoiled Gradient Echo pulse sequence, TR/TE/TI: 7/2/400 ms, flip angle 8 degrees, voxel size 0.94 × 0.94 × 1 mm, no inter-slice gaps, 160 axial slices ) and T2-weighted 2D dual-echo images (TR/TE2/TE1: 2000/80/20 ms, voxel size 0.47 × 0.47 × 3 mm, no inter-slice gaps, 44 axial slices).
MRI analysis-BWH
For the patients from BWH, the 1.5T 2D images were processed at BWH using a fully-automated in-house-developed TDS+ pipeline32,33 to derive cerebral T2 hyperintense lesion volume (T2LV) and brain parenchymal fraction (BPF), a normalized estimate of whole brain atrophy.34
MRI analysis-UCSF
For the patients from UCSF, 3T 3D images were processed at UCSF using an automated Structural Image Evaluation, using Normalization, of Atrophy (SIENAX) pipeline (https://fsl.fmrib.ox.ac.uk/fsl). While SIENAX is traditionally used to derive normalized brain parenchymal volume (BPV), to more closely harmonize the data from the two cohorts, we derived BPF from the UCSF images by dividing the SIENAX derived brain volume by (SIENAX brain volume + ventricular CSF). An experienced observer analyzed baseline and follow-up images concurrently using Jim software (v.7, Xinapse Systems, West Bergholt, UK; www.xinapse.com) to derive manually segmented T2LV; these masks were used as input for the SIENAX segmentation.
Statistical analysis
The baseline clinical and MRI characteristics of the two cohorts were compared using a two-sample t-test for continuous variables and a chi-squared test for dichotomous outcomes. Within each cohort, we estimated the association between the brain MRI measures (T2LV and BPF) and demographic and clinical measures (age, disease duration, EDSS at baseline, EDSS at 5 years and EDSS change) using Pearson’s correlation coefficients. For T2LV, we used a cube root transformation so that the variable was approximately normally distributed. We also treated EDSS as a continuous variable for all analyses given the moderate sample size in the two cohorts. We compared the Pearson’s correlation coefficients between cohorts using Fisher’s tests based on a z-transformation with the cocor library in R. In addition to these correlations, we fit a multivariable linear regression model with change in EDSS as the outcome and BPF, cube root transformed T2LV, age and sex as the predictors in all subjects and each cohort separately. Since linear regression and Pearson’s correlation coefficient assume underlying normality and this does not hold for the EDSS, we confirmed all analyses using a multilevel ordinal logistic regression model and Spearman’s correlation coefficient, and the results from these approaches were similar (data not shown). Finally, to determine if conversion of the SIENAX outputs to BPF instead of BPF led to inherent differences in the data, we estimated the correlations using BPF and BPV from SIENAX in the UCSF cohort.
Results
Demographic and clinical characteristics
As shown in Table 1, the BWH and UCSF patients were similar at baseline on sex (74 women each), age (mean±SD) (BWH: 39.5±8.4, UCSF: 39.5±8.0 years), disease duration (7.7±6.8, 7.2±6.6 years), and EDSS score (1.2±1.1, 1.4±1.2). The two groups were also similar on T2LV (BWH median 3.6 IQR 1.9–5.7 ml, UCSF 3.9, 1.4-8.1 ml, Table 1). At follow-up, four patients from BWH (4%) and seven patients from UCSF (7%) converted from RR to a diagnosis of SP MS (p=0.54). At follow-up, EDSS score increased to 1.6±1.5 in the BWH group and 2.3±1.4 in the UCSF group (p=0.001). We calculated the mean change over 5 years between EDSS scores in all subjects in the two groups separately, and then calculated the difference in the mean change between the two sites. For all subjects, the mean change was an increase of 0.69 (95% confidence interval: 0.51, 0.85). Within each site this was 0.46 (0.20, 0.72) at BWH and 0.91 (0.69, 1.13) at UCSF. The difference in change between sites was 0.45 (0.12, 0.78) (p=0.0086) and 0.0075 (0.0021, 0.013); p=0.0076 for the monthly rate of change. Thus, the UCSF cohort had higher on-study levels of progression of disability than the BWH cohort.
T2LV vs. clinical measures: correlation analysis
Univariate correlations are shown in Table 2, and Figures 1 and 2. Regarding T2LV vs. clinical measures, the UCSF cohort (n=100) showed a higher correlation than the BWH cohort (n=100) for the relationship between T2LV and disease duration (Table 2). For the correlation between T2LV and all EDSS variables, there were no significant differences between sites (Table 2). With the two cohorts pooled (n=200), the correlations resembled the UCSF site, with the relationships between T2LV and disease duration, EDSS at baseline, and EDSS at 5 years all reaching significance (Table 2).
Table 2.
MRI-clinical and MRI-MRI correlations
| All subjects (n=200) | BWH (n=100) | UCSF (n=100) | BWH vs. UCSF: p value | |
|---|---|---|---|---|
| Baseline T2LV vs: | ||||
| Disease duration | 0.272 (0.139, 0.396) p<0.001*** |
0.104 (−0.094, 0.295) p=0.303 |
0.395 (0.215, 0.549) p<0.001*** |
0.029* |
| EDSS at baseline | 0.194 (0.056, 0.324) p=0.006** |
0.178 (−0.02, 0.361) p=0.077 |
0.207 (0.011, 0.388) p=0.038* |
0.829 |
| EDSS at 5y | 0.257 (0.122, 0.382) p<0.001*** |
0.237 (0.043, 0.415) p=0.017* |
0.294 (0.103, 0.463) p=0.003** |
0.674 |
| EDSS 5y change | 0.121 (−0.018, 0.255) p=0.088 |
0.119 (−0.079, 0.309) p=0.237 |
0.130 (−0.068, 0.318) p=0.198 |
0.941 |
| Baseline BPF vs. | ||||
| Age | −0.188 (−0.318, −0.05) p=0.008** |
−0.202 (−0.383, −0.006) p=0.044* |
−0.523 (−0.653, −0.364) p<0.001*** |
0.009** |
| Disease duration | −0.111 (−0.246, 0.028) p=0.118 |
−0.220 (−0.399, −0.025) p=0.028* |
−0.331 (−0.495, −0.144) p=0.001** |
0.402 |
| EDSS at baseline | −0.235 (−0.362, −0.100) p=0.001** |
−0.174 (−0.358, 0.023) p=0.084 |
−0.371 (−0.528, −0.188) p<0.001*** |
0.137 |
| EDSS at 5y | −0.311 (−0.431, −0.180) p<0.001*** |
−0.072 (−0.264, 0.127) p=0.479 |
−0.366 (−0.525, −0.183) p<0.001*** |
0.029* |
| EDSS 5y change | −0.146 (−0.279, −0.007) p=0.039* |
0.070 (−0.129, 0.262) p=0.492 |
−0.037 (−0.232, 0.16) p=0.711 |
0.456 |
| Baseline MRI-MRI correlations: | ||||
| BPF vs. T2LV | −0.160 (−0.290, −0.020) p=0.023* |
−0.360 (−0.520, −0.180) p=0.0002** |
−0.230 (−0.410, −0.180) p=0.023* |
0.31 |
Key: Pearson correlation r values are shown with 95% confidence intervals. In the last column, the correlation coefficients between sites were compared by the approach of Fisher for the comparison of independent correlation coefficients. BWH = Brigham and Women’s Hospital; UCSF = University of California, San Francisco; BPF = brain parenchymal fraction; T2LV = global cerebral T2 hyperintense lesion volume (the cube root-transformed T2LV was used); EDSS = Expanded Disability Status Scale score;
p<0.05;
p<0.01;
p<0.001.
Figure 1.
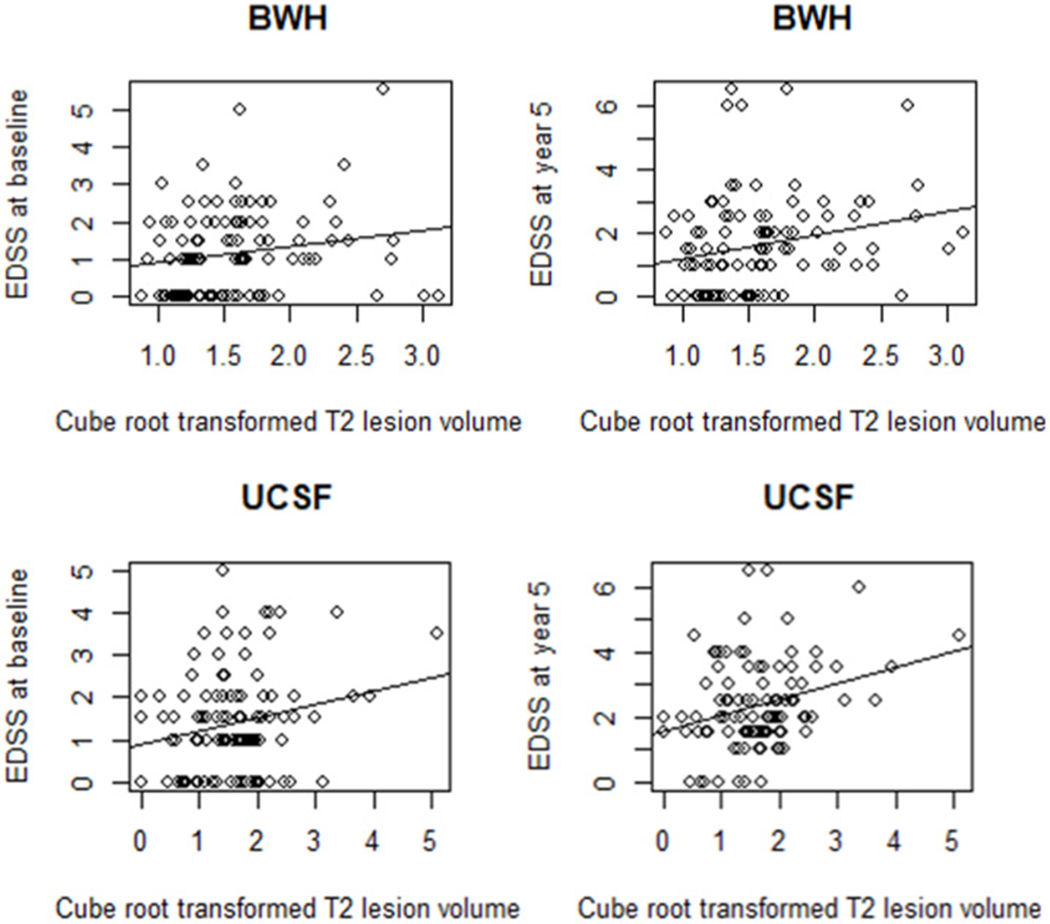
Baseline T2 lesion volume vs. EDSS score at baseline and 5-year follow-up
Global cerebral T2 hyperintense lesion volume (T2LV) at baseline vs. Expanded Disability Status Scale (EDSS) score at baseline and at 5-year follow-up from the Brigham and Women’s Hospital (BWH) (top) and the University of California, San Francisco (UCSF) (bottom) cohorts. Scatter plots with regression slopes illustrate the relationship between T2LV and EDSS score at baseline (BWH: Pearson r = 0.178, p=0.077, UCSF: r = 0.207, p=0.038), and at 5-year follow-up (BWH: Pearson r = 0.237, p=0.017, UCSF: r = 0.294, p=0.003). The correlation coefficients were not different between the two cohorts at either time point (p>0.05). See also Table 2.
Figure 2.

Baseline BPF vs. EDSS score at baseline and 5-year follow-up
Brain parenchymal fraction (BPF) at baseline vs. Expanded Disability Status Scale (EDSS) score at baseline and at 5-year follow-up from the Brigham and Women’s Hospital (BWH) (top) and the University of California, San Francisco (UCSF) (bottom) cohorts. Scatter plots with regression slopes illustrate the relationship between whole brain atrophy and EDSS score at baseline (BWH: Pearson r = −0.174, p=0.084, UCSF: r = −0.371, p<0.001), and at 5-year follow-up (BWH: Pearson r = −0.072, p=0.479, UCSF: r = −0.366, p<0.001). The correlation coefficients were different between the two cohorts at the 5-year time point (p<0.05). See also Table 2.
BPF vs. clinical measures: correlation analysis
Univariate correlations are shown in Table 2, and Figures 1 and 2. Regarding BPF vs. age, the UCSF cohort showed a higher inverse correlation with age than the BWH cohort (Table 2). Regarding BPF vs. clinical measures, the UCSF cohort showed a higher inverse correlation than BWH for the relationship between BPF and EDSS score at 5 years (Table 2). For the correlation between BPF and all EDSS variables and disease duration, there were no significant differences between sites (Table 2). With the two cohorts pooled, the correlations were significant for the relationships between BPF and age, EDSS at baseline, EDSS at 5 years, and EDSS 5-year change, but not for disease duration (Table 2).
BPF vs. T2LV: correlation analysis
Both the BWH and UCSF cohorts, and the pooled cohort, showed significant inverse correlations between BPF and T2LV, with no significant difference in the strength of correlation between sites (Table 2).
MRI prediction of 5-year disability change
Regression modeling assessed the multivariate relationships between baseline T2LV or BPF and change in EDSS score over the subsequent 5 years, adjusting for age, disease duration and sex. None of the baseline MRI parameters predicted the 5-year change in EDSS, considering each site separately (Table 3). However, with the pooled data from the two sites, a lower baseline BPF predicted disability worsening (p<0.05, Table 3).
Table 3.
Modeling the ability of MRI to predict disability change over 5 years
| All subjects (n=200) | BWH (n=100) | UCSF (n=100) | |
|---|---|---|---|
| BPF | −2.37 (−4.71, −0.02) p=0.048* |
3.30 (−4.03, 10.64) p=0.37 |
−0.72 (−6.18, 7.63) p=0.84 |
| T2LV | 0.16 (−0.10, 0.42) p=0.23 |
0.38 (−0.23, 0.98) p=0.22 |
0.17 (−0.12, 0.46) p=0.25 |
| Age | −0.009 (−0.031, 0.012) p=0.39 |
−0.011 (−0.044, 0.023) p=0.53 |
0.009 (−0.024, 0.042) p=0.60 |
| Sex | −0.19 (−0.59, 0.21) p=0.35 |
−0.16 (−0.78, 0.46) p=0.61 |
−0.11 (−0.62, 0.41) p=0.67 |
Key: These estimates with 95% confidence intervals are unstandardized regression coefficients from a linear regression with 5-year change in Expanded Disability Status Scale score as the outcome and these four predictors in the model together. BWH = Brigham and Women’s Hospital; UCSF = University of California, San Francisco; BPF = brain parenchymal fraction; T2LV = global cerebral T2 hyperintense lesion volume (the cube root-transformed T2LV was used);
p<0.05.
We also performed an exploratory analysis comparing the T2LV and BPF in subjects at 5-year follow-up who converted to SPMS (n=11) vs. those who did not (n=189). We used a linear regression model with conversion status, age, gender and cohort as a predictor. The estimated difference in cube root transformed T2LV comparing converters to non-converters was 0.51 (95% CI: 0.10, 0.92; p=0.014). The estimated difference in BPF was −0.006 (95% CI: −0.029, 0.017; p=0.60). We have chosen to not to present or discuss these results in further detail given the small sample size of the converter group.
Discussion
This study showed mixed results in comparing two cohorts of demographically and clinically baseline matched patients with RRMS on their clinical-MRI relationships in a 5-year study. The major finding was, despite the wide between-site differences in MRI acquisition (field strength, scanner vendor, voxel sizes, type of pulse sequences) and post-processing pipelines, most of the correlations were of similar direction and strength between cohorts.
However, a few of the comparisons showed clear differences, such as: 1) Age vs. BPF. It is possible that this result reflects a contribution of slight differences in disease duration, given that the latter is well known to influence brain atrophy in MS.35 2) Disease duration vs. lesion volume. This may also reflect differences in the disease severity of the cohorts, given that the UCSF patients clearly developed more disability and more of them developed progressive disease on-study. It is well known that cerebral lesion volume is related in part to disability change in MS.36 3) BPF vs. 5-year EDSS score. This may again reflect the different on-study change in disease severity of the cohorts. In addition, the weaker associations that were seen in the BWH cohort may have been a result of less precise volumetric assessment compared with the UCSF cohort given the differences in MRI acquisition (1.5T vs. 3T, T2-2D vs T1-3D sequences, and 3 mm vs. 1 mm slice thickness). However, a potential advantage of pooling data sets across centers is to increase statistical power due to larger sample size in the assessment of clinical-MRI relationships. This proved to be one benefit shown in our study, given that the pooled but not individual cohort data showed a link between baseline BPF and the subsequent 5-year change in EDSS score in addition to other stronger relationships in the data.
This works does not explore which factors make the most contribution to data heterogeneity, for example, whether the MRI field strength, type of sequence, resolution, or post-processing pipeline is the primary source of variability in the output metrics. The answer to these questions would only be possible if some of the data were acquired or processed in both centers, which we plan to test in future studies. Nonetheless, we present a useful benchmark for the limits of what is to be expected when data are pooled from multiple centers with vastly different data protocols.
A recurring theme throughout these results was that both T2 lesions and brain atrophy showed significant relationships to disease duration and EDSS scores. These results are supported by previous studies indicating that both white matter lesions and overall tissue destruction in the brain contribute to neurologic dysfunction in MS, and tend to provide complementary information on disease status and treatment efficacy.4,7,37 In RRMS, white matter lesions are thought to be largely the result of adaptive immunity responses, such as lymphocyte entry into the CNS.39 In contrast, brain atrophy, and the risk for developing secondary progressive disease, reflects a wide variety of processes not limited to the effect of destructive white matter lesions in the brain, but also gray matter factors such as cortical lesions, innate immunity, meningeal pathology, and neurodegeneration.39
We would conclude that there is a high degree of consistency in the information obtained from the MRI measures between sites, albeit with not perfect consistency, but clearly with more similarities than differences. These results provide an impetus to pool such data sets in the investigation of clinical-MRI relationships in large studies. Since the correlation coefficients are independent of the overall scaling of the metrics, these should be comparable if the metrics are related by a simple scaling factor – which they probably are for the MRI volumes.25 The differences in correlations where they do occur, may reflect differences in the clinical factors/disability of the cohorts. Despite the attempt to match at baseline, this study does not allow us to discern methodological vs. biological differences driving the differences in MRI data between cohorts. The differences in EDSS change on-study suggests that these may not be similar patients in terms of disease biology/severity. Indeed, the slightly higher EDSS score in the UCSF cohort at baseline suggests even at the study onset, that it may be a more severely affected group. It is well known that early EDSS measurements are related to subsequent changes in disability in MS.40
Table 4.
Whole brain volume measurement at the UCSF site: comparison of two methods
| BPF | BPV | |
|---|---|---|
| Disease duration | −0.331 (−0.495, −0.144) p=0.001** |
−0.332 (−0.496, −0.145) p=0.001** |
| EDSS at baseline | −0.371 (−0.528, −0.188) p<0.001*** |
−0.370 (−0.528, −0.187) p<0.001*** |
| EDSS at 5y | −0.366 (−0.525, −0.183) p<0.001*** |
−0.362 (−0.522, −0.178) p<0.001*** |
| EDSS 5y change | −0.037 (−0.232, 0.160) p=0.711 |
−0.033 (−0.228, 0.165) p=0.745 |
| T2LV | −0.227 (−0.406, −0.032) p=0.023* |
−0.231 (−0.409, −0.037) p=0.021* |
| Age | −0.523 (−0.653, −0.364) p<0.001*** |
−0.526 (−0.655, −0.367) p<0.001*** |
Key: Pearson correlations between the clinical/demographic variable in the first column and whole brain volume, with 95% confidence intervals. UCSF = University of California, San Francisco; BPF = brain parenchymal fraction; BPV=normalized brain parenchymal volume; EDSS = Expanded Disability Status Scale score; T2LV = global cerebral T2 hyperintense lesion volume (the cube root-transformed T2LV was used);
p<0.05;
p<0.01;
p<0.001.
Acknowledgments:
The following is a list of the members of the SUMMIT consortium who are co-authors of the work and approved the final version of the manuscript: Brigham and Women’s Hospital, Harvard Medical School (Boston, MA): Mark Anderson, Rohit Bakshi, Tanuja Chitnis, Brian Healy, Nikolaos Patsopoulos, Mariann Polgar-Turcsanyi, Francisco Quintana, Taylor Saraceno, Howard Weiner; University of California (San Francisco, CA): Jalayne Arias, Riley Bove, Refujia Gomez, Stephen Hauser, Roland Henry, Jill Hollenbach, Robin Lincoln, Jorge Oksenberg, Adam Renschen, Adam Santaniello. This work was presented in preliminary form at the 69th annual meeting of the American Academy of Neurology, Boston (April, 2017). This work was funded by the National Multiple Sclerosis Society and Biogen. The latter entity provided a medical accuracy review of this manuscript. The initial draft was prepared by the authors. The final draft was edited by, approved by, and fully under the control of the authors.
References
- 1.Ceccarelli A, Bakshi R, Neema M. MRI in multiple sclerosis: a review of the current literature. Curr Opin Neurol 2012;25:402–9. [DOI] [PubMed] [Google Scholar]
- 2.Kim G, Tauhid S, Dupuy SL, et al. An MRI-defined measure of cerebral lesion severity to assess therapeutic effects in multiple sclerosis. J Neurol 2016;263:531–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klawiter EC, Ceccarelli A, Arora A, et al. Corpus callosum atrophy correlates with gray matter atrophy in patients with multiple sclerosis. J Neuroimaging 2015;25:62–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tauhid S, Neema M, Healy BC, Weiner HL, Bakshi R. MRI phenotypes based on cerebral lesions and atrophy in patients with multiple sclerosis. J Neurol Sci 2014;346:250–4. [DOI] [PubMed] [Google Scholar]
- 5.Dell’Oglio E, Ceccarelli A, Glanz BI, et al. Quantification of global cerebral atrophy in multiple sclerosis from 3T MRI using SPM: The role of misclassification errors. J Neuroimaging 2015;25:191–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisniku LK, Brex PA, Altmann DR, et al. Disability and T2 MRI lesions: a 20-year follow-up of patients with relapse onset of multiple sclerosis. Brain 2008;131:808–17. [DOI] [PubMed] [Google Scholar]
- 7.Sormani MP, Arnold DL, De Stefano N, et al. Treatment effect on brain atrophy correlates with treatment effect on disability in multiple sclerosis. Ann Neurol 2014;75:43–9. [DOI] [PubMed] [Google Scholar]
- 8.Filippi M, Wolinsky JS, Comi G; CORAL Study Group. Effects of oral glatiramer acetate on clinical and MRI-monitored disease activity in patients with relapsing multiple sclerosis: a multicentre, double-blind, randomised, placebo-controlled study. Lancet Neurol 2006;5:213–20. [DOI] [PubMed] [Google Scholar]
- 9.Jovicich J, Czanner S, Han X, et al. MRI-derived measurements of human subcortical, ventricular and intracranial brain volumes: Reliability effects of scan sessions, acquisition sequences, data analyses, scanner upgrade, scanner vendors and field strengths. Neuroimage 2009;46:177–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu R, Tauhid S, Glanz BI, et al. Whole brain volume Measured from 1.5T versus 3T MRI in healthy subjects and patients with multiple sclerosis. J Neuroimaging 2016;26:62–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stankiewicz JM, Glanz BI, Healy BC, et al. Brain MRI lesion load at 1.5T and 3T versus clinical status in multiple sclerosis. J Neuroimaging 2011;21:e50–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leigh R, Ostuni J, Pham D, et al. Estimating cerebral atrophy in multiple sclerosis patients from various MR pulse sequences. Mult Scler 2002;8:420–9. [DOI] [PubMed] [Google Scholar]
- 13.Horsfield MA, Rovaris M, Rocca MA, et al. Whole brain atrophy in multiple sclerosis measured by two segmentation processes from various MRI sequences. J Neurol Sci 2003;216:169–77. [DOI] [PubMed] [Google Scholar]
- 14.Dupuy SL, Tauhid S, Kim G, Chu R, et al. MRI detection of hypointense brain lesions in patients with multiple sclerosis: T1 spin-echo vs. gradient-echo. Eur J Radiol 2015;84:1564–8. [DOI] [PubMed] [Google Scholar]
- 15.Biberacher V, Schmidt P, Keshavan A, et al. Intra- and interscanner variability of magnetic resonance imaging based volumetry in multiple sclerosis. Neuroimage 2016;142:188–97. [DOI] [PubMed] [Google Scholar]
- 16.Reid MW, Hannemann NP, York GE, et al. Comparing two processing pipelines to measure subcortical and cortical volumes in patients with and without mild traumatic brain injury. J Neuroimaging 2017;27:365–71. [DOI] [PubMed] [Google Scholar]
- 17.Ochs AL, Ross DE, Zannoni MD, Abildskov TJ, Bigler ED; Alzheimer’s Disease Neuroimaging Initiative. Comparison of automated brain volume measures obtained with NeuroQuant and FreeSurfer. J Neuroimaging 2015;25:721–7. [DOI] [PubMed] [Google Scholar]
- 18.Rajagopalan V, Pioro EP. Disparate voxel based morphometry (VBM) results between SPM and FSL softwares in ALS patients with frontotemporal dementia: which VBM results to consider? BMC Neurol 2015;15:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hemond CC, Chu R, Tummala S, Tauhid S, Healy BC, Bakshi R. Whole-brain atrophy assessed by proportional- versus registration-based pipelines from 3T MRI in multiple sclerosis. Brain Behav 2018;8:e01068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu R, Kim G, Tauhid S, Khalid F, Healy BC, Bakshi R. Whole brain and deep gray matter atrophy detection over 5 years with 3T MRI in multiple sclerosis using a variety of automated segmentation pipelines. PLoS One 2018;13:e0206939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gronenschild EH, Habets P, Jacobs HI, et al. The effects of FreeSurfer version, workstation type, and Macintosh operating system version on anatomical volume and cortical thickness measurements. PLoS One 2012;7:e38234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shinohara RT, Oh J, Nair G, et al. Volumetric analysis from a harmonized multisite brain MRI study of a single subject with multiple sclerosis. AJNR Am J Neuroradiol 2017;38:1501–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papinutto N, Bakshi R, Bischof A, et al. Gradient nonlinearity effects on upper cervical spinal cord area measurement from 3D T1 -weighted brain MRI acquisitions. Magn Reson Med 2018;79:1595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keshavan A, Paul F, Beyer MK, et al. Power estimation for non-standardized multisite studies. Neuroimage 2016;134:281–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones BC, Nair G, Shea CD, Crainiceanu CM, Cortese IC, Reich DS. Quantification of multiple-sclerosis-related brain atrophy in two heterogeneous MRI datasets using mixed-effects modeling. Neuroimage Clin 2013;3:171–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chua AS, Egorova S, Anderson MC, et al. Using multiple imputation to efficiently correct cerebral MRI whole brain lesion and atrophy data in patients with multiple sclerosis. Neuroimage 2015;119:81–8. [DOI] [PubMed] [Google Scholar]
- 27.Chua AS, Egorova S, Anderson MC, et al. Handling changes in MRI acquisition parameters in modeling whole brain lesion volume and atrophy data in multiple sclerosis subjects: Comparison of linear mixed-effect models. Neuroimage Clin 2015;8:606–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bove R, Chitnis T, Cree BA, et al. SUMMIT (Serially Unified Multicenter Multiple Sclerosis Investigation): creating a repository of deeply phenotyped contemporary multiple sclerosis cohorts. Mult Scler 2018;24:1485–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polman CH, Reingold SC, Banwell B,et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol 2011;69:292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurtzke JF. On the origin of EDSS. Mult Scler Relat Disord 2015;4:95–103. [DOI] [PubMed] [Google Scholar]
- 31.Lublin FD, Reingold SC, Cohen JA, et al. Defining the clinical course of multiple sclerosis: the 2013 revisions. Neurology 2014;83:278–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei X, Warfield SK, Zou KH, et al. Quantitative analysis of MRI signal abnormalities of brain white matter with high reproducibility and accuracy. J Magn Reson Imaging 2002;15:203–9. [DOI] [PubMed] [Google Scholar]
- 33.Wei X, Guttmann CR, Warfield SK, Eliasziw M, Mitchell JR. Has your patient’s multiple sclerosis lesion burden or brain atrophy actually changed? Mult Scler 2004;10:402–6. [DOI] [PubMed] [Google Scholar]
- 34.Sharma J, Sanfilipo MP, Benedict RHB, Weinstock-Guttman B, Munschauer FE, Bakshi R. Whole-brain atrophy in multiple sclerosis measured by automated versus semiautomated MR imaging segmentation. AJNR Am J Neuroradiol 2004;25:985–96. [PMC free article] [PubMed] [Google Scholar]
- 35.Eshaghi A, Marinescu RV, Young AL, et al. Progression of regional grey matter atrophy in multiple sclerosis. Brain 2018;141:1665–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uher T, Vaneckova M, Sobisek L, et al. Combining clinical and magnetic resonance imaging markers enhances prediction of 12-year disability in multiple sclerosis. Mult Scler 2017;23:51–61. [DOI] [PubMed] [Google Scholar]
- 37.Hemond CC, Healy BC, Tauhid S, et al. MRI phenotypes in MS: Longitudinal changes and miRNA signatures. Neurol Neuroimmunol Neuroinflamm 2019;6:e530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baecher-Allan C, Kaskow BJ, Weiner HL. Multiple sclerosis: mechanisms and immunotherapy. Neuron 2018;97:742–68. [DOI] [PubMed] [Google Scholar]
- 39.Zurawski J, Lassmann H, Bakshi R. Use of magnetic resonance imaging to visualize leptomeningeal inflammation in patients with multiple sclerosis: A Review. JAMA Neurol 2017;74:100–9. [DOI] [PubMed] [Google Scholar]
- 40.Tilling K, Lawton M, Robertson N, et al. Modelling disease progression in relapsing-remitting onset multiple sclerosis using multilevel models applied to longitudinal data from two natural history cohorts and one treated cohort. Health Technol Assess 2016;20:1–48. [DOI] [PMC free article] [PubMed] [Google Scholar]


