Abstract
Aims
The severe acute respiratory syndrome coronavirus 2, better known as COVID-19 has become the current health concern to the entire world. Initially appeared in Wuhan, China around December 2019, it had spread to almost 187 countries due to its high contagious nature. Precautionary measures remain the sole obliging tactic to cease the person to person transmissions till any effective method of treatment or vaccine is developed. Amidst the pandemic, research and development of new molecule is labour-intensive and tedious process. Drug repurposing is the concept of identifying therapeutically potent molecule from the library of pre-existing molecules.
Materials and methods
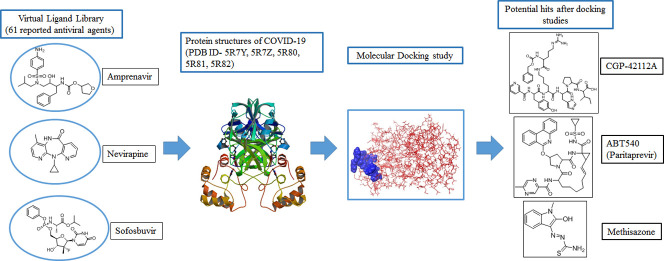
In the present study, 61 molecules that are already being used in clinics or under clinical scrutiny as antiviral agents are surveyed via docking study. Docking study was performed using Maestro interface (Schrödinger Suite, LLC, NY).
Key findings
Out of these 61 molecules, 37 molecules were found to interact with >2 protein structures of COVID-19. The docking results indicate that amongst the reported molecules, HIV protease inhibitors and RNA-dependent RNA polymerase inhibitors showed promising features of binding to COVID-19 enzyme. Along with these, Methisazone an inhibitor of protein synthesis, CGP42112A an angiotensin AT2 receptor agonist and ABT450 an inhibitor of the non-structural protein 3-4A might become convenient treatment option as well against COVID-19.
Significance
The drug repurposing approach provide an insight about the therapeutics that might be helpful in treating corona virus disease.
Keywords: Corona virus, COVID-19, SARS-CoV-2, MERS-CoV, Docking studies, Antiviral drugs
Graphical abstract
Highlights
-
•
Drug repurposing has become an emerging tactic to fight COVID-19.
-
•
Known antiviral agents used to study docking interaction with COVID-19 enzymes.
-
•
HIV protease inhibitors showed close interaction with COVID-19.
-
•
Methisazone, CGP42112A and ABT450 may become useful treatment against COVID-19.
1. Introduction
Novel corona virus disease (COVID-19) has become a pandemic threat to the public health. It is a respiratory malady causing fever, fatigue, dry cough, muscle aches, shortness of breath and some instances lead to pneumonia [1,2]. In serious conditions, it causes ARDS-Acute Respiratory Distress Syndrome i.e. a lung inflammation so severe that fluid builds up around and within the lungs which can cause septic shock due to dramatic fall in blood pressure and bodily organs are starved for oxygen. Incubation period of this corona virus is approximately 1 to 14 days. Symptoms and their severity vary from patient to patient. The elderly people, children below 6 years and patients with past medical history of asthma, diabetes, heart ailment are more vulnerable to this disease due to weaker or compromised immune systems. The epicenter of the outbreak was located in Wuhan, Hubei Province, China [2,3]. This outbreak was declared a Public Health Emergency of international concern on 30th January 2020 by WHO owing to is quick transmission with an estimated reproductive number (Ro) of 2.2. It has spread to nearly 187 countries worldwide with over 2,66,073 confirmed cases and over 11,184 confirmed deaths with a recorded case fatality rate (CFT) of 4.4 as of March 20, 2020 [4].
The causative agent for COVID-19 is SARS-CoV-2 (Severe acute respiratory syndrome coronavirus 2). Other similar agents previously known are Middle East respiratory syndrome (MERS) virus (MERS-CoV) and SARS-CoV [5,6]. They attack patient's lower respiratory system by invading the pulmonary epithelial cells, delivering their nucleocapsid and hijacking the cellular machinery to replicate in the cytoplasm. The virus family also affect heart, kidney, liver, gastrointestinal system and central nervous system. SARS-CoV-2 belongs to Coronaviridae family of enveloped single-stranded, positive-strand ribonucleic acid (RNA) structure. The structure of SARS-CoV-2 is in close resemblance to that of SARS-CoV. This SARS family contains 14 binding residues out of which 8 amino acids are specifically conserved for SARS-CoV-2. Importantly, the binding residues of this family interact with the ACE-2 (Angiotensin converting enzyme-2) directly [2,7].
Since the quick transmission of corona virus can be catastrophic for the entire world, the healthcare authorities have suggested certain preventive methods. Quarantining the infected patients, aggressive testing and rapid diagnosis of suspected victims, use of appropriate masks, frequent hand washing will help to counter and control the progression of this severe disease [8]. Currently, no drug or vaccine is available for coping this disease. Moreover, SARS-CoV-2 is far more contagious compared to other flu-viruses as one pre-symptomatic or asymptomatic person is capable to infect >2 healthy individuals. Researchers are now focusing on the repurpose strategy of existing drugs. Scientists working in this field have suggested the usage of some known broad-spectrum antiviral drugs like Nucleoside analogues and HIV-protease inhibitors as promising treatment methodology. RNA-dependent RNA polymerase (RdRp) and Angiotensin-converting enzyme 2 (ACE2) are also viable drug targets for COVID-19 treatment. Some antiviral drugs like Favinapir, Ritoavir, Oseltamivir, Lopinavir, Ganciclovir and Remdesivir are clinically tested against COVID-19 infection. Chloroquine, an antimalarial drug, has been proven to be effective in treatment of COVID-19 [2,9].
Until any accurate treatment methodology is available for COVID-19, the use of derivatives of previously known antiviral drugs is a useful strategy. In this study, docking studies were performed over binding pocket of COVID-19 to find the potential small molecule to combat life threatening corona virus disease.
2. Material and methods
2.1. Platform for molecular modelling
The computational investigations were performed using the Schrodinger software (Maestro 11.4, Schrodinger 2017-4).
2.2. Ligand preparation
Total 61 reported antiviral agents from the beginning of antiviral chemotherapy year 1960 to contemporary drugs in clinical trials were selected to perform the molecular docking studies to screen and identify the potent antiviral agents specifically for COVID-19 [10,11]. PubChem database was used to extract out the 3D chemical structures of the selected molecules. 3D and geometry optimizations with energy minimization of ligands were executed using algorithms monitored in Schrödinger Maestro v 11.4 [12]. LigPrep module (Schrodinger, LLC, NY, USA, 2009) was used from the Maestro builder panel to prepare ligand and generate 3D structure of the ligands by adding hydrogen atoms and removing salt and ionizing at pH (7 ± 2) [13]. Energy minimization was performed using OPLS_2005 force field by using the standard energy function of molecular mechanics and RMSD cut off 0.01 Ǻ to generate the low-energy ligand isomer [14].
2.3. Preparation of protein structures and grid generation
To combat the current situation of COVID-19 protein structure of COVID-19 main protease with co-crystallized structure (PDB IDs: 5R7Y, 5R7Z, 5R80, 5R81, 5R82, having resolution <2 Å, R-Value Free <0.30, R-Value Work <0.25) were selected and obtained from Protein Data Bank (http://www.rscb.org) with good resolutions [[15], [16], [17], [18], [19]]. Protein structure was prepared using protein preparation wizard in Maestro panel. During preparation of protein bond orders were assigned and hydrogen atoms were added as well. Water molecules were removed within 3 Ǻ of het groups [20]. Finally, OPLS-2005 force field was applied to minimize the structure of protein (Schrodinger, LLC, NY, USA, 2009) [21]. Further receptor grid boxes were generated using “Glide's Receptor Grid Generation” module at the active site (with the radius of 20 Å around the crystal structure) of co-crystallized ligand with the computing cubic box of 10 Ǻ × 10 Ǻ × 10 Ǻ [22].
2.4. Molecular docking
Molecular docking is a structure-based drug design approach to identify the essential amino acid interactions between the selected protein and generated ligands with low energy conformation [23]. Minimum interaction of the ligands characterized by the scoring function which used to foretell the binding affinity with the receptor. Glide Standard precision (SP), docking protocol was applied without smearing any constrain. Flexible docking with Glide Standard precision (SP) protocol was performed to predict the binding affinity and ligand efficiency as inhibitor of COVID-19 target [24]. Concluding energy assessment was done with the dock score. Visualization of docked ligands was done by Maestro interface (Schrödinger Suite, LLC, NY) [25].
where, a and b are co-efficient constant for vdW and Coul, respectively.
vdW = van der Waals energy; Coul = Coulomb energy; Hbond = Hydrogrn bonding with receptor; Metal = Binding with metal; Lipo = Constant term for lipophilic; BuryP = Buried polar group penalty; RotB = Rotatable bond penalty; Site = active site polar interaction [26].
3. Results and discussion
3.1. Docking studies
In order to find a potential candidate for treating COVID-19, molecular docking studies were performed over 61 antiviral agents on the binding pocket of enzyme COVID-19 (PDB ID: 5R7Y, 5R7Z, 5R80, 5R81 and 5R82). Some of these agents are marketed and some are under clinical development phase [27]. These molecules are active against different viral diseases viz. Hepatitis, HIV, Influenza, Herpes, Cytomegalovirus, Small Pox and Ebola virus. The mechanism of action of these compounds are also different like DNA polymerase inhibition, protease inhibition, inhibition of reverse-transcriptase, etc. The list of drugs tested for docking study is depicted in Table 1 .
Table 1.
List of antiviral agents docked against COVID-19 [27].
| No | Name | Structure | Active against | Mechanism |
|---|---|---|---|---|
| 1 | ABT450 | 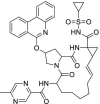 |
Hepatitis C | Inhibitor of the non-structural protein 3-4A (NS3/4A) serine protease |
| 2 | Acyclovir |  |
Cytomegalovirus infections | Inhibits viral DNA polymerase |
| 3 | Amprenavir |  |
HIV | Protease inhibitor |
| 4 | Arbidol (Umifenovir) | 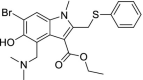 |
Influenza | Inhibits membrane fusion |
| 5 | Adefovir | 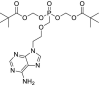 |
HIV | Reverse transcriptase inhibitor |
| 6 | Azidothimidine | 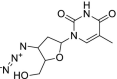 |
HIV | Inhibits reverse transcriptase |
| 7 | Asunaprevir | 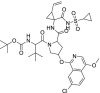 |
Hepatitis C | Protease inhibitor |
| 8 | Baricitinib |  |
Rheumatoid arthritis | Inhibits Janus kinase |
| 9 | Beclabuvir |  |
Hepatitis C | Inhibits non-structural protein 5B (NS5B) |
| 10 | Boceprevir |  |
Hepatitis C | NS3/4A protease inhibitor |
| 11 | Brivudin |  |
Herpes zoster | Blocks the action of DNA polymerases |
| 12 | Camostate |  |
Pancreatitis | Inhibits serine protease |
| 13 | Darunavir |  |
HIV | Inhibits HIV protease enzyme |
| 14 | Delavirdine |  |
HIV | Non-nucleoside reverse transcriptase inhibitor |
| 15 | Didanosine |  |
HIV | Nucleoside reverse transcriptase inhibitor |
| 16 | CGP42112A |  |
Vasodilation and blood pressure reduction | Angiotensin AT2 receptor agonist |
| 17 | Dasabuvir |  |
Hepatitis C | Inhibits the action of NS5B polymerase |
| 18 | Danoprevir |  |
Hepatitis C | NS3/4A protease inhibitor |
| 19 | Daclatasvir |  |
Hepatitis C | NS5A inhibitor |
| 20 | Elvitegravir | 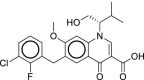 |
HIV | Integrase inhibitor |
| 21 | Favipiravir | 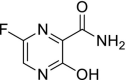 |
Influenza | Inhibits viral RNA-dependent RNA polymerase (RdRp) |
| 22 | Dihydroxy propyladenine | 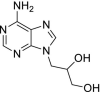 |
Herpes Virus | Inhibits viral replication |
| 23 | Efavirenz | 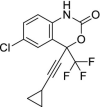 |
HIV | Inhibits non-nucleoside reverse transcriptase enzyme |
| 24 | Faldaprevir | 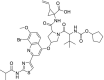 |
Hepatitis C | Protease inhibitor |
| 25 | Famciclovir | 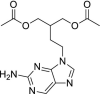 |
Hepatitis B | Inhibits viral DNA polymerase |
| 26 | Galidesivir | 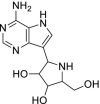 |
Ebola | RNA polymerase inhibitor |
| 27 | Entecavir | 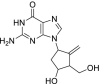 |
Hepatitis B | Inhibits reverse transcription |
| 28 | Elbasvir | 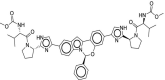 |
Hepatitis C | NS5A inhibitor |
| 29 | Ganciclovir | 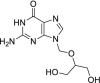 |
Cytomegalovirus | Inhibits viral DNA polymerases |
| 30 | Grazoprevir | 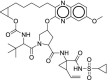 |
Hepatitis C | Blocks NS3 |
| 31 | Iodoxuridine | 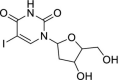 |
Herpes Simplex virus | Interferes viral DNA replication |
| 32 | Ritonavir | 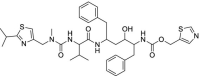 |
HIV | HIV protease inhibitor |
| 33 | Indinavir | 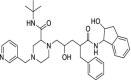 |
HIV | Protease inhibitor |
| 34 | Maraviroc | 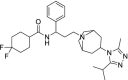 |
HIV | Negative allosteric modulator of the C—C chemokine receptor type 5 |
| 35 | Marboran/Methisazone | 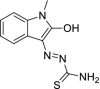 |
Small pox virus | Inhibits mRNA and protein synthesis |
| 36 | Lopinavir | 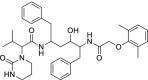 |
HIV | Protease inhibitor |
| 37 | Mericitabine | 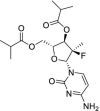 |
Hepatitis C | Inhibitor of RdRp |
| 38 | Nitrazoxanide | 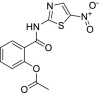 |
Broad-spectrum antiviral | Pyruvate:ferredoxin oxidoreductase (PFOR) enzyme |
| 39 | Radalbuvir | 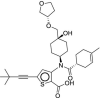 |
Hepatitis C | NS5B inhibitor |
| 40 | Remdesivir | 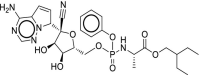 |
Ebola virus, Respiratory syncytial virus | Viral RNA polymerase |
| 41 | Raltegravir | 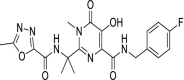 |
HIV | HIV-1 integrase inhibitor |
| 42 | Nevirapine | 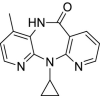 |
HIV | Non-nucleoside reverse transcriptase inhibitor |
| 43 | Foscarnet | 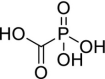 |
Herpes viruses, HIV, and hepatitis B virus | Inhibits the pyrophosphate binding site on viral DNA polymerases |
| 44 | Sequinavir |  |
HIV | Protease inhibitor |
| 45 | NSC306711 (Ferristatin II) |  |
Flavivirus | Degradation of Transferrin receptor- 1 |
| 46 | Amantadine |  |
Influenza A | Antagonism of the influenza virus A-M2 proton channel |
| 47 | Simeprevir |  |
Hepatitis C | Hepatitis C virus protease inhibitor |
| 48 | Ombitasvir | 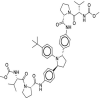 |
Hepatitis C | NS5A inhibitor |
| 49 | Sofosbuvir |  |
Hepatitis C | Inhibitor of viral RNA synthesis by inhibiting NS5B protein |
| 50 | Stavudine | 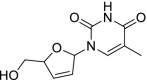 |
HIV | Inhibits HIV reverse transcriptase enzyme |
| 51 | Zanamivir | 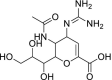 |
Influenza viruses | Neuraminidase inhibitor |
| 52 | Telbivudine | 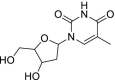 |
Hepatitis B | Reverse transcriptase inhibitor |
| 53 | Ravidasvir | 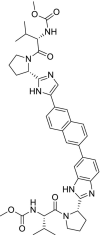 |
Hepatitis C | NS5A inhibitor |
| 54 | Zalcitabine | 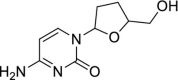 |
HIV | Nucleoside reverse transcriptase inhibitor |
| 55 | Tenofovir | 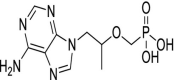 |
HIV | Inhibits the activity of HIV-1 reverse transcriptase |
| 56 | Telaprevir | 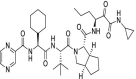 |
Hepatitis C | NS3/4a protease inhibitor |
| 57 | Velpatsvir |  |
Hepatitis C | NS5A protein inhibitor |
| 58 | Vedroprevir |  |
Hepatitis C | Inhibitor of the Hepatitis C virus (HCV) NS3 |
| 59 | Vaniprevir |  |
Hepatitis C | NS3/4A protease inhibitor |
| 60 | Uprifosbuvir |  |
Hepatitis C | NS5B polymerase inhibitor |
| 61 | Oseltamivir | 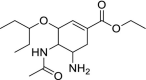 |
Influenza viruses A | Inhibits the neuraminidase enzyme |
All these 61 molecules were docked against the target enzyme COVID-19 and ranked based on their dock score. Compounds having dock score of −6.5 or less are considered better agent for inhibition of the COVID-19. A comparative analysis can be done by referring to Table 2 . This table represents the list of active molecules obtained after docking studies. These active molecules have dock score value of −6.5 or lower. Total 37 compounds showed binding interactions with different COVID-19 structures of PDB ID 5R7Y, 5R7Z, 5R80, 5R81 and 5R82.
Table 2.
Comparative docking study results on COVID-19 enzymes.
| Sr. No | Drug | Dock scorea |
||||
|---|---|---|---|---|---|---|
| 5R7Y | 5R7Z | 5R80 | 5R81 | 5R82 | ||
| 1. | NSC306711 (Ferristatin II) | −7.331 | – | – | −9.147 | – |
| 2. | Lopinavir | −6.834 | −6.968 | −7.331 | −8.44 | −7.58 |
| 3. | Elbasvir | – | – | – | −9.027 | – |
| 4. | Asunaprevir | – | −7.725 | – | −8.257 | −6.597 |
| 5. | Simeprevir | – | −7.778 | – | −7.784 | – |
| 6. | Remdesivir | – | −7.001 | −7.674 | – | −7.911 |
| 7. | CGP42112A | −7.108 | – | – | −7.521 | −7.243 |
| 8. | Indinavir | – | −7.058 | −7.157 | −6.834 | −6.796 |
| 9. | Ritonavir | −7.621 | – | −6.736 | −6.764 | −7.316 |
| 10. | ABT450 | – | −6.330 | – | −7.327 | −6.617 |
| 11. | Marboran/Methisazone | – | −7.542 | −6.829 | −6.928 | – |
| 12. | Galidesivir | −6.857 | – | – | – | −7.239 |
| 13. | Saquinavir | – | – | −7.227 | −7.632 | – |
| 14. | Baricitinib | – | – | −7.075 | −7.43 | – |
| 15. | Raltegravir | – | – | −7.057 | −7.257 | – |
| 16. | Delavirdine | – | −7.634 | – | – | – |
| 17. | Elvitegravir | – | −7.189 | – | – | – |
| 18. | Danoprevir | – | −6.956 | – | −6.83 | – |
| 19. | Galidesivir | – | −6.737 | −6.873 | −6.601 | – |
| 20. | Entecavir | −6.715 | −6.687 | – | – | – |
| 21. | Famciclovir | – | −6.521 | −6.965 | – | – |
| 22. | Uprifosbuvir | – | – | −7.558 | – | −6.805 |
| 23. | Oseltamivir | – | – | −6.891 | – | – |
| 24. | Azidothimidine | – | – | −6.873 | – | – |
| 25. | Sofosbuvir | – | – | −6.756 | – | −7.037 |
| 26. | Tenofovir | – | – | −6.644 | −6.739 | – |
| 27. | Mericitabine | – | – | −6.572 | – | – |
| 28. | Zanamivir | – | – | −6.548 | – | – |
| 29. | Didanosine | – | – | −6.526 | – | – |
| 30. | faldaprevir | – | – | – | −7.652 | −6.703 |
| 31. | Grazoprevir | – | – | – | −7.237 | – |
| 32. | Vedroprevir | – | – | – | −7.172 | – |
| 33. | Ravidasvir | – | – | – | −7.031 | – |
| 34. | Amprenavir | – | – | – | −6.583 | −6.744 |
| 35. | Efavirenz | – | – | – | −6.509 | −6.657 |
| 36. | Telaprevir | – | – | – | – | −7.083 |
| 37. | Daclatasvir | – | – | – | – | −6.881 |
– Indicates that dock score value is higher than −6.5.
Dock score value of −6.5 or lower is mentioned in table.
Out of these 37 molecules, Lopinavir, Asunaprevir, Remdesivir, CGP42112A, Indinavir, Ritonavir, ABT450, Marboran (Methisazone) and Galidesivir were found to interact with >2 protein structures of COVID-19. Importantly, amongst all the compounds anti-HIV drug Lopinavir binds to all the protein structures of COVID-19 with a dock score of less than −6.5. Moreover, Indinavir and Ritonavir also showed promising results in 4 out of 5 protein structures of COVID-19. Interestingly, all the HIV protease inhibitors showed excellent results in in silico docking studies.
Some of the HIV protease inhibitors are already tested for the infection of COVID-19. Lopinavir is more potent HIV protease inhibitor than ritonavir but it showed poor bioavalibility in vivo. Therefore a combination of these 2 drugs are tested against MERS-CoV and SARS-CoV. These drugs are found to be effective in silico, in vitro and in animal models but failed to impart effectiveness in clinical studies of COVID-19 patients [[28], [29], [30]]. An adenosine analog known as Remdesivir is a prodrug of Remdesivir-triphosphate. Mechanism of this drug is to inhibit RNA-dependent RNA polymerases (RdRps). It can terminate RNA synthesis by replacing ATP in the polymerization and thus known as chain terminator drug. Importantly, some studies suggested that ATP serves as the main substrate to NSP12 of SARS-CoV RdRp and SARS-CoV-2 RdRp shares 97% sequence similarity to SARS. Thus, Remdesivir can inhibit human corona viruses SARS-CoV, MERS-CoV, and SARS-CoV-2 strains that have ability to replicate in human epithelial cells [[30], [31], [32]].
Apart from this protease and RdRP inhibitors, researchers can focus on the other drugs like Methisazone, ABT450 and CGP42112A. Methisazone is an antiviral drug that works by inhibiting mRNA and protein synthesis in POX viruses [33] whereas CGP42112A is an angiotensin AT2 (Angiotensin receptor 2) receptor agonist that may alleviate the virus-induced lung injury [2]. ABT450 also known as Paritaprevir is an inhibitor of non-structural (NS) protein 3-4A serine protease for the treatment of hepatitis C [34].
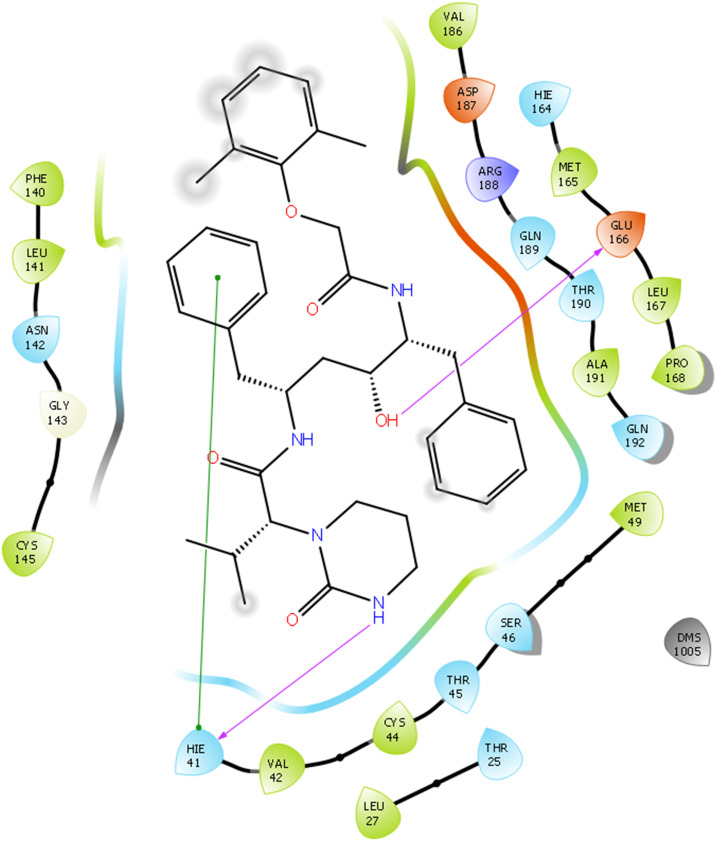
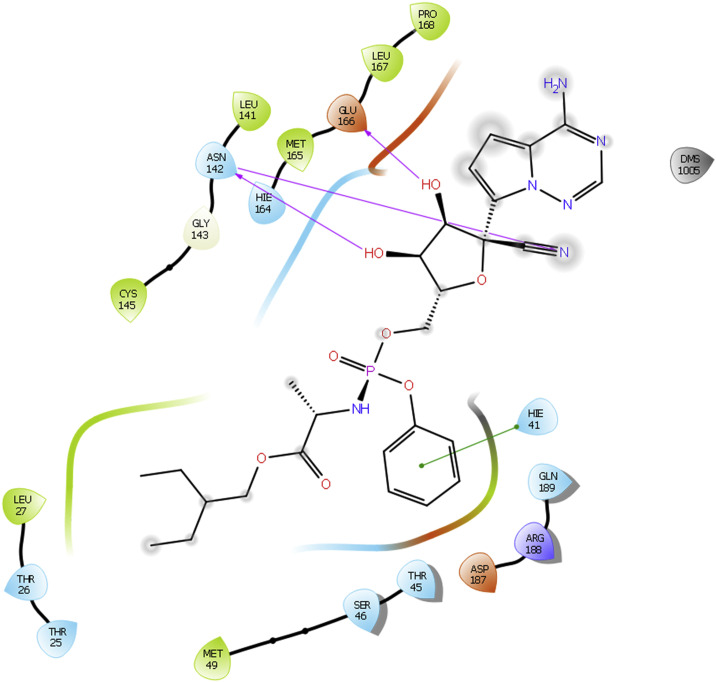
Docking interactions of some of active molecules based on docking studies are depicted in Fig. 1, Fig. 2, Fig. 3, Fig. 4 . The protease inhibitor Lopinavir interacted with all the protein structures of COVID-19. Binding interactions of Lopinavir with PDB ID 5R81 is presented in Fig. 1. Hydroxyl group of Lopinavir interacts by forming H-bond with amino acid Glu166. Furthermore, H-bond interaction is observed between amino acid Hie41 and ‘—NH’ group of tetrahydro pyrimidine ring scaffold. A pi-pi stacking interaction is also visible between amino acid Hie41 and phenyl ring. Although it can bind to the enzyme tightly, it is not proven effective in clinical studies. Whereas in case of Remdesivir, interactions with COVID-19 are same as that of Lopinavir but one additional strong H-bonding is observed for Remdesivir. Hydroxyl group of Remdesivir can form H-bond interaction with amino acid Glu166. Along with that, other hydroxyl groups have tight binding with amino acid Asn142. ‘N’ of cyano group also interacts with Asn142 by forming H-bonding. A phenyl group of Remdesivir forms pi-pi stacking interaction with amino acid Hie41. Based on our results, Remdesivir contains 5-cyano-3,4-dihydroxytetrahydrofuran ring scaffold which is responsible for enhanced binding affinity towards the COVID-19 enzymes and may be the reason for its good clinical activity against SARS CoV2.
Fig. 1.
Docking interactions of Lopinavir with 5R81.
Fig. 2.
Docking interactions of Remdesivir with 5R82.
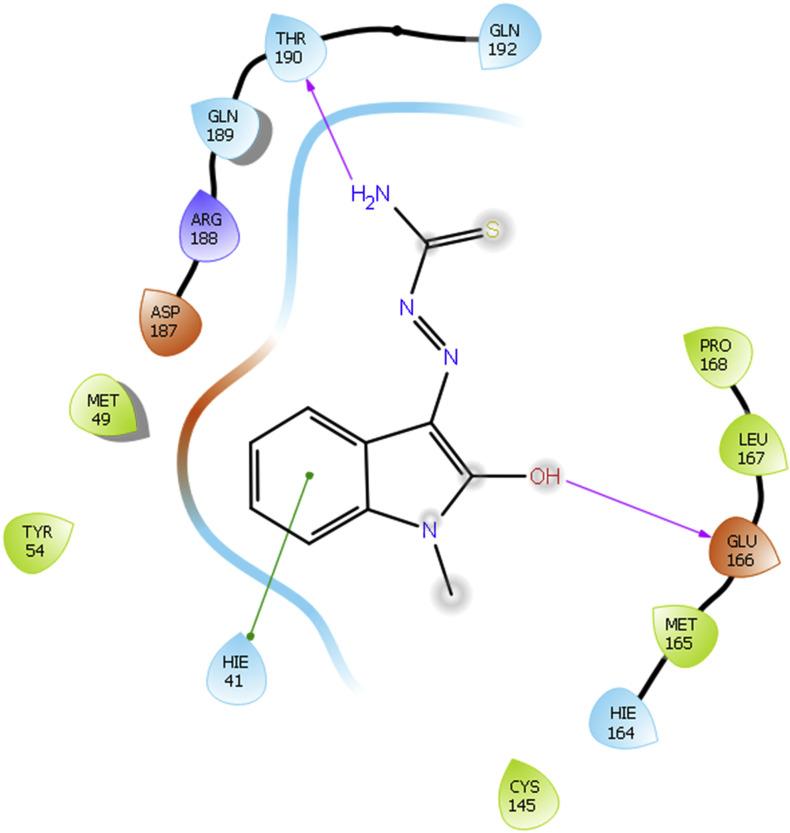
Fig. 3.
Docking interactions of Methisazone with 5R80.
Fig. 4.
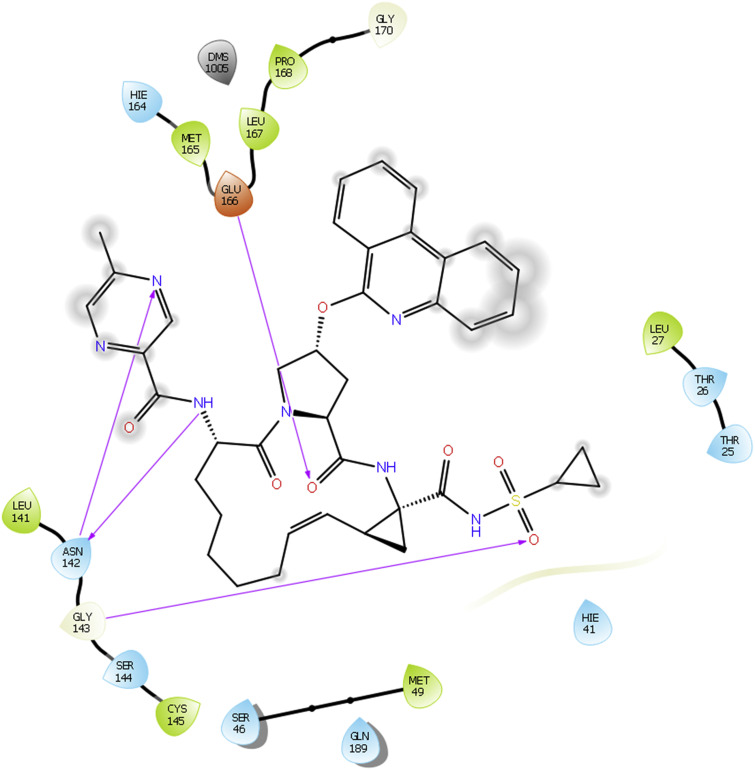
Docking interactions of ABT450 (Paritaprevir) with 5R82.
As Methisazone also showed higher binding score with COVID-19 enzyme, a representative binding image of Methisazone with 5R80 is given in Fig. 3.
Methisazone shows same binding interactions as that of Lopinavir. It interacts with protein structure of COVID-19 5R80. Hydroxyl group of Methisazone forms H-bonding interactions with amino acid Glu166. ‘—NH2’ functionality of Methisazone is responsible for H-bond formation with amino acid Thr190. Moreover, indole moiety of Methisazone is responsible for the pi-pi stacking interaction with amino acids Hie41.
Docking interaction diagram of one of the non-structural (NS) protein 3-4A serine protease inhibitor ABT450 is shown in Fig. 4 with PDB ID 5R82. ABT450 shows many H-bonding interactions with the enzyme same as that of Remdesivir. In this case, ‘NH’ of amide linkage and ‘N’ of pyrazine ring scaffold show H-bonding interactions with amino acid Asn142. Additionally, Glu166 forms H-bonding with ‘O’ of amide linkage. ‘O’ of sulfonyl group shows H-bond interaction with Gly143. These interactions may give rise to clinical potency of this compound against SARS CoV2.
Based on these docking results, it is estimated that compounds other than protease inhibitors can be beneficial as therapeutics for corona infection.
4. Conclusion
To combat the life-threatening corona virus infection, several studies are ongoing using antiviral drug therapies. HIV protease inhibitors have been suggested as one of the potential treatments of COVID-19. In this work, docking studies were performed on 61 molecules of known antiviral activities. In this study also many HIV protease inhibitors showed remarkable binding interactions with COVID-19 enzymes. These 4 protease inhibitors Lopinavir, Asunaprevir, Indinavir, and Ritonavir are found to be useful. Remdesivir acting on viral RNA polymerase also impart better activity in silico. Along with these, some new molecules emerged as COVID-19 inhibitors like Methisazone, ABT450 (Paritaprevir) and CGP42112A. Methisazone inhibits protein synthesis in POX virus and interacts with 5R7Z, 5R80 and 5R81 enzymes with dock score values of −7.542, −6.829 and −6.928, respectively. According to this study, angiotensin AT2 receptor will become a future drug target for COVID-19. One of the angiotensin AT2 receptors agonist CGP42112A shows potential characteristic of binding to COVID-19. The dock score values of CGP42112A with PDB ID 7R7Y, 5R81 and 5R82 are −7.108, −7.521 and −7.243, respectively. Additionally, a known drug treatment available for hepatitis C, Paritaprevir (ABT450) is found to be a useful candidate for corona disease. As per our study, along with protease inhibitors, researchers can also focus on the untouched drugs Methisazone, Paritaprevir and CGP42112A. These drugs may provide better drug therapies in the future.
Abbreviations
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- MERS
Middle East respiratory syndrome
- ACE-2
Angiotensin converting enzyme-2
- HIV
Human immunodeficiency virus
- NS3/4A
non-structural protein 3-4A
- RdRp
RNA-dependent RNA polymerase
Acknowledgments
Acknowledgements
Authors like to thank L.J. Institute of Pharmacy for research support. One of the authors would like to thank K. B. Institute of Pharmaceutical Education and Research for registration as research scholar.
Declaration of competing interest
The authors state no conflict of interest.
References
- 1.Hussin R., Siddappa N.B. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cynthia L., Qiongqiong Z., Yingzhu L. Research and Development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 2020 doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogoch A., Watts A., Thomas-Bachli C. Pneumonia of unknown etiology in Wuhan, China: potential for international spread via commercial air travel. J. Trav. Med. 2020 doi: 10.1093/jtm/taaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Organization WH . World Health Organization; 2020. Coronavirus Disease (COVID-2019) Situation Reports-61, 21 March 2020. [Google Scholar]
- 5.Elfiky Abdo A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248 doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wrapp D., NianShuang W., Kizzmekia S.C. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Sci. 2020 doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdul M.B., Areeba K., Usman A., Hira S. Evidence of the COVID-19 virus targeting the CNS: tissue distribution, host−virus interaction, and proposed neurotropic mechanisms. ACS Chem. Neurosci. 2020 doi: 10.1021/acschemneuro.0c00122. [DOI] [PubMed] [Google Scholar]
- 8.Catrin S., Sohrabi A.Z., Niamh O. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) Int. J. Surg. 2020;76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christian A.D., Jean-Marc R., Philippe C. New insights on the antiviral effects of chloroquine against coronavirus:what to expect for COVID-19? Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clercq E.D., Li G. Approved antiviral drugs over the past 50 years. Clin. Microbiol. Rev. 2016;29(3):695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.David M.J. Clinical trials of antiviral agents. J. Antimicrob. Chemother. 1992;29(2):97–103. doi: 10.1093/jac/29.2.97. [DOI] [PubMed] [Google Scholar]
- 12.Subramaniyan V., Palani M., Srinivasan P. Novel ligand-based docking; molecular dynamic simulations; and absorption, distribution, metabolism, and excretion approach to analyzing potential acetylcholinesterase inhibitors for Alzheimer's disease. J Pharm Anal. 2018;8:413–420. doi: 10.1016/j.jpha.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Modi P., Patel S., Chhabria M. Structure-based design, synthesis and biological evaluation of a newer series of pyrazolo [1, 5-a] pyrimidine analogues as potential anti-tubercular agents. Bioorg. Chem. 2019;87:240–251. doi: 10.1016/j.bioorg.2019.02.044. [DOI] [PubMed] [Google Scholar]
- 14.Pradeep H., Rajanikant G.K. A rational approach to selective pharmacophore designing: an innovative strategy for specific recognition of Gsk3b. Mol. Divers. 2012;16(3):553–562. doi: 10.1007/s11030-012-9387-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fearon D., Powell A.J., Douangamath A. Protein data Bank; 2020. PanDDA Analysis Group Deposition – Crystal Structure of COVID-19 Main Protease in Complex with Z45617795. [DOI] [Google Scholar]
- 16.Fearon D., Powell A.J., Douangamath A. Protein data Bank; 2020. PanDDA Analysis Group Deposition -- Crystal Structure of COVID-19 Main Protease in Complex with Z1220452176. [DOI] [Google Scholar]
- 17.Fearon D., Powell A.J., Douangamath A. Protein data Bank; 2020. PanDDA Analysis Group Deposition -- Crystal Structure of COVID-19 Main Protease in Complex with Z18197050. [DOI] [Google Scholar]
- 18.Fearon D., Powell A.J., Douangamath A. 2020. PanDDA Analysis Group Deposition -- Crystal Structure of COVID-19 Main Protease in Complex with Z1367324110, Protein Data Bank. [DOI] [Google Scholar]
- 19.Fearon D., Powell A.J., Douangamath A. Protein data Bank; 2020. PanDDA Analysis Group Deposition -- Crystal Structure of COVID-19 Main Protease in Complex with Z219104216. [DOI] [Google Scholar]
- 20.Sinha S., Patel S., Mohd A. Structure-based identification of novel sirtuin inhibitors against triple negative breast cancer: an in silico and in vitro study. Int. J. Biol. Macromol. 2019;140:454–468. doi: 10.1016/j.ijbiomac.2019.08.061. [DOI] [PubMed] [Google Scholar]
- 21.Halgren T.A., Murphy R.B., Friesner R.A. Glide: a new approach for rapid, accurate docking and scoring 2. Enrichment factors in database screening. J. Med. Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 22.Jasuja H., Chadha N., Kaur M., Silakari O. Dual inhibitors of Janus kinase 2 and 3 (JAK2/3): designing by pharmacophore- and docking-based virtual screening approach. Mol. Divers. 2014;18(2):253–267. doi: 10.1007/s11030-013-9497-z. [DOI] [PubMed] [Google Scholar]
- 23.Carlesso A., Chintha C., Gorman A.M. Merits and pitfalls of conventional and covalent docking in identifying new hydroxyl aryl aldehyde like compounds as human IRE1 inhibitors. Sci. Rep. 2019;9:3407. doi: 10.1038/s41598-019-39939-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hongbin H., Guigui Z., Yuquan Z. Reverse screening methods to search for the protein targets of chemopreventive compounds. Front. Chem. 2018;6:138. doi: 10.3389/fchem.2018.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maestro 11.4. Schrodinger, LLC; New York, NY: 2017. (4) [Google Scholar]
- 26.Bhuvaneshwari S., Sankaranarayanan K. Identification of potential CRAC channel inhibitors: pharmacophore mapping, 3D-QSAR modelling, and molecular docking approach. SAR QSAR Environ. Res. 2019;30(2):81–108. doi: 10.1080/1062936X.2019.1566172. [DOI] [PubMed] [Google Scholar]
- 27.Marrugo B., Loyd O. History and progress of antiviral drugs: from acyclovir to direct-acting antiviral agents (DAAs) for Hepatitis C. Medicina universitaria. 2015;17(68):165–174. [Google Scholar]
- 28.Bin C., Wang Y., Wen D., Liu Wen. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choy K., Wong A., Kaewpreedee P. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.T. Smith, J. Bushek, T. Prosser, COVID-19 Drug Therapy. Clinical Drug Information, Clinical Solutions.
- 31.Elfiky A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.L. Zhang, R. Zhou, Binding Mechanism of Remdesivir to SARS-CoV-2 RNA Dependent RNA Polymerase. Preprints. 10.20944/preprints202003.0267.v1. [DOI] [PubMed]
- 33.Yuriy P., V, Negrebetskaya E., Chernomaz A. Aromatic thiosemicarbazones, their antiviral action and interferon. 1. The decreasing of adenovirus type 1 resistance against interferon by methisazone in vitro. Биополимеры и клетка. 1996 [Google Scholar]
- 34.Ivan G., Borgia F., Riccardo Buonomo A. ABT-450: a novel protease inhibitor for the treatment of hepatitis C virus infection. Curr. Med. Chem. 2014;21(28):3261–3270. doi: 10.2174/0929867321666140706125950. [DOI] [PubMed] [Google Scholar]