Highlights
-
•
Validation studies of serologial antibody tests are needed.
-
•
Kinetics of SARS-CoV2 is a fundamental information in COVID-19 patients.
-
•
SARS-CoV-2 IgA response appears early and peaks at week 3.
Abstract
Validation studies of serological antibody tests must be properly designed for clinical, epidemiological and Public Health objectives such as confirmation of suspected COVID-19 cases, certification of seroconversion after infection, and epidemiological surveillance. We evaluated the kinetics of IgM, IgA and IgG SARS-CoV-2 antibodies in COVID-19 patients with confirmed (rRT-PCR) infection. We found that the IgA response appears and grows early, peaks at week 3, and it is stronger and more persistent than the IgM response. Further longitudinal investigations of virus-specific antibodies functions and of their protective efficacy over time are needed.
1. Introduction
Accurate diagnosis of COVID-19 is essential, not only to ensure appropriate patient care but also to facilitate identification of SARS-CoV-2 infected people, including asymptomatic carriers who need to be isolated to limit virus spread. Molecular testing to detect the SARS-CoV-2 RNA genome is widely employed to diagnose COVID-19 disease, asymptomatic infections and transmission chains [1]. However, there remains a great need for laboratory assays to measure antibody response and determine seroconversion. While such serological assays are not well suited to detect acute infections, they support a number of highly relevant applications. In fact, serological assays allows the study of immune response to SARS-CoV-2, and the identification of seroconversion; in addition, they may characterize COVID-19 course, and are essential for epidemiological studies and vaccine trials [2]. To provide “the right test at the right time” for the right target, the kinetics of the different antibody (Ab) isotypes production in COVID-19 patients must be thoroughly and preliminary investigated [3]. Aim of this paper is to describe the kinetics of SARS-CoV-2 IgA, and IgM in 19 COVID-19 patients using two different assays.
2. Methods
We used two different immunoassays to study the kinetics of SARS-CoV-2-specific antibodies (IgM, IgA, and IgG) for 6 weeks after the onset of symptoms (fever) in adult patients with confirmed (rRT-PCR) COVID-19. Tests were a chemiluminescent (CLIA) assay (MAGLUMI 2000 Plus), measuring SARS-CoV-2 specific IgM and IgG and an ELISA measuring specific IgG and IgA antibodies against SARS-CoV-2 (Euroimmun Medizinische Laboradiagnostika, Luebeck, Germany). Both assays have been performed according to the manufacturers’ instructions, as previously reported [4], [5]. The repeatability values (CV%) of CLIA assay for IgM are 3.06%, 1.84% and 4.05% at 0.61 kAU/L, 1.84 kAU/L and 4.39 kAU/L concentration levels, respectively; for IgG, CVs% are 5.69%, 3.86% and 3.18% at 0.48 kAU/L, 2.99 kAU/L and 10.59 kAU/L concentration levels, respectively. The SARS-CoV-2 IgM cut-off is 1.0 kAU/L, while for IgG the cut-off is 1.1 kAU/L [4]. The repeatability values (CV%) of ELISA for IgA range between 2.4% and 13.7% at a ratio of 1.03 and 0.20, respectively. For IgG, CVs % range between 3.9% and 16% at a ratio of 2.36 and 0.07, respectively. For both IgA and IgG the cut-off is ≥1.1. The study was submitted to the Ethical Committee of the University-Hospital of Padova (protocol number 23307).
3. Results
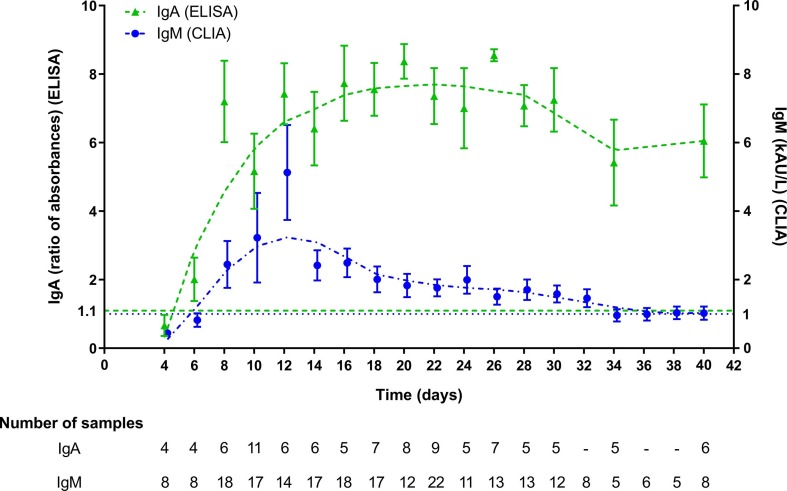
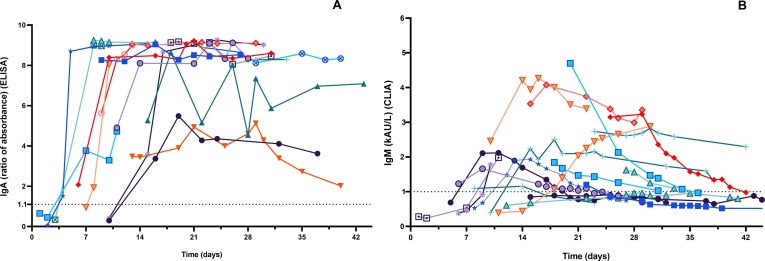
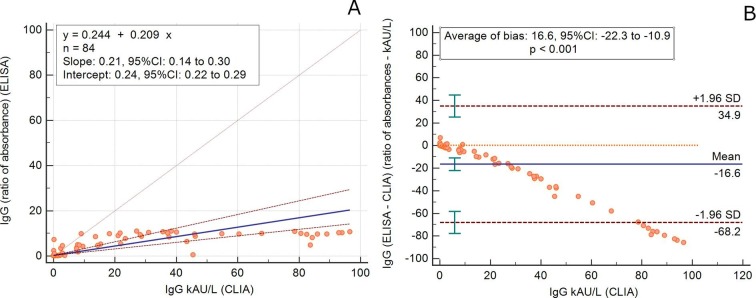
The kinetics of IgA-Abs were longitudinally tested in 19 patients (15 males, mean age 65.4 years, SD 14.5, range 22–81 y; 4 females, mean age 63.7 years, SD 7.8, range 53–70 y) for an average follow-up time of 7.5 days (SD 4.9). IgM-Abs kinetics was tested in 51 patients (37 males, men age 69.1 years, SD 13.5, range 22–89 y; 14 females, men age 62.6 years, SD 11.0, range 41–82 y) for 4.6 days (SD 4.0) (Fig. 1 ). Average levels of IgM and IgA antibodies increased since 6–8 days from the onset of COVID-19. Compared to IgM-Ab, IgA-Ab showed persistently higher levels for the whole observation period, with a peak level at 20–22 days. IgM-Ab levels peaked at 10–12 days and significantly declined after 18 days (Fig. 1). Fig. 2 shows the values of IgA-Ab and IgM-Ab in patients with more than 3 serial measurements (n = 18) that are heterogeneous in terms of onset and peak levels, but homogeneous for persistence. An IgA-Ab response to the S protein was detectable already in week 1 in 3/4 (75%) patients (Table 1 ). The values of IgG measured by the two assays was comparable and similar to the one already described with the same CLIA assay [4], being the clinical agreement 90.8% (number of patients = 84; Cohen’s K = 0.83; SE = 0.11) (Supplemental Fig. 1).
Fig. 1.
Kinetics of IgA (ELISA) and IgM (CLIA) of patients monitored from the onset of symptoms (fever).
Fig. 2.
Spaghetti plot of patients with more than 3 serial antibody determinations after the onset of symptoms (fever): A) IgA (n = 17 patients); B) IgM (n = 18 patients).
Table 1.
Descriptive statistics of IgA and IgM measurements, subdivided on the basis of each time point, up to 22–23 days (after the onset of fever).
| Time from the onset of fever | IgA (ratio of absorbances) | IgM (kAU/L) | |
|---|---|---|---|
| ≤5 days | n | 4 | 8 |
| Mean ± SD | 0.67 ± 0.62 | 0.44 ± 0.15 | |
| Median (IQR) | 0.55 (0.25–1.01) | 0.44(0.33–0.54) | |
| Min – Max | 0.05–1.52 | 0.24–0.69 | |
| n of Positive Tests (%) | 1/4 (25.0%) | 0/8 (0%) | |
| 6–7 days | n | 4 | 8 |
| Mean ± SD | 2.00 ± 1.27 | 0.82 ± 0.56 | |
| Median (IQR) | 1.64 (1.08–2.99) | 0.55 (0.48–1.07) | |
| Min-Max | 0.95–3.77 | 0.38–2.01 | |
| n of Positive Tests (%) | 3/4 (75.0%) | 2/8 (25.0%) | |
| 8–9 days | n | 6 | 18 |
| Mean ± SD | 7.20 ± 2.91 | 2.45 ± 2.90 | |
| Median (IQR) | 8.6 (5.63–9.21) | 1.02 (0.68–2.11) | |
| Min-Max | 1.93–9.25 | 0.38–8.57 | |
| n of Positive Tests (%) | 6/6 (100%) | 9/18 (50%) | |
| 10–11 days | n | 11 | 17 |
| Mean ± SD | 5.17 ± 3.64 | 3.22 ± 5.40 | |
| Median (IQR) | 4.90 (0.41–8.39) | 1.09 (0.7–2.12) | |
| Min-Max | 0.29–9.15 | 0.39–18.04 | |
| n of Positive Tests (%) | 8/11 (72.73%) | 9/17 (52.9%) | |
| 12–13 days | n | 6 | 14 |
| Mean ± SD | 7.42 ± 2.18 | 5.13 ± 5.18 | |
| Median (IQR) | 8.37 (6.29–9.00) | 2.81 (1.48–9.21) | |
| Min-Max | 3.48–9.06 | 0.44–16.03 | |
| n of Positive Tests (%) | 6/6 (100.0%) | 11/14 (78.6%) | |
| 14–15 days | n | 6 | 17 |
| Mean ± SD | 6.41 ± 2.63 | 2.41 ± 1.82 | |
| Median (IQR) | 6.68 (3.58–8.98) | 1.93 (1.04–3.53) | |
| Min-Max | 3.45–9.083 | 0.44–7.40 | |
| n of Positive Tests (%) | 6/6 (100.0%) | 13/17 (76.5%) | |
| 16–17 days | n | 5 | 18 |
| Mean ± SD | 7.74 ± 2.46 | 2.50 ± 1.78 | |
| Median (IQR) | 8.67 (8.48–9.04) | 1.86 (1.22–3.34) | |
| Min-Max | 3.37–9.13 | 0.75–7.69 | |
| n of Positive Tests (%) | 5/5 (100%) | 16/18 (88.9%) | |
| 18–19 days | n | 7 | 17 |
| Mean ± SD | 7.56 ± 2.04 | 2.01 ± 1.56 | |
| Median (IQR) | 8.29 (5.49–9.13) | 1.49 (1.12–2.09) | |
| Min-Max | 3.90–9.18 | 0.89–7.33 | |
| n of Positive Tests (%) | 7/7 (100%) | 16/17 (94.1%) | |
| 20–21 days | n | 8 | 12 |
| Mean ± SD | 8.38 ± 1.44 | 1.83 ± 1.18 | |
| Median (IQR) | 9.03 (8.30–9.09) | 1.36 (1.06–2.12) | |
| Min-Max | 4.94–9.22 | 0.78–4.70 | |
| n of Positive Tests (%) | 8/8 (100%) | 10/12 (83.3%) | |
| 22–23 days | n | 10 | 22 |
| Mean ± SD | 7.36 ± 2.60 | 1.76 ± 1.16 | |
| Median (IQR) | 8.70 (5.16–9.09) | 1.32 (0.92–2.26) | |
| Min-Max | 1.95–9.15 | 0.38–5.07 | |
| n of Positive Tests (%) | 10/10 (100%) | 15/22 (68.2%) |
n = number of samples obtained within the specified time period.
4. Discussion and conclusions
Detection of specific antibodies (IgM, IgA and IgG to SARS-CoV-2 spike protein) is useful to confirm SARS-CoV-2 infection in patients with PCR-positive COVID-19, essential in infected but asymptomatic subjects and in COVID-19 patients fist examined many week after the disease onset or in those with a low viral load. These serological assays are also essential to test the susceptibility or resistance to subsequent re-infection [6] and to perform epidemiological and surveillance studies. These tests are also being used to screen donor blood (convalescent plasma) to be transfused to patients with severe COVID-19 [7]. Timing at prescription is crucial for the interpretation of the test results and their rational and effective use for clinical decision. We have previously described the kinetics of IgM and IgG to SARS-CoV-2 using a chemiluminescent (CLIA) assay [4]. Here we show the peculiar characteristics of the kinetics of IgA antibodies in comparison to IgM, as well as their persistence over 38 days of follow-up from COVID-19 onset. The accuracy and reliability of serological methods is highly dependent from the choice of the targeted SARS-CoV-2 antigens and the assay format. The antigens used in the CLIA assay are the S-antigen and the N-protein, while the ELISA detects S1-specific IgA and IgG [3]. The differences observed between assays should be partially explained by differences in the targeted antigens. The spike (S) glycoprotein is densely glycosylated, with 66 N-linked glycosylation sites per trimer [8], a few of which only are the target of neutralizing antibodies [9]. Notwithstanding these fundamental differences, the IgG values measured with the two assays are comparable (Supplemental Figure). We acknowledge that we did not investigate any children and immunocompromised subjects, but only severely sick and adult patients; moreover, the follow-up period was not extended enough to properly test the antibody persistence over time. Nonetheless, given that the levels of spike-binding antibodies targeting S1 is highly correlated to those of neutralizing antibodies (NAbs), our results may be useful to design both, passive antibody therapy and vaccine development [10]. However, further longitudinal investigations of virus-specific antibodies functions and of their protective efficacy over time are needed.
Acknowledgement
We acknowledge the support of Euroimmun Medizinische Laboradiagnostika, Luebeck, Germany for kindly supplying the reagents without any influence in study design and data analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cca.2020.04.026.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary figure 1.
References
- 1.Mizumoto K., Kagaya K., Zarebski A., Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Euro Surveill. 2020;25(10) doi: 10.2807/1560-7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.T.F. Amana, T.H.O. Nguyen, V. Chromikova, S. Strohmeier, D. Stadblauer, Javier A et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. medRxiv http://doi.org/10.1101/2020.03.17.20037713. [DOI] [PMC free article] [PubMed]
- 3.Okba Nisreen M.A., Müller Marcel A., Li Wentao, Wang Chunyan, GeurtsvanKessel Corine H., Corman Victor M., Lamers Mart M., Sikkema Reina S., de Bruin Erwin, Chandler Felicity D., Yazdanpanah Yazdan, Le Hingrat Quentin, Descamps Diane, Houhou-Fidouh Nadhira, Reusken Chantal B.E.M., Bosch Berend-Jan, Drosten Christian, Koopmans Marion P.G., Haagmans Bart L. Severe Acute Respiratory Syndrome Coronavirus 2−Specific Antibody Responses in Coronavirus Disease 2019 Patients. Emerg. Infect. Dis. 2020;26(7) doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.A. Padoan, C. Cosma, L. Sciacovelli, D. Faggian, M. Plebani, Analytical performances of a chemiluminescence immunoassay for SARS-CoV-2 IgM/IgG and antibody kinetics. Clin. Chem. Lab. Med. 2020 Apr 16. pii:/j/cclm.ahead-of-print/cclm-2020-0443/cclm-2020-0443.xml. http://doi.org/10.1515/cclm-2020-0443. [Epub ahead of print]. [DOI] [PubMed]
- 5.R. Lassauniere, A. Frische, Z.B. Harboe, A.C.Y. Nielsen, A. Fomsgaard, K.A. Krogfelt et al. Evaluation of nine commercial SARS-CoV-2 immunoassays, medrxv http://doi.org/10.1101/2020.04.09.20056325.
- 6.L. Bao, W. Deng, H. Gao, C. Xiao, J. Liu, J. Xue et al. Reinfection could not occur in SARS-CoV-2 rhesus macaques. bioRxiv 2020:2020.03.13.990226. http://doi.org/10.1101/2020.03.13990226.
- 7.C. Shen, Z. Wang, F. Zhao, et al. Treatment of 5 Critically Ill Patients With COVID-19 With Convalescent Plasma, JAMA, Published online March 27, 2020. http://doi.org/10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed]
- 8.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Jet Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;S0092–8674:30262–30272. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.F. Wu, A. Wang, M. Liu, Q. Wang, J. Chen, S. Xia et al., Neutralising antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and they implications. medRxiv http://doi.org/10.1101/2020.03.30.20047365.