Abstract
The outbreak of coronavirus disease (COVID-19) caused by SARS-CoV-2 virus continually lead to worldwide human infections and deaths. Currently, there is no specific viral protein-targeted therapeutics. Viral nucleocapsid protein is a potential antiviral drug target, serving multiple critical functions during the viral life cycle. However, the structural information of SARS-CoV-2 nucleocapsid protein remains unclear. Herein, we have determined the 2.7 Å crystal structure of the N-terminal RNA binding domain of SARS-CoV-2 nucleocapsid protein. Although the overall structure is similar as other reported coronavirus nucleocapsid protein N-terminal domain, the surface electrostatic potential characteristics between them are distinct. Further comparison with mild virus type HCoV-OC43 equivalent domain demonstrates a unique potential RNA binding pocket alongside the β-sheet core. Complemented by in vitro binding studies, our data provide several atomic resolution features of SARS-CoV-2 nucleocapsid protein N-terminal domain, guiding the design of novel antiviral agents specific targeting to SARS-CoV-2.
Key words: COVID-19, Coronavirus, SARS-CoV-2, Nucleocapsid protein, RNA binding domain, Crystal structure, Antiviral targeting site
Graphical abstract
The crystal structure of the N-terminal RNA-binding domain of SARS-CoV-2 nucleocapsid protein was determined by X-ray. Compared with other previously reported coronavirus nucleocapsid protein N-terminal domains, the authors have identified a unique potential RNA-binding pocket that can guide the design of novel antiviral drugs targeting SARS-CoV-2.
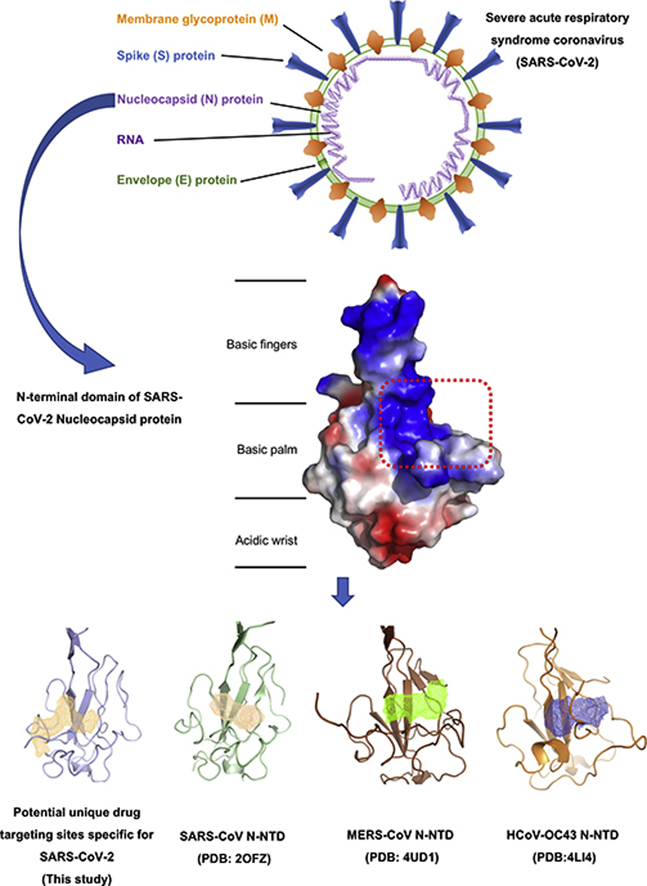
1. Introduction
The ongoing outbreak of coronavirus disease 2019 (COVID-19) is a new emerging human infectious disease caused by a novel coronavirus (severe acute respiratory syndrome coronavirus 2, SARS-CoV-2, previously known as 2019-nCoV). As of 12 March 2020, the novel coronavirus SARS-CoV-2 has spread to 118 countries and region. According to data from World Health Organization (WHO), the global confirmed case count of COVID-19 was 125,260, including 4613 deaths. The number of COVID-19 cases continues to soar, posing dramatic threat to health care system worldwide (COVID-2019 situation reports, World Health Organization, 12 March 2020)1. Despite remarkable efforts on containing the spread of the virus, there is no specific targeted therapeutic yet.
SARS-CoV-2 is a betacoronavirus with single-stranded RNA genomes, like MERS-CoV and SARS-CoV. The first two-thirds of the viral 30 kb RNA genome, mainly named as ORF1a/b region, translates into two polyproteins (pp1a and pp1ab) and encodes most of the non-structural proteins (nsp). The rest parts of the virus genome encode accessory proteins and four essential structural proteins, including spike (S) glycoprotein, small envelope (E) protein, matrix (M) protein, and nucleocapsid (N) protein2, 3, 4. Current antiviral drugs developed to treat coronavirus (CoV) infections primarily target S protein, the 3C-like (3CL), and papain-like (PLP) proteases5,6. Because mutant viruses in the S protein are prone to escape the targeted therapeutic with different host-cell receptor binding patterns6, there are limitations on targeting S protein for antiviral approaches. Antiviral protease inhibitors may nonspecifically act on the cellular homologous protease, resulting in host cell toxicity and severe side effects. Therefore, novel antiviral strategies are needed to combat acute respiratory infections caused by this novel coronavirus SARS-CoV-2.
The CoV N protein is a multifunctional RNA-binding protein necessary for viral RNA transcription and replication. It plays many pivotal roles in forming helical ribonucleoproteins during the packaging of the viral RNA genome, regulating viral RNA synthesis in replication/transcription, and modulating infected cell metabolism7, 8, 9. The primary functions of N protein are binding to the viral RNA genome and packing them into a long helical nucleocapsid structure or ribonucleoprotein (RNP) complex10,11. In vitro and in vivo experiments revealed that N protein bound to leader RNA, and maintained highly ordered RNA conformation suitable for replicating, and transcribing the viral genome8,12. More studies implicated that N protein regulated host–pathogen interactions, such as actin reorganization, host cell cycle progression, and apoptosis13, 14, 15. The N protein is also a highly immunogenic and abundantly expressed protein during infection, capable of inducing protective immune responses against SARS-CoV and SARS-CoV-216, 17, 18, 19.
The common domain architectures of coronavirus N protein consists of three distinct but highly conserved parts: an N-terminal RNA-binding domain (NTD), a C-terminal dimerization domain (CTD), and an intrinsically disordered central Ser/Arg (SR)-rich linker. Previous studies have revealed that the NTD is responsible for RNA binding, CTD for oligomerization, and (SR)-rich linker for primary phosphorylation, respectively20, 21, 22, 23, 24. The crystal structures of SARS-CoV N-NTD25, infectious bronchitis virus (IBV) N-NTD26,27, HCoV-OC43 N-NTD21, and mouse hepatitis virus (MHV) N-NTD28 have been solved. The CoVs N-NTD has been found to associate with the 3′ end of the viral RNA genome, possibly through electrostatic interactions. Additionally, several critical residues have been identified for RNA binding and virus infectivity in the N-terminal domain of coronavirus N proteins25,28, 29, 30. However, the structural and mechanistic basis for newly emerged novel SARS-CoV-2 N protein remains mostly unknown. Understanding these aspects will facilitate the discovery of agents that specifically block the coronavirus replication, transcription, and viral assembly31.
In this study, we report the crystal structure of SARS-CoV-2 nucleocapsid N-terminal domain (termed as SARS-CoV-2 N-NTD) as a model for understanding the molecular interactions that govern SARS-CoV-2 N-NTD binding to ribonucleotides. Compared with other solved CoVs N-NTD, we characterized the specificity surface electrostatic potential features of SARS-CoV-2 N-NTD. Additionally, we further demonstrated the potential unique nucleotide-binding pocket characteristics. Our findings will aid in the development of new drugs that interfere with viral N protein in SARS-CoV-2.
2. Materials and methods
2.1. Cloning, expression and purification
The SARS-CoV-2 N-FL plasmid was a gift from Guangdong Medical Laboratory Animal Center (Guangzhou, China). We designed several constructs, including SARS-CoV-2 N-FL (residues from 1 to 419), SARS-CoV-2 N-NTD domain (residues from 41 to 174), and SARS-CoV-2 N-NTD domain (residues from 33 to 180) depending on secondary structure predictions and sequence conservation characteristics. The constructs were cloned into the pRSF-Duet-1 vector with N-terminal His-SUMO tag and expressed in E. coli strain Rosetta. SARS-CoV-2 N-NTD (residues from 41 to 174, termed as SARS-CoV-2 N-NTD in the main text) was induced with 0.1 mmol/L isopropylthio-β-galactoside (IPTG) and incubated overnight at 16 °C in the Terrific Broth media (Sangon Biotech, Shanghai, China). After Nickel column chromatography followed by ulp1 protease digestion for tag removal, SARS-CoV-2 N-NTD proteins were further purified via size-exclusion chromatography (with the buffer consisting of 20 mmol/L Tris-HCl (pH 8.0), 150 mmol/L sodium chloride, 1 mmol/L dithiothreitol), and then concentrated by ultrafiltration to a final concentration of 22 mg/mL.
2.2. Crystallization and data collection
Crystals were grown from a solution containing 10 mmol/L sodium acetate, 50 mmol/L sodium cacodylate (pH 6.5), 13% PEG8000 (Sigma–Aldrich, St. Louis, MO, USA) by the hanging drop vapor diffusion method at 16 °C. Crystals were frozen in liquid nitrogen in reservoir solutions supplemented with 15% glycerol (v/v) as a cryoprotectant. X-ray diffraction data were collected at the South China Sea Institute of Oceanology, Chinese Academy of Sciences (Guangzhou, China) by Rigaku X-ray diffraction (XRD) instrument XtaLAB P200 007HF. The structure was solved by molecular replacement with the searching model (PDB: 2OG3)25 using PHENIX software suite32. The X-ray diffraction and structure refinement statistics are summarized in Table 1.
Table 1.
Data collection and refinement statistics.
| Item | SARS-CoV-2 N-NTDa |
|---|---|
| Protein Data Bank code | 6M3M |
| Wavelength (Å) | 1.5418 |
| Resolution range | 20.92–2.7 (2.796–2.7) |
| Space group | P 21 21 21 |
| Unit cell | |
| a, b, c (Å) | 58.88, 92.68, 97.32 |
| α, β, γ (°) | 90, 90, 90 |
| Total reflections | 98,913 (10,077) |
| Unique reflections | 15,133 (1481) |
| Multiplicity | 6.5 (6.8) |
| Completeness (%) | 99.55 (99.80) |
| Mean I/sigma (I) | 14.97 (2.9) |
| Wilson B-factor | 33.94 |
| R-mergeb | 0.1043 (0.3172) |
| R-measc | 0.1135 (0.3436) |
| R-pimd | 0.04388 (0.1303) |
| CC1/2 | 0.991 (0.96) |
| CC | 0.998 (0.99) |
| Reflections used in refinement | 15,126 (1481) |
| Reflections used for R-free | 1514 (138) |
| R-worke | 0.2578 (0.3551) |
| R-freef | 0.2934 (0.4058) |
| CC (work) | 0.908 (0.692) |
| CC (free) | 0.851 (0.635) |
| Number of non-hydrogen atoms | 3952 |
| Macromolecules | 3822 |
| Solvent | 130 |
| Protein residues | 499 |
| RMS (bonds) (Å) | 0.004 |
| RMS (angles) (°) | 0.72 |
| Ramachandran favored (%) | 96.48 |
| Ramachandran allowed (%) | 3.52 |
| Ramachandran outliers (%) | 0.00 |
| Rotamer outliers (%) | 0.00 |
| Clashscore | 11.15 |
| Average B-factor | 31.60 |
| Macromolecules | 31.86 |
| Solvent | 24.03 |
Statistics for the highest-resolution shell are shown in parentheses.
R-merge=∑hkl∑i|Ii(hkl)–<I(hkl)>|/∑hkl∑iIi(hkl), where Ii(hkl) is the intensity measured for the i th reflection and < I(hkl) > is the average intensity of all reflections with indices hkl.
R-work=∑hkl||Fobs(hkl)|–|Fcalc(hkl)||/∑hkl|Fobs(hkl)|.
R-free is calculated in an identical manner using 10% of randomly selected reflections that were not included in the refinement.
2.3. Surface plasmon resonance analysis
Surface plasmon resonance (SPR) analysis was performed using a Biacore T200 with the CM5 sensor chip (GE Healthcare Life Sciences, Pittsburgh, PA, USA) at room temperature (25 °C). SARS-CoV-2 N-NTD was exchanged into PBS buffer via gel-filtration. The CM5 chip surface was activated for 10 min using 0.2 mol/L EDC/0.05 mol/L NHS. After the injection of 30 μg/mL protein in 10 mmol/L sodium acetate (pH 5.5) for three times to immobilize on one of channels of the CM5 chip up to ∼5800 response units, 10 μL of 1 mol/L ethanolamine (pH 8.0) was used for blocking the remaining activated groups. Each of the analytes (AMP, GMP, UMP, and CMP) was dissolved in PBS (pH 7.4, 0.05% NP-20) and flowed through the chip surface at a flow rate of 30 μL/min at 25 °C. Analytes (30 μL) were injected for affinity analysis with 60 s dissociation time. To understanding the dose-dependent affinity of analytes and SARS-CoV-2 N-NTD, we tested nine dilutions of analytes from 0.15625 to 10 mmol/L. A blank flow channel was used as a reference channel to correct the bulk refractive index by subtracting the response signal of the reference channel from the signals of protein immobilized cells. The dissociation constant (KD) for analytes binding to SARS-CoV-2 N-NTD was determined from the association and dissociation curves of the sensorgrams, using the BIA evaluation program (Biacore).
2.4. Biolayer interferometry assays
Biolayer interferometry assays (BLI) experiments were performed using an Octet RED96e instrument from ForteBio (Fremont, CA, USA). All assays were run at 25 °C with continuous 100 rpm shaking. PBS with 0.01% Tween-20 used as the assay buffer. Biotinylated SARS-CoV-2 N-NTD proteins were tethered on super streptavidin (SSA) biosensors (ForteBio) by dipping sensors into 100 μg/mL protein solution. Average saturation response levels of 10–15 nm were achieved in 15 min for all samples. Sensors with protein tethered were washed in assay buffer for 10 min to eliminate nonspecifically bound protein molecules, and establish stable baselines before starting association–dissociation cycles with AMP/GMP/UMP/CMP. Raw kinetic data collected were processed in the data analysis software provided by the manufacturer using double reference subtraction in which both 0.01% Tween-20 only reference and inactive reference were subtracted. Resulting data were analyzed based on the 1:1 binding model from which Kon and Koff values were obtained, and then KD values were calculated.
3. Results
3.1. Sequence features of SARS-CoV-2 nucleocapsid protein
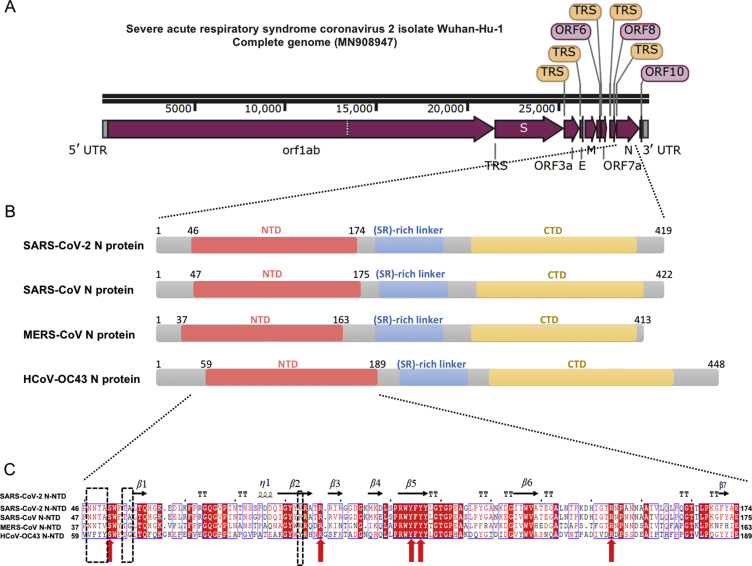
It has been reported that the complete genome of SARS-CoV-2 (GenBank: MN908947, Wuhan-Hu-1 coronavirus) is 29.9 kb in length, similarly to 27.9 kb SARS-CoV and 30.1 kb MERS-CoV genome33,34 (Fig. 1A). Nucleocapsid (N) protein is translated from the 3′ end structural ORF35, 36, 37. According to Virus Variation Resource in National Center for Biotechnology Information databank38, SARS-CoV-2 N protein-encoding regions are conserved among the known NCBI 103 genome datasets. Only a few variations (S194L in virus strain Foshan/20SF207/2020, K249I in virus strain Wuhan/IVDC-HB-envF13-21/2020, P344S in virus strain Guangzhou/20SF206/2020) in N protein are found in public genomic epidemiology. Overall domain architecture of N protein among four coronaviruses (SARS-CoV-2, SARS-CoV, MERS-CoV, and HCoV-OC43) is shown in Fig. 1B, which indicates that SARS-CoV-2 shares typical characteristics with other coronaviruses. Zooming into the completed genomic sequence of SARS-CoV-2 N protein-encoding region, we found that the sequence identities between SARS-CoV-2 with SARS-CoV, MERS-CoV, and HCoV-OC43 were 89.74%, 48.59%, 35.62%, respectively (Supporting Information Fig. S1)39,40. Since full-length SARS-CoV-2 N protein aggregated status were found in our expression and purification studies (Supporting Information Fig. S2), as well as previously reported data on other coronavirus nucleocapsid protein, we next investigated the structural studies on N-terminal region of SARS-CoV-2 N protein (termed as SARS-CoV-2 N-NTD).
Figure 1.
Sequence features of SARS-CoV-2 nucleocapsid protein. (A) The complete whole genomic features of SARS-CoV-2 isolate Wuhan-Hu-1 (Genebank: MN908947). UTR: untranslated region; orf/ORF: open reading frame; TRS: transcriptional regulatory sequences; S: spike glycoprotein encoding region; E: envelope protein encoding region; M: membrane protein encoding region; N: nucleocapsid protein encoding region. The figure is illustrated by SnapGene Viewer. (B) Domain architectures of coronavirus nucleocapsid protein. NTD: N-terminal RNA-binding domain; CTD: C-terminal dimerization domain. (C) Multiple sequence alignment of SARS-CoV-2 N-NTD with SARS-CoV N-NTD (UniProtKB: P59595), MERS-CoV N-NTD (UniProtKB: R9UM87), HCoV-OC43 N-NTD (UniProtKB: P33469). Red arrows indicate conserved residues for ribonucleotide binding sites, and dash boxes indicate residues variably in the structural comparisons.
3.2. Crystal structure of SARS-CoV-2 N-NTD
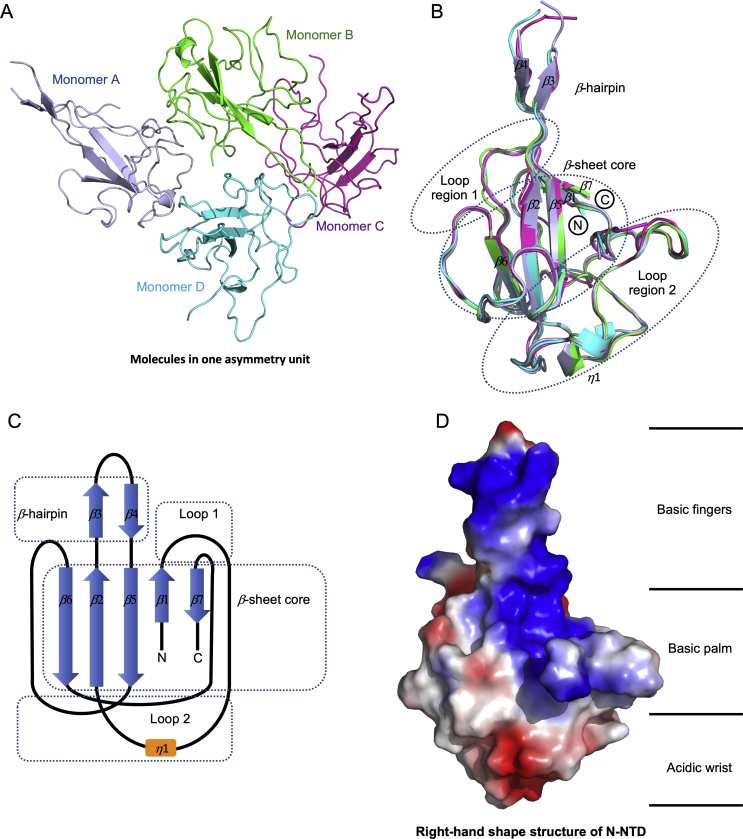
In order to obtain the atomic information of SARS-CoV-2 N-NTD, we solved the structure at 2.7 Å resolution using X-ray crystallography technology. Briefly, 47–173 residues of SARS-CoV-2 N protein were constructed, expressed, and purified as described protocol (Section 2 Materials and methods). The structure of SARS-CoV-2 N-NTD was determined by molecular replacement using the SARS-CoV N-NTD structure (PDB:2OfG3) as the search model25. The final structure was refined to R-factor and R-free values of 0.26 and 0.29, respectively. The complete statistics for data collection, phasing and refinement are presented in Table 1. Unlike to SARS-CoV N-NTD crystals packing modes (monoclinic form at 2OFZ, cubic form at 2OG3)25, SARS-CoV-2 N-NTD crystal shows orthorhombic crystal packing form with four N-NTD monomers in one asymmetry unit (Fig. 2A). In a previous study, Saikatendu et al.25 found that SARS-CoV N-NTD had two different packing modes in distinct crystal forms. The symmetry molecules in the monoclinic crystal form pack in a head-to-head linear 3D array, with most of the interfacial interactions being made by residues of the positively charged β-hairpin (Supporting Information Fig. S3A). In the cubic crystal form, the SARS-CoV N-NTD packs in a helical tubules array (Fig. S3B). In our study, SARS-CoV-2 N-NTD packs into orthorhombic crystal form, where the interfacial interactions are formed by residues of β-hairpin fingers and palm regions (Fig. S3C). Although evidence for real RNP organization in the mature virions is lacking, the differences in the crystal packing patterns may implicate other potential contacts in SARS-CoV-2 RNP formation process. All four monomers in one asymmetric unit of the SARS-CoV-2 N-NTD crystal structure shared similar right-handed (loops)-(β-sheet core)-(loops) sandwiched fold, as conserved among the CoVs N-NTD (Fig. 2B). The β-sheet core consists of five antiparallel β-strands with a single short 310 helix just before strand β2, and a protruding β-hairpin between strands β2 and β5. The β-hairpin is functionally essential for CoV N-NTD, implicated in mutational analysis of amino acid residues for RNA binding30 (Fig. 2C). The SARS-CoV-2 N-NTD is enriched in aromatic and basic residues, folding into a right-hand shape, which resembles with a protruded basic finger, a basic palm, and an acidic wrist (Fig. 2D).
Figure 2.
Structural overview of SARS-CoV-2 N-NTD. (A) Ribbon representation of SARS-CoV-2 N-NTD molecules in one asymmetric unit. The four molecules are highlighted with different colors, respectively. (B) Superimpositions of four molecules in one asymmetric unit. The dash circles indicate the sandwiched structure composed of Loop region 1, β-sheet core, and Loop region 2. The β-strand is labeled with β1 to β7, and the 310 helix is labeled with 1. (C) Topological style illustration of SARS-CoV-2 N-NTD structure. (D) Electrostatic surface of the SARS-CoV-2 N-NTD. Blue denotes positive charge potential, while red indicates negative charge potential. The potential distribution was calculated by Pymol. The values range from –6kT (red) to 0 (white) and to +6kT (blue), where k is the Boltzmann constant, and T is the temperature.
3.3. Comparison of SARS-CoV-2 N-NTD with related viral N-NTD structures
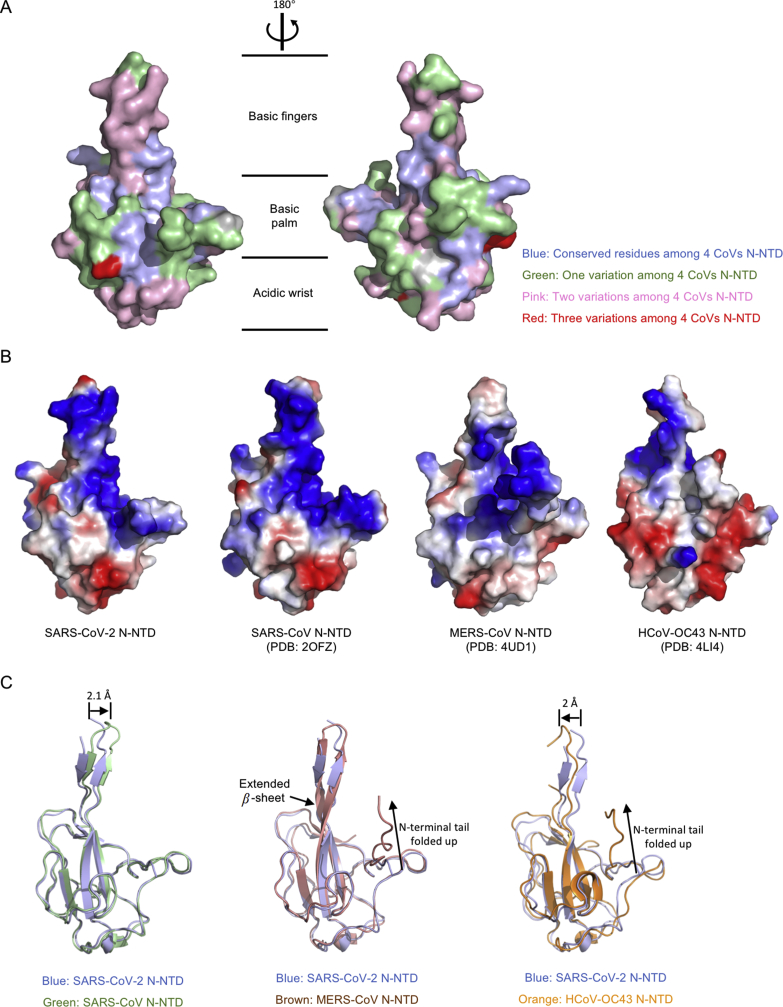
To obtain more specific information, we first mapped the conserved residues between SARS-CoV-2 N-NTD with SARS-CoV N-NTD, MERS-CoV N-NTD, and HCoV-OC443 N-NTD, respectively (Fig. 3A). The most conserved residues distribute on the basic palm region (Fig. 3A, blue and green area), while the less conserved residues locate in basic fingers and acidic wrist regions (Fig. 3A, pink and red area). The available CoVs N-NTD crystal structures allowed us to compare the electrostatic potential on the surface. As shown in Fig. 3B, although CoV N-NTDs all adapt similar overall organizations, the surface charge distribution patterns are different. Consistently with our observations, the previous modeling of related coronaviral N-NTDs also showed markedly differ in surface charge distributions25. The superimposition of SARS-CoV-2 N-NTD with three kinds of CoVs N-NTD is shown in Fig. 3C. Compared with SARS-CoV N-NTD, SARS-CoV-2 N-NTD shows a 2.1 Å movement in the β-hairpin region forward to nucleotide binding site (Fig. 3C, left panel). While compared with MERS-CoV N-NTD, SARS-CoV-2 N-NTD shows a less extended β-hairpin region, and a distinct relax N-terminal tail (Fig. 3C, middle panel). Inconsistently, SARS-CoV-2 N-NTD shows a distinct loosen N-terminal tail, and a 2 Å movement in the β-hairpin region backward to the opposite side of nucleotide binding site when the structure is compared with HCoV-OC43 N-NTD (Fig. 3C, right panel). These differences dramatically change the surface characterizations of the protein, which may result in the RNA-binding cleft being adaptive to its RNA genome.
Figure 3.
Comparison of SARS-CoV-2 N-NTD with related viral N-NTD structures. (A) Mapping the conserved surfaces of four CoV N-NTDs in SARS-CoV-2 N-NTD structure. The multiple sequence alignment used for mapping is shown in Fig. 1C. Blue denotes conserved residues among 4 CoVs N-NTD; green denotes one variation among 4 CoVs N-NTD; pink denotes two variations among 4 CoVs N-NTD; red denotes three variations among 4 CoVs N-NTD. (B) Electrostatic surface of the SARS-CoV-2 N-NTD, SARS-CoV N-NTD, MERS-CoV N-NTD, HCoV-OC43 N-NTD. Blue denotes positive charge potential, while red indicates negative charge potential. (C) Overall structural comparison of SARS-CoV-2 N-NTD with related viral N-NTD structures. Left: superimposition of SARS-CoV-2 N-NTD (blue) to SARS-CoV N-NTD (green); middle: superimposition of SARS-CoV-2 N-NTD (blue) to MERS-CoV N-NTD (brown); right: superimposition of SARS-CoV-2 N-NTD (blue) to HCoV-OC43 N-NTD (orange).
3.4. A potential unique drug targeting pocket in SARS-CoV-2 N-NTD
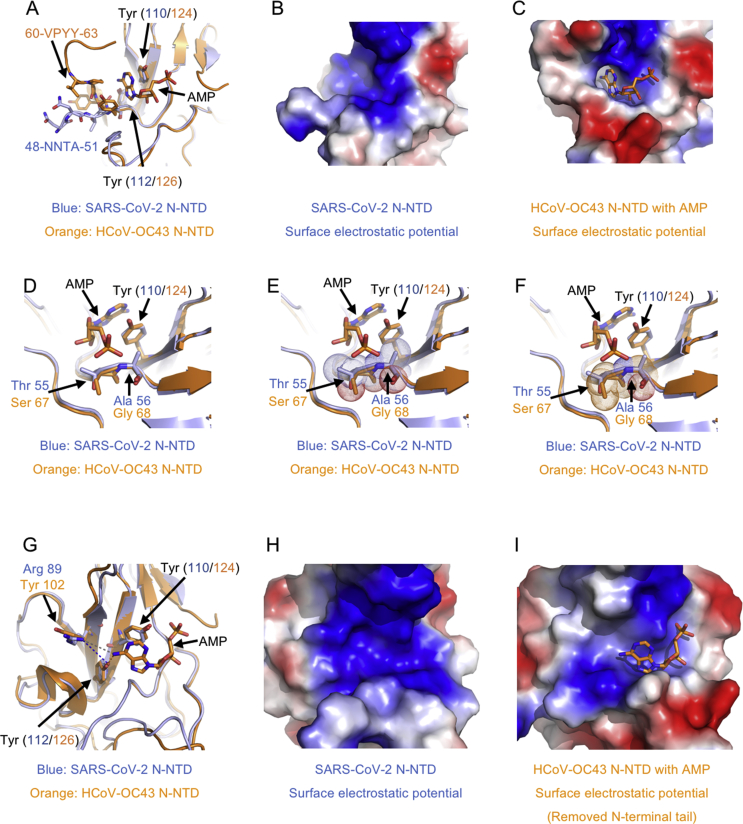
Although there are several CoV N-NTDs structures solved, the structural basis for ribonucleoside 5′-monophosphate binding of N protein had only been described in HCoV-OC43, a relative type typically causing mild cold symptoms41. Since the surface characterizations of N-NTD between SARS-CoV-2 with HCoV-OC43 are distinct, we next explored the differences of RNA binding mechanistic basis with superimposition analysis. Previous studies have shown that HCoV-OC43 N-NTD contained adenosine monophosphate (AMP)/uridine monophosphate (UMP)/cytosine monophosphate (CMP)/guanosine monophosphate (GMP) binding site alongside the middle two β-strands of its β-sheets core41. In the complex structure of HCoV-OC43 N-NTD with ribonucleotides, the phosphate group was bound by Arg122 and Gly68 via ionic interactions, the pentose sugar ribose 2′-hydroxyl group was recognized by Ser64 and Arg164, the nitrogenous base was inserted into a hydrophobic pocket consisting of Phe57, Pro61, Tyr63, Tyr102, Tyr124, and Tyr126, mainly interacted with Tyr124 via π−π stacking forces (Supporting Information Fig. S4). It is proposed that this ribonucleotide binding mechanism is essential for all coronavirus N proteins, applying to develop CoV N-NTD-target agents.
To obtain the structure information of SARS-CoV-2 N-NTD ribonucleotide binding site, we made a superimposition of SARS-CoV-2 N-NTD with HCoV-OC43 N-NTD-AMP complex. As expected, the root mean square deviation (RMSD) between these two structure coordinates is 1.4 Å over 136 superimposed Cα atoms. However, several differences around the ribonucleotide binding site were shown as the superimposition of SARS-CoV-2 N-NTD with HCoV-OC43 N-NTD. The major difference is the N-terminal tail of N-NTD with sequence variation (SARS-CoV-2: 48 NNTA 51 versus HCoV-OC43: 60 VPYY 63). In HCoV-OC43 N-NTD, the tail folded up to compose a nitrogenous base binding channel, whereas this region extended outward in SARS-CoV-2 (Fig. 4A). The N-terminal tail movement contributed to the change of N-NTD surface charge distribution, at which the nucleotide binding cavity became easier to access in SARS-CoV-2 N-NTD (Fig. 4B and C). The second difference is on phosphate group binding site, where SARS-CoV-2 N-NTD has larger sidechain residues (55 TA 56) compared with HCoV-OC43 N-NTD equivalents (67 SG 68) (Fig. 4D). Structural superimposition suggested additional polar properties of Thr55 and Ala56 in SARS-CoV-2 N-NTD may increase the steric clash with ribonucleotide phosphate moiety (Fig. 4E and F). The third difference is on the edge of the nitrogenous base recognized hydrophobic pocket, where SARS-CoV-2 N-NTD had Arg89 residues compared with HCoV-OC43 N-NTD Tyr102 equivalents (Fig. 4G). The change of these residue sidechain may lead to dramatic decreasing of non-polar properties and increasing of polar features in the nitrogenous base binding site (Fig. 4H and I). To evaluate these different observations in our structure, we next performed surface plasmon resonance (SPR) analysis experiments to assess the binding affinity between SARS-CoV-2 N-NTD with all four kinds of ribonucleotide AMP/UMP/CMP/GMP. Intriguingly, all 5′-monophosphate ribonucleotides, excepted for GMP (KD value is 8 mmol/L), show little binding signals in assays (Supporting Information Fig. S5). Similar results were shown in the Bio-Layer Interferometry (Octet RED96e, Fortebio) interaction analysis for SARS-CoV-2 N-NTD with all four kinds of ribonucleotide AMP/UMP/CMP/GMP (Supporting Information Fig. S6). To elucidate the binding of SARS-CoV-2 N-NTD with RNA, we next used SPR to determine the dissociation constants. A viral RNA derived intergenic sequence, 5′-(AAGUUCGUUU)-3′, was used in the SPR experiments. As shown in Supporting Information Fig. S7, the dissociation constants (KD) for RNA binding to wild type (WT) SARS-CoV-2 N-NTD is 140 μmol/L, around 2000-fold smaller than the value of Y110A mutant (KD = 330 mmol/L). This implies that WT SARS-CoV-2 N-NTD has a higher binding affinity to viral derived RNA than mutant in the binding pocket. Altogether, the above results suggest potential distinct ribonucleotide-binding patterns between SARS-CoV-2 N protein with HCoV-OC43 N protein.
Figure 4.
A potential unique drug target pocket in SARS-CoV-2 N-NTD. (A) Detailed view of ribonucleotide binding pocket in superimposition structures between SARS-CoV-2 N-NTD with HCoV-OC43 N-NTD AMP complex. AMP, interacting residues and equivalents are highlighted with stick representation. (B) Electrostatic surface of the potential ribonucleotide binding pocket on SARS-CoV-2 N-NTD. (C) Electrostatic surface of the ribonucleotide binding pocket on HCoV-OC43 N-NTD. (D) Detailed view of phosphate group binding site in superimposition structures between SARS-CoV-2 N-NTD with HCoV-OC43 N-NTD AMP complex. (E) Dot representation of SARS-CoV-2 residues Thr55 and Ala56, which indicates potential steric clashes with the ribonucleotide phosphate group. (F) Dot representation of HCoV-OC43 N-NTD residues Ser67 and Gly68. (G) Detailed view of the nitrogenous base binding site in superimposition structures between SARS-CoV-2 N-NTD with HCoV-OC43 N-NTD AMP complex. (H) Electrostatic surface of the potential ribonucleotide nitrogenous base binding pocket on SARS-CoV-2 N-NTD. (I) Electrostatic surface of the ribonucleotide nitrogenous base binding pocket on HCoV-OC43 N-NTD. In electrostatic surface potential panels, blue denotes positive charge potential, while red indicates negative charge potential. The potential distribution was calculated by Pymol. The values range from –6kT (red) to 0 (white) and to +6kT (blue), where k is the Boltzmann constant, and T is the temperature.
To calculate the pocket volume, we input the coordinate files of SARS-CoV-2 N-NTD and other CoVs N-NTD into the online web server DoGSiteScorer42. The server predicted potential pockets, described them through descriptors, and queried the model for druggability estimations. A druggability score between 0 and 1 was returned. The higher the score, the more druggable the pocket is estimated to be. As shown in Supporting Information Fig. S8A, a pocket volume of 279 Å3 is predicted on the SARS-CoV-2 N-NTD. SARS-CoV N-NTD and MERS-CoV N-NTD pocket volumes are similar as SARS-CoV-2 N-NTD's volume (Fig. S8B and S8C), whereas mild type coronavirus HCoV-OC43 has a larger pocket with a volume of 352 Å3 (Fig. S8D). Their pocket surfaces are distinct among SARS-CoV-2 N-NTD, SARS-CoV N-NTD, and MERS-CoV N-NTD, with different enclosure ratio and surface/volume values (Fig. S8E).
4. Discussion
Structure-based drug discovery has been shown to be a promising approach for the development of new therapeutics. Many ongoing studies are developed to treat COVID-19 primarily targeting the spike protein, viral proteases (3C-like protease, and papain-like protease). However, there is little effective targeted therapeutic currently. Recent studies demonstrated that N proteins would be an excellent drug-targeting candidate in other CoVs since they had several critical functions, such as RNA genomic packing, viral transcription and assembly, in the infectious cell11. However, the molecular basis of SARS-CoV-2 N protein is yet to be elucidated. Here, we present the 2.7 Å crystal structure of SARS-CoV-2 N protein N-terminal domain, revealing the specific surface charge distributions which may facilitate drug discovery specifically to SARS-CoV-2 N protein ribonucleotide binding domain.
On the structural basis of SARS-CoV-2 N-NTD, several residues in the ribonucleotide binding domain were found to recognize the CoV RNA substrates distinctly. The N-terminal tail residues (Asn48, Asn49, Thr50, and Ala51) is found to be more flexible and extended outward compared with equivalent residues in HCoV-OC43 N-NTD, possibly opening up the binding pocket into fitting with viral RNA genomic high order structure. Residues Arg89, instead of HCoV-OC43 N-NTD Tyr102, may contribute to guanosine base recognition despite the overall ribonucleotide binding that may be excluded by residues Thr55 and Ala56 in the phosphate moiety recognition site. In a previous study in which crystal structures of the CoV N-NTD in complex with five ligands (AMP, CMP, GMP, UMP, and inhibitor PJ34, respectively) were reported, Lin et al.41 demonstrated several general features for developing the hCoV-OC43 N-NTD targeting agents. The first feature is a polycyclic aromatic core (nitrogenous base in ribonucleotides), which is required to enable π−π stacking with the Tyr102 residues. The second feature is hydrogen-bond-forming moieties to the aromatic core mediates specific interactions with the N-NTD (nitrogenous base in ribonucleotides). Third, attaching a branching moiety (or moieties) that fits the ribonucleotide-binding pocket can enhance the drug affinity and specificity. In our SARS-CoV-2 N-NTD structure, the observed differences affect the above features in the pocket. For example, the Arg89 in SARS-CoV-2 N-NTD will decrease the hydrogen-bond-forming moieties to the aromatic core (this effect may be weakened for guanosine base recognition as the arginine–guanosine interactions are the most abundant contacts found in the amino acid–nucleotide interactions). Additionally, the branching phosphate group binding site in SARS-CoV-2 N-NTD structure is different from hCoV-OC43 N-NTD one, at which Thr55 and Ala56 in SARS-CoV-2 N-NTD may increase the steric clash with ribonucleotide phosphate moiety. Therefore, the above results suggest that the introduction of a hydrogen-bond-forming moiety (a guanosine base like moiety) at the base recognition site and avoiding the steric clash at the branching phosphate group binding site would benefit the high-affinity ligand development.
Until now, seven coronaviruses have been identified as human-susceptible viruses, among which HCoV-229E, HCoV-NL63, HCoV-HKU1, and HCoV-OC43 with low pathogenicity cause mild respiratory symptoms similar as the common cold. In contrast, the other three betacoronaviruses (SARS-CoV-2, SARS-CoV, and MERS-CoV) lead to severe and potentially fatal respiratory tract infections33,43,44. A previous study reported the structural basis of HCoV-OC43 N-NTD with AMP, GMP, UMP, CMP, and a virtual screening-base compound PJ34. However, our data suggested that SARS-CoV-2 employed a unique pattern for binding ribonucleotides with atomic resolution information. The structure not only helps us to understand the ribonucleotide-binding mechanisms between severe infectious coronavirus with mild infectious one, but also guide the design of novel antiviral agents specific targeting to SARS-CoV-2.
Acknowledgments
We thank Guangdong Medical Laboratory Animal Center for providing the N-protein encoding gene plasmids, Dr. Yongzhi Lu from Guangzhou Institutes of Biomedicine and Health (Chinese Academy of Sciences) for the initial crystals X-ray diffraction screening, supports of Dr. Xuan Ma from South China Sea Institute of Oceanology (Chinese Academy of Sciences) for home source X-ray diffraction facility. This work was supported by National Natural Science Foundation of China (31770801) and Special Fund for Scientific and Technological Innovation Strategy of Guangdong Province of China (2018B030306029 and 2017A030313145) to Shoudeng Chen, National Natural Science Foundation of China (81430041, 81620108017) and National Key Basic Research Program, China (SQ2018YFC090075) to Hong Shan, National Natural Science Foundation of China (81870019) to Yan Yan.
Author contributions
Shoudeng Chen and Hong Shan conceived the project. Shoudeng Chen, Hong Shan and Sisi Kang made the constructs, performed crystallographic, made figures and wrote the paper. Sisi Kang purified proteins, carried out the in vitro binding experiments, and the biochemical experiments. Liping Zhang and Changsheng Zhang helped X-ray diffraction data collection and analysis. Mei Yang and Suhua He helped with protein purification, carried out the in vitro binding experiments, and the biochemical experiments. Zhaoxia Huang, Xiaoxue Chen, Ziliang Zhou, Zhechong Zhou, Qiuyue Chen, Yan Yan and Zhongsi Hong analyzed data.
Conflicts of interest
The authors declare no conflict of interest.
Data availability statement
The structure in this paper is deposited to the Protein Data Bank with 6M3M access code.
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2020.04.009.
Contributor Information
Hong Shan, Email: shanhong@mail.sysu.edu.cn.
Shoudeng Chen, Email: chenshd5@mail.sysu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.World Health Organization . 12 March 2020. COVID-2019 situation reports.https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports Available from: [Google Scholar]
- 2.Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr Top Microbiol Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo Y., Cao Q., Hong Z., Tan Y., Chen S., Jin H. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—an update on the status. Mil Med Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramajayam R., Tan K.P., Liang P.H. Recent development of 3C and 3CL protease inhibitors for anti-coronavirus and anti-picornavirus drug discovery. Biochem Soc Trans. 2011;39:1371–1375. doi: 10.1042/BST0391371. [DOI] [PubMed] [Google Scholar]
- 6.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson G.W., Stohlman S.A., Tahara S.M. High affinity interaction between nucleocapsid protein and leader/intergenic sequence of mouse hepatitis virus RNA. J Gen Virol. 2000;81:181–188. doi: 10.1099/0022-1317-81-1-181. [DOI] [PubMed] [Google Scholar]
- 8.Stohlman S.A., Baric R.S., Nelson G.N., Soe L.H., Welter L.M., Deans R.J. Specific interaction between coronavirus leader RNA and nucleocapsid protein. J Virol. 1988;62:4280–4287. doi: 10.1128/jvi.62.11.4288-4295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cong Y.Y., Ulasli M., Schepers H., Mauthe M., V'kovski P., Kriegenburg F. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J Virol. 2020;94 doi: 10.1128/JVI.01925-19. pii.e01925-01919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masters P.S., Sturman L.S. Background paper: functions of the coronavirus nucleocapsid protein. Adv Exp Med Biol. 1990;276:235–238. doi: 10.1007/978-1-4684-5823-7_32. [DOI] [PubMed] [Google Scholar]
- 11.McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang T.K., Wu M.P.J., Chen S.T., Hou M.H., Hong M.H., Pan F.M. Biochemical and immunological studies of nucleocapsid proteins of severe acute respiratory syndrome and 229E human coronaviruses. Proteomics. 2005;5:925–937. doi: 10.1002/pmic.200401204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du L., Zhao G., Lin Y., Chan C., He Y., Jiang S. Priming with rAAV encoding RBD of SARS-CoV S protein and boosting with RBD-specific peptides for T cell epitopes elevated humoral and cellular immune responses against SARS-CoV infection. Vaccine. 2008;26:1644–1651. doi: 10.1016/j.vaccine.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Surjit M., Liu B., Chow V.T., Lal S.K. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J Biol Chem. 2006;281:10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh P.K., Chang S.C., Huang C.C., Lee T.T., Hsiao C.W., Kou Y.H. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J Virol. 2005;79:13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12:E254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu S.J., Leng C.H., Lien S.P., Chi H.Y., Huang C.Y., Lin C.L. Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine. 2006;24:3100–3108. doi: 10.1016/j.vaccine.2006.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shang B., Wang X.Y., Yuan J.W., Vabret A., Wu X.D., Yang R.F. Characterization and application of monoclonal antibodies against N protein of SARS-coronavirus. Biochem Biophys Res Commun. 2005;336:110–117. doi: 10.1016/j.bbrc.2005.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Y., Shen X., Yang R.F., Li Y.X., Ji Y.Y., He Y.Y. Identification of an epitope of SARS-coronavirus nucleocapsid protein. Cell Res. 2003;13:141–145. doi: 10.1038/sj.cr.7290158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo Y.S., Lin S.Y., Wang S.M., Wang C.T., Chiu Y.L., Huang T.H. Oligomerization of the carboxyl terminal domain of the human coronavirus 229E nucleocapsid protein. FEBS Lett. 2013;587:120–127. doi: 10.1016/j.febslet.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen I.J., Yuann J.M.P., Chang Y.M., Lin S.Y., Zhao J., Perlman S. Crystal structure-based exploration of the important role of Arg106 in the RNA-binding domain of human coronavirus OC43 nucleocapsid protein. Biochim Biophys Acta. 2013;1834:1054–1062. doi: 10.1016/j.bbapap.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang C.K., Chen C.M.M., Chiang M.H., Hsu Y.L., Huang T.H. Transient oligomerization of the SARS-CoV N protein—implication for virus ribonucleoprotein packaging. PloS One. 2013;8:e65045. doi: 10.1371/journal.pone.0065045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang C.K., Sue S.C., Yu T.H., Hsieh C.M., Tsai C.K., Chiang Y.C. Modular organization of SARS coronavirus nucleocapsid protein. J Biomed Sci. 2006;13:59–72. doi: 10.1007/s11373-005-9035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wootton S.K., Rowland R.R., Yoo D. Phosphorylation of the porcine reproductive and respiratory syndrome virus nucleocapsid protein. J Virol. 2002;76:10569–10576. doi: 10.1128/JVI.76.20.10569-10576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saikatendu K.S., Joseph J.S., Subramanian V., Neuman B.W., Buchmeier M.J., Stevens R.C. Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein. J Virol. 2007;81:3913–3921. doi: 10.1128/JVI.02236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jayaram H., Fan H., Bowman B.R., Ooi A., Jayaram J., Collisson E.W. X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation. J Virol. 2006;80:6612–6620. doi: 10.1128/JVI.00157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan H., Ooi A., Tan Y.W., Wang S., Fang S., Liu D.X. The nucleocapsid protein of coronavirus infectious bronchitis virus: crystal structure of its N-terminal domain and multimerization properties. Structure. 2005;13:1859–1868. doi: 10.1016/j.str.2005.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grossoehme N.E., Li L., Keane S.C., Liu P., Dann C.E., Iii, Leibowitz J.L. Coronavirus N protein N-terminal domain (NTD) specifically binds the transcriptional regulatory sequence (TRS) and melts TRS-cTRS RNA duplexes. J Mol Biol. 2009;394:544–557. doi: 10.1016/j.jmb.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keane S.C., Lius P., Leibowitzs J.L., Giedroc D.P. Functional transcriptional regulatory sequence (TRS) RNA binding and helix destabilizing determinants of murine hepatitis virus (MHV) nucleocapsid (N) protein. J Biol Chem. 2012;287:7063–7073. doi: 10.1074/jbc.M111.287763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan Y.W., Fang S., Fan H., Lescar J., Liu D.X. Amino acid residues critical for RNA-binding in the N-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 2006;34:4816–4825. doi: 10.1093/nar/gkl650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin S.M., Lin S.C., Hsu J.N., Chang C.K., Chien C.M., Wang Y.S. Structure-based stabilization of non-native protein–protein interactions of coronavirus nucleocapsid proteins in antiviral drug design. J Med Chem. 2020;63:3131–3141. doi: 10.1021/acs.jmedchem.9b01913. [DOI] [PubMed] [Google Scholar]
- 32.Liebschner D., Afonine P.V., Baker M.L., Bunkoczi G., Chen V.B., Croll T.I. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu L.L., Malik Peiris J.S. Pathogenesis of severe acute respiratory syndrome. Curr Opin Immunol. 2005;17:404–410. doi: 10.1016/j.coi.2005.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan Y.J., Lim S.G., Hong W. Characterization of viral proteins encoded by the SARS-coronavirus genome. Antivir Res. 2005;65:69–78. doi: 10.1016/j.antiviral.2004.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hatcher E.L., Zhdanov S.A., Bao Y., Blinkova O., Nawrocki E.P., Ostapchuck Y. Virus variation resource—improved response to emergent viral outbreaks. Nucleic Acids Res. 2017;45:D482–D490. doi: 10.1093/nar/gkw1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W.Z. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin S.Y., Liu C.L., Chang Y.M., Zhao J.C., Perlman S., Hou M.H. Structural basis for the identification of the N-terminal domain of coronavirus nucleocapsid protein as an antiviral target. J Med Chem. 2014;57:2247–2257. doi: 10.1021/jm500089r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Volkamer A., Kuhn D., Rippmann F., Rarey M. DoGSiteScorer: a web server for automatic binding site prediction, analysis and druggability assessment. Bioinformatics. 2012;28:2074–2075. doi: 10.1093/bioinformatics/bts310. [DOI] [PubMed] [Google Scholar]
- 43.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evans P.R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D Biol Crystallogr. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 47.Weiss M.S. Global indicators of X-ray data quality. J Appl Crystallogr. 2001;34:130–135. [Google Scholar]
- 48.Diederichs K., Karplus P.A. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat Struct Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structure in this paper is deposited to the Protein Data Bank with 6M3M access code.