Highlights
-
•
Gnotobiotic (Gn) calves were susceptible to infection with PDCoV, but not with PEDV.
-
•
Cytopathic effects (CPE) were identified in two primary bovine mesenchymal cell types inoculated with PDCoV or PEDV.
-
•
High PDCoV or PEDV RNA titers and PDCoV or PEDV antigens were also detected in inoculated cells.
-
•
The in vitro observations only partially coincided with the corresponding in vivo data from Gn calves.
Keywords: Primary bovine cells, PDCoV, PEDV, Interspecies transmission, Virus, Pig
Abstract
Unlike porcine epidemic diarrhea virus (PEDV) that infects only pigs, porcine deltacoronavirus (PDCoV) has the capacity to infect different animal species. In vivo gnotobiotic calves were previously confirmed to be susceptible to infection with PDCoV, but not with PEDV. We next investigated in vitro whether primary bovine cells are susceptible to PDCoV or PEDV infection. We conducted quantification of viral RNA in cell culture supernatants and immunofluorescent staining for the detection of PDCoV or PEDV antigen in two primary bovine cell types inoculated with the PDCoV strain OH-FD22 or PEDV strain PC22-P40 grown in LLC-PK or Vero cells, respectively, and supplemented with 1.25∼5 μg/mL of trypsin in the cell culture medium. The primary cells were isolated from the kidney or heart of a gnotobiotic calf, and both cell types were vimentin-positive, but E-Cadherin-negative, resembling mesenchymal cells. Similar to the previous in vivo observation, cytopathic effects (CPE) that consisted of enlarged and rounded cells, followed by cell shrinkage and detachment, were identified in the two primary cell types inoculated with PDCoV. Unexpectedly, similar CPE was also identified in the two cell types inoculated with PEDV. High PDCoV or PEDV RNA titers and PDCoV or PEDV antigens were detected in the cell culture supernatants and CPE-positive cells, respectively. Our study revealed that primary bovine mesenchymal cells are susceptible to infection with PDCoV and PEDV. The in vitro observation partially coincided with the corresponding in vivo data from gnotobiotic calves.
1. Introduction
Coronaviruses are enveloped, single-stranded RNA viruses of positive-sense polarity. Their genomes range from approximately 26–32 kb in size (Jung et al., 2016; Jung and Saif, 2015). The family Coronaviridae of the order Nidovirales is divided into the four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus. Bats are the projected host reservoir for Alphacoronaviruses and Betacoronaviruses, while birds are the suspect host for Gammacoronaviruses and Deltacoronaviruses (Jung et al., 2016; Jung and Saif, 2015). Porcine epidemic diarrhea virus (PEDV) (genus Alphacoronavirus) and porcine deltacoronavirus (PDCoV) (genus Deltacoronavirus) share similar clinical and pathological features, including acute diarrhea, dehydration and mortality in neonatal piglets (Jung et al., 2016; Jung and Saif, 2015).
For the last four decades since the first discovery of PEDV in 1977, PEDV has been found only within the pig population (Jung and Saif, 2015). However, PDCoV-induced pig diarrhea recently emerged in the United States in 2014 (Jung et al., 2016). PDCoV is a novel enteropathogenic coronavirus in pigs and was previously reported in wild birds, although the mechanisms of interspecies transmission of deltacoronaviruses between pigs and birds are unclear (Jung et al., 2016). PDCoV may be incompletely adapted to pigs and as such may have a capacity to infect different animal species (Jung et al., 2016), as also suggested by previous in vitro or in vivo observations (Boley et al., 2020; Li et al., 2018; Liang et al., 2019). Our previous in vivo study also revealed that gnotobiotic calves were susceptible to infection with the newly emerging PDCoV, but not with PEDV (Jung et al., 2017). In prior studies, we isolated various primary cells from gnotobiotic calves to test growth of the GIII.2 bovine norovirus strain CV186-OH that did not grow in regular bovine cell lines (Oka et al., 2018). Additionally, two different primary bovine cells, morphologically and molecularly (vimentin-positive, but E-Cadherin-negative) similar to mesenchymal cells, were isolated and successfully propagated in vitro. Therefore, we attempted to use the two primary bovine cell types to investigate: i) whether they are also susceptible to infection with the cell culture-adapted PDCoV OH-FD22, but not with the cell culture-adapted PEDV PC22-P40, similar to the previous in vivo observations in gnotobiotic calves (Jung et al., 2017); ii) if or how in vitro observations from bovine cells coincide with the in vivo counterparts from gnotobiotic calves; and iii) based on that, to which extent the cell culture-based observations may be useful to predict the corresponding in vivo expectations.
2. Materials and methods
2.1. Virus
The PDCoV OH-FD22-P8 (passage 8) virus was serially passaged in LLC porcine kidney (LLC-PK) (ATCC CL-101) cells supplemented with 10 μg/mL of trypsin in the cell culture medium for a total of 8 passages, as described previously (Hu et al., 2015). After the 6th passage, the virus was purified once by a plaque assay and then further serially passaged (Hu et al., 2015). The viral RNA titer of the OH-FD22-P8 used in this study was 10.5 log10 genomic equivalents (GE)/mL, and the infectious titer was 8.6 log10 plaque forming units (PFU)/mL (Jung et al., 2018). The Vero cell-culture grown PEDV PC22-P40 (passage 40) virus was also used to infect primary bovine cells, as described previously (Jung et al., 2017; Oka et al., 2014). The PEDV PC22-P40 retained its pathogenic characteristics, similar to the parent virus (Lin et al., 2017).
2.2. Isolation of primary bovine cells from the kidney or heart of a gnotobiotic calf
Two primary bovine cell types were isolated from the kidney and heart of a 7-day-old gnotobiotic calf. After removing the pericardial or renal capsule, approximately 10 g of the cardiac or renal parenchyma were collected. Cells were dissociated mechanically from tissues using sterile metal mesh screens (Sigma-Aldrich). Isolated cells were propagated and passaged in the following growth medium: Dulbecco’s modified eagle medium/F12 (DMEM/F12) (Gibco, USA) supplemented with 10 % fetal bovine serum (FBS), 2% penicillin/streptomycin (Gibco), 1% insulin-transferrin-sodium selenite (Roche), and 10 ng/mL of human epidermal growth factor (Invitrogen). Cells were passaged 3–4 times before use, and growth of cells and bacterial contamination were monitored during each passage. Virus was inoculated onto 2 to 3-day-old, 100 % confluent cell monolayers. All animal-related experimental or euthanasia protocols were approved by the Ohio State University Institutional Animal Care and Use Committee.
2.3. Immunofluorescent staining for the detection of E-Cadherin or vimentin in the primary bovine cells isolated
The primary bovine cells isolated from the kidney were spindle-shaped, whereas the cells isolated from the heart were polygonal to slightly elongate. We tested the cell monolayers by immunofluorescent (IF) staining to determine if they were positive or negative for E-Cadherin (a marker for epithelial cells) and/or vimentin (a marker of mesenchymal cells, such as fibroblasts and smooth muscle cells). The cell monolayers in 6-well plates were fixed with 100 % ethanol at 4 °C overnight and tested in duplicate by IF staining, as described previously (Hu et al., 2015; Jung et al., 2018), for the detection of E-Cadherin or vimentin antigen, using monoclonal antibodies against human E-Cadherin (Invitrogen, CA, USA) or human vimentin (Novus Biologicals, Littleton, CO, USA). Monoclonal antibodies were diluted 1 in 25−50 in phosphate-buffered saline (PBS).
2.4. Infection of primary bovine cells with PDCoV or PEDV
The cell culture conditions tested to infect primary bovine cells with a multiplicity of infection (MOI) 1 of PDCoV or PEDV in 6-well plates were as follows: Washing of cells with maintenance medium (DMEM/F12 supplemented with 2% penicillin/streptomycin) (MM) twice to remove FBS, virus adsorption for one hour, and then removal of the inoculum and the addition of 2 mL MM supplemented with 1.25–2.5 μg/ml and 2.5−5 μg/mL of trypsin (Gibco) (MMT) for PDCoV and PEDV, respectively. A higher concentration of trypsin in MMT was used for growth of PEDV compared with PDCoV based on our previous observations (Hu et al., 2015; Jung et al., 2018; Oka et al., 2014). Overall, the cell culture conditions for viral infections were similar to the previous methods used in Vero cells and LLC-PK or IPEC-J2 (pig small intestinal epithelial cell line) cells for PEDV and PDCoV, respectively (Hu et al., 2015; Jung et al., 2018; Oka et al., 2014). However, only lower concentrations of trypsin were added to MMT (Hu et al., 2015; Jung et al., 2018; Oka et al., 2014), because primary bovine cells were expected to be less tolerant to trypsin than the immortalized cell lines used previously.
2.5. Analysis of PDCoV or PEDV RNA in cell culture supernatants
Infection of primary bovine cells with PDCoV or PEDV was conducted in duplicate. One-hundred microliters of cell culture supernatants were collected from PDCoV- or PEDV-inoculated and non-inoculated cells in 6-well plates at 18 and 42 h post-inoculation (hpi). The RNA was extracted from 50 μl of cell culture supernatants centrifuged at 2000 × g for 30 min at 4 °C, using the Mag-MAX Viral RNA Isolation Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. The detection limit of qRT-PCR for PDCoV and PEDV was 10 GE per reaction, corresponding to 5.6 and 5.8 log10 GE/mL of PDCoV and PEDV in cell culture supernatants, respectively (Jung et al., 2017). Log10-transformed viral RNA titers (log10 GE/mL) were expressed as the means ± standard deviation of the means (SD).
2.6. Immunofluorescent staining for the detection of PDCoV or PEDV antigen in primary bovine cells
When PDCoV- or PEDV-inoculated cells showed more than 50 % CPE in 6-well plates, they were washed with PBS and fixed with 100 % ethanol at 4 °C overnight. The infected cells were prepared in duplicate and tested by IF staining for the detection of PDCoV or PEDV antigen, using hyperimmune gnotobiotic pig antiserum against PDCoV OH-FD22 strain or monoclonal antibody 6C8-1 against the nucleocapsid protein of PEDV, respectively, as described previously (Jung et al., 2017). Trypsin alone-treated cells were tested as negative controls for IF staining.
3. Results
3.1. The isolated primary bovine cells appeared to be mesenchymal cells positive for vimentin, but negative for E-Cadherin
After the first 3–4 passages, the isolated primary bovine cells were 100 % confluent at approximately 2–3 days after seeding approximately 2 × 105 cells in 6-well plates. The primary cells isolated from the kidney appeared to be homogeneous cells that were spindle-shaped (Fig. 1 A–B), whereas the other primary cells isolated from the heart appeared to be pleomorphic cells that were polygonal to slightly elongate (Fig. 2 A–B). By IF staining, the primary cells isolated from the kidney were positive for vimentin but negative for E-Cadherin. Based on the morphologic and molecular characteristics, the isolated cells were considered to be fibroblast- or smooth muscle-like cells that belong to mesenchymal cells. In our study, the Primary Bovine Kidney mesenchymal cells were referred to as “PBK cells”. Similarly, the primary cells isolated from the heart were positive for vimentin but negative for E-Cadherin. Because of the limited morphologic and molecular characteristics, the cells were considered to be only a type of mesenchymal cells of heart origin. In our study, the Primary Bovine Heart mesenchymal cells were also referred to as “PBH cells”.
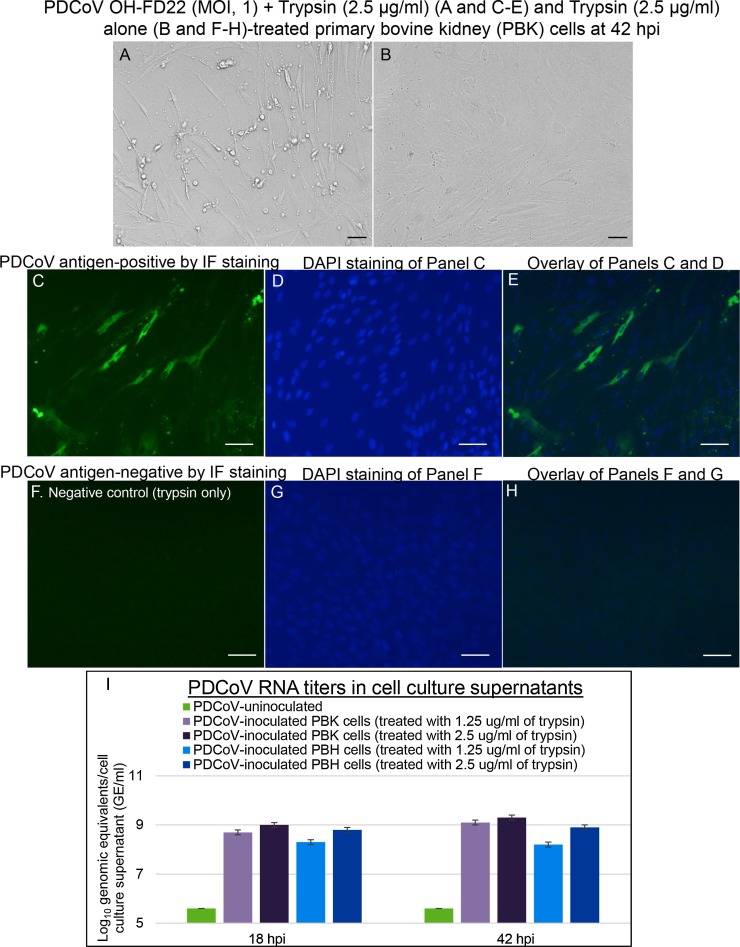
Fig. 1.
Cytopathic effects (CPE) and localization of PDCoV antigens by immunofluorescent (IF) staining in primary bovine kidney (PBK) cells inoculated with the PDCoV strain OH-FD22 (MOI, 1), and supplemented with 2.5 μg/mL of trypsin in the cell culture medium. (A) PDCoV OH-FD22-inoculated PBK cells at 42 h post-inoculation (hpi), showing CPE that consists of enlarged, rounded, and densely granular cells that occurred singly or in clusters, often forming cell clumps, followed by cell shrinkage and detachment. (B) PDCoV-uninoculated PBK cells supplemented with 2.5 μg/mL of trypsin showing normal spindle-shaped cells. Note that most of PBK cells are spindle-shaped. (C) IF staining of the inoculated PBK cells at 42 hpi, showing that the enlarged, rounded or spindle-shaped cells are positive for PDCoV antigen (green staining). (D) Blue-fluorescent 4′, 6-diamidino-2-phenylindole dihydrochloride (DAPI) staining of Panel C to counterstain nuclear DNA. (E) Overlay of Panels C and D. (F) IF staining of PDCoV-uninoculated PBK cells treated with 2.5 μg/mL of trypsin at 42 hpi, showing no cells positive for PDCoV antigen. (G) DAPI staining of Panel F. (H) Overlay of Panels F and G. Scale bars =50 μm (A–H). (I) PDCoV RNA titers by qRT-PCR in cell culture supernatants of PDCoV-inoculated PBK and primary bovine heart cells supplemented with 1.25 to 2.5 μg/mL of trypsin at 18 and 42 hpi. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
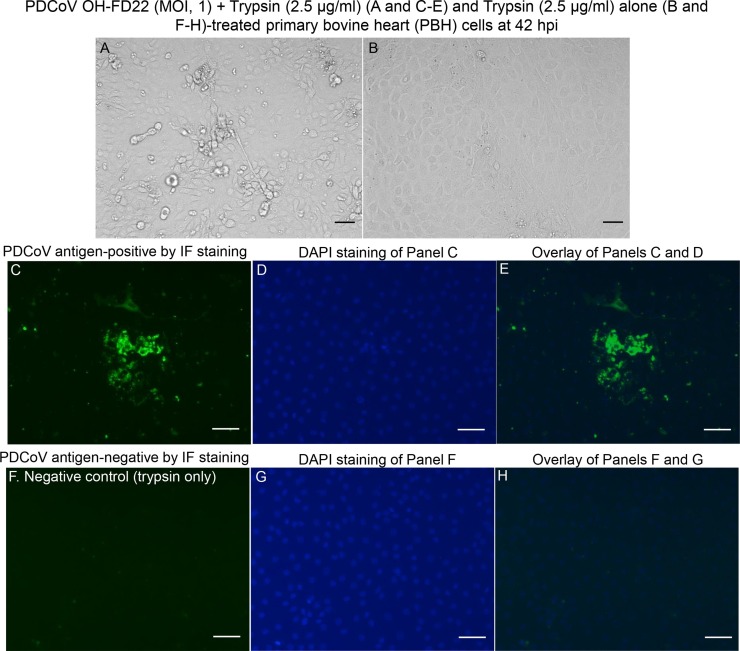
Fig. 2.
Cytopathic effects (CPE) and localization of PDCoV antigens by immunofluorescent (IF) staining in primary bovine heart (PBH) cells inoculated with the PDCoV strain OH-FD22 (MOI, 1), and supplemented with 2.5 μg/mL of trypsin in the cell culture medium. (A) PDCoV OH-FD22-inoculated PBH cells at 42 h post-inoculation (hpi), showing CPE that consists of enlarged, rounded, and densely granular cells that occurred singly or in clusters, often forming cell clumps, followed by cell shrinkage and detachment. (B) PDCoV-uninoculated PBH cells supplemented with 2.5 μg/mL of trypsin showing normal cells. (C) IF staining of the inoculated PBH cells at 42 hpi, showing that the enlarged, rounded, and clustered cells are positive for PDCoV antigen (green staining). (D) Blue-fluorescent 4′, 6-diamidino-2-phenylindole dihydrochloride (DAPI) staining of Panel C to counterstain nuclear DNA. (E) Overlay of Panels C and D. (F) IF staining of PDCoV-uninoculated PBH cells treated with 2.5 μg/mL of trypsin at 42 hpi, showing no cells positive for PDCoV antigen. (G) DAPI staining of Panel F. (H) Overlay of Panels F and G. Scale bars =50 μm (A–H). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
3.2. Cytopathic effects and PDCoV antigens were identified in PDCoV-inoculated primary bovine cells
At 18 hpi, in PDCoV-inoculated PBK and PBH cells treated with 1.25–2.5 μg/ml of trypsin, approximately 30–40 % of cells showed cytopathic effects (CPE) such as enlarged and rounded cells that occurred singly or in clusters, followed by cell shrinkage and detachment. At 42 hpi, PDCoV-inoculated PBK and PBH cells treated with 1.25–2.5 μg/ml of trypsin exhibited approximately 50–60 % of CPE (Figs. 1A–B and 2 A–B). By IF staining, low numbers (approximately 20–25 % of the cells remained on the plates) of the single or clustered cells that showed CPE in PBK (Fig. 1C–E) and PBH cells (Fig. 2C–E) at 42 hpi, were positive for PDCoV antigen, regardless of the different trypsin concentrations. No IF-positive cells were found in the PDCoV-uninoculated, trypsin-treated negative controls (Figs. 1F–H and 2 F–H).
3.3. Cytopathic effects and PEDV antigens were also identified in PEDV-inoculated primary bovine cells
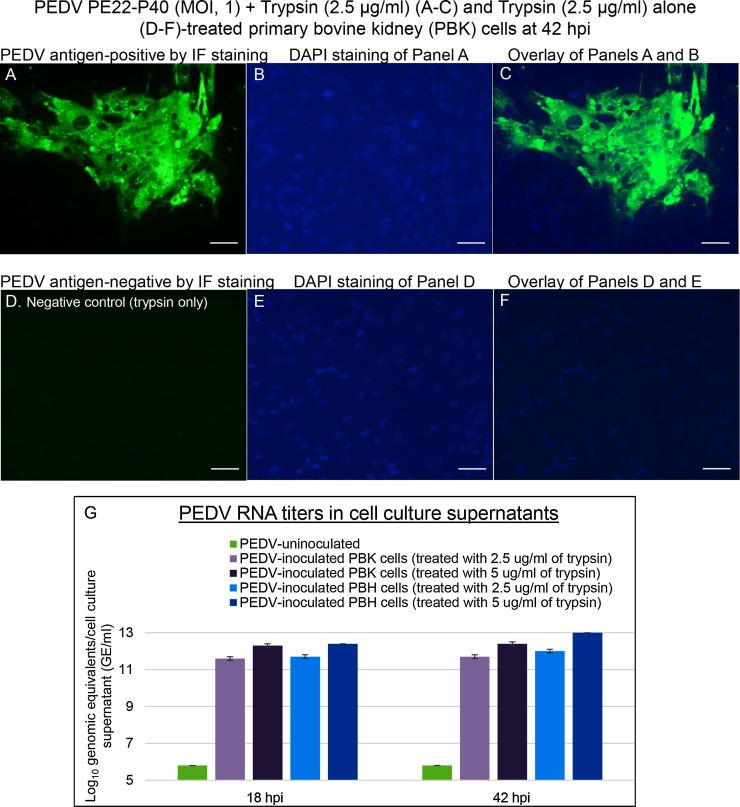
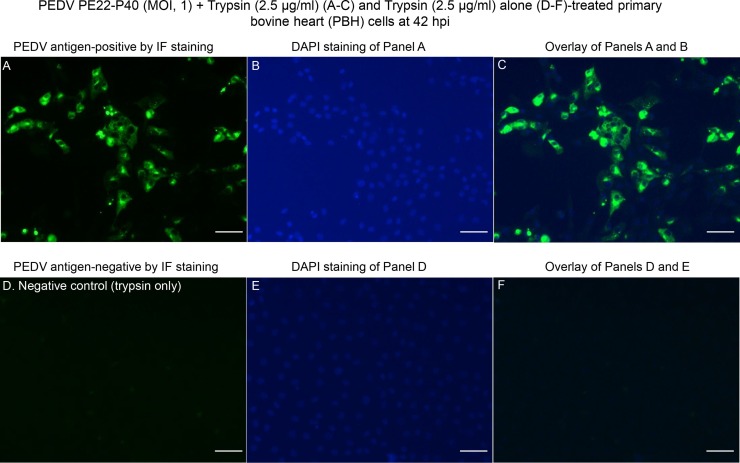
At 18 hpi, there was no significant CPE observed in PEDV-inoculated PBK and PBH cells treated with 2.5−5 μg/mL of trypsin. At 42 hpi, however, PEDV-inoculated PBK and PBH cells treated with 2.5−5 μg/mL of trypsin exhibited more than 50%–100% of CPE. Similar to CPE observed in PDCoV-inoculated cells (Figs. 1A and 2 A), CPE consisted of enlarged, rounded, and densely granular cells that occurred singly or in clusters, often forming cell clumps, followed by cell shrinkage and detachment. Relative to PEDV-inoculated PBK cells treated with 2.5−5 μg/mL of trypsin or PEDV-inoculated PBH cells treated with the lower concentration of trypsin, 2.5 μg/mL, PEDV-inoculated PBH cells treated with the higher concentration of trypsin, 5 μg/mL, showed complete cell detachment at 42 hpi. By IF staining, low numbers (approximately 20–25 % of the cells remained on the plates) of the single or clustered cells that showed CPE in PBK (Fig. 3 A–C) and PBH cells (Fig. 4 A–C) at 42 hpi, were positive for PEDV antigen, regardless of the different trypsin concentrations, except for the completely detached, PEDV-inoculated PBH cells treated with 5 μg/mL of trypsin at 42 hpi. No IF-positive cells were found in the PEDV-uninoculated, trypsin-treated negative controls (Figs. 3D–F and 4 D–F).
Fig. 3.
Localization of PEDV antigens by immunofluorescent (IF) staining in primary bovine kidney (PBK) cells inoculated with the PEDV strain PE22-P40 (MOI, 1), and supplemented with 2.5 μg/mL of trypsin in the cell culture medium. (A) IF staining of the inoculated PBK cells at 42 h post-inoculation (hpi), showing that the enlarged, rounded, and clustered cells are positive for PEDV antigen (green staining). (B) Blue-fluorescent 4′, 6-diamidino-2-phenylindole dihydrochloride (DAPI) staining of Panel A to counterstain nuclear DNA. (C) Overlay of Panels A and B. Note that the entire clustered cells positive for PEDV antigen (arrowheads) appeared to be syncytial or multinucleated cells. (D) IF staining of PEDV-uninoculated PBK cells treated with 2.5 μg/mL of trypsin at 42 hpi, showing no cells positive for PEDV antigen. (E) DAPI staining of Panel D. (F) Overlay of Panels D and E. Scale bars =50 μm (A–F). (G) PEDV RNA titers by qRT-PCR in cell culture supernatants of PEDV-inoculated PBK and primary bovine heart cells supplemented with 2.5–5 μg/mL of trypsin at 18 and 42 hpi. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
Fig. 4.
Localization of PEDV antigens by immunofluorescent (IF) staining in primary bovine heart (PBH) cells inoculated with the PEDV strain PE22-P40 (MOI, 1), and supplemented with 2.5 μg/mL of trypsin in the cell culture medium. (A) IF staining of the inoculated PBH cells at 42 h post-inoculation (hpi), showing that the enlarged, rounded, and clustered cells are positive for PEDV antigen (green staining). (B) Blue-fluorescent 4′, 6-diamidino-2-phenylindole dihydrochloride (DAPI) staining of Panel A to counterstain nuclear DNA. (C) Overlay of Panels A and B. (D) IF staining of PEDV-uninoculated PBH cells treated with 2.5 μg/mL of trypsin at 42 hpi, showing no cells positive for PEDV antigen. (E) DAPI staining of Panel D. (F) Overlay of Panels D and E. Scale bars =50 μm (A–F). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
3.4. High PDCoV RNA titers were observed in the cell culture supernatants of trypsin-treated, PDCoV-inoculated PBK and PBH cells at 18 and 42 hpi
Mean PDCoV RNA titers (SD) in PDCoV-inoculated PBK cells treated with 2.5 μg/mL of trypsin were 9.0 (0.1) log10 GE/mL and 9.3 (0.1) log10 GE/mL at 18 and 42 hpi, respectively, whereas viral RNA titers in the lower concentration trypsin (1.25 μg/mL)-treated, PDCoV-inoculated PBK cells were lower, such as 8.7 (0.1) log10 GE/mL and 9.1 (0.1) log10 GE/mL at 18 and 42 hpi, respectively (Fig. 1I). Similarly, mean PDCoV RNA titers (SD) in PDCoV-inoculated PBH cells treated with 2.5 μg/mL of trypsin were 8.8 (0.1) log10 GE/mL and 8.9 (0.1) log10 GE/mL at 18 and 42 hpi, respectively, whereas viral RNA titers in the lower concentration trypsin (1.25 μg/mL)-treated, PDCoV-inoculated PBH cells were lower, such as 8.3 (0.1) log10 GE/mL and 8.2 (0.1) log10 GE/mL at 18 and 42 hpi, respectively. There was also a trend toward higher PDCoV RNA titers in the cell culture supernatants of PDCoV-inoculated PBK cells (of kidney origin) compared with the PBH counterpart (of heart origin) (Fig. 1I). None of the cell culture supernatants from the PDCoV-uninoculated, trypsin-treated negative controls was positive for PDCoV RNA at 18−42 hpi.
3.5. High PEDV RNA titers were also observed in the cell culture supernatants of trypsin-treated, PEDV-inoculated PBK and PBH cells at 18 and 42 hpi
Mean PEDV RNA titers (SD) in PEDV-inoculated PBK cells treated with 5 μg/mL of trypsin were 12.3 (0.1) log10 GE/mL and 12.4 (0.1) log10 GE/mL at 18 and 42 hpi, respectively, whereas viral RNA titers in the lower concentration trypsin (2.5 μg/mL)-treated, PEDV-inoculated PBK cells were lower, such as 11.6 (0.1) log10 GE/mL and 11.7 (0.1) log10 GE/mL at 18 and 42 hpi, respectively (Fig. 3G). Similarly, mean PEDV RNA titers (SD) in PEDV-inoculated PBH cells treated with 5 μg/mL of trypsin were 12.4 (0) log10 GE/mL and 13.0 (0) log10 GE/mL at 18 and 42 hpi, respectively, whereas viral RNA titers in the lower concentration trypsin (2.5 μg/mL)-treated, PEDV-inoculated PBH cells were lower, such as 11.7 (0.1) log10 GE/mL and 12.0 (0.1) log10 GE/mL at 18 and 42 hpi, respectively. Dissimilar to the observation from PDCoV-inoculated cells, there was a trend toward higher PEDV RNA titers in the cell culture supernatants of PEDV-inoculated PBH cells (of heart origin) compared with the PBK counterpart (of kidney origin) (Fig. 3G). None of the cell culture supernatants from the PEDV-uninoculated, trypsin-treated negative controls was positive for PEDV RNA at 18−42 hpi.
4. Discussion
Based on our data from two different primary cells of kidney and heart origin, bovine mesenchymal cells were susceptible to infection with PDCoV and PEDV, that coincided with the in vivo susceptibility of gnotobiotic calves to infection with PDCoV, as reported previously (Jung et al., 2017). However, the observation is discrepant with the in vivo data from gnotobiotic calves inoculated orally with a similar strain of PEDV (Jung et al., 2017). The previous study revealed that PDCoV-inoculated gnotobiotic calves developed acute subclinical infection without histologic lesions and no detectable PDCoV antigen in the intestines and other major organs, including kidney and heart, but with persisting fecal viral RNA shedding and seroconversion (Jung et al., 2017). On the other hand, no PEDV-inoculated gnotobiotic calves exhibited fecal virus shedding, seroconversion, histopathology or detectable PEDV antigens in the corresponding tissues. The data indicate that gnotobiotic calves were susceptible to infection with the emerging PDCoV, but not with PEDV. Therefore, our in vitro observation from the primary bovine cell cultures only partially coincided with the corresponding in vivo data from gnotobiotic calves.
PEDV has been found only within the pig population (Jung and Saif, 2015). In contrast to the epidemiologic features of PEDV, the virus also replicated in cell lines from different animal species, such as bats (bat lung cells, Tb1-Lu), humans (human hepatocellular carcinoma cells, Huh7) (Liu et al., 2015) and birds (duck intestinal epithelial cells, MK-DIEC) (Khatri, 2015). These observations could partially support the discrepant in vivo and in vitro results from our previous and current studies, respectively (Jung et al., 2017). However, it is also noteworthy that unlike our previous and current studies, none of the previous reports showed any evidence to support whether the in vitro observations from the bat or bird cell cultures coincide with in vivo data from PEDV-inoculated bats or birds (Khatri, 2015; Liu et al., 2015).
Relative to PEDV, PDCoV is a novel enteropathogenic CoV in pigs (Jung et al., 2016). PDCoV may be incompletely adapted to pigs and as such may have a capacity to infect different animal species, such as calves, chickens, and turkeys (Boley et al., 2020; Jung et al., 2017; Liang et al., 2019). Our previous in vitro study demonstrated that a variety of cell lines of different animal species or organs origin were susceptible to infection with PDCoV, including human (Huh7 and human cervical cancer cells, HeLa), monkey (African green monkey kidney cells, Vero-CCL81), avian (chicken embryo fibroblast cells, DF-1), and canine (Madin-Darby canine kidney cells, MDCK), implicating its zoonotic or interspecies transmission potential (Li et al., 2018). We also identified aminopeptidase N (APN) as a major cell entry receptor for PDCoV in the variety of cells of the different animal species origin. Structural analyses of the APN ectodomain revealed four independently folded domains, termed domains I through IV (Chen et al., 2012), which were highly conserved among different animal species, including bovine (Bos tarus) (Li et al., 2018). Therefore, similar to the cells of human, monkey, avian or canine origin that permitted PDCoV infection (Li et al., 2018), the spike 1 protein of PDCoV also seemed to engage APN via the conserved domain II for entry into the primary bovine cells of kidney or heart origin. However, it is noteworthy that in our previous in vivo study, no PDCoV-inoculated calves exhibited PDCoV antigen in the small and large intestines, kidney, or heart (Jung et al., 2017). On the other hand, the mechanisms by which PEDV entered into the primary bovine cells remain unknown as does the receptor for PEDV.
Our previous study revealed limited infectivity of PDCoV in gnotobiotic calves (Jung et al., 2017), and likewise, in our subsequent studies, we found that gnotobiotic calf-passaged strains of PDCoV had no increased infectivity in gnotobiotic calves after serial calf passage, indicating incomplete adaptation of the virus to calves. Therefore, in our current study, we further attempted to verify if the previous in vivo finding could present an abortive infection with PDCoV in gnotobiotic calves. However, we did not verify which primary bovine mesenchymal cell type was more susceptible to PDCoV or PEDV, by comparing the infectivity and replication rates between the two cell types. Our current study demonstrated that PDCoV or PEDV previously grown in other swine or monkey cell cultures such as LLC-PK or Vero cells, respectively, can infect and replicate in vitro in primary bovine mesenchymal cells. Compared with PDCoV RNA titers, ranging from 8.2 to 9.3 log10 GE/mL, in the cell culture supernatants of PBK or PBH cells treated with 1.25–2.5 μg/ml of trypsin at 18−42 hpi, higher PEDV RNA titers, ranging from 11.6 to 13.0 log10 GE/mL, were observed in the cell culture supernatants of PBK or PBH cells treated with 2.5−5 μg/mL of trypsin during the same period. The higher PEDV RNA titers might be a result of greater replication and release of PEDV in the PBK or PBH cells treated with the higher concentrations of trypsin, although the detailed mechanisms need to be investigated further. In addition, supplementation of 1.25–2.5 μg/ml and 2.5−5.0 μg/mL of trypsin in the cell culture maintenance medium for PDCoV and PEDV, respectively, appeared to be beneficial for both viral growth and induction of CPE in the two primary bovine cell types, although further studies are needed to confirm the absolute necessity of trypsin in the cell culture maintenance medium for PDCoV and PEDV replication. The results were similar to the beneficial effect of the exogenous trypsin on both growth and induction of CPE of these two viruses in the pig or monkey cell lines (Hu et al., 2015; Jung et al., 2018; Oka et al., 2014; Shi et al., 2017).
5. Conclusion
We demonstrated that primary bovine mesenchymal cells were susceptible to infection with PDCoV and PEDV. The in vitro observation only partially coincided with the corresponding in vivo data from gnotobiotic calves that were susceptible to infection with PDCoV, but not with PEDV. Based on our data, cell culture-based observations on growth of coronaviruses may not be comprehensive enough to postulate their in vivo infectivity, although unlike PEDV, PDCoV showed the capacity to infect and replicate in both gnotobiotic calves in vivo and in bovine primary cells in vitro. However, further studies are still needed to investigate whether bovine intestinal epithelial cells are also susceptible to PDCoV and PEDV.
Declaration of Competing Interest
None of the authors of this paper has a financial or personal relationship with other people or organizations that could inappropriately influence or bias the content of the paper.
Acknowledgements
We thank Dr. Juliette Hanson, Ronna Wood, and Jeffery Ogg for assistance with animal care. Salaries and research support were provided by state and federal funds appropriated to the Ohio Agricultural Research and Development Center, The Ohio State University, Wooster, Ohio, USA.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.vetmic.2020.108660.
Contributor Information
Kwonil Jung, Email: jung.221@osu.edu.
Linda J. Saif, Email: saif.2@osu.edu.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Boley P.A., Alhamo M.A., Lossie G., Yadav K.K., Vasquez-Lee M., Saif L.J., Kenney S.P. Porcine Deltacoronavirus Infection and Transmission in Poultry, United States. Emerging Infect. Dis. 2020;26:255–265. doi: 10.3201/eid2602.190346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Lin Y.L., Peng G., Li F. Structural basis for multifunctional roles of mammalian aminopeptidase N. Proc. Natl. Acad. Sci. U. S. A. 2012;109:17966–17971. doi: 10.1073/pnas.1210123109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Jung K., Vlasova A.N., Chepngeno J., Lu Z., Wang Q., Saif L.J. Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J. Clin. Microbiol. 2015;53:1537–1548. doi: 10.1128/JCM.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Saif L.J. Porcine epidemic diarrhea virus infection: Etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet. J. 2015;204:134–143. doi: 10.1016/j.tvjl.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Saif L.J. Porcine deltacoronavirus infection: Etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res. 2016;226:50–59. doi: 10.1016/j.virusres.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Hu H., Saif L.J. Calves are susceptible to infection with the newly emerged porcine deltacoronavirus, but not with the swine enteric alphacoronavirus, porcine epidemic diarrhea virus. Arch. Virol. 2017;162:2357–2362. doi: 10.1007/s00705-017-3351-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Miyazaki A., Hu H., Saif L.J. Susceptibility of porcine IPEC-J2 intestinal epithelial cells to infection with porcine deltacoronavirus (PDCoV) and serum cytokine responses of gnotobiotic pigs to acute infection with IPEC-J2 cell culture-passaged PDCoV. Vet. Microbiol. 2018;221:49–58. doi: 10.1016/j.vetmic.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri M. Porcine epidemic diarrhea virus replication in duck intestinal cell line. Emerg Infect. Dis. 2015;21:549–550. doi: 10.3201/eid2103.141658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Hulswit R.J.G., Kenney S.P., Widjaja I., Jung K., Alhamo M.A., van Dieren B., van Kuppeveld F.J.M., Saif L.J., Bosch B.J. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc. Natl. Acad. Sci. U. S. A. 2018;115:5135–5143. doi: 10.1073/pnas.1802879115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q., Zhang H., Li B., Ding Q., Wang Y., Gao W., Guo D., Wei Z., Hu H. Susceptibility of chickens to porcine deltacoronavirus infection. Viruses. 2019;11:573. doi: 10.3390/v11060573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.M., Hou Y., Marthaler D.G., Gao X., Liu X., Zheng L., Saif L.J., Wang Q. Attenuation of an original US porcine epidemic diarrhea virus strain PC22A via serial cell culture passage. Vet. Microbiol. 2017;201:62–71. doi: 10.1016/j.vetmic.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Tang J., Ma Y., Liang X., Yang Y., Peng G., Qi Q., Jiang S., Li J., Du L., Li F. Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J. Virology. 2015;89:6121–6125. doi: 10.1128/JVI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T., Saif L.J., Marthaler D., Esseili M.A., Meulia T., Lin C.M., Vlasova A.N., Jung K., Zhang Y., Wang Q. Cell culture isolation and sequence analysis of genetically diverse US porcine epidemic diarrhea virus strains including a novel strain with a large deletion in the spike gene. Vet. Microbiol. 2014;173:258–269. doi: 10.1016/j.vetmic.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T., Stoltzfus G.T., Zhu C., Jung K., Wang Q., Saif L.J. Attempts to grow human noroviruses, a sapovirus, and a bovine norovirus in vitro. PLoS One. 2018;13 doi: 10.1371/journal.pone.0178157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Jia S., Zhao H., Yin J., Wang X., Yu M., Ma S., Wu Y., Chen Y., Fan W., Xu Y., Li Y. Novel approach for isolation and identification of porcine epidemic diarrhea virus (PEDV) strain NJ using porcine intestinal epithelial cells. Viruses. 2017;9:19. doi: 10.3390/v9010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.