Abstract
The coronavirus disease 2019 (COVID-19) pandemic has focused attention on the need to develop effective therapies against the causative agent, SARS-CoV-2, and also against other pathogenic coronaviruses (CoV) that have emerged in the past or might appear in future. Researchers are therefore focusing on steps in the CoV replication cycle that may be vulnerable to inhibition by broad-spectrum or specific antiviral agents. The conserved nature of the fusion domain and mechanism across the CoV family make it a valuable target to elucidate and develop pan-CoV therapeutics. In this article, we review the role of the CoV spike protein in mediating fusion of the viral and host cell membranes, summarizing the results of research on SARS-CoV, MERS-CoV, and recent peer-reviewed studies of SARS-CoV-2, and suggest that the fusion mechanism be investigated as a potential antiviral target. We also provide a supplemental file containing background information on the biology, epidemiology, and clinical features of all human-infecting coronaviruses, along with a phylogenetic tree of these coronaviruses.
Keywords: Middle east respiratory syndrome, Severe acute respiratory syndrome, SARS-CoV-2, COVID-19, Spike protein, Fusion peptide
Highlights
-
•
SARS-CoV, MERS-CoV, and SARS-CoV-2 entry (receptor binding and membrane fusion) is governed by the viral spike (S) protein.
-
•
A predicted furin cleavage in SARS-CoV-2 differentiates it from SARS-CoV, and may affect its entry and transmissibility.
-
•
The proposed SARS-CoV-2 FP using a pairwise sequence alignment with SARS-CoV shows 93% sequence homology.
-
•
S protein can be activated for early plasma membrane or late endosomal membrane entry depending on protease availability.
-
•
The fusion peptide is well conserved across the CoV family, making it a good target for pan coronavirus antivirals.
1. Introduction
Coronavirus (CoV) cell infection begins with viral entry, in which the viral particle recognizes a host cell receptor and fuses its membrane with the host cell membrane (Belouzard et al., 2012). These two steps are mediated by the coronavirus spike (S) protein. In addition to mediating entry, the S protein is the principal antigenic determinant and the target of neutralizing antibodies (Walls et al., 2016; Yuan et al., 2017; Zhang et al., 2004). This makes the S protein a valuable target in vaccine and antiviral efforts, as summarized in (Du et al., 2017, 2009; Xia et al., 2014). As the urgency to develop effective therapeutics against coronaviruses rises, it is important to also consider broad-spectrum properties of such therapies, as there will likely be future outbreaks for which we will need to be prepared for a rapid response.
Of the two functions that the S protein mediates, there have been considerably more efforts studying therapeutics against receptor binding, as it contains major antigenic determinants (Ying et al., 2015). However, across the coronavirus family, the receptor binding domain is poorly conserved (Belouzard et al., 2012; Graham et al., 2013; Li, 2015), and so therapeutics that target the receptor binding function have low potential as a pan-coronavirus solution. In fact, it was observed that monoclonal antibodies that recognized the SARS-CoV receptor binding domain did not recognize the SARS-CoV-2 receptor binding domain, highlighting the low cross-reactivity in this region (Wrapp et al., 2020). On the other hand, the membrane fusion domain is amongst the most conserved areas in the S protein (Lai et al., 2017; Madu et al., 2009b), and so, targeting membrane fusion may have greater cross-functional success against future coronavirus outbreaks (Xia et al., 2019).
In this article, we review the S protein structure, function, and its role in mediating fusion, and notable therapeutic strategies for blocking fusion, focusing on results obtained on SARS-CoV, MERS-CoV, and recent peer-reviewed studies on SARS-CoV-2. We also provide a supplemental file containing background information on the biology, epidemiology, and clinical features of all human infecting coronaviruses, along with a phylogenetic tree of these coronaviruses.
2. S protein structure and function
As discussed, the CoV S protein has an important role in an infection as it regulates viral entry into host cells and is also the major antigenic determinant, which is usually targeted by the host antibody response (Bosch et al., 2003; Walls et al., 2016; Yuan et al., 2017). These roles emphasize the importance of this protein in both viral entry and interaction with the host's immune system. The S protein is one of 4 structural proteins encoded by the CoV single-stranded, positive-sense RNA genome. In the viral membrane, the S protein participates in two key events: binding to the cellular receptor and inducing fusion between the viral and the cellular membranes (Belouzard et al., 2012). The accomplishment of these two events will drive the release of the viral RNA genome in the host cell and the subsequent start of the viral replication cycle (Masters and Perlman, 2013).
Following viral genome release into the host cell, translation of the ORF produces 16 non-structural proteins that form the viral replicase-transcriptase complex (Fig. 1 A). This complex aids in viral genome replication and subgenomic transcription. The subgenomic RNAs encode four structural proteins, spike (S), envelope (E), membrane (M), and nucleocapsid (N), along with several accessory proteins. Upon translation, the S, E, and M structural proteins are inserted into the rough endoplasmic reticulum. From there, the proteins travel along the secretory pathway to the endoplasmic reticulum-Golgi apparatus intermediate compartment (ERGIC), the site of CoV particle assembly. Afterwards, the virus is released from the cell via exocytosis (de Haan and Rottier, 2005).
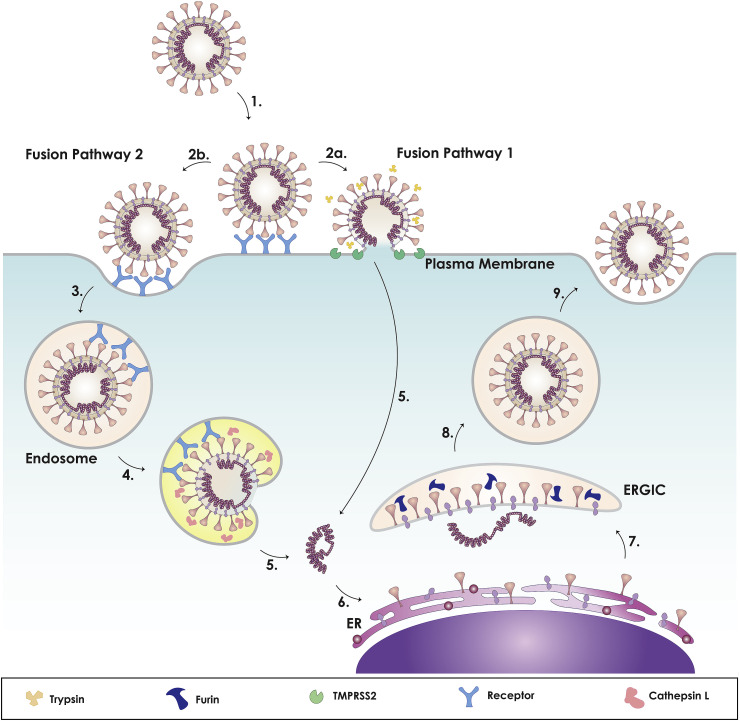
Fig. 1.
Coronavirus spike (S) protein. A. Cartoon figure of the CoV particle (top) and complete CoV viral genome (bottom). CoVs have a lipid envelope with three structural transmembrane proteins: spike (S), membrane (M), and envelope (E). The virus interior contains the viral genome encapsulated by the nucleocapsid (N) protein. The CoV single stranded genome encodes for 16 non-structural proteins, including the papain-like protease (PLpro), 3C-like protease (3CLpro), RNA-dependent RNA polymerase (RdRp), helicase (Hel), and exonuclease (ExoN). The subgenomic RNAs encode four structural proteins: spike (S; dark pink), envelope (E; dark blue), membrane (M; purple), and nucleocapsid (N; magenta) and a number of accessory proteins (Chan et al., 2020; de Wit et al., 2016). B. Cartoon figure of the CoV S protein trimer. C. The CoV S gene denoting the functional components of the protein. The CoV S protein is composed of the two subunits: S1 and S2, encompassing the major functional components: SP (signal peptide, pink); NTD (N-terminal domain; green), CTD (C-terminal domain; light blue), FP (fusion peptide; red), HR1 (heptad repeat 1; purple), HR2 (heptad repeat 2; orange), TM (transmembrane; yellow), and CP (cytoplasmic; dark blue). The S protein has two cleavage sites denoted with dark purple (S1/S2) and pink (S2’) arrows. D. Sequence alignment of S1/S2 cleavage site (dark purple arrow) and S2’ cleavage site (pink) between MERS-CoV, SARS-CoV, and SARS-CoV-2. E. Within the genome, the fusion peptide is highlighted, denoting the sequences from MERS-CoV FP and SARS-CoV FP. Red denotes the conserved residues between MERS-CoV, SARS-CoV, and SARS-CoV-2 FP sequences; blue denotes the SARS-CoV and SARS-CoV-2 FP conserved residues; green denotes the SARS-CoV and MERS-CoV FP conserved residues; purple denotes the MERS-CoV and SARS-CoV-2 conserved residues. The fusion peptide sequence of SARS-CoV-2 was determined by performing a pairwise alignment with MUSCLE through Geneious (version 2020.0.5). Amino acid sequence of the spike proteins was obtained from NCBI Genbank based on the following: SARS-CoV-2 (MN908947.3), MERS-CoV (AFS88936.1), SARS-CoV (AAP13441.1).
In the ERGIC, the S protein monomer is also extensively modified via N-glycosylation and trimerizes (Heald-Sargent and Gallagher, 2012). For some coronaviruses (i.e. MERS-CoV, but not SARS-CoV), the S protein can also be partially processed by furin and furin-like proteases during protein biosynthesis in the Golgi apparatus depending on the host cell type (Kleine-Weber et al., 2018; Millet and Whittaker, 2014; Yang et al., 2014). In some coronaviruses, the S protein that is not incorporated into the viral particle could be transferred to the cell surface (Fehr and Perlman, 2015).
Structurally, the S protein is a ~180–200 kDa type I transmembrane protein, with the N-terminus facing the extracellular space, held in the viral membrane via its transmembrane domain, with a short C-terminal segment facing the intracellular space (Bosch et al., 2003). The extracellular domain is split into two subunits or domains, S1 and S2, which mediate receptor binding and membrane fusion, respectively (Fig. 1B and C). Visually, the S protein trimers form a characteristic bulbous, crown-like halo surrounding the viral particle, for which coronaviruses were named. Structural modeling of coronavirus S protein monomers show that the S1 and S2 subunit form the bulbous head and stalk region, respectively. Aspects of the structure will be referred to throughout the article (Fig. 2 ).
Fig. 2.
MERS-CoV (A), SARS-CoV (B), and SARS-CoV-2 (C) protein models. Models were built to show the predicted structure of the S1/S2, the S2′ cleavage site and the FP, which are not solved in cryo-EM structures. Trimers and monomers were modeled using SARS-CoV (PDB# 5X58) and MERS-CoV (PDB# 6Q05) structures using the methodology described in (Jaimes et al., 2020b). Color scheme is as described for Fig. 1.
The S1 subunit contains two subdomains, the N-terminal domain (NTD) and the C-terminal domain (CTD) (Fig. 1C). One or both subdomains can serve as the receptor-binding domain (RBD) with the piece of the RBD that directly contacts the receptor termed the receptor-binding motif (RBM). A general rule of thumb is that the NTD mediates viral binding to sugar-based receptors, whereas the CTD mediates binding to protein-based receptor, though there are exceptions, for example the mouse hepatitis virus NTD recognizes proteinaceous CEACAM1 as its receptor (Peng et al., 2011).
Both SARS-CoV and SARS-CoV-2 utilize the CTD to bind angiotensin-converting enzyme 2 (ACE2) (Li et al., 2003; Zhou et al., 2020), abundantly detected on lung and small intestine cells (Hamming et al., 2004). The amino acid differences in the RBM (~50% sequence homology) across SARS-CoV and SARS-CoV-2 have been hypothesized to strengthen or weaken RBM-ACE2 associations, though it is unclear if the overall effect of all these differences results in stronger recognition of SARS-CoV-2 for ACE2 (Yan et al., 2020). It is reported that SARS-CoV-2 S does bind ACE2 with 10- to 20-fold higher affinity than SARS-CoV S binding to ACE2 (Wrapp et al., 2020). However, another study reported that purified SARS-CoV-2 and SARS-CoV RBD binds ACE2 with similar affinities (Walls et al., 2020), so further experiments should be conducted to determine the impact of residue changes on ACE2 binding.
MERS-CoV utilizes the CTD to bind proteinaceous dipeptidyl peptidase 4 (DPP4) (Raj et al., 2013), detected on lung and kidney cells (van Doremalen et al., 2014). The well-conserved nature of DPP4 across a wide variety of species (e.g. bats, dromedaries, humans) provides more insight into the zoonotic capabilities of MERS-CoV. Although MERS-CoV does not infect murine cells since MERS-CoV S does not bind to mouse DPP4, it is possible to infect mice by replacing blades 4 & 5 of mouse DPP4 with blades from animals susceptible to MERS-CoV S binding, forming chimeric DPP4s (Barlan et al., 2014). With the ability to interchange blades 4 & 5, it was determined that MERS-CoV S preferentially binds to human, horse, camel, goat, and bat DPP4, listed in decreasing order (Barlan et al., 2014). Another contributing factor to the zoonotic potential of MERS-CoV comes from reports that MERS-CoV S has the ability to bind sialic acid receptors in addition to DPP4 (Li et al., 2017). Recent work further revealed that sialic acid receptors play a role in transmissibility between species, as they reaffirmed the binding affinity reported by the Barlan group (Widagdo et al., 2019).
Significant progress has been made to understand the RBD-receptor interaction, and crystal structures of the SARS-CoV S (Li et al., 2005; Song et al., 2018), MERS-CoV S (Wang et al., 2013) and SARS-CoV-2 S (Yan et al., 2020) in complex with its receptor have been determined. Similar studies have revealed that receptor binding requires one of the trimers to be in the “up” position for SARS-CoV and SARS-CoV-2, but not for MERS-CoV (Gui et al., 2017; Song et al., 2018; Yan et al., 2020; Yuan et al., 2017).
The CoV S protein is also classified as a class I viral fusion protein, based on the structure of its fusion subunit (White et al., 2008). Within this class, the fusion subunit is largely composed of α-helical secondary structures (Fig. 2), and its function is regulated through proteolytical priming or cleavage at specific sites to induce the fusion-competent state of the S protein (White and Whittaker, 2016). The S2 or fusion subunit contains a variety of motifs (Fig. 1C), starting with the fusion peptide (FP) which is the functional fusogenic element of the S protein. The FP describes a short segment (15–25 amino acids), conserved across the viral family that is composed of mostly hydrophobic residues, such as glycine (G) or alanine (A), which inserts in the host cell membrane to trigger the fusion event (Epand, 2003). Fusion peptides tend to be sensitive to point mutations, in that a single mutation can negate fusion (Madu et al., 2009b). However, the fusion peptide is loosely defined, as these requirements are not absolute but rather serve as guidelines to identify the fusion peptide region.
For SARS-CoV, several regions have been suggested as the FP. Using a Wimley and White interfacial hydrophobicity scale to identify regions with a higher propensity to insert into membranes, the region 770–788 was identified and a peptide corresponding to this region was shown to induce fusion and membrane leakage in large unilamellar vesicles (Sainz et al., 2005). Further work identified regions 873–888 and 1185–1202 as strong membrane interacting regions and proposed that these regions, in conjunction with 770–788, work synergistically to mediate fusion (Guillén et al., 2008a, 2008b). Separately, the region 798–835 was also identified as a fusion peptide, since single point mutagenesis studies demonstrated its importance in fusion, with an extremely conserved region, SFIEDLLFNKV (798–808) (Madu et al., 2009b, 2009a). An excellent review that discusses the journey of identifying the SARS-CoV FP and key findings is (Millet and Whittaker, 2018).
Research specifically focusing on the MERS-CoV FP has been rather limited. Sequence alignment, using MUSCLE software from the Geneious bioinformatic software platform, based on the SARS-CoV FP suggested that the highly conserved SFIEDLLFNKV motif is similar within MERS-CoV and a potential region of the MERS-CoV FP was revealed (Lai et al., 2017). The sequence region 888–898, RSARSAIEDLLFDKV, was strongly suggested to comprise the MERS-CoV FP based on single point mutagenesis screening with giant unilamellar vesicles identifying the critical hydrophobic residues for syncytium-forming ability (Alsaadi et al., 2019).
Based on current understanding of the SARS-CoV FP, we suggest here a preliminary SARS-CoV-2 FP using a pairwise sequence alignment. The suggested FP has 93% sequence homology with SARS-CoV FP, displaying strong conservation across these two viruses. The current understanding of FP residues are as described in Fig. 1E.
Downstream of the FP are the heptad repeat regions (HR1 and HR2), with HR1 more N-terminal and longer than HR2. Both HR1 and HR2 are composed of repetitive heptapeptide, HPPHCPC, in which H represents hydrophobic or traditionally bulky residues, P are polar or hydrophilic residues, and C are other charged residues (Chambers et al., 1990). This allows the HR region to adopt an α-helix secondary structure with a hydrophobic interface to drive membrane fusion (White et al., 2008). Following the HR2 region is the transmembrane (TM) domain, which anchors the S protein in the viral membrane. Lastly, at the C-terminal end of the S protein is the cytoplasmic tail.
3. Proteolytic activation of CoV S
As reiterated, the CoV S drives receptor binding and membrane fusion. However, in order to catalyze the membrane fusion reaction, the S protein needs to be primed by an appropriate protease at the S1 and S2 interface (S1/S2) and triggered immediately upstream of the FP (S2’) (Fig. 1D). What is fascinating about this triggering event is that several different proteases can trigger, and it is the protease requirements that drive viral tropism. Both SARS-CoV and MERS-CoV S can be triggered to fuse at either the plasma membrane or the endosomal membrane, and the route of entry is dependent on protease availability (Fig. 3 ). The emerging SARS-CoV-2 also utilizes both pathways (Hoffmann et al., 2020). If the plasma membrane-route proteases are available, the virus can fuse via an “early pathway” at the plasma membrane, but if not, the virus can fuse via a “late pathway” at the endosomal membrane. Intriguingly, the activation of coronavirus fusion varies, depending on the protease in the local environment, highlighting the flexibility of coronavirus S proteins to respond to the particular chemical cues available. In this section, we will review both routes, as well as the proteases involved in activating S.
Fig. 3.
Model of coronavirus dual entry pathway. This model depicts the two methods of viral entry: early pathway and late pathway. As the virus binds to its receptor (1), it can achieve entry via two routes: plasma membrane or endosome. For SARS-CoV: The presence of exogeneous and membrane bound proteases, such as trypsin and TMPRSS2, triggers the early fusion pathway (2a). Otherwise, it will be endocytosed (2b, 3). For MERS-CoV: If furin cleaved the S protein at S1/S2 during biosynthesis, exogeneous and membrane bound proteases, such as trypsin and TMPRSS2, will trigger early entry (2a). Otherwise, it will be cleaved at the S1/S2 site (2b) causing the virus to be endocytosed (3). For both: Within the endosome, the low pH activates cathepsin L (4), cleaving S2′ site, triggering the fusion pathway and releasing the CoV genome. Upon viral entry, copies of the genome are made in the cytoplasm (5), where components of the spike protein are synthesized in the rough endoplasmic reticulum (ER) (6). The structural proteins are assembled in the ER-Golgi intermediate compartment (ERGIC), where the spike protein can be pre-cleaved by furin, depending on cell type (7), followed by release of the virus from the cell (8, 9). For SARS-CoV-2: Studies currently show that SARS-CoV-2 can utilize membrane bound TMPRSS2 or endosomal cathepsin L for entry and that the S protein is processed during biosynthesis. Other factors that can influence the viral entry pathway are calcium and cholesterol (not shown).
3.1. Plasma membrane route (“early pathway”)
An early indication that SARS-CoV can utilize the plasma membrane pathway came from cell-cell fusion studies that found that HEK293T cells transiently expressing the SARS-CoV S protein mediated cell-cell fusion with target Vero E6 cells when treated with a low concentration of trypsin, whereas a similar setup utilizing a low pH pulse in the absence of trypsin did not mediate any fusion (Simmons et al., 2004). Likewise, trypsin was also found to induce MERS-CoV and SARS-CoV-2 S protein mediated cell-cell fusion of Vero E6 cells and 293T cells, respectively (Ou et al., 2020; Shirato et al., 2013). Subsequent studies using retroviral pseudoparticles expressing SARS-CoV S protein (SARSpp) or MERS-CoV S protein (MERSpp) revealed that trypsin treatment of these pseudoparticles after binding to their respective receptor resulted in effective infection at the plasma membrane surface (Qian et al., 2013; Simmons et al., 2005). In fact, pretreating the S protein prior to receptor binding was found to abrogate infection, and this is hypothesized to occur because trypsin treatment causes the S protein to undergo irreversible conformational changes so that it cannot mediate fusion (Matsuyama et al., 2005; Park et al., 2016; Simmons et al., 2005). This phenomenon has yet to be investigated for SARS-CoV-2. Another comprehensive study investigating the impact of a panel of additional exogenous proteases on SARS-CoV and MERS-CoV live virus infection of Vero E6 cells revealed that thermolysin and elastase also efficiently mediate cellular entry at the plasma membrane after binding (Belouzard et al., 2010; Matsuyama et al., 2005; Shirato et al., 2013). The finding that elastase could mediate entry is clinically important, since elastase is produced by inflammatory cells in the lungs during SARS-CoV infection and could thus promote the progression of SARS-CoV infection.
Although these exogeneous proteases are capable of activating fusion, they do not provide insights into the fusion mechanism in the human respiratory tract, a major target for coronaviruses. This is because coronaviruses should only be cleaved after receptor engagement and not before, but this timing would be difficult to control if exogenous proteases were the in vivo activator. Thus, transmembrane proteases are of interest as they are localized on the plasma membrane surface where the virus encounters the receptor. The type II transmembrane serine proteases (TTSPs) are a family of such proteases anchored in the cellular membrane and have already been implicated in influenza virus infection (Choi et al., 2009). In particular, the transmembrane protease/serine subfamily member 2 (TMPRSS2) and TMPRSS4 were found to activate and enable influenza virus spread, even in the absence of extracellular trypsin (Bertram et al., 2010; Böttcher et al., 2006; Chaipan et al., 2009).
Inspired by the impact of TTSP on influenza virus spread, researchers have also sought to investigate whether similar TTSPs could impact SARS-CoV and MERS-CoV fusion. An initial study demonstrated that a soluble form of a TTSP protease, TMPRSS11a, could cleave and activate SARS-CoV S protein for fusion (Kam et al., 2009). Subsequent studies investigated whether membrane-bound TTSP proteases were capable of activating the SARS-CoV S protein also. It was determined that cell lines transiently expressing TMPRSS2 support both SARSpp and live SARS-CoV infection at the plasma membrane without exogeneous or late-pathway proteases (Glowacka et al., 2011; Matsuyama et al., 2010; Shulla et al., 2011). Similar studies with MERSpp and live MERS-CoV also identified membrane-bound TMPRSS2 as an activator of MERS-CoV infection (Gierer et al., 2013; Qian et al., 2013; Shirato et al., 2013). SARS-CoV-2 pseudoparticles (SARS2pp) and live SARS-CoV-2 also utilize membrane-bound TMPRSS2 for plasma membrane entry (Hoffmann et al., 2020; Matsuyama et al., 2020), much like SARS-CoV and MERS-CoV.
Other TTSP proteases have been found to support, in various degrees, CoV infection at the plasma membrane surface. Utilizing MERSpp, it was shown that TMPRSS11a and TMPRSS11e can also activate MERS-CoV S for viral infection (Zmora et al., 2018). However, the TTSP family is not a blanket activator of coronavirus S mediated fusion. Although TMPRSS4 was found to activate SARS-CoV and MERS-CoV S mediated cell-cell fusion, it was ineffective in promoting viral infection (Glowacka et al., 2011; Qian et al., 2013). Transient expression of TMPRSS2, 4, 11a, 11d, and 11e were shown to enhance SARS-CoV-2 S-mediated cell-cell fusion, though it is unknown if these proteases activate S for infection (Ou et al., 2020). The discrepancies in these findings could result from high S expression and/or interaction of large surface areas in cell-cell fusion assays that encourage more fusion than would be possible in viral-cell fusion (Glowacka et al., 2011). More intriguingly, TTSP activation can also differ between SARS-CoV and MERS-CoV. TMPRSS11d, also known as human airway trypsin-like protease (HAT), is unable to activate SARS-CoV S infection, but can activate MERS-CoV S infection (Bertram et al., 2011; Zmora et al., 2018). While differences between SARS-CoV and MERS-CoV activation will be further discussed in the following paragraph, it is noteworthy to mention that the coronavirus S has evolved to have flexibility in a variety of proteases that can cleave and activate it, providing a possible rationale for their zoonotic potential.
Although a high degree of sequence conservation exists between the SARS-CoV and MERS-CoV S2 membrane fusion domains, there are key differences in their fusion mechanism for plasma membrane entry. For one, MERS-CoV S contains a furin cleavage site at S1/S2, whereas SARS-CoV S does not (Fig. 1D) (Millet and Whittaker, 2014). Furin or furin-like proteases found in the trans-Golgi network (TGN) can encounter and act on such cleavage sites in coronavirus S as it is being synthesized (Millet and Whittaker, 2015). MERS-CoV has been observed to be cleaved during biosynthesis at the S1/S2 position (Millet and Whittaker, 2014), and this pre-cleavage was shown to promote subsequent MERS-CoV cleavage by TMPRSS2 at the S2′ position to activate plasma membrane fusion, whereas if there was no pre-cleavage, immediate entry via TMPRSS2 was not observed (Park et al., 2016). Furin activation of MERS-CoV at the S2′ position for entry has also been observed, based on the presence of specific basic residues (Millet and Whittaker, 2014), but the exact role is still unclear (Matsuyama et al., 2018). There are no indications that SARS-CoV or SARS-CoV-2 S can be cleaved by furin at S2’.
SARS-CoV S is not cleaved during biosynthesis and does not require a S1/S2 pre-cleavage event for plasma membrane fusion. While it is suspected that the S1/S2 cleavage event can cause conformational changes that further exposes the S2’ site for immediate plasma membrane fusion, it is noted that there can be alternative ways to cause these conformational changes, such as receptor binding (Park et al., 2016). Indeed, it is observed that SARS-CoV S binds its receptor with a 10- to 20- fold higher affinity compared to that of MERS-CoV S with its receptor, so stronger receptor binding can perhaps compensate for uncleaved S1/S2, at least in the case for SARS-CoV to support plasma membrane entry. It has been suggested that the S1/S2 site may enable these coronaviruses to spread and infect new organisms with low receptor affinity. Interestingly, TMPRSS2 overexpression was observed to allow uncleaved MERS-CoV to infect cells, bypassing the required S1/S2 cleavage event (Kleine-Weber et al., 2018), once again highlighting the adaptability of the coronavirus S protein.
Unexpectedly, SARS-CoV-2 S also possesses a potential furin cleavage site at the S1/S2 region, which is unique for SARS-like CoVs (Coutard et al., 2020; Walls et al., 2020; Wrapp et al., 2020; Jaimes et al., 2020a). Western blots have shown that S proteins in SARS2pp can be processed at the S1/S2 boundary during biosynthesis, similar to what was reported for MERS-CoV (Walls et al., 2020). However, the impact of the potential furin cleavage site and the accompanying extended structural loop at S1/S2 remains to be determined.
3.2. Endosomal route (“late pathway”)
In the absence of exogenous or membrane-bound proteases that enable entry at the plasma membrane surface, coronaviruses can be internalized via clathrin- and non-clathrin-mediated endocytosis (Inoue et al., 2007; Wang et al., 2008). A caveat is that if MERS-CoV is not pre-cleaved at the S1/S2 site during biosynthesis, then it will also be endocytosed, regardless of the presence of plasma membrane proteases (Park et al., 2016). As the virus is shuttled along the endocytic pathway towards the cell interior, the pH in the endosome decreases. For some viruses (e.g. influenza, vesicular stomatitis virus), the presence of low pH triggers fusion. As such, treating the cells with lysosomotropic agents to prevent endosomal acidification abrogates infection of these pH-dependent viruses (Ochiai et al., 1995). Initial experiments infecting Vero E6 cells with SARSpp in the presence of lysosomotropic agents suggested that SARSpp was sensitive to low pH conditions (Simmons et al., 2004). However, since a pH pulse could not mediate cell-cell fusion and SARSpp exposure to low pH prior to binding did not reduce infectivity, there were grounds to suspect other factors may be involved in CoV endosomal fusion.
The low pH environment also activates endosomal proteases, such as cathepsins, a family of cysteine proteases. Of interest are cathepsin B and cathepsin L, which become active in the early and late endosome, respectively, and are known activators of other members in the CoV family (Qiu et al., 2006; Regan et al., 2008). Indeed, subsequent studies demonstrated that the cathepsin B/L inhibitor MDL28170 drastically reduced SARSpp (Huang et al., 2006; Simmons et al., 2005) and MERS-CoV entry into MRC-5 cells (Gierer et al., 2013). Studies on SARS-CoV-2 have echoed similar themes; SARS2pp are sensitive to cathepsin B/L inhibitors (Hoffmann et al., 2020; Ou et al., 2020). Confirmation that all these coronaviruses are activated by cathepsin L for fusion in the late endosome was determined using inhibitors specific for either cathepsin B or L (Ou et al., 2020; Qian et al., 2013; Simmons et al., 2005), along with direct observation of SARS-CoV trafficking to late endosomes (Mingo et al., 2015). Thus, it is believed that SARS-CoV, MERS-CoV, and SARS-CoV-2 dependency on low pH in the endosomal route is indirect; acidic conditions are required to activate cathepsin L protease, which in turn then act on S, resulting in a virus primed to undergo subsequent fusion steps. To further confirm the independence of these coronaviruses entry from pH, it is important to reiterate that both viruses can undergo plasma membrane fusion, setting a precedence for neutral pH fusion.
3.3. SARS-CoV and MERS-CoV cleavage sites
Although both the SARS-CoV and the MERS-CoV S protein can be activated by a similar array of proteases, it is crucial to mention that these proteases act on the S protein at different sites, which may lead to slightly different activities (Table 1 ). TMPRSS2 has been reported to cleave both SARS-CoV and MERS-CoV at the S2’ site to activate plasma membrane fusion (Kleine-Weber et al., 2018; Reinke et al., 2017). However, for this activation, MERS-CoV must be pre-cleaved by furin at the S1/S2 site during S biosynthesis (Kleine-Weber et al., 2018; Millet and Whittaker, 2014; Park et al., 2016), whereas SARS-CoV does not need cleavage at the S1/S2 for activation (Reinke et al., 2017). Although likely, it is yet to be formally determined if SARS-CoV-2 also needs to be pre-cleaved at the S1/S2 site for plasma membrane fusion.
Table 1.
SARS-CoV and MERS-CoV activating proteases, locations, and sites. Both SARS-CoV and MERS-CoV S protein contain two sites (S1/S2 and S2’) that can be cleaved by various proteases. For each protease, this table details its location in the cell (TGN: trans-Golgi network, plasma membrane bound, endosome, exogenously found), the fusion pathway it triggers, and its cleavage sites. Since cleavage at a particular site may not necessarily activate the S protein for fusion, this table distinguishes between sites that are just cleaved versus sites that result in S activation.
| Protease | Location | Pathway | SARS-CoV Sites | MERS-CoV Sites |
|---|---|---|---|---|
| Furin | TGN | Biosynthesis | Does not activate | Cleaves at RSVR (S1/S2) (Kleine-Weber et al., 2018; Millet and Whittaker, 2014) |
| Trypsin | Exogeneous | Plasma Membrane | Activates at R667 (S1/S2), R797 (S2′) sequentially (Belouzard et al., 2009) | Activates at unidentified sites (Shirato et al., 2013) |
| Elastase | Exogeneous | Plasma Membrane | Activates at T795 (S2′) (Belouzard et al., 2009) | Activates at unidentified sites (Shirato et al., 2013) |
| Thermolysin | Exogeneous | Plasma Membrane | Activates at unidentified sites (Matsuyama et al., 2005) | Activates at unidentified sites (Shirato et al., 2013) |
| TMPRSS2 | Membrane Bound | Plasma Membrane | Cleaves at R667 (S1/S2), activates at R797 (S2′) (Reinke et al., 2017) | Activates at RSAR (S2′) Needs prior S1/S2 cleavage (Kleine-Weber et al., 2018) |
| Cathepsin L | Late endosome | Endosomal | Cleaves S1/S2 at T678 (Bosch et al., 2008), activates at unidentified sites | Activates at RSAR (S2′) and unidentified sites (Kleine-Weber et al., 2018) |
The role of the furin cleavage site in the S1/S2 position as it relates to viral tropism and pathogenicity is of interest to the scientific community. In the case of influenza virus, low-pathogenicity influenza strains contain a single basic residue cleavage site, whereas highly pathogenic influenza strains have a polybasic furin cleavage site (Sun et al., 2010). It has been suggested that the presence of furin cleavage sites in influenza HA expands viral tropism, as furin and furin-like proteases are ubiquitously expressed in most cell lines. Thus, it has been hypothesized that the additional furin site present at S1/S2 may also allow SARS-CoV-2 to spread more efficiently than other SARS-like-CoVs (Coutard et al., 2020; Walls et al., 2020), though experimental work will need to be done to show whether this is the case. Of interest is to also determine how SARS-CoV-2 acquired this cleavage site, whether from animal-to-human or human-to-human transmission (Andersen et al., 2020), in order to determine the pathogenicity of emerging coronaviruses.
Trypsin, an exogeneous activator of plasma membrane fusion, has different cleavage requirements than TMPRSS2. For SARS-CoV, trypsin shows elements of a two-step activation process; it first cleaves arginine at the SARS-CoV S1/S2 site (R667), followed by the S2′ site (R797) for fusion (Belouzard et al., 2009), whereas TMPRSS2 does not require the S1/S2 cleavage site. This aspect highlights the flexibility of the CoV FP as it can be cleaved by different proteases resulting in slightly different activities. As further evidence of CoV FP flexibility, elastase cleaves SARS-CoV S at the S2′ site at T795 (Belouzard et al., 2010), which is two residues upstream of the trypsin cleavage site at R797, suggesting that the S protein can even accommodate cleavages at different sites. However, mutagenesis studies have demonstrated that elastase mediates maximal fusion when cleaving at the 797 site, showing that while the S protein is flexible in cleavage location, it has preferred sites. For MERS-CoV and SARS-CoV-2, the exact sites at S1/S2 and S2’ position for exogeneous protease cleavage have yet to be determined and it would be interesting to observe if small variations in the sites exist across different proteases in these cases as well.
Regarding endosomal pathway-related cleavage, it has been suggested that cathepsin L can cleave SARS-CoV at a site slightly downstream of the S1/S2 site at T678, though fusion functional studies have yet to be conducted (Bosch et al., 2008). For MERS-CoV, functional studies have shown that S1/S2 and S2′ sites are dispensable, though cathepsin L activation of MERS-CoV is slightly reduced when the S2′ is mutated (Kleine-Weber et al., 2018). This seems to indicate that cathepsin L may be using auxiliary S1/S2 and/or S2’ sites for activation. It is difficult to screen for these sites since cathepsin has a relatively indiscriminate recognition motif, though studies have shown that cathepsin L prefers aromatic residues at the P2 location (Biniossek et al., 2011). While it seems that SARS-CoV, MERS-CoV, and SARS-CoV-2 exhibit similar behavior when entering via the endocytic route, it is important to emphasize that the cathepsin L cleavage sites are unknown and it is possible that the location or the number of sites vary and could in turn, impact entry efficiency of MERS-CoV or SARS-CoV-2 endocytic route. All in all, the extensive range of proteases that activate SARS-CoV, MERS-CoV, and SARS-CoV-2 attest to their promiscuity in infecting a wide range of cell types that present these proteases and may explain the expanded tropism of these viruses.
3.4. CoV entry remarks
As described above, SARS-CoV, MERS-CoV, and SARS-CoV-2 can enter cells using an early pathway or a late pathway, depending on protease availability and cell type (Fig. 3). If membrane-bound proteases, most notably TMPRSS2, are available on the host cell, they can cleave both viruses for early fusion at the plasma membrane surface after receptor binding. As noted, MERS-CoV also requires the S1/S2 cleavage during biosynthesis for plasma membrane fusion (this has yet to be determined for SARS-CoV-2). Otherwise, the virus is endocytosed and the increasingly acidic conditions can activate cathepsin L to trigger fusion at the endosomal membrane. Table 2 summarizes cell lines susceptible to SARS-CoV and MERS-CoV infection, as well as their infection pathways. Initial studies of SARS-CoV-2-susceptible cell lines can be found in (Hoffmann et al., 2020; Ou et al., 2020). A general rule of thumb is that these coronaviruses will utilize the plasma membrane route in lung cells as TMPRSS is commonly expressed in lung cells and the endosomal route for all other cell types.
Table 2.
SARS-CoV and MERS-CoV entry pathways in commonly used cell lines. For each cell line, the pathway that SARS-CoV or MERS-CoV uses to infect that cell line is described. If the pathway is unknown, it is noted if the cell line is susceptible to either SARS-CoV or MERS-CoV or if the cell line has been tested.
| Cell Type | Cell Type | SARS-CoV Infection Notes | MERS-CoV Infection Notes |
|---|---|---|---|
| 16HBE | Human bronchial epithelia | Likely using early pathway (Kam et al., 2009) | Not tested |
| A549 | Human alveolar basal epithelial carcinoma | Lacks ACE2, does not infect (Kam et al., 2009; Simmons et al., 2004) | Weak/No infection (Gierer et al., 2013; Qian et al., 2013) |
| BEAS-2B | Human bronchial epithelial | Lacks ACE2, does not infect (Kam et al., 2009) | Not tested |
| BSC-1 | African Green Monkey respiratory epithelial | Late pathway (Mingo et al., 2015) | Not tested |
| Caco-2 | Human colon epithelial adenocarcinoma | Infects (Mossel et al., 2005) | Early pathway (Gierer et al., 2013; Kleine-Weber et al., 2018) Could use both pathways (Park et al., 2016) |
| Calu-3 | Human bronchial epithelial adenocarcinoma | Early pathway (Kawase et al., 2012) | Early pathway (Park et al., 2016; Shirato et al., 2013) |
| COS-7 | African Green Monkey kidney fibroblast | Lacks ACE2, does not infect (Simmons et al., 2004) Late pathway with ACE2 transfection (Inoue et al., 2007) |
Not tested |
| EA-HY | Human endothelium | Not tested | Does not infect (Gierer et al., 2013) |
| HEK293T | Human embryonic kidney | Late pathway (Simmons et al., 2004) Late pathway with ACE2 transfection (Bertram et al., 2011; Huang et al., 2006; Shulla et al., 2011; Wang et al., 2008) |
Late pathway with DPP4 transfection (Gierer et al., 2013) |
| HeLa | Human cervix epithelial adenocarcinoma | Late pathway with ACE2 transfection (Kawase et al., 2012) | Lacks DPP4, does not infect (Shirato et al., 2013) |
| HepG2 | Human liver carcinoma | Late pathway (Inoue et al., 2007) | Not tested |
| HOS | Human bone, fibroblast/epithelial osteosarcoma | Does not infect (Simmons et al., 2004) | Infects (Gierer et al., 2013) |
| HT1080 | Human fibrosarcoma | Infects (Simmons et al., 2004) | Not tested |
| Huh-7 | Human liver carcinoma | Infects (Simmons et al., 2004) | Late pathway (Millet and Whittaker, 2014; Park et al., 2016) |
| Human Airway Epithelial | Primary | Not tested | Early pathway (Park et al., 2016) |
| LLCMK2 | Rhesus Macaque kidney epithelial | Not tested | Late pathway (Qian et al., 2013) |
| MRC-5 | Human lung fibroblast | Not tested | Likely late pathway (Gierer et al., 2013; Millet and Whittaker, 2014; Shirato et al., 2013) |
| NHBE | Primary normal human bronchial epithelial cells | Not tested | Late pathway (Millet and Whittaker, 2014) |
| RPE | Human retina epithelial | Not tested | Does not infect (Gierer et al., 2013) |
| U373 | Human glioblastoma | Not tested | Does not infect (Gierer et al., 2013) |
| Vero E6 | African Green Monkey kidney epithelial | Late pathway (Glowacka et al., 2011; Huang et al., 2006; Matsuyama et al., 2010; Simmons et al., 2004) | Late pathway (Kleine-Weber et al., 2018; Millet and Whittaker, 2014; Qian et al., 2013; Shirato et al., 2013) |
| WI-38 | Human lung fibroblast | Not tested | Likely late pathway (Millet and Whittaker, 2014; Shirato et al., 2013) |
Although a variety of triggers exist to dictate the pathway of coronavirus entry and fusion, it is worth considering whether these pathways are unique or redundant. Early studies seem to suggest that the plasma membrane pathway is the preferred route, as it enables the virus to directly enter the cells and disseminate. In fact, it has been shown that exogeneous trypsin treatment enhanced SARS-CoV replication in Vero cells by 100-fold when entering via the plasma membrane route as opposed to the endosomal route (Matsuyama et al., 2005). The delayed viral growth kinetics associated with the endosomal route could be attributed to the 30-minute lag time the virus requires to traffic from the cellular membrane to the endosomal membrane.
Moreover, the plasma membrane pathway has also been implicated as the clinically relevant pathway (Shulla et al., 2011). SARS-CoV, MERS-CoV, and SARS-CoV-2 replicate rapidly in the lungs during disease progression, and lung epithelial cells have been reported to have high TMPRSS2 expression (Donaldson et al., 2002), suggesting that these viruses can employ TMPRSS2 for entry. This view is further supported by observations that SARS-CoV receptor, ACE2, was found to colocalize with TMPRSS2 so that after receptor binding, SARS-CoV can immediately be processed by TMPRSS2 for entry (Shulla et al., 2011). It will be of interest to observe if TMPRSS2 colocalization with ACE2 can also immediately process SARS-CoV-2 entry, and likewise for MERS-CoV, if the MERS-CoV receptor, DPP4, can also colocalize with TMPRSS2 to support plasma membrane fusion.
Further support for the relevance of the plasma membrane pathway comes from experiments with Calu-3 cells, an immortalized human lung cell line reported to have mRNA expression levels of ACE2, DPP4, TMPRSS2, and cathepsin L similar to the human lung, so that they may serve as a good representative model for SARS-CoV, MERS-CoV, and SARS-CoV-2 susceptibility. Using Calu-3 cells, researchers have shown that both SARS-CoV and MERS-CoV infection are mitigated using a TMPRSS2 inhibitor, camostat, but not with a cathepsin inhibitor, EST (Kawase et al., 2012; Shirato et al., 2013). This implies that TMPRSS2-mediated entry is the preferred pathway, since cathepsin inhibitors have minimal impact on entry when the TMPRSS2 route is available. In fact, mice treated with a TMPRSS2 inhibitor had a higher survival rate (60%) than mice treated with a cathepsin inhibitor (10%) when challenged with a lethal dose of SARS-CoV (Zhou et al., 2015). Similar studies also allude to the importance of TMPRSS2 in SARS-CoV-2 infection as camostat significantly reduced both SARS2pp and live SARS-CoV-2 infection of Calu-3 cells (Hoffmann et al., 2020). Interestingly, treatment of Calu-3 cells with both camostat and EST drastically reduced SARS-CoV, but not MERS-CoV infection, much more so than with camostat alone (Kawase et al., 2012; Shirato et al., 2013). This suggests that SARS-CoV can more easily employ the late pathway than MERS-CoV.
These studies bring into question the importance of the late pathway, especially as cathepsin L is not as highly expressed as TMPRSS2 in respiratory cells (Park et al., 2016). Furthermore, for other human coronaviruses, it has been suggested that replication in cell culture may select for cathepsin L activation (Kleine-Weber et al., 2018). Intriguingly, usage of non-lung cell culture lines that support SARS-CoV and MERS-CoV infection, such as Vero E6 (SARS-CoV/SARS-CoV-2) or Huh-7 (MERS-CoV) have demonstrated that the cathepsin L pathway is much more prominent as these cells express minimal TMPRSS2 and infection is strongly reduced in the presence of cathepsin inhibitors (Hoffmann et al., 2020; Park et al., 2016; Simmons et al., 2005). While the TMPRSS2 route seems to hold higher clinical significance, it is important to consider that in the later stages of SARS-CoV and MERS-CoV infection, the virus may spread beyond the lungs, especially as MERS-CoV infection may result in kidney failure. Thus, studies of other cellular systems are highly valued to enable comprehensive understanding SARS-CoV and MERS-CoV entry pathways and how it depends on cell type.
4. S protein membrane fusion
Viral membrane fusion is the process by which enveloped viruses merge their membrane with the host cell membrane so that the virus can deliver its genome in the cell, resulting in the eventual production of new virions (Harrison, 2015, 2008). Coronavirus membrane fusion occurs after receptor binding, so that both its membrane and the host cell membrane are proximal. However, membrane fusion is not a spontaneous process, as there are high energy requirements to bring the membranes close together (Cohen and Melikyan, 2004; Martens and McMahon, 2008). It is the role of the viral fusion protein to serve as the catalyst by providing the energy requirement to drive this reaction.
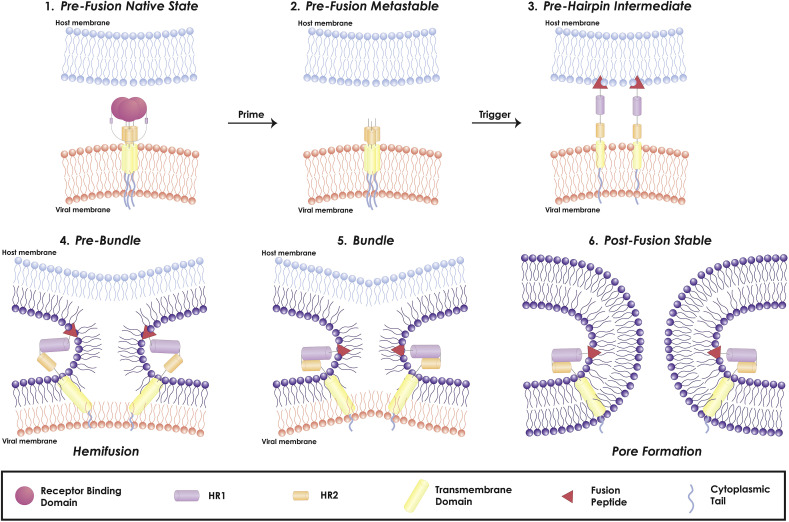
Viral fusion proteins are grouped into three distinct classes (I, II and III) based on their structure and function (White et al., 2008; White and Whittaker, 2016). As mentioned previously, the coronavirus S protein is a class I fusion protein, due to the structural characteristics of its fusion domain, the need for protease cleavage to be fusion-competent and the presence of heptad repeats that fold into a six-helix bundle (Bosch et al., 2003). Class I fusion proteins catalyze the membrane fusion reaction though a sequence of states: (1) pre-fusion native state, (2) pre-fusion metastable state, (3) pre-hairpin intermediate state, (4) post-fusion stable state (Fig. 4 ). As the S protein is synthesized, it adopts the pre-fusion native state. Proteolytic processing at the S1/S2 will cause the S protein to adopt a pre-fusion metastable state. This priming event generates separate S1 and S2 domains, which are then non-covalently associated. (Tripet et al., 2004).
Fig. 4.
Coronavirus viral fusion pathway model based on class I fusion protein understanding. The captions above the figure describe the state of the fusion protein, the captions below describe the state of the membranes. The S protein starts in the pre-fusion native state (1) and undergoes priming of the S1 subunit by relevant proteases to achieve the pre-fusion metastable state (2). Subsequent triggering by relevant proteases will enable the FP to insert in the host membrane and allow the S protein to form the pre-hairpin intermediate (3). The pre-hairpin begins to fold back on itself due to HR1 and HR2 interactions forming the pre-bundle (4), bundle (5), and eventual post-fusion stable (6) states. During the S protein foldback, the two membranes will approach each other until the outer leaflets merge (hemifusion) and eventually the inner leaflets merge (pore formation). Adapted from (White and Whittaker, 2016).
In this metastable state, the fusion protein must overcome a kinetic barrier to transition to the next state. The energy to overcome this barrier can be provided by a trigger that will interact with the fusion protein, resulting in a series of conformational changes that will enable the fusion protein to insert its FP into the host membrane, forming a pre-hairpin intermediate state. The triggering event(s) are usually environmental cues that inform the virus about its microenvironment. As an example, the influenza virus fusion protein is triggered by low pH; as the virus is trafficked through the endosome, the increasingly acidic conditions eventually destabilize its fusion protein, so that the fusion peptide is able to insert into the endosomal membrane and commence the fusion process (Carr and Kim, 1993). Since the triggering step initiates the fusion cascade, it is very well regulated to ensure that the virus fuses in an appropriate location. This is especially important as class I viral fusion proteins are generally not reversible; they should not be triggered to fuse at a non-optimal condition (i.e. not bound to a cell). Furthermore, the triggering event also exposes the influenza fusion peptide, as it is buried within the subunit prior to trigger to protect its hydrophobic nature from surrounding aqueous environment. However, for SARS-CoV and MERS-CoV, recently determined cryo-EM structures have shown that both of their FPs are partially exposed at the surface (Yuan et al., 2017). This seems to be a feature of coronaviruses, as the murine hepatitis virus fusion peptide is also exposed in its prefusion state (Walls et al., 2016).
After insertion of the FP, the three HR1 regions assemble into a coiled-coil trimer and three HR2 regions bind to the hydrophobic grooves of the HR1 trimer in an antiparallel manner. (Guillén et al., 2005). The assembly of the HR1 and HR2 domains is known as the fusion core or six-helix bundle, and it is this conformational rearrangement that pulls the viral and host cell membrane into proximity so that they can fuse, bringing the fusion protein into a stable post-fusion state. The process of membrane fusion itself is composed of two stages: hemifusion and pore formation (Fig. 4). The hemifusion stage is defined as the merging of only the outer leaflets of the opposing membranes, with still distinct inner leaflets and no content mixing. The hemifusion stage is considered a transient intermediate that will either dissociate and form two separate vesicles or proceed to pore formation (Lentz et al., 2000). In pore formation, both outer and inner leaflets are merged, forming a connection between the interior of the virus and the host cell cytoplasm, so that the virus can transfer its genetic material through the newly formed pore. Additional reviews on the mechanics of (viral) membrane fusion are (Chernomordik and Kozlov, 2003, 2008; Harrison, 2015).
4.1. Role of cholesterol in fusion
The membrane fusion reaction is dependent on the lipid composition of the viral and/or cellular membranes (Chernomordik and Kozlov, 2008). In both of them, sphingolipids and cholesterol molecules tend to pack together and form microdomains termed “lipid rafts” floating within the "sea" of phospholipids (Simons and Ikonen, 1997). Sphingolipids possess long, saturated acyl chains that have reduced Van der Waals interactions with the unsaturated acyl chains of phospholipids in membranes, due to the inability to align the unsaturated carbon chains. Cholesterol can fill gaps between associating sphingolipids, leading to tighter packing and ordering of cholesterol and sphingolipids that gives lipids rafts detergent-resistance above their non-raft counterparts. The high lipid tail ordering in rafts also influences the distribution of protein and other lipids (Munro, 2003). As an example, GPI-anchored or palmitoylated proteins tend to be raft-associated (Levental et al., 2010) and sphingolipids like GM1 tend to also be enriched in lipid rafts.
Studies on the role of lipid rafts in viral membrane fusion have shown two major themes: (1) viral transmembrane receptors may be concentrated within rafts and serve as "hotspots" for viral entry, and (2) cholesterol in rafts promotes fusion by reducing the energy needed to form fusion intermediates (Yang et al., 2016). For human immunodeficiency virus (HIV), membrane fusion requires multiple fusion protein and receptor interactions, and rafts have been proposed as a means to enrich fusion proteins and receptors for viral entry. Biophysical studies have shown HIV fusion protein interactions with raft domain lipids (Hammache et al., 1999). In vitro studies treating HIV particles with a cholesterol sequestering drug, β-cyclodextrin, was shown to inactivate the virus by rendering it incapable of membrane fusion (Liao et al., 2001), highlighting a potential fusion inhibition strategy.
Attempts to understand CoV interactions with lipid rafts have met with limited success. For SARS-CoV, the location of its ACE2 receptor in rafts is still controversial. Earlier studies showed that ACE2 was found in non-raft fractions in CHO cells transiently expressing it and Vero E6 cells endogenously expressing it (Li et al., 2007; Warner et al., 2005). Other studies have observed that ACE2 colocalizes with established raft proteins caveolin-1, flotillin-2, and ganglioside GM1 in Vero E6 cells (Glende et al., 2008; Lu et al., 2008), with the discrepancies between these results attributed to different experimental techniques (Glende et al., 2008). For the study with transiently expressed ACE2 in CHO cells, overexpression could cause ACE2 to also partition into non-raft areas. For the studies with Vero E6 cells, endogenous expression of ACE2 and its distribution depend on the time point following cell seeding and it is possible that the different groups might have measured ACE2 location at different time points. Further studies on the influence of cholesterol on SARS-CoV infectivity have strengthened the notion that ACE2 is a raft protein (Glende et al., 2008; Lu et al., 2008). Treating Vero cells with a cholesterol-chelating drug, methyl-β-cyclodextrin (mβCD) that disrupts raft formation, has shown that SARS-CoV can still infect in the absence of lipid rafts. It was observed that SARSpp infection was inhibited 60–90% in cells following mβCD treatment. A possible explanation is that ACE2 is a raft protein. By disrupting raft formation, ACE2 is no longer concentrated in microdomains, and this reduced receptor availability lessens SARS-CoV docking and binding efficiency. It will be of interest to observe if SARS-CoV-2 infectivity is similarly inhibited by this drug treatment.
In summary, it is suggested that ACE2 is a raft protein and SARS-CoV entry requires cholesterol. However, there remains a number of questions on the mechanism by which cholesterol is influencing infection. Rather than simply organizing receptors into rafts, it is possible that cholesterol is directly influencing membrane fusion dynamics, by encouraging fusion intermediate formation, but formal evidence for this role for coronaviruses has not been obtained. With regard to MERS-CoV, its DPP4 receptor has been reported to partition into the raft fraction, in Jurkat cells, a human T cell line (Ishii et al., 2001). It would be interesting to observe if DPP4 also partitions into the raft phase in MERS-CoV susceptible cell lines and to determine the dependence of MERS-CoV entry on cholesterol.
4.2. Role of calcium in fusion
As previously discussed, the extracellular environment heavily influences CoV membrane fusion. Extracellular proteases and pH both serve direct and/or indirect roles in enabling SARS-CoV and MERS-CoV fusion. Recently, the ion content of the cellular environment has been investigated for its role in viral fusion, as ions have been shown to be crucial for other membrane fusion processes, such as synaptic vesicle fusion (Martens and McMahon, 2008). Initial studies have discovered that the rubella virus fusion machinery coordinates with calcium ions (Ca2+) for proper orientation and insertion into the host membrane (Dubé et al., 2014). This coordination/requirement was specific to Ca2+ as both fusion assays and infectivity experiments demonstrated that magnesium, manganese, and zinc cations did not enable fusion when supplemented at the same concentration as Ca2+ for rubella virus. Later it was shown by (Nathan et al., 2019) that the Ebola virus fusion machinery also coordinates with Ca2+ in entry, further corroborated by (Das et al., 2020).
Inspired by these studies, we investigated whether the infectivity of SARS-CoV and MERS-CoV was also influenced by Ca2+ and if this is also correlated to the entry steps. By employing extracellular and/or intracellular calcium chelating compounds, it is possible to infect cells in Ca2+ depleted conditions and probe resulting infectivity (Dubé et al., 2016). Infectivity assays with SARSpp and MERSpp demonstrated that SARS-CoV and MERS-CoV entry into Vero e6 and Huh7 cell lines was reduced when intracellular calcium was chelated (Lai et al., 2017; Straus et al., 2019). These results suggest that SARS-CoV and MERS-CoV entry into cells using the endocytic route has aspects of calcium dependence. Ca2+ interactions crucial for CoV entry were further explored with biophysical techniques. Using electron spin resonance (ESR), it was observed that both SARS-CoV and MERS-CoV FP induces the ordering of the lipid acyl chains upon interaction, termed membrane ordering. Increased membrane ordering aids fusion by increasing the negative curvature of the bilayer as it bends during the fusion process, which will reduce the repulsive energy between two opposing membranes (Ge and Freed, 2009). In the presence of Ca2+, the FPs are able to induce greater membrane ordering, indicating that Ca2+ may promote fusion by stabilizing a structure of the FP that organizes the lipids in a manner that facilities membrane merging.
Although both SARS-CoV and MERS-CoV FP interact with calcium, there are differences in their interactions; the SARS-CoV FP was seen to inducer greater membrane-ordering effects than the MERS-CoV FP (Straus et al., 2019). This aspect was manifested in infectivity experiments showing that SARSpp infection was reduced more compared to MERSpp infection when Ca2+ was chelated. Further evidence highlighting the differences between these CoVs comes from isothermal calorimetry titration (ITC) experiments demonstrating that MERS-CoV FP binds one Ca2+ ion, whereas the SARS-CoV FP binds two. After partitioning the SARS-CoV FP domain into two separate domains, FP1 and FP2, it was determined using ESR experiments that each of these domains induces greater membrane order in the presence of Ca2+, suggesting that SARS-CoV FP binds Ca2+ in both the FP1 and FP2 domain. In contrast, mutagenesis experiments conducted on the MERS-CoV FP suggested only one negatively-charged amino acid (E891) in the FP1 region that could bind Ca2+. Although these experiments suggest that MERS-CoV FP can only bind one Ca2+ in its FP1 region, while SARS-CoV FP can bind two, one in each of its FP1 and FP2 regions, further experiments should be conducted to complete this data set. ESR experiments should verify that only the MERS-CoV FP1, not FP2, has a membrane-ordering effect dependent on Ca2+ and mutagenesis experiments should find sites in the SARS-CoV FP1 and FP2 domain that bind Ca2+. Regardless, it is remarkable that despite exhibiting such high sequence conservation, there are subtle differences between SARS-CoV and MERS-CoV entry requirements.
5. SARS-CoV and MERS-CoV current antifusogenics
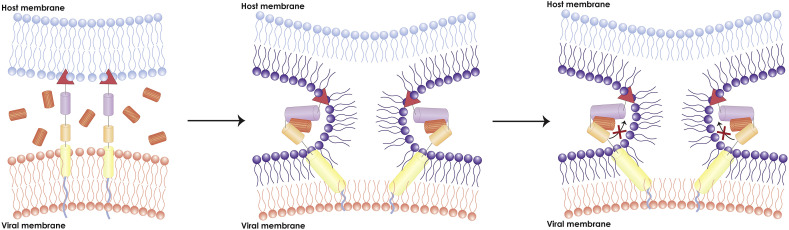
As previously noted, after insertion of the viral fusion peptide in the target cellular membrane, the more N terminal heptad repeat region (HR1) folds into a trimeric helical coiled-coil structure. Subsequently, the C-terminal heptad repeat region (HR2) dissociates into monomers and packs against the grooves of the HR1 trimer in an antiparallel manner (Peisajovich and Shai, 2003). This packing is driven by hydrophobic interactions between HR1 and HR2 and forms the viral fusion core, also known as the six-helix bundle. The formation of the fusion core brings the viral and target membranes together so membrane fusion can commence. Thus, disrupting fusion core formation would seem to be a valid anti-fusion strategy (Fig. 5 ).
Fig. 5.
Model of major antiviral inhibitor pathway. This model depicts the inhibitory mechanism of a major CoV inhibitory peptide: HR2 peptide. Exogeneous HR2 peptides present during the CoV membrane fusion can competitively bind with CoV HR1. This prevents CoV HR2 from locking with HR1 and arrests the membrane fusion reaction, subsequently preventing pore formation.
In the early 1990s, two groups discovered that peptides derived from the HIV-1 gp41 HR2 region could bind to the HR1 region in the pre-intermediate state, preventing the gp41 HR2 from binding and forming the fusion core (Jiang et al., 1993; Wild et al., 1994). By competitively binding to the HR1 region, these HR2 peptides potently inhibited HIV-1 infection at nanomolar concentrations. One of these peptides, DP-178, was tested in proof of principle clinical studies and demonstrated that patients receiving peptide treatment displayed viral load reduction. Further clinical studies established the long-term safety and efficiency of DP-178 and eventually it was approved by the US Food and Drug Administration for HIV/AIDS treatment named Fuzeon (enfuvirtide) as the first fusion inhibitor drug (Matthews et al., 2004).
As these coronaviruses utilize the fusion core to drive membrane fusion, it is worth considering whether CoV fusion could be arrested using HR2-derived peptides. Fuzeon's inhibition potential is specific to HIV-1, as studies with the related HIV-2 were not as promising (Wild et al., 1994). This is due to sequence variation in the HR regions between these two viruses and so CoV specific HR peptides are required for inhibiting CoV fusion. Shortly after the SARS outbreak in 2003, three independent groups designed peptides based on the SARS-CoV HR2 region and found that they were inhibitory (Bosch et al., 2004; Liu et al., 2004; Yuan et al., 2004). Circular-dichroism (CD), a technique that reports on the structural and disordered content of a peptide, showed that HR2 peptides bound to viral HR1 regions have more alpha-helical content than only HR2, suggesting that HR2 peptides form a stable structure when bound to HR1 (Bosch et al., 2004; Liu et al., 2004). These results demonstrate that HR2 peptides can compete with the virus fusion protein's own HR2 region to bind HR1 and prevent fusion. A similar tale can be told for MERS-CoV HR2-based peptides. Two groups investigated MERS-CoV fusion core formation and found that an analogous HR2 peptide inhibited infection (Gao et al., 2013; Lu et al., 2014). CD experiments proved that the HR2 peptide bound to HR1, suggesting that the inhibition mechanism is identical to that of SARS-CoV HR2 and HIV HR2 peptides (Lu et al., 2014).
Sequence alignment of HR1 and HR2 region between SARS-CoV and SARS-CoV-2 show 92.6% and 100% sequence homology, respectively, suggesting that HR2 peptides may also inhibit SARS-CoV-2. Preliminary studies with analogous SARS-CoV-2 HR2 peptides have displayed similar inhibitory behavior in blocking SARS2pp infection of ACE2 expressing cells. CD experiments further confirmed SARS-CoV-2 HR1 and HR2 interact, exhibiting alpha-helical content characteristic of six-helix bundle formation (Xia et al., 2020). Detailed information regarding the peptide sequence and its half maximal inhibitory concentration values (IC50) or related values can be found in Table 3 .
Table 3.
SARS-CoV and MERS-CoV HR2 based inhibitory peptides data. The inhibition data describes IC or analogous values reported for the various inhibitors in blocking infection of SARS-CoV or MERS-CoV infection. As the procedures to test inhibition varied among different groups, the different viral strains and cell lines are noted. The * for P21S10 refers to the position of the residue used to form the staple.
| Virus | Peptide Name | Sequence | Inhibition Data | References |
|---|---|---|---|---|
| SARS-CoV | HR2-8 | ELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYEQYIK | EC50: 17 μM (SARS-CoV Strain 5688 infection of Vero E6 cells) | Bosch et al. (2004) |
| SARS-CoV | CP-1 | GINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYE | IC50: 19 μM (SARS-CoV Strain WHU infection of Vero E6 cells) | Liu et al. (2004) |
| SARS-CoV | HR1-1 | NGIGVTQNVLYENQKQIANQFNKAISQIQESLTTTSTA | EC50: 0.14 μM (HIV pseudotyped SARS-CoV S infection of VeroE6 cells) EC50: 3.68 μM (SARS-CoV Strain BJ01 infection of Vero E6 cells) |
Yuan et al. (2004) |
| SARS-CoV | HR2-18 | IQKEIDRLNEVAKNLNESLIDLQELGK | EC50: 1.19 μM (HIV pseudotyped SARS-CoV S infection of VeroE6 cells) EC50: 5.22 μM (SARS-CoV Strain BJ01 infection of Vero E6 cells) |
Yuan et al. (2004) |
| SARS-CoV | P6 | YQDVNCTDVSTAIHADQLTP | IC90: 113 μM (SARS-CoV Strain GZ50 infection of FRhK-4 cells) | Zheng et al. (2005) |
| SARS-CoV | P8 | QYGSFCTQLNRALSGIAAEQ | IC90: 24.9 μM (SARS-CoV Strain GZ50 infection of FRhK-4 cells) | Zheng et al. (2005) |
| SARS-CoV | P10 | IQKEIDRLNEVAKNLNESLI | IC90: 73.5 μM (SARS-CoV Strain GZ50 infection of FRhK-4 cells) | Zheng et al. (2005) |
| SARS-CoV | P6+P8+P10 | IC90: 0.9 μM (SARS-CoV Strain GZ50 infection of FRhK-4 cells | Zheng et al. (2005) | |
| SARS-CoV | SR9 | ISGINASVVNIQKEIRLNEVAKNLNESLIDLQEL | EC50: <100 nM (SARS-CoV infection of Vero E6 cells with trypsin treatment) | Ujike et al. (2008) |
| SARS-CoV | HR2P | DISGINASVVNIQKEIDRLNEVAKNLNESLIDLQEL | IC50: 2.81 μM (HIV pseudotyped SARS-CoV S infection of ACE2/293T cells) | Xia et al. (2019) |
| MERS-CoV | P1 | LTQINTTLLDLTYEMLSLQQVVKALNESYIDLKEL | IC50: 3 μM (HIV pseudotyped MERS-CoV S infection of Huh7 cells) | Gao et al. (2013) |
| MERS-CoV | HR2P | SLTQINTTLLDLTYEMLSLQQVVKALNESYIDLKEL | IC50: 0.6 μM (MERS-CoV EMC/2012 infection of Huh7 cells) IC50: 0.6 μM (MERS-CoV EMC/2012 infection of Calu-3 cells) IC50: 13.9 μM (MERS-CoV EMC/2012 infection of HFL cells) |
Lu et al. (2014) |
| MERS-CoV | HR2P-M2 | SLTQINTTLLDLEYEMKKLEEVVKKLEESYIDLKEL | IC50: 0.55 μM (MERS-CoV S mediated HEK293T and Huh7 cell-cell fusion) | Lu et al. (2014) |
| MERS-CoV | P21S10 | LDLTYEMLSLQQVV K*LNE*Y | EC50: 3.03 μM (HIV pseudotyped MERS-CoV S infection of Huh7 cells) | Wang et al. (2018) |
| SARS-CoV-2 | 2019-nCoV-HR2P | DISGINASVVNIQKEIDRLNEVAKNLNESLIDLQEL | IC50: 0.98 μM (Pseudotyped SARS-CoV-2 S infection of ACE2/293T cells) | Xia et al. (2020) |
| Pan-CoV | EK1 | SLDQINVTFLDLEYEMKLEEAIKLEESYIDLKEL | IC50: 0.26 μM (HIV pseudotyped MERS-CoV S infection of Huh7) IC50: 0.62 μM (MERS-CoV Strain EMC/2012 infection of Calu-3 cells) IC50: 2.23 μM (HIV pseudotyped SARS-CoV S infection of ACE2/293T cells) IC50: 2.38 μM (Pseudotyped SARS-CoV-2 S infection of ACE2/293T cells) |
(Xia et al., 2020, 2019) |
Although the inhibition mechanism is identical, it is observed that MERS-CoV HR2 peptides inhibited MERS-CoV replication in Vero cells 32-fold more than their SARS-CoV counterpart (Lu et al., 2014). Crystal structures of the SARS-CoV and MERS-CoV fusion cores showed that residue differences in the HR1 and HR2 region enabled MERS-CoV to form more hydrogen bonds in the fusion core, suggesting that MERS-CoV HR2 peptides have a greater binding affinity for the HR1 region compared with SARS-CoV HR2 peptides. Another explanation focuses on the differences in the fusion pathways between the two viruses. In the cell lines used, MERS-CoV is hypothesized to infect via the plasma membrane route, whereas SARS-CoV infects via the endosomal route. Since the peptides are likely more available in the extracellular space than in an endosome, this could explain why MERS-CoV is more sensitive to HR2 inhibitors. In fact, one study observed that the SARS-CoV HR2 peptide did block SARS-CoV infection of Vero E6 cells more potently when trypsin treatment caused SARS-CoV to utilize the plasma membrane pathway, rather than the endosomal pathway (Ujike et al., 2008). To be more conclusive, it would be important to study the efficacy of these peptides in inhibiting infection of lung-derived cell lines, such as Calu-3, since they could more accurately recapitulate in vivo infection. This would also shed light on the viral entry pathway that these peptides inhibit and how efficient they block viral entry at the plasma membrane pathway versus the endosomal pathway.
A common theme amongst these studies and those of other enveloped viruses is that only the HR2 peptide can bind viral HR1 in an inhibitory manner; analogous HR1 peptides exhibit no antiviral capacity. This is because singular HR1 peptides tend to form self-associated oligomers and aggregate in solution (Lu et al., 1995). However, by noting that the major HR1 helical content resulting in oligomerization was located in the downstream sections of HR1, one group was able to report success with a SARS-CoV HR1 peptide derived from upstream sections of HR1 (Yuan et al., 2004). CD experiments confirmed that this peptide has low α-helical content, preventing self-oligomerization.
These studies also demonstrated that the SARS-CoV and MERS-CoV HR2 peptides are not cross reactive: SARS-CoV HR2 peptides do not inhibit MERS-CoV infection, and vice versa (Gao et al., 2013; Lu et al., 2014). Recently, one group challenged this notion and screened HR2-derived peptides from all human-infecting CoV (HCoV), including SARS-CoV, MERS-CoV, HCoV-229E, HCoV-NL63, HCoV-OC43 (OC43), and HCoV-HKU1 (Xia et al., 2019). They found that the HR2 peptide from HCoV-OC43 inhibited all these aforementioned CoVs. The unique pan inhibition property can be attributed to key amino acid differences that strengthen HR1-HR2 interactions. As an example, the crystal structures of MERS-CoV and SARS-CoV HR1 interaction with the OC43 HR2 peptide showed that the OC43 HR2 peptide has a valine that fits neatly into the hydrophobic pockets of MERS-CoV and SARS-CoV HR1. The corresponding SARS-CoV and MERS-CoV HR2 residues, alanine and threonine, fit the pocket loosely, or not at all. This study also found that the OC43 peptide inhibits SARS-like CoV, which have reservoir in bats and recognize human receptors (Ge et al., 2013; Menachery et al., 2016). This suggests that the OC43 peptide exhibits promise as a treatment during coronavirus outbreaks.
While there is great potential towards developing CoV HR2 peptides, especially the OC43 peptide as a CoV therapeutic, there remains work to improve the IC50 values, which are roughly three order of magnitude higher than the IC50 values reported for HIV HR2 peptides (Du et al., 2009). Strategies aimed at improving IC50 and pharmacokinetic values have included introducing mutations or adding conjugates. It has been previously reported that introducing negatively and positively charged amino acids Glu (E) and Lys (K) with three or four residues in between them can form E-K or K-E salt bridges that enhance the peptide's stability, solubility, and antiviral activity (Lu et al., 2014; Marqusee and Baldwin, 1987; Otaka et al., 2002). Combining these mutations with others not involved in HR1 binding on the OC43 HR2 peptide resulted in decreased IC50 and increased solubility values. This mutated peptide, named EK1, was found to be more inhibitory than the native HR2 peptide for SARS-CoV (IC50 values of 2.23 and 2.81, respectively) and MERS-CoV (IC50 values of 0.26 and 1.06, respectively) pseudovirus infection. EK1 was also evaluated for its ability to protect mice against live MERS-CoV challenge. Infection was uniformly lethal for untreated mice, while those treated either prophylactically or therapeutically had 100% and 75% survival, respectively (Xia et al., 2019). Most promisingly, EK1 also significantly inhibits SARS-CoV-2 pseudoparticle infection, though slightly less potent than its native HR2 peptide (IC50 values of 2.38 and 0.98, respectively) (Xia et al., 2020). Regardless, these results highlight fusion inhibitory compounds as a promising strategy to block coronavirus infections.
Additional efforts sought to decrease HR2 IC50 values by improving peptide localization to the virus. One group conjugated a MERS-CoV HR2 peptide, HR2P-M2, to a monoclonal antibody that recognizes the MERS-CoV S RBD region. This showed potent synergism in blocking both receptor binding and membrane fusion by inhibiting MERS-CoV S-mediated cell-cell fusion as well as MERSpp infection (Wang et al., 2019). Furthermore, since CoV receptors and proteases tend to partition in lipid-enriched domains, another group tested the possibility of directing HR2 peptides towards these lipid-enriched domains where CoV entry occurs. Conjugating palmitate, which partitions into lipid rafts, to HR2P-M2 was found to increase inhibition of SARSpp and MERSpp cell entry (Park and Gallagher, 2017). On the other hand, conjugating tocopherol, which partitions into non-raft domains, to HR2P-M2 had minimal impact on cell entry. An intriguing aspect of this work is that these palmitate-conjugated peptides also strongly inhibited CoV entry via the endosomal route, which is believed to be relatively resistant to the inhibitory action of these peptides. Indeed, immunofluorescence microscopy showed that lipid-tagged peptides were detected intracellularly, whereas non-lipid-tagged peptides were not detected on cells.
Additional efforts sought to improve the pharmacokinetic values which describe the living organism's response to a drug of HR2P-M2 and can include parameters such as drug bioavailability, metabolism, or half-life. Peptide stapling is one technique, which generates a "brace" that locks a specific conformation for the peptide, resulting in increased target affinity, cell permeability, and serum stability (Frank et al., 2014; Hojo et al., 2016). For HR2 peptides, peptide stapling can stabilize the α-helical nature of the peptide, which is critical to their potency. An all-hydrocarbon peptide stapling of HR2P-M2, named P21S10, resulted in a 27-fold increase in the area under the plasma concentration-time curve value, which measures the total systemic exposure to a drug, while maintaining a similar EC50 and in vivo half-life when compared to HR2P-M2 (Wang et al., 2018). An alternative strategy includes conjugating peptides to gold nanorods. Nanorods have emerged as a promising drug delivery system for improving pharmacokinetic values. One group conjugated HR2-based peptides to a gold nanorod with a polyethylene glycol coating (PIH-AuNR), which demonstrated a 10-fold inhibition increase of MERS-CoV S mediated cell-cell fusion and longer inhibitory activity over its non-conjugated counterpart. In vivo studies demonstrated that PIH-AuNR are biocompatible with mice; over 12 days, there were no significant differences in body weight and behavioral abnormalities between PIH-AuNR treated and control groups (Huang et al., 2019).
An alternative strategy of using peptides to target non-HR regions has also been studied, albeit to a limited degree. While screening a variety of peptide fragments spanning the SARS-CoV S protein for SARS-CoV infection inhibitory activity, researchers identified inhibitory peptides P6, P8, and P10. P10 was derived from the HR2 region, and thus its inhibitory capabilities results from blocking 6-HB formation (Zheng et al., 2005). Intriguingly, P6 and P8 were derived from regions slightly upstream of the S1/S2 and S2’ cleavage sites, which suggest that non-HR regions may be of interest as an anti-fusogenic strategy. The authors hypothesized that the antiviral activity of P6 could result from interference with S1/S2 cleavage, preventing conformational changes for fusion.
Although these studies demonstrate that anti-fusogenic peptides can inhibit pathogenic coronavirus infection, they have not been evaluated for use in humans. There are in vivo mouse trials with the HR2P-M2 peptide (Channappanavar et al., 2015; Jiang et al., 2015), though only time will tell if these peptides will become FDA-approved. An alternative strategy is to screen FDA-approved drugs for their ability to inhibit CoV membrane fusion because the time frame and monetary cost associated with repurposing approved drugs is generally less than developing novel drugs. An initial screen identified imatinib, an Abelson kinase inhibitor (Dyall et al., 2014). Abelson kinases are tyrosine kinases that regulate a variety of cellular pathways and have been previously shown to inhibit viral replication of other viruses, such as Ebola virus, coxsackie virus, and vaccinia virus, at different stages. Subsequent studies demonstrated that imatinib inhibits SARS-CoV and MERS-CoV pseudovirus fusion with BSC-1 and Huh7 endosomal membranes, respectively (Coleman et al., 2016). As Abelson kinases are involved in pathways that enable cytoskeleton rearrangements, it is hypothesized that they inhibit fusion by preventing key cytoskeleton rearrangements for fusion (Sisk et al., 2018).
6. Next steps in research
SARS-CoV, MERS-CoV, and the emerging SARS-CoV-2 cell entry is governed by the S protein. The conserved nature of the fusion mechanism, as well as the fusion subunit sequence, makes it a potentially valuable target for developing a pan-CoV therapeutic. Increased understanding of the fusion mechanism is necessary to know which critical areas to target.
While the surface route is believed to be more clinically relevant, as lung cells express TMPRSS2, this will need to be verified with in vivo experiments. TMPRSS2 knockout mice demonstrated less severe immunopathology when infected with SARS-CoV or MERS-CoV, however, at least for SARS-CoV, viral spread was still detected in the alveoli (Iwata-Yoshikawa et al., 2019). This suggest that other proteases, such as cathepsin L, could be activating the virus, and that the endosomal route may hold clinical relevance. Thus, it is important to expand research efforts to understand the role of these other proteases. As a starting point, identifying the elusive cathepsin L cleavage sites would provide greater insight into additional fusion-relevant domains within the S protein. Furthermore, both MERS-CoV and SARS-CoV-2 have predicted cleavage sites at the S1/S2 and S2’ positions (Coutard et al., 2020; Millet and Whittaker, 2014; Walls et al., 2020; Wrapp et al., 2020). Although furin has been shown to process the MERS-CoV S1/S2 site during biosynthesis, the role of furin or furin-like proteases for SARS-CoV-2 entry is currently unknown, but will likely fundamentally affect viral entry. It is also important to note that key in vitro results should be corroborated with in vivo experiments, as knowledge of clinically relevant routes, proteases, and S domains can guide therapeutic development to block viral entry. In a similar manner, studies to determine the impact of cholesterol and Ca2+ on SARS-CoV-2 would be of interest to guide novel antiviral strategies.
Current studies on anti-CoV fusogenics have identified HR2 peptides as a promising countermeasure. Studies with the EK1 peptide have demonstrated its potential effectiveness as a pan-CoV solution, as it inhibited SARS-CoV, MERS-CoV, and SARS-CoV-2 pseudoparticle infection with IC50 values around 2 μM (Xia et al., 2019, 2020). Furthermore, in vivo studies have shown that EK1 protects mice from MERS-CoV challenge when administered either prophylactically or therapeutically, with 100% or 75% survival, respectively. As the treatment group was administered EK1 peptides 30 min post-infection, it would be of interest to observe the effectiveness of EK1 on a clinical timescale, perhaps several days after infection, to assess its therapeutic potential.
To address the pandemic caused by SARS-CoV-2 and future coronaviruses rapidly, it is important to continue studies on their entry mechanism, which can guide novel therapeutic efforts. Many aspects of the viral fusion reaction (S domains, proteases, lipid compositions, ionic environments) may be valuable targets in arresting membrane fusion by a variety of agents (small molecules, antibodies, peptides). The research community is poised to see what novel therapeutics can address CoV infections.
Acknowledgements
The authors express their gratitude to members of the Daniel and Whittaker groups for their helpful discussions. We have received financial support for coronavirus-related projects from National Institutes of Health research grant R01AI35270. TT is supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1650441.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2020.104792.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Alsaadi E.A.J., Neuman B.W., Jones I.M. A fusion peptide in the spike protein of MERS coronavirus. Viruses. 2019;11:825. doi: 10.3390/v11090825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;89:44–48. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlan A., Zhao J., Sarkar M.K., Li K., McCray P.B., Perlman S., Gallagher T. Receptor variation and susceptibility to Middle East respiratory syndrome coronavirus infection. J. Virol. 2014;88:4953–4961. doi: 10.1128/JVI.00161-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. Unit. States Am. 2009;106:5871–5876. doi: 10.1073/PNAS.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Madu I., Whittaker G.R. Elastase-mediated activation of the severe acute respiratory syndrome coronavirus spike protein at discrete sites within the S2 domain * □ S downloaded from. J. Biol. Chem. 2010;285:22758–22763. doi: 10.1074/jbc.M110.103275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R., Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Glowacka I., Blazejewska P., Soilleux E., Allen P., Danisch S., Steffen I., Choi S.-Y., Park Y., Schneider H., Schughart K., Pöhlmann S. TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in caco-2 cells. J. Virol. 2010;84:10016–10025. doi: 10.1128/JVI.00239-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Glowacka I., Müller M.A., Lavender H., Gnirss K., Nehlmeier I., Niemeyer D., He Y., Simmons G., Drosten C., Soilleux E.J., Jahn O., Steffen I., Pöhlmann S. Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease. J. Virol. 2011;85:13363–13372. doi: 10.1128/JVI.05300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biniossek M.L., Nägler D.K., Becker-Pauly C., Schilling O. Proteomic identification of protease cleavage sites characterizes prime and non-prime specificity of cysteine cathepsins B, L, and S. J. Proteome Res. 2011;10:5363–5373. doi: 10.1021/pr200621z. [DOI] [PubMed] [Google Scholar]
- Bosch B.J., Bartelink W., Rottier P.J.M. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 2008;82:8887–8890. doi: 10.1128/jvi.00415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch J., Martina B.E.E., Van Der Zee R., Lepault J., Haijema B.J., Versluis C., Heck A.J.R., De Groot R., Osterhaus A.D.M.E., Rottier P.J.M. 2004. Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) Infection Inhibition Using Spike Protein Heptad Repeat-Derived Peptides, PNAS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch J., Van Der Zee R., De Haan C.A.M., Rottier P.J.M. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex downloaded from. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher E., Matrosovich T., Beyerle M., Klenk H.-D., Garten W., Matrosovich M. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 2006;80:9896–9898. doi: 10.1128/JVI.01118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr C.M., Kim P.S. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-W. [DOI] [PubMed] [Google Scholar]
- Chaipan C., Kobasa D., Bertram S., Glowacka I., Steffen I., Tsegaye T.S., Takeda M., Bugge T.H., Kim S., Park Y., Marzi A., Pöhlmann S. Proteolytic activation of the 1918 influenza virus hemagglutinin. J. Virol. 2009;83:3200–3211. doi: 10.1128/JVI.02205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers P., Pringle C.R., Easton A.J. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J. Gen. Virol. 1990;71:3075–3080. doi: 10.1099/0022-1317-71-12-3075. [DOI] [PubMed] [Google Scholar]
- Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Lu L., Xia S., Du L., Meyerholz D.K., Perlman S., Jiang S. Protective effect of intranasal regimens containing peptidic Middle East respiratory syndrome coronavirus fusion inhibitor against MERS-CoV infection. J. Infect. Dis. 2015;212:1894–1903. doi: 10.1093/infdis/jiv325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik L.V., Kozlov M.M. Protein-lipid interplay in fusion and fission of biological membranes. Annu. Rev. Biochem. 2003;72:175–207. doi: 10.1146/annurev.biochem.72.121801.161504. [DOI] [PubMed] [Google Scholar]
- Chernomordik L.V., Kozlov M.M. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 2008;15:675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.Y., Bertram S., Glowacka I., Park Y.W., Pöhlmann S. Type II transmembrane serine proteases in cancer and viral infections. Trends Mol. Med. 2009;15:303–312. doi: 10.1016/j.molmed.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Cohen F.S., Melikyan G.B. The energetics of membrane fusion from binding, through hemifusion, pore formation, and pore enlargement. J. Membr. Biol. 2004;199:1–14. doi: 10.1007/s00232-004-0669-8. [DOI] [PubMed] [Google Scholar]
- Coleman C.M., Sisk J.M., Mingo R.M., Nelson E.A., White J.M., Frieman M.B. Abelson kinase inhibitors are potent inhibitors of severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus fusion. J. Virol. 2016;90:8924–8933. doi: 10.1128/jvi.01429-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das D.K., Bulow U., Diehl W.E., Durham N.D., Senjobe F., Chandran K., Luban J., Munro J.B. Conformational changes in the Ebola virus membrane fusion machine induced by pH, Ca2+, and receptor binding. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan C.A.M., Rottier P.J.M. Advances in Virus Research. 2005. Molecular interactions in the assembly of coronaviruses; pp. 165–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson S.H., Hirsh A., Li D.C., Holloway G., Chao J., Boucher R.C., Gabriel S.E. Regulation of the epithelial sodium channel by serine proteases in human airways. J. Biol. Chem. 2002;277:8338–8345. doi: 10.1074/jbc.M105044200. [DOI] [PubMed] [Google Scholar]
- Du L., He Y., Zhou Y., Liu S., Zheng B.-J., Jiang S. The spike protein of SARS-CoV — a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Yang Y., Zhou Y., Lu L., Li F., Jiang S. MERS-CoV spike protein: a key target for antivirals. Expert Opin. Ther. Targets. 2017;21:131–143. doi: 10.1080/14728222.2017.1271415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubé M., Etienne L., Fels M., Kielian M. Calcium-dependent rubella virus fusion occurs in early endosomes. J. Virol. 2016;90:6303–6313. doi: 10.1128/jvi.00634-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubé M., Rey F.A., Kielian M. Rubella virus: first calcium-requiring viral fusion protein. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall J., Coleman C.M., Hart B.J., Venkataraman T., Holbrook M.R., Kindrachuk J., Johnson R.F., Olinger G.G., Jahrling P.B., Laidlaw M., Johansen L.M., Lear-Rooney C.M., Glass P.J., Hensley L.E., Frieman M.B. Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob. Agents Chemother. 2014;58:4885–4893. doi: 10.1128/AAC.03036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epand R.M. Fusion peptides and the mechanism of viral fusion. Biochim. Biophys. Acta Biomembr. 2003;1614:116–121. doi: 10.1016/S0005-2736(03)00169-X. [DOI] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Coronaviruses Methods Protoc. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank A.O., Vangamudi B., Feldkamp M.D., Souza-Fagundes E.M., Luzwick J.W., Cortez D., Olejniczak E.T., Waterson A.G., Rossanese O.W., Chazin W.J., Fesik S.W. Discovery of a potent stapled helix peptide that binds to the 70N domain of replication protein A. J. Med. Chem. 2014;57:2455–2461. doi: 10.1021/jm401730y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Lu G., Qi J., Li Y., Wu Y., Deng Y., Geng H., Li H., Wang Q., Xiao H., Tan W., Yan J., Gao G.F. Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S protein of Middle East respiratory syndrome coronavirus. J. Virol. 2013;87:13134–13140. doi: 10.1128/jvi.02433-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge M., Freed J.H. Fusion peptide from influenza hemagglutinin increases membrane surface order: an electron-spin resonance study. Biophys. J. 2009;96:4925–4934. doi: 10.1016/j.bpj.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.-Y., Li J.-L., Yang X.-L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., Zhang Y.-J., Luo C.-M., Tan B., Wang N., Zhu Y., Crameri G., Zhang S.-Y., Wang L.-F., Daszak P., Shi Z.-L. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierer S., Bertram S., Kaup F., Wrensch F., Heurich A., Kramer-Kuhl A., Welsch K., Winkler M., Meyer B., Drosten C., Dittmer U., von Hahn T., Simmons G., Hofmann H., Pohlmann S. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can Be activated by TMPRSS2, and is targeted by neutralizing antibodies. J. Virol. 2013;87:5502–5511. doi: 10.1128/jvi.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glende J., Schwegmann-Wessels C., Al-Falah M., Pfefferle S., Qu X., Deng H., Drosten C., Naim H.Y., Herrler G. Importance of cholesterol-rich membrane microdomains in the interaction of the S protein of SARS-coronavirus with the cellular receptor angiotensin-converting enzyme 2. Virology. 2008;381:215–221. doi: 10.1016/J.VIROL.2008.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Müller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., Niemeyer D., Schneider H., Drosten C., Pöhlmann S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Publ. Gr. 2013;11 doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui M., Song W., Zhou H., Xu J., Chen S., Xiang Y., Wang X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén J., Kinnunen P.K.J., Villalaín J. Membrane insertion of the three main membranotropic sequences from SARS-CoV S2 glycoprotein. Biochim. Biophys. Acta Biomembr. 2008;1778:2765–2774. doi: 10.1016/J.BBAMEM.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén J., Pérez-Berná A.J., Moreno M.R., Villalaín J. A second SARS-CoV S2 glycoprotein internal membrane-active peptide. Biophysical characterization and membrane interaction. Biochemistry. 2008;47:8214–8224. doi: 10.1021/bi800814q. [DOI] [PubMed] [Google Scholar]
- Guillén J., Pérez-Berná A.J., Moreno M.R., Villalaín J. Identification of the membrane-active regions of the severe acute respiratory syndrome coronavirus spike membrane glycoprotein using a 16/18-mer peptide scan: implications for the viral fusion mechanism. J. Virol. 2005;79:1743–1752. doi: 10.1128/JVI.79.3.1743-1752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammache D., Yahi N., Maresca M., Rard G.É., Roni P., Fantini J. Human erythrocyte glycosphingolipids as alternative cofactors for human immunodeficiency virus type 1 (HIV-1) entry: evidence for CD4-induced interactions between HIV-1 gp120 and reconstituted membrane microdomains of glycosphingolipids (Gb3 and GM3) J. Virol. 1999;73(6):5244–5248. doi: 10.1128/jvi.73.6.5244-5248.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L.C., Lely A.T., Navis G.J., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S.C. Viral membrane fusion. Virology. 2015;479–480:498–507. doi: 10.1016/J.VIROL.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S.C. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald-Sargent T., Gallagher T. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses. 2012;4:557–580. doi: 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Mü M.A., Drosten C., Pö S., Krü N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Pö Hlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor article SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:1–10. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojo K., Hossain M.A., Tailhades J., Shabanpoor F., Wong L.L.L., Ong-Pålsson E.E.K., Kastman H.E., Ma S., Gundlach A.L., Rosengren K.J., Wade J.D., Bathgate R.A.D. Development of a single-chain peptide agonist of the relaxin-3 receptor using hydrocarbon stapling. J. Med. Chem. 2016;59:7445–7456. doi: 10.1021/acs.jmedchem.6b00265. [DOI] [PubMed] [Google Scholar]
- Huang I.C., Bosch B.J., Li F., Li W., Kyoung H.L., Ghiran S., Vasilieva N., Dermody T.S., Harrison S.C., Dormitzer P.R., Farzan M., Rottier P.J.M., Choe H. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 2006;281:3198–3203. doi: 10.1074/jbc.M508381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Li M., Xu Y., Zhang J., Meng X., An X., Sun L., Guo L., Shan X., Ge J., Chen J., Luo Y., Wu H., Zhang Y., Jiang Q., Ning X. Novel gold nanorod-based HR1 peptide inhibitor for Middle East respiratory syndrome coronavirus. ACS Appl. Mater. Interfaces. 2019;11 doi: 10.1021/acsami.9b04240. 19799–19807. [DOI] [PubMed] [Google Scholar]
- Inoue Y., Tanaka N., Tanaka Y., Inoue S., Morita K., Zhuang M., Hattori T., Sugamura K. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J. Virol. 2007;81:8722–8729. doi: 10.1128/JVI.00253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T., Ohnuma K., Murakami A., Takasawa N., Kobayashi S., Dang N.H., Schlossman S.F., Morimoto C. 2001. CD26-mediated Signaling for T Cell Activation Occurs in Lipid Rafts through its Association with CD45RO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J. Virol. JVI.01815-18. 2019;93:1815–1833. doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes J.A., Andre N.M., Chappie J.S., Millet J.K., Whittaker G.R. J. Molec. Biol.; 2020. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically-sensitive activation loop. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes J.A., Millet J.K., Stout A.E., André N.M., Whittaker G.R. A tale of two viruses: the distinct spike glycoproteins of feline coronaviruses. Viruses. 2020;12:83. doi: 10.3390/v12010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Lin K., Strick N., Neurath A.R. HIV-1 inhibition by a peptide [3] Nature. 1993;365:113. doi: 10.1038/365113a0. [DOI] [PubMed] [Google Scholar]
- Jiang S., Xia S., Yu F., Lu L., Jiang Shibo, Tao X., Garron T., Du L., Lu Lu, Tseng C.-T.K. 2015. Intranasally Administered Peptidic Viral Fusion Inhibitor Protected hDPP4 Transgenic Mice from MERS-CoV Infection. [DOI] [Google Scholar]
- Kam Y.-W., Okumura Y., Kido H., Ng L.F.P., Bruzzone R., Altmeyer R. Cleavage of the SARS coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro. 2009. [DOI] [PMC free article] [PubMed]
- Kawase M., Shirato K., van der Hoek L., Taguchi F., Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol. 2012;86:6537–6545. doi: 10.1128/jvi.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Weber H., Elzayat M.T., Hoffmann M., Pöhlmann S. Functional analysis of potential cleavage sites in the MERS-coronavirus spike protein. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-34859-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A.L., Millet J.K., Daniel S., Freed J.H., Whittaker G.R. The SARS-CoV fusion peptide forms an extended bipartite fusion platform that perturbs membrane order in a calcium-dependent manner. J. Mol. Biol. 2017;429:3875–3892. doi: 10.1016/j.jmb.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz B.R., Malinin V., Haque M.E., Evans K. Protein machines and lipid assemblies: current views of cell membrane fusion. Curr. Opin. Struct. Biol. 2000;10:607–615. doi: 10.1016/s0959-440x(00)00138-x. [DOI] [PubMed] [Google Scholar]
- Levental I., Lingwood D., Grzybek M., Coskun Ü., Simons K. Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc. Natl. Acad. Sci. U.S.A. 2010;107:22050–22054. doi: 10.1073/pnas.1016184107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. 2015. Receptor Recognition Mechanisms of Coronaviruses: a Decade of Structural Studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structural biology: structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science (80- 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li G.M., Li Y.G., Yamate M., Li S.M., Ikuta K. Lipid rafts play an important role in the early stage of severe acute respiratory syndrome-coronavirus life cycle. Microb. Infect. 2007;9:96–102. doi: 10.1016/j.micinf.2006.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Hulswit R.J.G., Widjaja I., Raj V.S., McBride R., Peng W., Widagdo W., Tortorici M.A., Van Dieren B., Lang Y., Van Lent J.W.M., Paulson J.C., De Haan C.A.M., De Groot R.J., Van Kuppeveld F.J.M., Haagmans B.L., Bosch B.J. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc. Natl. Acad. Sci. U.S.A. 2017;114:E8508–E8517. doi: 10.1073/pnas.1712592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Z., Cimakasky L.M., Hampton R., Nguyen D.H., Hildreth J.E.K. Lipid rafts and HIV pathogenesis: host membrane cholesterol is required for infection by HIV type 1. AIDS Res. Hum. Retrovir. 2001;17:1009–1019. doi: 10.1089/088922201300343690. [DOI] [PubMed] [Google Scholar]
- Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R., Xiong H., Farmar J., Debnath A.K., Tien P., Jiang S. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Liu Q., Zhu Y., Chan K.-H., Qin L., Li Y., Wang Q., Fuk-Woo Chan J., Du L., Yu F., Ma C., Ye S., Yuen K.-Y., Zhang R., Jiang S. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat. Commun. 2014;5 doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Blacklow S.C., Kim P.S. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Mol. Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- Lu Y., Liu D.X., Tam J.P. Lipid rafts are involved in SARS-CoV entry into Vero E6 cells. Biochem. Biophys. Res. Commun. 2008;369:344–349. doi: 10.1016/J.BBRC.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madu I.G., Belouzard S., Whittaker G.R. SARS-coronavirus spike S2 domain flanked by cysteine residues C822 and C833 is important for activation of membrane fusion. Virology. 2009;393:265–271. doi: 10.1016/j.virol.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madu I.G., Roth S.L., Belouzard S., Whittaker G.R. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J. Virol. 2009;83:7411–7421. doi: 10.1128/jvi.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marqusee S., Baldwin R.L. Helix stabilization by Glu-...Lys+ salt bridges in short peptides of de novo design. Proc. Natl. Acad. Sci. U.S.A. 1987;84:8898–8902. doi: 10.1073/pnas.84.24.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens S., McMahon H.T. Mechanisms of membrane fusion: disparate players and common principles. Nat. Rev. Mol. Cell Biol. 2008;9:543–556. doi: 10.1038/nrm2417. [DOI] [PubMed] [Google Scholar]
- Masters P.S., Perlman S. Coronaviridae. Fields Virology. 2013:825–858. [Google Scholar]
- Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F., Sakata M., Tahara M., Kutsuna S., Ohmagari N., Kuroda M., Suzuki T., Kageyama T., Takeda M. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. Unit. States Am. 2020;202002589 doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Shirato K., Kawase M., Terada Y., Kawachi K., Fukushi S., Kamitani W. Middle East respiratory syndrome coronavirus spike protein is not activated directly by cellular furin during viral entry into target cells. J. Virol. 2018;92 doi: 10.1128/jvi.00683-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Ujike M., Morikawa S., Tashiro M., Taguchi F. Proceedings of the National Academy of Sciences of the United States of America. 2005. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews T., Salgo M., Greenberg M., Chung J., DeMasi R., Bolognesi D. Enfuvirtide: the first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nat. Rev. Drug Discov. 2004;3(3):215–225. doi: 10.1038/nrd1331. [DOI] [PubMed] [Google Scholar]
- Menachery V.D., Yount B.L., Sims A.C., Debbink K., Agnihothram S.S., Gralinski L.E., Graham R.L., Scobey T., Plante J.A., Royal S.R., Swanstrom J., Sheahan T.P., Pickles R.J., Corti D., Randell S.H., Lanzavecchia A., Marasco W.A., Baric R.S. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. U.S.A. 2016;113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Physiological and molecular triggers for SARS-CoV membrane fusion and entry into host cells. Virology. 2018;517:3–8. doi: 10.1016/J.VIROL.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U.S.A. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingo R.M., Simmons J.A., Shoemaker C.J., Nelson E.A., Schornberg K.L., D’souza R.S., Casanova J.E., White J.M. 2015. Ebola Virus and Severe Acute Respiratory Syndrome Coronavirus Display Late Cell Entry Kinetics: Evidence that Transport to NPC1+ Endolysosomes Is a Rate-Defining Step. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossel E.C., Huang C., Narayanan K., Makino S., Tesh R.B., Peters C.J. Exogenous ACE2 expression allows refractory cell lines to support severe acute respiratory syndrome coronavirus replication. J. Virol. 2005;79:3846–3850. doi: 10.1128/jvi.79.6.3846-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro, S. Lipid rafts: elusive or illusive? 2003 Nov 14;115(4):377-88. [DOI] [PubMed]
- Nathan L., Lai A.L., Millet J.K., Straus M.R., Freed J.H., Whittaker G.R., Daniel S. Calcium ions directly interact with the Ebola virus fusion peptide to promote structure–function changes that enhance infection. ACS Infect. Dis. acsinfecdis.9b00296. 2019;6(2):250–260. doi: 10.1021/acsinfecdis.9b00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai H., Sakai S., Hirabayashi T., Shimizu Y., Terasawa K. Inhibitory effect of bafilomycin A1, a specific inhibitor of vacuolar-type proton pump, on the growth of influenza A and B viruses in MDCK cells. Antivir. Res. 1995;27:425–430. doi: 10.1016/0166-3542(95)00040-S. [DOI] [PubMed] [Google Scholar]
- Otaka A., Nakamura M., Nameki D., Kodama E., Uchiyama S., Nakamura S., Nakano H., Tamamura H., Kobayashi Y., Matsuoka M., Fujii N. Remodeling of gp41-C34 peptide leads to highly effective inhibitors of the fusion of HIV-1 with target cells we thank dr. Terrence R. Burke, jr., NCI, NIH, frederick, MD 21702-1201, for proofreading the manuscript and providing useful comments. This research was supported in part by a grant-in-aid for scientific research from the ministry of education, culture, sports, science and technology, Japan, the Japan society for the promotion of science, and the Japan Health science foundation. Angew. Chem. Int. Ed. 2002;41:2937. doi: 10.1002/1521-3773(20020816)41:16<2937::AID-ANIE2937>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.E., Gallagher T. Lipidation increases antiviral activities of coronavirus fusion-inhibiting peptides. Virology. 2017;511:9–18. doi: 10.1016/j.virol.2017.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.E., Li K., Barlan A., Fehr A.R., Perlman S., McCray P.B., Gallagher T. Proteolytic processing of middle east respiratory syndrome coronavirus spikes expands virus tropism. Proc. Natl. Acad. Sci. U.S.A. 2016;113:12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisajovich S.G., Shai Y. Viral fusion proteins: multiple regions contribute to membrane fusion. Biochim. Biophys. Acta Biomembr. 2003;1614:122–129. doi: 10.1016/S0005-2736(03)00170-6. [DOI] [PubMed] [Google Scholar]
- Peng G., Sun D., Rajashankar K.R., Qian Z., Holmes K.V., Li F. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10696–10701. doi: 10.1073/pnas.1104306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z., Dominguez S.R., Holmes K.V. Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. PloS One. 2013;8 doi: 10.1371/journal.pone.0076469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z., Hingley S.T., Simmons G., Yu § Christopher, Das Sarma J., Bates † Paul, Weiss S.R. Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry. J. Virol. 2006;80:5768–5776. doi: 10.1128/JVI.00442-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H.W., Müller M.A., Dijkman R., Muth D., Demmers J.A.A., Zaki A., Fouchier R.A.M., Thiel V., Drosten C., Rottier P.J.M., Osterhaus A.D.M.E., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan A.D., Shraybman R., Cohen R.D., Whittaker G.R. Differential role for low pH and cathepsin-mediated cleavage of the viral spike protein during entry of serotype II feline coronaviruses. Vet. Microbiol. 2008;132:235–248. doi: 10.1016/j.vetmic.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke L.M., Spiegel M., Plegge T., Hartleib A., Nehlmeier I., Gierer S., Hoffmann M., Hofmann-Winkler H., Winkler M., Pöhlmann S. Different residues in the SARS-CoV spike protein determine cleavage and activation by the host cell protease TMPRSS2. PloS One. 2017;12 doi: 10.1371/journal.pone.0179177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz B., Rausch J.M., Gallaher W.R., Garry R.F., Wimley W.C. Identification and characterization of the putative fusion peptide of the severe acute respiratory syndrome-associated coronavirus spike protein downloaded from. J. Virol. 2005;79:7195–7206. doi: 10.1128/JVI.79.11.7195-7206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Kawase M., Matsuyama S. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J. Virol. 2013;87:12552–12561. doi: 10.1128/jvi.01890-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry downloaded from. J. Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U.S.A. 2005 doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Reeves J.D., Rennekamp A.J., Amberg S.M., Piefer A.J., Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- Sisk J.M., Frieman M.B., Machamer C.E. Coronavirus S protein-induced fusion is blocked prior to hemifusion by Abl kinase inhibitors. J. Gen. Virol. 2018;99:619–630. doi: 10.1099/jgv.0.001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus M.R., Tang T., Lai A.L., Flegel A., Bidon M., Freed J.H., Daniel S., Whittaker G.R. Ca2+ ions promote fusion of Middle East Respiratory Syndrome coronavirus with host cells and increase infectivity. Journal of Virology. 2019;2019 doi: 10.1101/2019.12.18.881391. 12.18.881391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Tse L.V., Ferguson A.D., Whittaker G.R. Modifications to the hemagglutinin cleavage site control the virulence of a neurotropic H1N1 influenza virus. J. Virol. 2010;84:8683–8690. doi: 10.1128/jvi.00797-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripet B., Howard M.W., Jobling M., Holmes R.K., Holmes K.V., Hodges R.S. Structural characterization of the SARS-coronavirus spike S fusion protein core. J. Biol. Chem. 2004;279:20836–20849. doi: 10.1074/JBC.M400759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujike M., Nishikawa † Hiroki, Otaka A., Yamamoto Naoki, Yamamoto Norio, Matsuoka M., Kodama E., Fujii N., Taguchi F. Heptad repeat-derived peptides block protease-mediated direct entry from the cell surface of severe acute respiratory syndrome coronavirus but not entry via the endosomal pathway. J. Virol. 2008;82:588–592. doi: 10.1128/JVI.01697-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Miazgowicz K.L., Milne-Price S., Bushmaker T., Robertson S., Scott D., Kinne J., McLellan J.S., Zhu J., Munster V.J. Host species restriction of Middle East respiratory syndrome coronavirus through its receptor, dipeptidyl peptidase 4. J. Virol. 2014;88:9220–9232. doi: 10.1128/jvi.00676-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. 2020. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Bosch B.-J., Frenz B., Rottier P.J.M., Dimaio F., Rey F.A., Veesler D. 2016. Cryo-electron Microscopy Structure of a Coronavirus Spike Glycoprotein Trimer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Hua C., Xia S., Li W., Lu L., Jiang S. Combining a fusion inhibitory peptide targeting the MERS-CoV S2 protein HR1 domain and a neutralizing antibody specific for the S1 protein receptor-binding domain (RBD) showed potent synergism against pseudotyped MERS-CoV with or without mutations in RBD. 2019. Viruses 11. [DOI] [PMC free article] [PubMed]
- Wang C., Xia S., Zhang P., Zhang T., Wang W., Tian Y., Meng G., Jiang S., Liu K. Discovery of hydrocarbon-stapled short α-helical peptides as promising Middle East respiratory syndrome coronavirus (MERS-CoV) fusion inhibitors. J. Med. Chem. 2018;61 doi: 10.1021/acs.jmedchem.7b01732. 2018–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang P., Liu K., Guo F., Zhang Y., Zhang G., Jiang C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008;18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Shi X., Jiang L., Zhang S., Wang D., Tong P., Guo D., Fu L., Cui Y., Liu X., Arledge K.C., Chen Y.-H., Zhang L., Wang X. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner F.J., Lew R.A., Smith A.I., Lambert D.W., Hooper N.M., Turner A.J. Angiotensin-converting enzyme 2 (ACE2), but not ACE, is preferentially localized to the apical surface of polarized kidney cells. J. Biol. Chem. 2005;280:39353–39362. doi: 10.1074/jbc.M508914200. [DOI] [PubMed] [Google Scholar]
- White J.M., Delos S.E., Brecher M., Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.M., Whittaker G.R. Fusion of enveloped viruses in endosomes. Traffic. 2016;17:593–614. doi: 10.1111/tra.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widagdo W., Okba N.M.A., Li W., de Jong A., de Swart R.L., Begeman L., van den Brand J.M.A., Bosch B.-J., Haagmans B.L. Species-specific colocalization of Middle East respiratory syndrome coronavirus attachment and entry receptors. J. Virol. 2019;93 doi: 10.1128/jvi.00107-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild C.T., Shugars D.C., Greenwell T.K., McDanal C.B., Matthews T.J. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. U.S.A. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Liu Q., Wang Q., Sun Z., Su S., Du L., Ying T., Lu L., Jiang S. Middle East respiratory syndrome coronavirus (MERS-CoV) entry inhibitors targeting spike protein. Virus Res. 2014;194:200–210. doi: 10.1016/j.virusres.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Yan L., Xu W., Agrawal A.S., Algaissi A., Tseng C.-T.K., Wang Q., Du L., Tan W., Wilson I.A., Jiang S., Yang B., Lu L. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019;5:eaav4580. doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., Ying T., Liu S., Shi Z., Jiang S., Lu L. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 2020 doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020;80– doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.-T., Kreutzberger A.J.B., Lee J., Kiessling V., Tamm L.K. The role of cholesterol in membrane fusion. Chem. Phys. Lipids. 2016;199:136–143. doi: 10.1016/j.chemphyslip.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Du L., Liu C., Wang L., Ma C., Tang J., Baric R.S., Jiang S., Li F. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc. Natl. Acad. Sci. U.S.A. 2014;111:12516–12521. doi: 10.1073/pnas.1405889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying T., Li H., Lu L., Dimitrov D.S., Jiang S. Development of human neutralizing monoclonal antibodies for prevention and therapy of MERS-CoV infections. Microb. Infect. 2015;17:142–148. doi: 10.1016/j.micinf.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K., Yi L., Chen J., Qu X., Qing T., Rao X., Jiang P., Hu J., Xiong Z., Nie Y., Shi X., Wang W., Ling C., Yin X., Fan K., Lai L., Ding M., Deng H. Suppression of SARS-CoV entry by peptides corresponding to heptad regions on spike glycoprotein. Biochem. Biophys. Res. Commun. 2004;319:746–752. doi: 10.1016/J.BBRC.2004.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y., Zhang X., Gao G.F. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Wang G., Li J., Nie Y., Shi X., Lian G., Wang W., Yin X., Zhao Y., Qu X., Ding M., Deng H. Identification of an antigenic determinant on the S2 domain of the severe acute respiratory syndrome coronavirus spike glycoprotein capable of inducing neutralizing antibodies. J. Virol. 2004;78:6938–6945. doi: 10.1128/JVI.78.13.6938-6945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B.J., Guan Y., He M.L., Sun H., Du L., Zheng Y., Wong K.L., Chen H., Chen Y., Lu L., Tanner J.A., Watt R.M., Niccolai N., Bernini A., Spiga O., Woo P.C.Y., Kung H.F., Yuen K.Y., Huang J.D. Synthetic peptides outside the spike protein heptad repeat regions as potent inhibitors of SARS-associated coronavirus. Antivir. Ther. 2005;10:393–403. [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Vedantham P., Lu K., Agudelo J., Carrion R., Nunneley J.W., Barnard D., Pöhlmann S., McKerrow J.H., Renslo A.R., Simmons G. Protease inhibitors targeting coronavirus and filovirus entry. Antivir. Res. 2015;116:76–84. doi: 10.1016/J.ANTIVIRAL.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmora P., Hoffmann M., Kollmus H., Moldenhauer A.S., Danov O., Braun A., Winkler M., Schughart K., Stefan Pöhlmann X. TMPRSS11A activates the influenza A virus hemagglutinin and the MERS coronavirus spike protein and is insensitive against blockade by HAI-1. J. Biol. Chem. 2018;293:13863–13873. doi: 10.1074/jbc.RA118.001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.