Summary
Interferon-induced transmembrane protein 3 (IFITM3) is a key interferon effector that broadly prevents infection by diverse viruses. However, the cellular factors that control IFITM3 homeostasis and antiviral activity have not been fully elucidated. Using site-specific photo-crosslinking and quantitative proteomic analysis, here we present the identification and functional characterization of VCP/p97 AAA-ATPase as a primary interaction partner of IFITM3. We show that IFITM3 ubiquitination at lysine 24 is crucial for VCP binding, trafficking, turnover, and engagement with incoming virus particles. Consistently, pharmacological inhibition of VCP/p97 ATPase activity leads to defective IFITM3 lysosomal sorting, turnover, and co-trafficking with virus particles. Our results showcase the utility of site-specific protein photo-crosslinking in mammalian cells and reveal VCP/p97 as a key cellular factor involved in IFITM3 trafficking and homeostasis.
Keywords: chemical proteomics, photo-crosslinking, unnatural amino acid, protein-protein interaction, IFITM3, VCP/p97
Graphical Abstract
Highlights
-
•
Photo-crosslinking proteomics identify VCP/p97 as an IFITM3-interacting protein
-
•
Ubiquitination of IFITM3 is crucial for interaction with VCP
-
•
Lysine 24 ubiquitination regulates IFITM3 trafficking and turnover
-
•
Depletion or inhibition of VCP leads to delayed turnover and accumulation of IFITM3
Using site-specific photo-crosslinking and quantitative proteomic analysis, Wu et al. report the identification of VCP/p97 AAA-ATPase as a primary interaction partner of IFITM3, a crucial host restriction factor for limiting virus infections. Further functional studies demonstrate that VCP/p97 regulates IFITM3 lysosomal trafficking and turnover, revealing an important regulatory mechanism of IFITM3 in immune system.
Introduction
Interferons (IFNs) mediate the first-line host innate immune defense against viral infection by inducing the expression of hundreds of IFN-stimulated genes (ISGs) (Muller et al., 1994, Schoggins and Rice, 2011, Schoggins et al., 2011, MacMicking, 2012). Among these ISGs, the IFN-induced transmembrane protein (IFITM) family has been shown to be responsible for a significant portion of the IFN-mediated antiviral activity (Brass et al., 2009, Bailey et al., 2014). In recent years, extensive studies have shown that IFITM3, the most active isoform of IFITM family (Brass et al., 2009, Gorman et al., 2016), provides potent antiviral activity in mammalian cells against many pathogenic viruses, including influenza virus, hepatitis C virus, dengue virus, West Nile virus, vesicular stomatitis virus, human immunodeficiency virus, SARS coronavirus, and Ebola virus (Brass et al., 2009, Weidner et al., 2010, Huang et al., 2011, Lu et al., 2011, Schoggins et al., 2011, Perreira et al., 2013, Bailey et al., 2014). In line with cellular studies, Ifitm3 homozygous knockout mice are more susceptible to influenza virus infection (Bailey et al., 2012, Everitt et al., 2012). More importantly, a significant percentage of human patients hospitalized by seasonal influenza virus infection carries a genetic polymorphism expressing partial loss-of-function alleles of IFITM3 (Zhang et al., 2013, Wang et al., 2014, Zani and Yount, 2018). Therefore, IFITM3 appears to be a key IFN-induced host effector restricting viral infection in mammals.
Over the years, many studies have explored the biochemical properties and antiviral mechanism of IFITM3. IFITM3 is largely localized to intracellular late endolysosomes (Amini-Bavil-Olyaee et al., 2013, Desai et al., 2014, Weston et al., 2014), and may traffic from the plasma membrane to intracellular compartments via an N-terminal YxxΦ sorting motif (Jia et al., 2012, Jia et al., 2014, Chesarino et al., 2014a). IFITM3 is further regulated by post-translational modifications in mammalian cells (Chesarino et al., 2014b). We previously discovered that S-palmitoylation at conserved membrane-proximal cysteine residues regulates IFITM3 membrane targeting and antiviral activity (Yount et al., 2010, Percher et al., 2016), and that ubiquitination of lysine residues controls its turnover and stability (Yount et al., 2012). IFITM3 does not block the binding or uptake of viruses into host cells, but instead restricts deposition of viral contents into cytosol (Feeley et al., 2011) by preventing virus-cell fusion (Liao et al., 2019). Studies have initially suggested that IFITM3 blocks viral membrane hemifusion (Li et al., 2013b), but then implied that IFITM3 inhibits fusion pore formation at a post-hemifusion stage (Desai et al., 2014, Suddala et al., 2019) through directly changing membrane fluidity and/or curvature (Lin et al., 2013, Chesarino et al., 2017) or by indirectly altering the lipid concentration and/or composition of vesicle membranes (Amini-Bavil-Olyaee et al., 2013). In addition, IFITM3 was shown to incorporate into nascent virions during viral assembly to limit viral entry (Compton et al., 2014, Tartour et al., 2014). Furthermore, IFITM3 may directly suppress viral protein synthesis to restrict virus replication (Lee et al., 2018). Nevertheless, there is still no clear consensus on the precise antiviral mechanism of IFITM3 (Diamond and Farzan, 2012, Liao et al., 2019).
Our laboratories have been focusing on characterization of IFITM3 antiviral properties and mechanisms (Yount et al., 2010, Yount et al., 2012, Peng and Hang, 2015, Peng and Hang, 2016, Percher et al., 2016, Spence et al., 2019). We developed a site-specific fluorescence-labeling method for IFITM3 (Peng and Hang, 2016), which integrated amber suppression technology (Wang et al., 2006, Chin, 2017, Young and Schultz, 2018) for site-specific incorporation of a cycloalkene unnatural amino acid (UAA) into the protein with bioorthogonal labeling for fluorophore conjugation. Live-cell imaging of IFITM3 using this method has revealed that IFITM3 directly engages virus-containing vesicles (Spence et al., 2019).
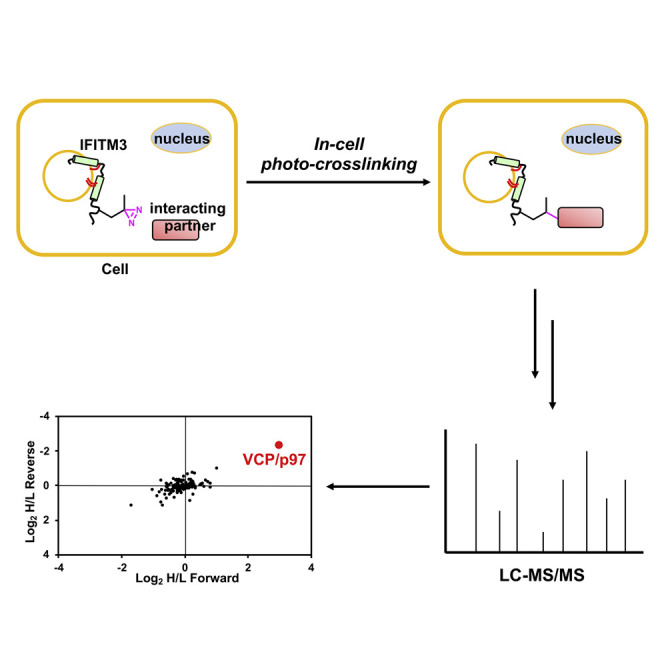
To further characterize the biochemical and cellular properties of IFITM3, we sought to identify its interacting proteins by mass spectrometry (MS) analysis. Profiling of IFITM3-interacting proteins has been challenging with standard co-immunoprecipitation (coIP)-MS approach. Indeed, due to the “stickiness” of IFITM3 as a membrane protein, such analyses previously conducted by us and others mainly recovered IFITM3 homo- or hetero-oligomerizations (John et al., 2013) as well as many “false-positive” membrane-associated proteins (Fu et al., 2017, Hubel et al., 2019). Moreover, membrane protein interactions may only be retained under a native lipid environment, which is often disrupted by detergents during cell lysis (Daley, 2008). To overcome this technical issue, we turned our attention to in-cell photo-crosslinking, which generates a covalent bond between interacting proteins upon photoirradiation (Tanaka et al., 2008, Pham et al., 2013, Peng and Hang, 2015). As a result of the covalent complex formation, stringent or even denaturing conditions for cell lysis and protein isolation can be utilized to eliminate non-specific false-positives. More importantly, photo-crosslinking in live cells allows trap of protein interactions under native conditions. In this regard, photo-crosslinkable UAAs have been site-specifically incorporated into proteins of interest via amber suppression technology (Chin et al., 2002, Hino et al., 2005, Tippmann et al., 2007, Ai et al., 2011, Chou et al., 2011, Xie et al., 2017), which enabled covalent capture of protein-protein interactions in live cells and identification by MS analysis (Zhang et al., 2011, Zhang et al., 2017, Yang et al., 2016, He et al., 2017, Kleiner et al., 2018).
Herein, we present the application of a photo-crosslinking proteomics approach that relies on amber suppression-mediated site-specific incorporation of a diazirine-containing amino acid and quantitative MS analysis to identify IFITM3-interacting proteins (Figure 1 ). The VCP/p97 AAA-ATPase was retrieved from the proteomics study and validated to interact with IFITM3. We show that VCP, in complex with the UBXD1 cofactor, mainly associates with ubiquitinated IFITM3. The ubiquitination-deficient lysine 24 mutant of IFITM3 exhibits delayed turnover, impaired steady-state distribution in lysosomes, and altered co-trafficking with incoming virus particles. In line with these results, depletion of the VCP-UBXD1 complex leads to abnormal turnover and accumulation of IFITM3. Moreover, we demonstrate that chemical inhibition of VCP ATPase activity results in accumulation of ubiquitinated IFITM3, delayed turnover, and impaired trafficking of IFITM3 to lysosomes and incoming virus particles. Collectively, these results identify a functional interaction of IFITM3 with VCP, and characterize a key regulatory mechanism in immune system for IFITM3 trafficking, turnover, and homeostasis.
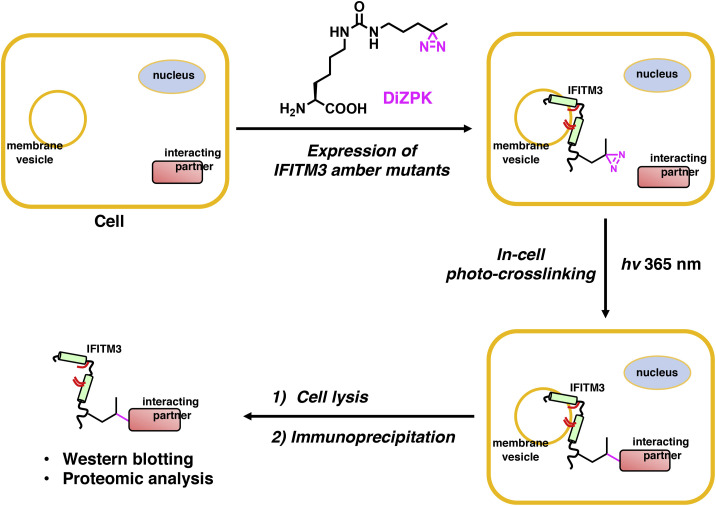
Figure 1.
Site-Specific Photo-Crosslinking Strategy for Studying IFITM3-Interacting Proteins in Live Cells
The amber suppression technology mediates site-specific incorporation of the diazirine-containing DiZPK into IFITM3, which enables in-cell photo-crosslinking with interacting proteins. The photo-crosslinked complexes were analyzed by western blot and identified by quantitative proteomics after IP.
Results
Site-Specific Incorporation of Photo-Crosslinkable Amino Acid into IFITM3 in Mammalian Cells
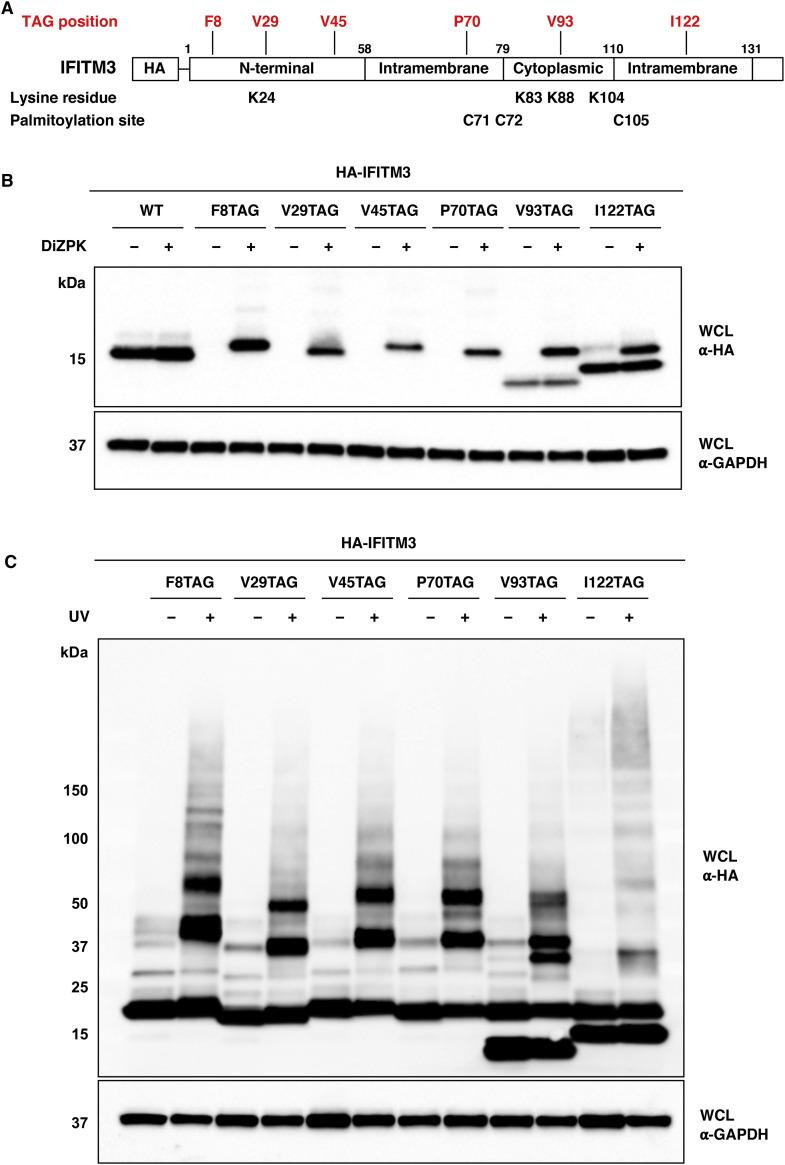
To capture IFITM3-interacting proteins in live cells, we chose to use amber suppression technology to site-specifically incorporate DiZPK (Zhang et al., 2011), a UAA that contains a photo-activatable diazirine functionality and facilitates covalent photo-crosslinking of protein-protein interactions (Figure 1). We selected several amino acid positions on IFITM3 that are situated in different domains of the protein (Figure 2 A). These residues are not essential for IFITM3 activity (John et al., 2013), free of post-translational modifications, and likely accessible for protein-protein interactions. We then mutated them individually into the amber codon TAG for DiZPK incorporation. To evaluate the amber suppression efficiency in mammalian cells, HEK293T cells were co-transfected with two plasmids encoding an aminoacyl-tRNA synthetase/tRNA pair and an HA-tagged IFITM3-TAG mutant, i.e., pCMV-DiZPK-PylRS (Zhang et al., 2011) and pCMV-HA-IFITM3-TAG, respectively, in the absence or presence of DiZPK. Western blot analysis of whole-cell lysates indicated that all IFITM3-TAG mutants afforded full-length protein expressions with varying levels that were dependent on the presence of DiZPK (Figure 2B). Notably, IFITM3-TAG mutants with the amber codon positions near the C terminus also generated a large portion of truncated proteins, suggesting suboptimal amber suppression efficiency at these near C-terminal positions. It is possible that truncated proteins were also generated for other IFITM3 mutants, but degraded more efficiently than longer truncated proteins observed for C-terminal IFITM3 mutants.
Figure 2.
DiZPK Is Incorporated into IFITM3 via Amber Suppression and Enables Photo-Crosslinking in Live Cells
(A) Domain structure of IFITM3 protein showing amino acid residues mutated to amber codons and locations of lysine and cysteine residues.
(B) Western blot analysis of HEK293T cells transfected with HA-IFITM3-TAG mutants in the absence or presence of DiZPK.
(C) Western blot analysis of HEK293T cells transfected with HA-IFITM3-TAG mutants in the presence of DiZPK with or without UV irradiation. High-molecular-weight bands that appeared in UV-irradiated samples indicate putative photo-crosslinked IFITM3 complexes.
We then examined whether the incorporation of DiZPK could induce protein photo-crosslinking of IFITM3 in live cells. For this purpose, cells expressing DiZPK-modified IFITM3 were subjected to 365-nm photoirradiation and lysed for western blot analysis. As expected, a series of slower-migrating bands indicative of higher-molecular-weight photo-crosslinked complexes were observed for all IFITM3-TAG mutants only when cells were treated with UV irradiation (Figure 2C). Interestingly, IFITM3 mutants with varying DiZPK positions showed distinct photo-crosslinking patterns and efficiency (Figure 2C), suggesting that IFITM3 photo-crosslinking induced by DiZPK is largely dependent on the location and environment of the UAA. Moreover, the photo-crosslinked complexes can be efficiently isolated by anti-HA IP (Figure S1A). Taken together, these results show that photo-activatable DiZPK is site-specifically incorporated into IFITM3 and allows photo-crosslinking of IFITM3-interacting proteins.
Photo-Crosslinking Proteomics Identify IFITM3-Interacting Partners
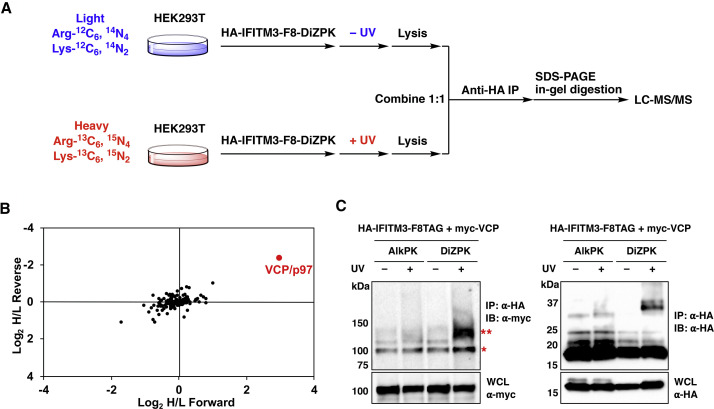
Having validated the expression and photo-crosslinking of IFITM3 by western blot, we then proceeded to profile IFITM3 protein interactions using stable isotope labeling by amino acids in cell culture (SILAC)-based quantitative proteomics (Ong et al., 2002). In this regard, we cultured HEK293T cells with SILAC media before transfection to express DiZPK-modified HA-IFITM3 and then treated the cells with UV irradiation, which was grouped as the “photo-crosslinking sample”; as the “non-photo-crosslinking control,” cells cultured in corresponding SILAC medium were not treated with UV. After cell lysis, the photo-crosslinking sample and non-photo-crosslinking control were equally mixed, subjected to anti-HA affinity purification, and analyzed by MS for protein identification and quantification (Figure 3 A). It is worth mentioning that we used very harsh denaturing conditions (i.e., 4% SDS) for cell lysis to break non-covalent IFITM3 interactions and favor identification of covalent complexes. Moreover, to eliminate potential false-positives and increase identification reliability, we performed two sets of proteomics experiments by switching the SILAC labels and UV treatment conditions (termed “forward” and “reverse” SILAC experiments); i.e., in the forward SILAC experiment, only heavy-labeled cells were UV irradiated (Figure 3A), whereas only light-labeled cells were UV irradiated in the reverse SILAC experiment. We initially focused on IFITM3 modified with DiZPK at position 8 for photo-crosslinking proteomics studies, as this mutant exhibited optimal photo-crosslinking efficiency. The forward and reverse SILAC experiments robustly recovered one protein, i.e., VCP (valosin-containing protein, also called p97), that was selectively more enriched in the photo-crosslinking sample versus the non-photo-crosslinking control (Figure 3B; Table S1), thus indicating that VCP is crosslinked to IFITM3 in response to UV. Notably, the proteomics analysis also identified many other proteins (Figure 3B, shown in black) that were not enriched in photo-crosslinking samples, including IFITM3-interacting proteins, such as VAPA (Table S1) (Amini-Bavil-Olyaee et al., 2013), and non-specific binders, such as ribosomal proteins.
Figure 3.
Quantitative Proteomics Enable Identification of Photo-Crosslinked IFITM3-Interacting Proteins
(A) Scheme for “forward” SILAC quantitative proteomic analysis of photo-crosslinked interaction partners of IFITM3. In a complementary “reverse” SILAC proteomic analysis, light cells were UV irradiated, while heavy cells were not.
(B) Normalized SILAC ratios for forward and reverse SILAC proteomic analysis with DiZPK-modified IFITM3 at position 8. VCP, highly enriched in photo-crosslinking samples, is marked in red. Other identified proteins that were not enriched in photo-crosslinking samples are shown in black and represent non-covalent IFITM3-interacting proteins or non-specifically binding proteins.
(C) Validation of IFITM3-VCP photo-crosslinking complex. HEK293T cells expressing AlkPK or DiZPK-modified HA-IFITM3 and myc-VCP were UV irradiated and subjected to anti-HA IP for western blot analysis. The red asterisk indicates co-immunoprecipitated VCP and the double red asterisks indicate photo-crosslinked IFITM3-VCP complex.
To validate the interaction of IFITM3 with VCP and the formation of a photo-crosslinked complex, we co-expressed HA-IFITM3-F8TAG in the presence of AlkPK (Li et al., 2013a), a lysine derivative without the diazirine group serving as a negative control, or DiZPK with myc-tagged VCP in HEK293T cells for photo-crosslinking, anti-HA IP, and western blot analysis. As shown in Figure 3C, VCP indeed interacts with IFITM3 as revealed by the co-immunoprecipitated VCP bands. More importantly, a slower-migrating band with molecular weight equal to the summed mass of VCP and IFITM3 was observed only for DiZPK-modified IFITM3 in the presence of UV, confirming the formation of photo-crosslinked IFITM3-VCP complex (Figure 3C). We also performed a similar photo-crosslinking experiment on other IFITM3-TAG mutants. Intriguingly, IFITM3-TAG mutants with DiZPK incorporated at the N terminus (e.g., positions 8, 29, and 45) showed relatively stronger photo-crosslinked bands with VCP, whereas DiZPK at position 93 and 122 almost failed to induce crosslinking (Figure S1B), again confirming that the photo-crosslinking efficiency is dependent on DiZPK position. Collectively, these results demonstrate that DiZPK-modified IFITM3 is robustly photo-crosslinked with VCP.
VCP Interaction with IFITM3 Is Largely Dependent on Ubiquitination
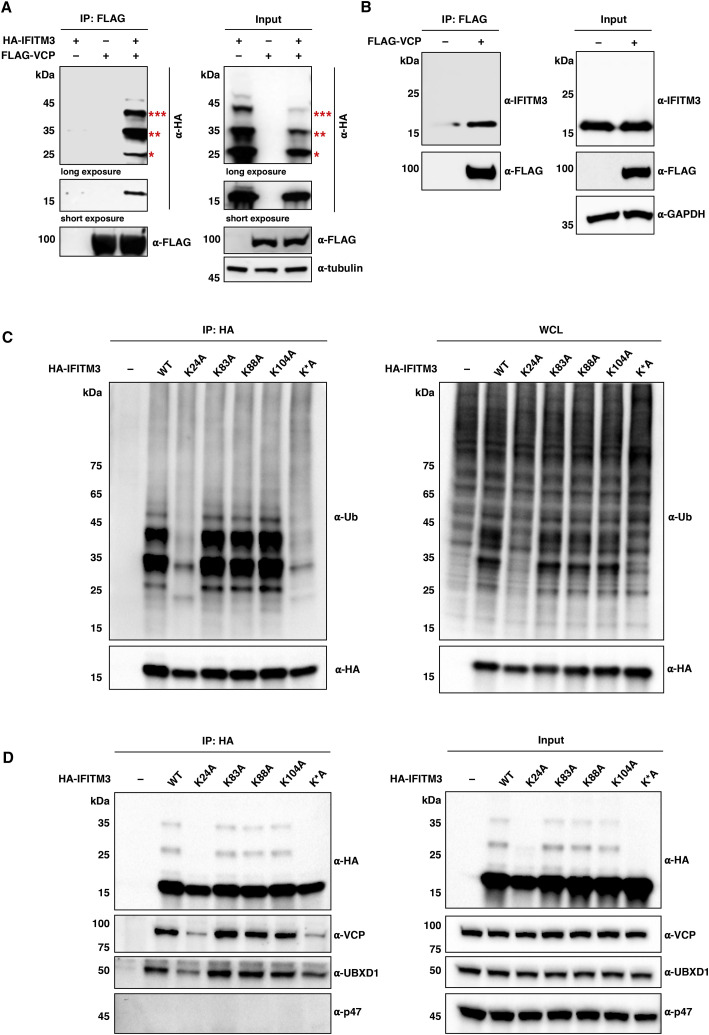
As the interaction between IFITM3 and VCP had not been characterized previously, we pursued the significance of this interaction. VCP is a hexameric AAA-type ATPase involved in diverse cellular processes, ranging from endoplasmic reticulum-associated degradation (ERAD) to membrane fusion, nuclear factor κB (NF-κB) activation, and chromatin-associated degradation (Meyer et al., 2012, Stach and Freemont, 2017, Ye et al., 2017). Additional coIP experiments with FLAG-tagged VCP further confirmed that both exogenously introduced and endogenously expressed IFITM3 associated with VCP (Figures 4A and 4B). We observed that ubiquitinated IFITM3 co-immunoprecipitated with VCP (Figure 4A), consistent with the common feature of VCP-involved processes that all require ubiquitinated substrates (Stach and Freemont, 2017). To determine whether ubiquitination of IFITM3 is essential for interaction with VCP, we analyzed IFITM3 single-lysine mutants and a quadruple-lysine mutant, in which all four lysines were mutated to alanine (i.e., K∗A). Our results showed that K24 was the major ubiquitination site, and that K24A and K∗A mutants of IFITM3 were both nearly non-ubiquitinated (Figure 4C), consistent with our previous studies on mouse Ifitm3 (Yount et al., 2012). Moreover, coIP revealed that binding of IFITM3 K24A or K∗A mutant to VCP was indeed greatly diminished (Figure 4D), implying that IFITM3 ubiquitination at N-terminal K24 is essential for interacting with VCP. Because VCP requires different cofactors to fulfill distinct roles (Stach and Freemont, 2017), we probed the anti-HA immunoprecipitates with specific antibodies against well-known VCP cofactors, such as p47 (also known as NSFL1C) and UBXD1 (also known as UBXN6). These experiments showed that the interaction complex of IFITM3 and VCP involved UBXD1 as the cofactor (Figure 4D). Taken together, these data suggest that IFITM3 interacts with VCP largely in a ubiquitination-dependent manner, with UBXD1 as the cofactor.
Figure 4.
IFITM3 Ubiquitination Is Important for Interaction with VCP
(A) CoIP of HA-IFITM3 with FLAG-VCP. HEK293T cells were transfected with HA-IFITM3 and/or FLAG-VCP, and cell lysates were immunoprecipitated with anti-FLAG resin for western blot analysis. The red asterisks indicate IFITM3 ubiquitination bands with high molecular weights.
(B) CoIP of endogenous IFITM3 with FLAG-VCP. HeLa cells were transfected with FLAG-VCP, and cell lysates were immunoprecipitated with anti-FLAG resin for western blot analysis.
(C) Analysis of ubiquitination status of IFITM3 and lysine mutants. HEK293T cells were transfected with HA-IFITM3, single lysine mutants (i.e., K24A, K83A, K88A, and K104A), or the quadruple lysine mutant K∗A, lysed for anti-HA IP, and analyzed by western blot.
(D) CoIP analysis of HA-IFITM3 and lysine mutants with endogenous VCP and its cofactors. The anti-HA immunoprecipitates were prepared as in (C) and analyzed by western blot.
K24 Ubiquitination Regulates IFITM3 Trafficking, Turnover, and Interaction with Incoming Virus Particles
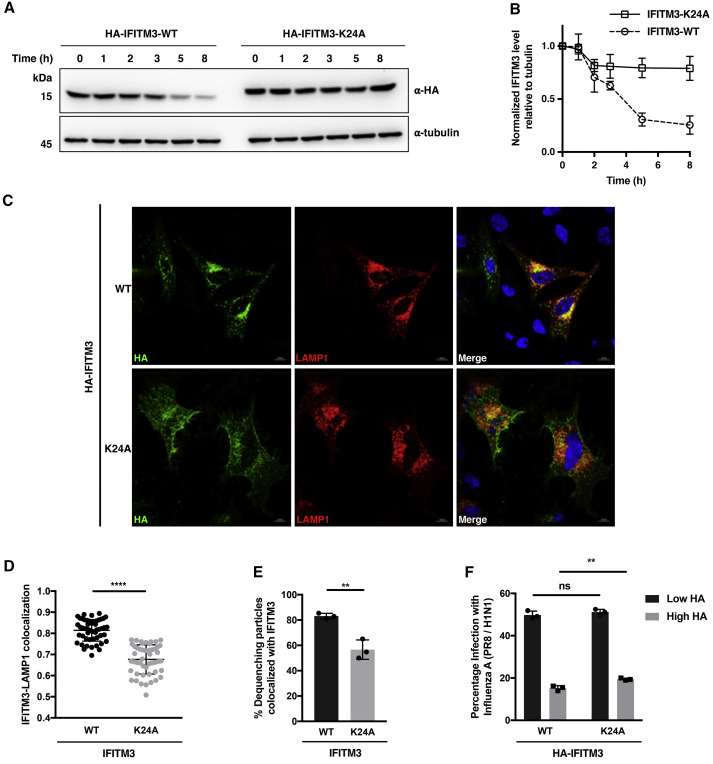
To investigate the functions of IFITM3 ubiquitination and interaction with VCP, we took advantage of the IFITM3-K24A mutant. As both ubiquitination and VCP interaction are implicated in protein degradation, we first monitored the degradation of IFITM3-K24A mutant in parallel with wild-type. Cells expressing HA-IFITM3 were treated with cycloheximide (CHX) for different time periods and analyzed by western blot. We observed that the ubiquitination-deficient K24A variant was degraded much more slowly compared with wild-type IFITM3 (Figures 5A and 5B), indicating that K24 ubiquitination is an important regulator of IFITM3 turnover and stability.
Figure 5.
K24 Ubiquitination Regulates IFITM3 Turnover, Localization, and Co-trafficking with Incoming Virus Particles
(A) Analysis of turnover of IFITM3 and K24A mutant. HeLa cells expressing HA-IFITM3 or K24A mutant were treated with CHX (25 μg/mL) for the indicated times and lysed for anti-HA western blot analysis.
(B) Quantification of IFITM3 levels normalized to tubulin levels shown in (A). Data are represented as mean ± SD, n = 3.
(C) Immunofluorescence analysis of IFITM3 and K24A mutant. HeLa cells were transfected with indicated HA-IFITM3 construct and mCherry-LAMP1, and processed for immunofluorescence with an Alexa Fluor 488-conjugated anti-HA antibody. Scale bars, 10 μm.
(D) Quantification of Pearson coefficients for the IFITM3-LAMP1 colocalization shown in (C). Data are represented as mean ± SD, n = 50 cells. ∗∗∗∗p < 0.0001 calculated by Student's t test.
(E) Relative percentage of DiD-IAV particles colocalized with IFITM3 and IFITM3-K24A at the time of dequenching. HeLa IFITM2/3-KO cells expressing BODIPY-labeled IFITM3 were infected with DiD-IAV particles and monitored for DiD dequenching and IFITM3 trafficking by time-lapse imaging. Data are represented as mean ± SD of three independent experiments. ∗∗p < 0.01 calculated by Student's t test.
(F) Antiviral activity of IFITM3 and K24A mutant. HEK293T cells expressing HA-IFITM3 or HA-IFITM3-K24A were infected with IAV (PR8/H1N1) at an MOI of 2.5 for 6 h. Cells were fixed and stained with anti-HA and anti-influenza NP antibodies to measure IFITM3 expression and virus infection, respectively, with flow cytometry. “Low HA” and “High HA” indicate cell populations expressing low and high levels of HA-IFITM3, respectively. Data are represented as mean ± SD of three independent experiments. ∗∗p < 0.01, n.s. indicates p > 0.05 calculated by Student's t test.
Fluorescence imaging experiments were then performed to examine the localization and trafficking of IFITM3. Immunofluorescence studies demonstrated that the K24A mutant exhibited more diffuse intracellular signals compared with wild-type IFITM3 (Figure 5C). Although wild-type IFITM3 colocalized frequently with lysosome marker protein LAMP1 as reported previously (Desai et al., 2014, Peng and Hang, 2016), overlapping of the K24A mutant and LAMP1 was significantly reduced (Figures 5C and 5D), accounting for the impaired turnover of K24A mutant. In addition to steady-state distribution, we applied a previously developed live-cell fluorescence imaging method (Peng and Hang, 2016) to study the dynamic trafficking of K24A mutant during influenza A virus (IAV) infection. The method involves site-specific incorporation of a cycloalkene UAA into IFITM3 and subsequent bioorthogonal labeling with BODIPY-tetrazine in live cells. We performed the live-cell imaging in HeLa IFITM2/3-KO cells that were infected with 1,1′-dioctactadecyl-3,3,3′,3′-tetramethylindodicarbocyanine (DiD)-labeled IAV particles (Spence et al., 2019). Disruption of ubiquitination by K24 mutation did not significantly affect virus-trafficking dynamics or total percentage of DiD-labeled virus particles that underwent lipid mixing (Figure S2). However, we observed an apparent reduction in the percentage of virus particles colocalized with IFITM3-K24A or K24R mutant at the time of lipid mixing compared with wild-type IFITM3 (Figures 5E and S2G), indicating that K24 ubiquitination is required for maximal incorporation of IFITM3 into virus-containing vesicles at the time of viral fusion. We also assessed the antiviral activity of IFITM3-K24A mutant against IAV infection. Interestingly, although the K24A mutant was still protective against IAV, its activity was slightly decreased compared with wild-type IFITM3 (Figure 5F). Together, these results demonstrate that site-specific K24 ubiquitination not only determines IFITM3 turnover and steady-state distribution in lysosomes but also regulates IFITM3 dynamic trafficking to incoming virus particles.
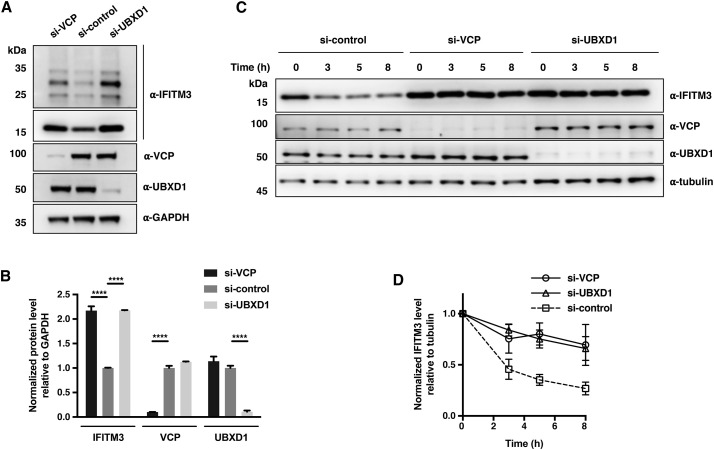
Depletion of VCP or UBXD1 Leads to Delayed Turnover and Accumulation of IFITM3
To investigate the regulatory role of VCP on IFITM3, we used small interfering RNA (siRNA) oligonucleotides to interfere with VCP expression. As UBXD1 was shown to be specifically involved in the VCP-IFITM3 interaction complex (Figure 4D), we also included UBXD1 siRNA in these studies. Western blot analysis of whole-cell lysates confirmed specific and efficient interference of VCP or UBXD1 expression after treatment with respective siRNA (Figure 6 A). We then monitored IFITM3 levels, ubiquitination status, and turnover rate after cellular depletion of VCP or UBXD1. As shown in Figures 6A and 6B, the endogenous levels of non-ubiquitinated IFITM3 in HeLa cells were upregulated by knockdown of VCP or UBXD1. Also, the IFITM3 ubiquitination levels were significantly increased in VCP- or UBXD1-depleted samples (Figure 6A). In line with these results, we repeatedly observed decreased ubiquitination levels of IFITM3, as well as downregulation of unmodified IFITM3, when VCP was overexpressed (Figures 4A and S3), suggesting that VCP facilitates the recycling of IFITM3. More importantly, consistent with the above results, the turnover rate of endogenous IFITM3 was greatly reduced when VCP or UBXD1 was knocked down (Figures 6C and 6D). These results demonstrate that knockdown of VCP-UBXD1 complex impairs IFITM3 turnover and leads to accumulation in lysosomes, consistent with our K24A mutant data.
Figure 6.
Depletion of VCP or UBXD1 Leads to Accumulation of IFITM3 and Delayed Turnover
(A) Analysis of endogenous IFITM3 levels upon VCP or UBXD1 knockdown. HeLa cells were transfected with indicated siRNA for 72 h and lysed for western blot analysis.
(B) Quantification of non-ubiquitinated IFITM3, VCP, and UBXD1 levels shown in (A). Data are represented as mean ± SD, n = 3. ∗∗∗∗p < 0.0001 calculated by two-way ANOVA test.
(C) Analysis of endogenous IFITM3 turnover upon VCP or UBXD1 knockdown. HeLa cells were transfected with indicated siRNA for 72 h, treated with CHX (25 μg/mL) for the indicated times, and lysed for western blot analysis.
(D) Quantification of IFITM3 levels normalized to tubulin levels shown in (C). Data are represented as mean ± SD, n = 3.
VCP/p97 ATPase Activity Is Crucial for IFITM3 Trafficking, Turnover, and Antiviral Activity
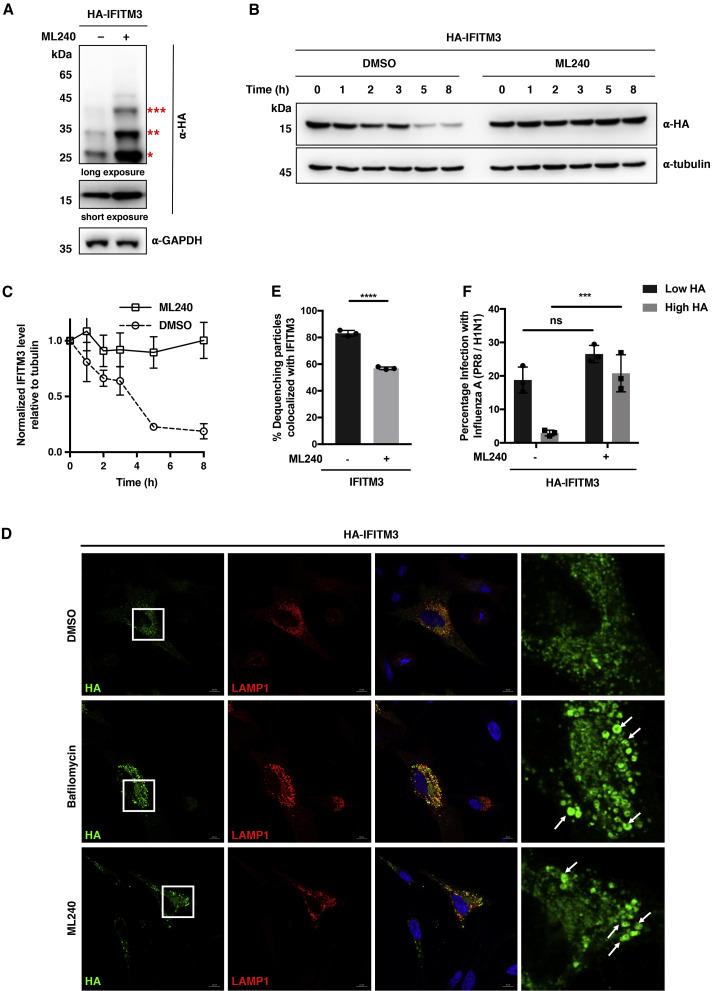
To further explore the regulation of IFITM3 by VCP, we used a pharmacological approach using ML240 (Chou et al., 2013), a specific VCP inhibitor, to deactivate VCP ATPase activity. Western blot analysis of exogenous IFITM3 demonstrated accumulation of both non-ubiquitinated and ubiquitinated IFITM3 after long-term treatment of cells with ML240 (Figures 7A, S4A, and S4B). Similar results were observed for endogenous IFITM3 in ML240-treated samples (Figures S4C–S4E). More importantly, consistent with these data, inhibition of VCP ATPase activity with ML240 significantly delayed the degradation of both overexpressed and endogenous IFITM3 (Figures 7B, 7C, S4F, and S4G), consistent with the delayed turnover of K24A mutant and indicative of the involvement of VCP ATPase activity in IFITM3 degradation.
Figure 7.
Chemical Inhibition of VCP ATPase Activity with ML240 Impairs IFITM3 Turnover, Lysosomal Sorting, and Trafficking
(A) Analysis of IFITM3 and ubiquitination levels upon treatment with ML240. HEK293T cells were transfected with HA-IFITM3 for 12 h, treated with DMSO or ML240 (2.5 μM) for another 12 h, and then lysed for western blot analysis. The red asterisks indicate IFITM3 ubiquitination bands with high molecular weights.
(B) Analysis of IFITM3 turnover upon treatment with ML240. HeLa cells expressing HA-IFITM3 were treated with CHX (25 μg/mL) for the indicated times in the presence of DMSO or ML240 (2.5 μM) and lysed for western blot analysis.
(C) Quantification of IFITM3 levels normalized to tubulin levels shown in (B). Data are represented as mean ± SD, n = 3.
(D) Immunofluorescence analysis of HA-IFITM3 in the presence of bafilomycin A1 or ML240. HeLa cells were transfected with HA-IFITM3 and mCherry-LAMP1, treated with DMSO, bafilomycin A1 (100 ng/mL), or ML240 (2.5 μM) for 12 h, and processed for immunofluorescence with an Alexa Fluor 488-conjugated anti-HA antibody. Scale bars, 10 μm. The right panels show magnified squared regions in the corresponding left panels. White arrows indicate enlarged IFITM3-containing compartments.
(E) Relative percentage of DiD-IAV particles colocalized with IFITM3 at the time of dequenching upon ML240 treatment. HeLa IFITM2/3-KO cells expressing BODIPY-labeled IFITM3 were infected with DiD-IAV particles, acutely treated with ML240 (1 μM), and monitored for DiD dequenching and IFITM3 trafficking by time-lapse imaging. Data are represented as mean ± SD of three independent experiments. ∗∗∗∗p < 0.0001 calculated by Student's t test.
(F) Antiviral activity of IFITM3 in the presence of ML240. HeLa IFITM2/3-KO cells were transfected with HA-IFITM3 and infected with IAV (PR8/H1N1) at an MOI of 2.5 for 2 h. Cells were then treated with ML240 (2.5 μM) for 4 h, fixed, and stained with anti-HA and anti-influenza NP antibodies to measure IFITM3 expression and virus infection, respectively, with flow cytometry. “Low HA” and “High HA” indicate cell populations expressing low and high levels of HA-IFITM3, respectively. Data are represented as mean ± SD of three independent experiments. ∗∗∗p < 0.001, n.s. indicates p > 0.05 calculated by Student's t test.
To examine the degradation pathway of IFITM3, we compared the effects of ML240 with those of proteasome and lysosome inhibitors. Treatment with lysosomal acidification inhibitors, such as chloroquine and bafilomycin A1 led to obvious accumulation of both ubiquitinated and non-ubiquitinated IFITM3 (Figure S5A), as observed in ML240 treatment. In contrast, proteasome inhibition with MG132 had almost no effect on IFITM3 and its ubiquitination levels, although MG132 did cause a general increase in cellular ubiquitination (Figure S5A). In addition, similar to ML240, bafilomycin A1 was also shown to slow down the turnover of IFITM3 (Figure S5B). Meanwhile, our immunofluorescence studies confirmed that IFITM3 accumulated in LAMP1-positive lysosomal compartments upon treatment with bafilomycin A1 or ML240 (Figure 7D). More strikingly, both bafilomycin A1 and ML240 treatment led to significant enlargement of IFITM3-containing vesicles. The magnified images demonstrate that IFITM3 was restricted to the limiting membrane of these vesicles, pointing to a defect in sorting of IFITM3 into lysosomes for degradation (Figure 7D). These results suggest that IFITM3 is turned over via the lysosome degradation pathway, which relies on VCP ATPase activity for sorting into lysosomes.
We also applied the live-cell fluorescence imaging method to investigate the dynamic co-trafficking of IFITM3 with incoming IAV particles upon ML240 treatment. Long-term treatment of cells with ML240 (e.g., 12–24 h) was shown to increase steady-state IFITM3 levels, which may simply result in less virus infection due to high cellular IFITM3 concentration. To minimize the chronic effects of ML240 on IFITM3 levels and decouple them from the short-term effects of ML240 on virus infection, we applied acute treatment of cells with ML240 immediately after infection with DiD-labeled IAV. Although ML240 did not significantly change the virus fusion rate or the total percentage of fusing particles (Figure S6), suggesting that the endosomal milieu was largely unaffected, the probability of IFITM3 colocalization with fusing virus particles was reduced by ML240 treatment (Figure 7E). Furthermore, in antiviral activity assays, we observed that acute administration of ML240 immediately after virus infection significantly increased IAV infection efficiency in HeLa cells (Figure S7) as well as in HeLa IFITM2/3-KO cells expressing HA-IFITM3 (Figure 7F). By contrast, ML240 exhibited negligible effects on virus infection in IFITM3-deficient cells (Figures S7 and 7F). Collectively, these pharmacological intervention studies demonstrate that VCP ATPase activity plays a critical role in IFITM3 trafficking, turnover, and interaction with incoming virus particles.
Discussion
Over the past decade, numerous research efforts have been made to characterize the antiviral activity and mechanism of action of IFITM3. As our continued endeavors in studying IFITM3 biochemical and antiviral properties, we integrated the amber suppression-enabled photo-crosslinking strategy with quantitative SILAC proteomics to identify IFITM3-interacting proteins. The unique advantage of photo-crosslinking to covalently stabilize protein interactions has indeed allowed reliable identification of VCP/p97 as an interaction partner of IFITM3. Notably, during preparation of this manuscript, a coIP-MS analysis on IFITM3 has been reported to identify over 70 IFITM3-interacting candidate proteins, including VCP as one of the confident hits (Hubel et al., 2019). However, the IFITM3-VCP interaction and its functional significance were not validated. It is noteworthy that VAPA as a known IFITM3-interacting protein (Amini-Bavil-Olyaee et al., 2013) was also identified in our proteomics analysis, but not enriched in the photo-crosslinking samples. This is reasonable as VAPA specifically interacts with IFITM3 transmembrane II domain (Amini-Bavil-Olyaee et al., 2013), which is far away from the photo-activatable UAA. Other known IFITM3 interactors, such as tyrosine kinase FYN (Jia et al., 2012) and E3 ubiquitin ligase NEDD4 (Chesarino et al., 2015), were not identified in our proteomics analysis, probably due to low cellular abundance. Consistently, these interacting proteins were also absent from the aforementioned coIP-MS study on IFITM3 (Hubel et al., 2019). As demonstrated, the photo-crosslinking efficiency was highly dependent on DiZPK position, another unique feature of photo-crosslinking that only occurs between proteins in close proximity due to the short half-life of photo-generated reactive species. Therefore, photo-crosslinking proteomics on other positions should generate distinct IFITM3-interacting proteins.
We validated the interaction of IFITM3 and VCP by coIP and discovered that this interaction is largely dependent on IFITM3 K24 ubiquitination. Previous studies have shown that IFITM3 is ubiquitinated by NEDD4 E3 ubiquitin ligase, which is recruited via the PPxY motif (17-PPNY-20 in IFITM3) (Chesarino et al., 2015). The lysine residue closest to the PPxY motif, K24 at the N terminus of IFITM3, was indeed confirmed to be exclusively ubiquitinated. It was previously discovered that ubiquitination controls the turnover and stability of IFITM3 (Yount et al., 2012). However, the exact regulatory mechanism remains unclear. Our present data provide evidence that, as a functional consequence of binding with ubiquitinated IFITM3, the VCP-UBXD1 complex regulates the trafficking and sorting of IFITM3 to lysosomes for turnover. This notion is supported by the observations that (1) mutation of K24 to Ala prevents interaction with the VCP-UBXD1 complex and, more importantly, interferes with IFITM3 transport to lysosomes for degradation (Figure 5); (2) depletion of the VCP-UBXD1 complex consistently leads to delayed turnover and accumulation of IFITM3 (Figure 6); and (3) long-term inhibition of VCP ATPase activity with ML240 results in enlargement of IFITM3-containing vesicles, delayed turnover, and accumulation of IFITM3 (Figure 7), similar to inhibition of lysosomal acidification. Therefore, we propose that the VCP-UBXD1 complex acts downstream of IFITM3 ubiquitination and directs IFITM3 transport and/or sorting to lysosomal compartments for degradation.
In addition, our present data also suggest that K24 ubiquitination plays an important role in directing IFITM3 transport to virus-bearing vesicles, likely through interaction with VCP. To support this, we demonstrated that IFITM3-K24A mutant was less colocalized with incoming IAV particles (Figure 5E), and that acute treatment with ML240 decreased the colocalization probability of IFITM3 with IAV (Figure 7E).
Dissection of the IFITM3 degradation pathway provides important insights into the intrinsic regulatory mechanism for IFITM3. IFITM3 is expressed at a low basal level in most cell types but becomes highly expressed upon IFN stimulation. Besides inhibiting virus infection, IFITM3 has recently been shown to be involved in other biological processes, for example, shuttling IFN regulatory factor 3, a critical transcription factor for activating IFN production, to lysosomes for degradation (Jiang et al., 2018), and blocking syncytin-mediated fusion to impair syncytiotrophoblast formation (Buchrieser et al., 2019, Zani et al., 2019), which are surprisingly deleterious to immune defense and fetal development, respectively. Moreover, IFITM3 is highly expressed in many cancers and has been implicated in cancer pathogenesis (Schoenherr et al., 2017, Min et al., 2018, Liu et al., 2019). These findings suggest that temporal control of IFITM3 expression levels is crucial for its physiological functions. Our study thus highlights the importance of the VCP-UBXD1 complex in modulating the expression levels and biological consequences of IFITM3. It is, however, still worth mentioning that other pathways are not excluded for IFITM3 degradation, especially under stress conditions. For example, it was recently shown that rapamycin-induced degradation of IFITM3 partially requires the ESCRT machinery (Shi et al., 2018).
Although the exact molecular function of VCP in controlling IFITM3 trafficking remains unclear, it is most likely that the VCP-UBXD1 complex is recruited to IFITM3 to disassemble IFITM3 oligomers or membrane complexes. As IFITM3 oligomerization is believed to directly affect membrane fluidity and curvature (John et al., 2013), disassembly of IFITM3 oligomers is likely to be required for transport of IFITM3 from limiting membrane to intraluminal vesicles. Notably, VCP, as well as its homolog Cdc48, has emerged as a universal segregase protein that binds to ubiquitinated proteins and uses energy from ATP hydrolysis to extract them from lipid membranes or macromolecular complexes (Cooney et al., 2019, Twomey et al., 2019). Such substrate proteins include, for example, ERAD substrates, NF-κB inhibitor alpha, caveolin-1, and so on (Stach and Freemont, 2017, Ye et al., 2017). Our characterization of IFITM3-VCP interaction thus further expands the VCP client list.
Is the VCP-IFITM3 interaction related to IFITM3 antiviral activity? We observed that the interaction-less K24A mutant was less efficient to restrict IAV infection compared with wild-type IFITM3 (Figure 5F). It was noted that most of the antiviral activity of IFITM3-K24A mutant was still retained. This may be explained by the residual interaction of K24A mutant with VCP (Figure 4D) and/or enhanced stability of this mutant (Figures 5A and 5B) at steady state contributing to virus restriction (Yount et al., 2012). As long-term treatment of cells with VCP inhibitor ML240 (e.g., 12–24 h) leads to increased steady-state IFITM3 levels, we applied acute ML240 treatment (i.e., 4 h) of virus-infected cells to differentiate its short-term effects on IFITM3 antiviral activity from the prolonged effects on IFITM3 levels. Interestingly, our pharmacological study showed that acute treatment with ML240 antagonized the antiviral capability of IFITM3 (Figure 7F), implying that VCP may be required for its complete antiviral activity. It is important to note that ML240 appeared to be quite specific to VCP, showing little activity against protein kinases and receptors (Chou et al., 2013). Only limited kinases, such as PIP5K3 and DNAPK (Chou et al., 2013) were shown to be slightly inhibited by ML240. As they have not been implicated in IFITM3 function and host immunity against viruses from previous genome-wide screenings (Shah et al., 2015, Puschnik et al., 2017), these potential off-targets of ML240 do not likely affect interpretation of our results. Nevertheless, it is still possible that unknown ML240 off-target effects (Bastola and Chien, 2018) would contribute to the phenotypes on IFITM3 we observed. Unfortunately, the multiple functions and targets of VCP, as well as its cofactor UBXD1, have precluded us from using siRNA interference in live-cell trafficking and antiviral activity assays, as VCP and UBXD1 are involved in general endolysosomal sorting and trafficking (Ritz et al., 2011, Haines et al., 2012, Papadopoulos et al., 2017, Schoenherr et al., 2017), virus uptake and infectivity (Panda et al., 2013, Watanabe et al., 2014, Bhowmick et al., 2017, Hu et al., 2017, Zhang et al., 2019), and regulation of other antiviral proteins (Wang et al., 2013, Hao et al., 2015). Knockdown of VCP or UBXD1 is therefore expected to affect multiple targets and pathways besides IFITM3. In addition, the increased IFITM3 level upon VCP or UBXD1 depletion (Figure 6) may further confound interpretation of virus infection data. Nevertheless, our studies imply a possible functional relevance of VCP with IFITM3 antiviral activity.
To summarize, the photo-crosslinking proteomics study described herein has revealed interaction of IFITM3 with VCP and uncovered key functional roles of the interaction on modulating IFITM3 trafficking, homeostasis, and engagement with incoming virus particles, which greatly advances our understanding on the regulatory mechanisms of this IFN effector. Similar photo-crosslinking proteomic analyses on other IFITM3-TAG mutants in future should enable profiling of the complete IFITM3 interactome. In addition, the site-specific protein photo-crosslinking approach described here should broadly facilitate functional analysis of other host effectors involved in immune response.
Significance
IFITM3 is a crucial host restriction factor for limiting the infection of multiple viruses in humans and mice. Characterization of the antiviral properties and regulatory mechanisms of IFITM3 is critical for advancing our understanding of innate immune response as well as host-pathogen interactions. Facing this biological question, we apply a chemical biology approach that integrates site-specific protein modification via amber suppression with photo-crosslinking and SILAC quantitative proteomics to profile the interacting proteins of IFITM3 in live cells. Retrieved as the top hit from the proteomic analysis, the VCP/p97 AAA-ATPase is validated to interact with IFITM3 and functionally characterized to be a regulator for IFITM3 lysosomal distribution, turnover, and trafficking to incoming virus particles. Our study, therefore, not only demonstrates the utility of this integrative site-specific photo-crosslinking proteomics strategy to profile protein interactions in live cells, but also provides important insights into the regulatory mechanisms of IFITM3 as a critical host immune factor.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-IFITM3 antibody | Proteintech | Cat#11714-1-AP; RRID: AB_2295684 |
| Rabbit polyclonal anti-UBXD1 antibody | Proteintech | Cat#14706-1-AP; RRID: AB_2210488 |
| Rabbit polyclonal anti-p47 antibody | Proteintech | Cat#15620-1-AP |
| Rabbit polyclonal anti-GAPDH antibody HRP-conjugated | Proteintech | Cat#HRP-60004; RRID: AB_2737588 |
| Rabbit polyclonal anti-Tubulin antibody HRP-conjugated | Proteintech | Cat#HRP-66031 |
| VCP monoclonal antibody (5) | ThermoFisher Scientific | Cat#MA3-004; RRID: AB_2214638 |
| Mono- and polyubiquitinylated conjugates, mAb (FK2H) (HRP conjugate) antibody | Enzo Life Sciences | Cat#BML-PW0150; RRID: AB_10554460 |
| Anti-HA-Peroxidase; Rat monoclonal antibody (clone 3F10) conjugated with peroxidase | Roche | Cat#12013819001; RRID: AB_390917 |
| Anti-c-myc-Peroxidase; Mouse monoclonal antibody (clone 9E10) conjugated to peroxidase | Roche | Cat#11814150001; RRID: AB_390910 |
| Monoclonal ANTI-FLAG® M2 antibody | Sigma-Aldrich | Cat#F1804; RRID: AB_262044 |
| Mouse anti-Influenza A virus nucleoprotein monoclonal antibody (clone AA5H) | Abcam | Cat#ab20343; RRID: AB_445525 |
| Peroxidase-AffiniPure goat anti-mouse IgG (H + L) antibody | Jackson ImmunoResearch | Cat#115-035-003; RRID: AB_10015289 |
| Peroxidase-AffiniPure goat anti-rabbit IgG (H+L) antibody | Jackson ImmunoResearch | Cat#111-035-003; RRID:AB_2313567 |
| Peroxidase-AffiniPure goat anti-mouse IgG, light chain specific antibody | Jackson ImmunoResearch | Cat#115-035-174; RRID: AB_2338512 |
| Peroxidase IgG fraction monoclonal mouse anti-rabbit IgG, light chain specific | Jackson ImmunoResearch | Cat#211-032-171 |
| HA-Tag (6E2) mouse mAb (Alexa Fluor 488 conjugate) | Cell Signaling Technology | Cat#2350 |
| Bacterial and Virus Strains | ||
| Influenza A/PR/8/34 (H1N1) | Charles River Laboratories | Cat#10100374 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DiZPK | (Zhang et al., 2011) | N/A |
| AlPK | (Li et al., 2013a) | N/A |
| 2’-aTCOK | (Peng and Hang, 2016) | N/A |
| BODIPY-Tz | (Peng and Hang, 2016) | N/A |
| Bafilomycin A1 | Abcam | Cat#ab120497 |
| Cycloheximide (CHX) | Cell Signaling Technology | Cat#2112 |
| MG-132 | Selleck | Cat#S2619 |
| Chloroquine (CQ) | Selleck | Cat#S4157 |
| ML240 | Sigma-Aldrich | Cat#SML1071 |
| Benzonase | Sigma-Aldrich | Cat#E1014 |
| Protease Inhibitor Cocktail | Roche | Cat#11873580001 |
| DiD | ThermoFisher Scientific | Cat#V22887 |
| 12C6-L-lysine | Sigma-Aldrich | Cat#L8662 |
| 12C6-L-arginine | Sigma-Aldrich | Cat#A6969 |
| 13C6,15N2-L-lysine | Cambridge Isotope | Cat#CNLM-291-H-PK |
| 13C6,15N4-L-arginine | Cambridge Isotope | Cat#CNLM-539-H-PK |
| DMEM | ThermoFisher Scientific | Cat#11995065 |
| DMEM for SILAC | ThermoFisher Scientific | Cat#88364 |
| Fetal Bovine Serum (FBS) | ThermoFisher Scientific | Cat#26140079 |
| Fetal Bovine Serum (FBS), dialyzed | ThermoFisher Scientific | Cat#26400044 |
| Opti-MEM | ThermoFisher Scientific | Cat#31985070 |
| FluoroBrite DMEM | ThermoFisher Scientific | Cat#A1896701 |
| ProLong Gold Antifade Mountant with DAPI | ThermoFisher Scientific | Cat#P36935 |
| ViaFect Transfection Reagent | Promega | Cat#E4983 |
| Lipofectamine 3000 | ThermoFisher Scientific | Cat#L3000015 |
| Xtremegene 9 | Roche | Cat#6365809001 |
| DharmaFECT Transfection Reagent | Dharmacon | Cat#T-2001-03 |
| Clarity Western ECL Substrate | Bio-Rad | Cat#1705060 |
| Amersham ECL Select Reagent | GE Healthcare | Cat#RPN2235 |
| Critical Commercial Assays | ||
| AlexaFluor-647 Antibody Labeling Kit | ThermoFisher Scientific | Cat#A20186 |
| BCA assay | ThermoFisher Scientific | Cat#23228 |
| QuikChange II Site-Directed Mutagenesis Kit | Agilent Technologies | Cat#200523 |
| Deposited Data | ||
| Proteomics database search results | This study; Mendeley Data | https://doi.org/10.17632/tds8by96gc.1 |
| Experimental Models: Cell Lines | ||
| HEK293T | ATCC | Cat#CRL-11268 |
| HeLa | ATCC | Cat#CCL-2 |
| IFITM2/3 KO Hela cells | (Spence et al., 2019) | N/A |
| Oligonucleotides | ||
| Control siRNA (CGTACGCGGAATACTTCGA) | (Kirchner et al., 2013) | N/A |
| VCP siRNA (AACAGCCAUUCUCAAACAGAATT) | (Kirchner et al., 2013) | N/A |
| UBXD1 siRNA (CCAGGUGAGAAAGGAACUUTT) | (Kirchner et al., 2013) | N/A |
| Primers for cloning and mutagenesis; See Table S2 | This study | N/A |
| Recombinant DNA | ||
| pCMV-DiZPK-PylRS | (Zhang et al., 2011) | Addgene Plasmid #91706 |
| LAMP1-mCherry | (Van Engelenburg and Palmer, 2010) | Addgene Plasmid #45147 |
| Human VCP cDNA | Open Biosystems | Clone #7415 |
| Human IFITM3 cDNA | Open Biosystems | Clone #10410 |
| pCMV-HA vector | TaKaRa | Cat#635690 |
| pCMV-myc vector | TaKaRa | Cat#635689 |
| pCMV-Tag2B vector | Agilent Technologies | Cat#211172 |
| pCMV-HA-IFITM3; See Table S2 | This study | N/A |
| pCMV-HA-IFITM3-TAG mutants; See Table S2 | This study | N/A |
| pCMV-myc-VCP; See Table S2 | This study | N/A |
| pCMV-FLAG-VCP; See Table S2 | This study | N/A |
| Software and Algorithms | ||
| ImageLab | Bio-Rad | http://www.bio-rad.com/en-us/product/image-lab-software?ID=KRE6P5E8Z |
| Graphpad Prism 8 | Graphpad | https://www.graphpad.com/ |
| ZEN blue 2012 | Zeiss | https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html |
| Volocity | PerkinElmer | https://www.perkinelmer.com/category/image-analysis-software |
| FlowJo | FlowJo, LLC | https://www.flowjo.com/ |
| MaxQuant v1.6.3.4 | Max Planck Institute of Biochemistry | https://www.maxquant.org/maxquant/ |
| Andromeda | Max Planck Institute of Biochemistry | http://coxdocs.org/doku.php?id=maxquant:andromeda:start |
| Perseus | Max Planck Institute of Biochemistry | https://www.maxquant.org/perseus/ |
| Other | ||
| EZview™ Red anti-HA affinity gels | Sigma-Aldrich | Cat#E6779 |
| EZview™ Red anti-Flag affinity gels | Sigma-Aldrich | Cat#F2426 |
| Pierce™ anti-HA magnetic beads | ThermoFisher Scientific | Cat#88836 |
Lead Contact and Materials Availability
Further information and requests for reagents should be directed to and will be fulfilled by the Lead Contact, Tao Peng (tpeng@pku.edu.cn). All unique/stable reagents generated in this study are available from the Lead Contact.
Experimental Model and Subject Details
HEK293T (Sex: Female) and HeLa (Sex: Female) cells used in this study were purchased from ATCC. IFITM2/3 KO Hela cells were previously generated in the laboratory (Spence et al., 2019). HEK293T, HeLa, and IFITM2/3 KO Hela cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; ThermoFisher Scientific) supplemented with 10% fetal bovine serum (FBS; ThermoFisher Scientific). All cells were grown in a humidified, 5% CO2 incubator at 37°C.
For SILAC-based proteomics studies, HEK293T cells were cultured in arginine and lysine-deficient DMEM (DMEM for SILAC; ThermoFisher Scientific) supplemented with 10% dialyzed FBS (ThermoFisher Scientific). For culture of light labeled cells, SILAC medium containing 12C6-L-lysine (Lys0; 0.80 mM; Sigma-Aldrich) and 12C6-L-arginine (Arg0; 0.40 mM; Sigma-Aldrich) was used. For culture of heavy labeled cells, SILAC medium containing 13C6,15N2-L-lysine (Lys8; 0.80 mM; Cambridge Isotope) and 13C6,15N4-L-arginine (Arg10; 0.80 mM; Cambridge Isotope) was used (Ong and Mann, 2006). After seven cell doublings, the incorporation of the heavy isotopes was estimated to be >98% as determined by LC-MS/MS analysis.
Influenza virus A/PR/8/34 (H1N1) was from Charles River Laboratories (10100374) and stored at −80°C until use.
Method Details
General Reagents
Anti-IFITM3 (11714-1-AP), UBXD1 (14706-1-AP), p47 (15620-1-AP), GADPH (HRP-60004), and Tubulin (HRP-66031) antibodies were ordered from Proteintech. VCP antibody (MA3-004) was from ThermoFisher Scientific. Ubiquitin antibody (FK2; BML-PW0150) was purchased from Enzo Life Sciences. Anti-HA (3F10; 12013819001) and anti-Myc (9E10; 11814150001) antibodies conjugated to HRP were purchased from Roche. Anti-FLAG antibody (M2; F1804) was from Sigma-Aldrich. The secondary antibodies HRP-conjugated goat anti-mouse IgG (115-035-003) and HRP-conjugated goat anti-rabbit IgG (111-035-003) were purchased from Jackson ImmunoResearch. Secondary antibodies goat anti-mouse IgG (Jackson ImmunoResearch, 115-035-174) and mouse anti-rabbit IgG (Jackson ImmunoResearch, 211-032-171) specific to light chain and conjugated to HRP were used in western blot of co-immunoprecipitates. For immunoprecipitation, anti-HA (E6779) and anti-Flag (F2426) affinity gels were purchased from Sigma-Aldrich. For immunofluorescence, anti-HA antibody conjugated to Alexafluor-488 (2350) was purchased from Cell Signaling Technology. Influenza A virus nucleoprotein antibody (AA5H; ab20343) was from Abcam. AlexaFluor-647 Antibody Labeling Kit (A20186) was ordered from ThermoFisher Scientific.
For pharmacological treatments, Bafilomycin A1 (ab120497) was from Abcam. ML240 (SML1071) was from Sigma-Aldrich. Cycloheximide (CHX; 2112) was purchased from Cell Signaling Technology. MG-132 (S2619) and chloroquine (S4157) were from Selleck.
For UAA incorporation, DiZPK (Zhang et al., 2011) and AlkPK (Li et al., 2013a) were synthesized in the laboratory according to published procedures. 2’-aTCOK was previously synthesized in the laboratory (Peng and Hang, 2016).
For live cell imaging, DiD was from ThermoFisher Scientific (V22887). BODIPY-Tz was previously synthesized in the laboratory (Peng and Hang, 2016, Spence et al., 2019).
Plasmid, Transfection, and siRNA Knockdown
pCMV-DiZPK-PylRS plasmid encoding a DiZPK-aminoacyl-tRNA synthetase/tRNA pair was kindly provided by Professor Peng R. Chen at Peking University (Zhang et al., 2011) and is available on Addgene (plasmid #91706). Human IFITM3 cDNA was purchased from Open Biosystems and PCR cloned into pCMV-HA vector (TaKaRa) using EcoRI and SalI as the restriction sites. All mutants of IFITM3 were generated by QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies, 200523) using primers listed in Table S2. VCP cDNA was purchased from Open Biosystems and PCR cloned into pCMV-Tag2B vector (Agilent Technologies) using BamHI and HindIII as the restriction sites and pCMV-myc vector (TaKaRa) using SalI and KpnI as the restriction sites. LAMP1-mCherry plasmid (Van Engelenburg and Palmer, 2010) was obtained from Addgene (plasmid #45147).
ViaFect transfection reagent purchased from Promega or Lipofectamine 3000 from ThermoFisher Scientific was used for transfection of Hela cells, and Xtremegene 9 purchased from Roche was used for transfection of HEK293T cells. For RNA interference, siRNAs were synthesized by Genepharma and transfected into cells using DharmaFECT transfection reagent (Dharmacon).
siRNA sequences are as follows:
Control: CGTACGCGGAATACTTCGA;
VCP: AACAGCCAUUCUCAAACAGAATT (Kirchner et al., 2013);
UBXD1: CCAGGUGAGAAAGGAACUUTT (Kirchner et al., 2013).
Expression of DiZPK-Modified IFITM3 and Photo-Crosslinking
HEK293T cells were grown in normal growth media in 6-well plates. Before transfection, cell media were exchanged to fresh complete growth media containing 1 mM DiZPK (Zhang et al., 2011). Cells were then transfected with pCMV-DiZPK-PylRS and IFITM3-TAG mutant plasmids using Xtremegene 9 (Roche; 6 μL Xtremegene 9 + 1 μg of pCMV-DiZPK-PylRS + 1 μg of IFITM3-TAG mutant per well in a 6-well plate). Cells were grown for additional 16–24 h before collected and lysed with 4% SDS lysis buffer (4% SDS, 150 mM NaCl, 50 mM triethanolamine, pH 7.4) for western blot to assess the expression of IFITM3. For in-cell photocrosslinking, cells expressing DiZPK-modified protein were covered with PBS (1 mL) and subjected to UV irradiation at 365 nm on ice for 10 min using a Spectrolinker XL-1000 UV crosslinker (Spectronics) at a distance of 3 cm. Cells were then collected and lysed for western blot.
Western Blot Analysis
Cells were generally lysed with 4% SDS lysis buffer containing Roche protease inhibitor cocktail and benzonase. Cell lysates were clarified by centrifugation at 12,000× g for 20 min and quantified by the BCA assay (ThermoFisher Scientific). Equal amounts of cell lysates were diluted with 4X reducing SDS-loading buffer and β-mercaptoethanol at 1 mg/mL concentration. The samples were heated for 10 min at 95°C and loaded onto 4−20% Tris-HCl gels (Bio-Rad) for SDS-PAGE separation. Proteins on the gel were transferred to nitrocellulose membrane using Trans-Blot Turbo Transfer System (Bio-Rad). The membranes were incubated with indicated antibody in PBST (0.1% Tween-20 in PBS) containing 5% nonfat milk at 4°C overnight and washed with PBST. Blots were then developed using Clarity Western ECL substrate (Bio-Rad) or Amersham ECL Select (GE Healthcare) and imaged with a Bio-Rad ChemiDoc MP Imager. Western blots were analyzed by calculating the densitometry of protein bands on ImageLab software (Bio-Rad). The densitometry of protein of interest band relative to that of corresponding loading control was quantified.
Co-immunoprecipitation
Cells were harvested after 20 h transfection and washed with PBS twice. The cell pellets were then lysed on ice with Triton X-100 lysis buffer (1% Triton X-100, 150 mM NaCl, 50 mM Tris, pH 7.4) supplemented with Roche protease inhibitor cocktail. After 30 min, the cell lysates were centrifuged at 12,000× g for 20 min at 4°C. The cleared cell lysates were quantified using the BCA assay (ThermoFisher Scientific). Equal amounts of cell lysates were incubated with EZview Red anti-HA or anti-FLAG affinity gel (Sigma-Aldrich) at 4°C for 3 h with end-over-end rotation. The beads were washed five times with Triton X-100 lysis buffer and eluted with 1% SDS buffer for western blot.
Analysis of IFITM3 Turnover
HEK293T cells were cultured to 80% confluency and transfected with HA-IFITM3 for 20 h. Cells were then treated with CHX and ML240 for indicated time and lysed with 4% SDS for western blot analysis. Alternatively, Hela cells were transfected with VCP or UBXD1 siRNA for 72 h and treated with CHX for indicated time before analysis with western blot.
Immunofluorescence
Hela cells grown on glass coverslips in 6-well plates were co-transfected with HA-IFITM3 constructs and LAMP1-mCherry for 16 h, and treated with Bafilomycin A1 or ML240 for 12 h. Cells were then fixed with 3.7% (w/v) paraformaldehyde, permeabilized with 0.1% (w/v) Triton X-100, and blocked with 5% (w/v) BSA in PBS. Cells were incubated with anti-HA antibody conjugated to AlexaFluor-488 (1:800) in 0.05% Triton X-100 in PBS at 4°C overnight and washed with PBS three times. Finally, the coverslips were mounted onto slides using ProLong Gold Antifade Mountant with DAPI (ThermoFisher Scientific) and examined by an inverted LSM 780 laser scanning confocal microscope (Zeiss) with a Zeiss Plan-Apochromatic 63×/1.4 N.A. oil immersion objective. Images were acquired and analyzed with the ZEN blue 2012 software (Zeiss). Pearson's correlation coefficients were calculated in ZEN blue 2012 software (Zeiss).
Live Cell Imaging of IFITM3 and Virus Particle Trafficking
For live cell imaging studies, IFITM 2/3 KO HeLa cells were seeded onto fibronectin-coated 35 mm glass coverslip dishes (MatTek) 24 h prior to experiments. On the next day, cells were transfected with HA-IFITM3-F8TAG constructs (1 μg per dish) and pCMV-DiZPK-PylRS plasmid (1 μg per dish) using 6 μL Viafect (Promega) in complete cell growth media containing 2′-aTCOK. After overnight incubation, cell media were changed into fresh cell growth media without UAA. After another 2 h culture at 37°C/5% CO2, cells were labeled with BODIPY-Tz (250 nM) in FluoroBrite DMEM (ThermoFisher Scientific) for 0.5 h at 37°C and washed with complete cell growth media 3 times over 1 h (Peng and Hang, 2016). Cells were then chilled on ice before spinoculation of DiD-labeled virus onto monolayers at 1,500× g and 6°C for 20 min. Purified influenza virus A/PR/8/34 (H1N1) particles were labeled with self-quenching concentrations of the lipophilic dye DiD (ThermoFisher Scientific) as previously reported (Spence et al., 2019). Unbound particles were removed by five washes with cold PBS, and 500 μL cold imaging buffer (140 mM NaCl, 2.5 mM KCl, 1.8 mM MgCl2, 20 mM HEPES, 5 mM sucrose, 2 μM Hoechst 33342, and 2% FBS) was added to cover the cells. The dish was immediately mounted on the microscope objective and focused. The coverslip dish was then flooded with 1.5 mL warm imaging buffer to mark the start of experiments (t = 0). Images were acquired every 10 s over the duration of the experiments using a single Z-section. Live-cell microscopy was performed with an AxioObserver.Z1 wide-field epifluorescence microscope (Zeiss) equipped with a 40×/1.3N.A. objective, DAPI/GFP/Texas Red/Cy5 filter set, and heated environmental enclosure maintained at 37°C, as previously described (Spence et al., 2019). Image analysis and single-particle tracking were performed using Volocity software (PerkinElmer). Mean measurements (± standard deviations) were derived from three separate experiments, unless otherwise indicated.
Influenza Virus Infection Assay
For the infection assay, IFITM 2/3 KO HeLa cells were seeded in 12 well plates and cultured overnight. Cells were transfected with HA-IFITM3 constructs (1 μg/well) using 3 μL Lipofectamine 3000 in 1 mL of complete cell growth media. The cells were infected with influenza virus A/PR/8/34 virus (H1N1) with MOI of 2.5. After 2 h of infection, DMSO or ML240 was added. After 4 h, cells were washed with PBS, trypsinized, and collected in cluster tubes. Cells were washed again with PBS and then fixed with 240 μL of 4% PFA in PBS for 5 min. The fixed cells were permeabilized with 200 μL of 0.2% saponin in PBS for 10 min and then blocked with 200 μL of 0.2% BSA and 0.2% saponin in PBS for 10 min. Cells were treated with anti-influenza NP antibody conjugated to AlexaFluor-647 (1:250) and anti-HA antibody conjugated to AlexaFluor-488 (1:300) in 0.02% saponin in PBS. After three washes with 0.02% saponin in PBS, cells were resuspended in 100 μL PBS containing 0.2% BSA and 0.02% saponin. The samples were analyzed by flow cytometry (BD LSRII). Data analysis was performed using FlowJo software. All samples were first gated by IFITM3-positive staining, indicating successful transfection, and then NP-positive percentage of NP-positive staining, indicated successful infection.
Isolation of Photo-Crosslinked IFITM3 Complexes and Proteomic Analysis
HEK293T cells were cultured in 150 mm dishes to ~80% confluency, and transfected with HA-IFITM3-F8TAG and pCMV-DiZPK-PylRS using Xtremegene 9 (Roche) in the presence of 1 mM DiZPK for 20 h. Cells were then irradiated with UV at 365 nm for 10 min on ice before harvested and washed with PBS twice. In “forward” SILAC experiment, heavy-labeled cells were irradiated with UV while light-labeled cells were not. Alternatively, in “reverse” SILAC experiment, light-labeled cells were irradiated with UV while heavy-labeled cells were not. The cell pellets were lysed with 4% SDS lysis buffer supplemented with Roche protease inhibitor cocktail and benzonase. Cell lysates were centrifuged at 12,000× g for 20 min to remove cellular debris. Protein concentrations were determined by the BCA assay (ThermoFisher Scientific). Heavy-labeled cell lysates were mixed with light-labeled cell lysates in 1:1 ratio according to protein concentrations. The mixed cell lysates were then diluted with Triton X-100 buffer (1% Triton X-100, 150 mM NaCl, 50 mM Tris, pH 7.4) until the SDS concentration is less than 0.1%. The diluted cell lysates were incubated with anti-HA magnetic beads (ThermoFisher Scientific) at 4°C for 1.5 h with end-over-end rotation. The beads were then washed five times with chilled RIPA buffer (1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 50 mM triethanolamine pH 7.4) and eluted with 0.5 M NH4OH twice. The pooled eluates were dried, resuspended in 1X SDS-PAGE loading buffer, reduced with 10 mM DTT for 30 min, and then alkylated with 50 mM iodoacetamide in the dark for another 30 min. The resulting samples were separated by SDS-PAGE and subjected to in-gel digestion as previously reported for LC-MS/MS analysis (Shevchenko et al., 2006).
All LC-MS/MS analysis was carried out at the Proteomics Resource Center at The Rockefeller University, New York, NY, USA. LC-MS analysis was performed with a Dionex 3000 nano-HPLC coupled to an Orbitrap XL mass spectrometer (ThermoFisher Scientific). A C18 reverse-phase column (75 μm diameter, 15 cm length) was used for sample loading with a 180-minute gradient increasing from 95% buffer A (water with 0.1% formic acid) and 5% buffer B (acetonitrile with 0.1% formic acid) to 75% buffer B in 133 minutes at 0.2 μL/min. The mass spectrometer was operated in top-8-CID-mode with a resolution of 60,000@m/z 400. One full MS scan (300–2000 MW) was followed by three data-dependent scans of the nth most intense ions with dynamic exclusion enabled. Peptides fulfilling a Percolator calculated 1% false discovery rate (FDR) threshold were reported. Acquired tandem MS spectra were extracted and quantified using MaxQuant software (Cox and Mann, 2008). Trypsin was set as the digestion enzyme with two missed cleavages allowed. Carbamidomethylation of cysteine was set as a fixed modification, while methionine oxidation and N-terminal acetylation were set as variable modifications. Mass deviation for MS/MS peaks was set at a maximum of 0.5 m/z and maximum false discovery rates (FDR) were set to 0.01 both at the peptide and at the protein levels. Mass spectra were searched with Andromeda against Uniprot complete human database concatenated with common known contaminants. Only unique and razor peptides were used for quantification with minimal two ratio counts. “Re-quantify” feature of MaxQuant was used to correct quantification of proteins. Known contaminants and reverse hits were removed before data processing. The search results were analyzed by Perseus (Tyanova et al., 2016).
Quantification and Statistical Analysis
Data were presented as mean ± standard deviation determined from biological replicates. Statistical analysis was performed with GraphPad Prism. The method for determining error bars and significance is indicated in the corresponding figure legends with biological replicate number indicated. For significance, ns = non-significant (p > 0.05), ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Data and Code Availability
The proteomics database search results have been deposited to the Mendeley database (https://doi.org/10.17632/tds8by96gc.1).
Acknowledgments
We thank Prof. Peng R. Chen at Peking University for the pCMV-DiZPK-PylRS plasmid. We also thank the Proteomics Resource Center at The Rockefeller University for assistance in proteomic analysis. T.D. and X.Y. acknowledge the Tri-Institutional Program in Chemical Biology at The Rockefeller University. K.C. acknowledges support from NIH-NIAID R01AI134824. H.C.H. acknowledges support from NIH-NIGMS R01M087544. T.P. acknowledges support from National Natural Science Foundation of China (21778010), Shenzhen Science and Technology Innovation Committee (JCYJ20170412150832022), and Shenzhen Peacock Plan (KQTD2015032709315529).
Author Contributions
H.C.H. and T.P. conceived and supervised the study. T.P., X.W., J.S.S., and T.D. performed the experiments. X.Y., C.C., Y.Z., Y.L., and Y.S. generated reagents. X.W., J.S.S., T.D., K.C., H.C.H., and T.P. interpreted the data. H.C.H. and T.P. wrote the manuscript with input from all authors.
Declaration of Interests
The authors declare no competing interests.
Published: April 2, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.chembiol.2020.03.004.
Contributor Information
Howard C. Hang, Email: hhang@mail.rockefeller.edu.
Tao Peng, Email: tpeng@pku.edu.cn.
Supplemental Information
References
- Ai H.-W., Shen W., Sagi A., Chen P.R., Schultz P.G. Probing protein–protein interactions with a genetically encoded photo-crosslinking amino acid. ChemBioChem. 2011;12:1854–1857. doi: 10.1002/cbic.201100194. [DOI] [PubMed] [Google Scholar]
- Amini-Bavil-Olyaee S., Choi Y.J., Lee J.H., Shi M., Huang I.C., Farzan M., Jung J.U. The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe. 2013;13:452–464. doi: 10.1016/j.chom.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey C.C., Huang I.C., Kam C., Farzan M. Ifitm3 limits the severity of acute influenza in mice. PLoS Pathog. 2012;8:e1002909. doi: 10.1371/journal.ppat.1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey C.C., Zhong G., Huang I.-C., Farzan M. IFITM-family proteins: the cell's first line of antiviral defense. Annu. Rev. Virol. 2014;1:261–283. doi: 10.1146/annurev-virology-031413-085537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastola P., Chien J. Co-selected mutations in VCP: a novel mechanism of resistance to VCP inhibitors. Cell Death Dis. 2018;9:35. doi: 10.1038/s41419-017-0049-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick S., Chakravarty C., Sellathamby S., Lal S.K. The influenza A virus matrix protein 2 undergoes retrograde transport from the endoplasmic reticulum into the cytoplasm and bypasses cytoplasmic proteasomal degradation. Arch. Virol. 2017;162:919–929. doi: 10.1007/s00705-016-3153-8. [DOI] [PubMed] [Google Scholar]
- Brass A.L., Huang I.C., Benita Y., John S.P., Krishnan M.N., Feeley E.M., Ryan B.J., Weyer J.L., van der Weyden L., Fikrig E. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchrieser J., Degrelle S.A., Couderc T., Nevers Q., Disson O., Manet C., Donahue D.A., Porrot F., Hillion K.-H., Perthame E. IFITM proteins inhibit placental syncytiotrophoblast formation and promote fetal demise. Science. 2019;365:176–180. doi: 10.1126/science.aaw7733. [DOI] [PubMed] [Google Scholar]
- Chesarino N.M., Compton A.A., McMichael T.M., Kenney A.D., Zhang L., Soewarna V., Davis M., Schwartz O., Yount J.S. IFITM3 requires an amphipathic helix for antiviral activity. EMBO Rep. 2017;18:1740–1751. doi: 10.15252/embr.201744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarino N.M., McMichael T.M., Hach J.C., Yount J.S. Phosphorylation of the antiviral protein IFITM3 dually regulates its endocytosis and ubiquitination. J. Biol. Chem. 2014;289:11986–11992. doi: 10.1074/jbc.M114.557694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarino N.M., McMichael T.M., Yount J.S. Regulation of the trafficking and antiviral activity of IFITM3 by post-translational modifications. Future Microbiol. 2014;9:1151–1163. doi: 10.2217/fmb.14.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarino N.M., McMichael T.M., Yount J.S. E3 ubiquitin ligase NEDD4 promotes influenza virus infection by decreasing levels of the antiviral protein IFITM3. PLoS Pathog. 2015;11:e1005095. doi: 10.1371/journal.ppat.1005095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin J.W. Expanding and reprogramming the genetic code. Nature. 2017;550:53–60. doi: 10.1038/nature24031. [DOI] [PubMed] [Google Scholar]
- Chin J.W., Martin A.B., King D.S., Wang L., Schultz P.G. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C., Uprety R., Davis L., Chin J.W., Deiters A. Genetically encoding an aliphatic diazirine for protein photocrosslinking. Chem. Sci. 2011;2:480–483. [Google Scholar]
- Chou T.-F., Li K., Frankowski K.J., Schoenen F.J., Deshaies R.J. Structure–activity relationship study reveals ML240 and ML241 as potent and selective inhibitors of p97 ATPase. ChemMedChem. 2013;8:297–312. doi: 10.1002/cmdc.201200520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton A.A., Bruel T., Porrot F., Mallet A., Sachse M., Euvrard M., Liang C., Casartelli N., Schwartz O. IFITM proteins incorporated into HIV-1 virions impair viral fusion and spread. Cell Host Microbe. 2014;16:736–747. doi: 10.1016/j.chom.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney I., Han H., Stewart M.G., Carson R.H., Hansen D.T., Iwasa J.H., Price J.C., Hill C.P., Shen P.S. Structure of the Cdc48 segregase in the act of unfolding an authentic substrate. Science. 2019;365:502–505. doi: 10.1126/science.aax0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Daley D.O. The assembly of membrane proteins into complexes. Curr. Opin. Struct. Biol. 2008;18:420–424. doi: 10.1016/j.sbi.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Desai T.M., Marin M., Chin C.R., Savidis G., Brass A.L., Melikyan G.B. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog. 2014;10:e1004048. doi: 10.1371/journal.ppat.1004048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond M.S., Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 2012;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt A.R., Clare S., Pertel T., John S.P., Wash R.S., Smith S.E., Chin C.R., Feeley E.M., Sims J.S., Adams D.J. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeley E.M., Sims J.S., John S.P., Chin C.R., Pertel T., Chen L.-M., Gaiha G.D., Ryan B.J., Donis R.O., Elledge S.J. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog. 2011;7:e1002337. doi: 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu B., Wang L., Li S., Dorf M.E. ZMPSTE24 defends against influenza and other pathogenic viruses. J. Exp. Med. 2017;214:919–929. doi: 10.1084/jem.20161270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman M.J., Poddar S., Farzan M., Diamond M.S. The interferon-stimulated gene Ifitm3zrestricts West Nile virus infection and pathogenesis. J. Virol. 2016;90:8212–8225. doi: 10.1128/JVI.00581-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines D.S., Lee J.E., Beauparlant S.L., Kyle D.B., den Besten W., Sweredoski M.J., Graham R.L.J., Hess S., Deshaies R.J. Protein interaction profiling of the p97 adaptor UBXD1 points to a role for the complex in modulating ERGIC-53 trafficking. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.016444. M111.016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Q., Jiao S., Shi Z., Li C., Meng X., Zhang Z., Wang Y., Song X., Wang W., Zhang R. A non-canonical role of the p97 complex in RIG-I antiviral signaling. EMBO J. 2015;34:2903–2920. doi: 10.15252/embj.201591888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D., Xie X., Yang F., Zhang H., Su H., Ge Y., Song H., Chen P.R. Quantitative and comparative profiling of protease substrates through a genetically encoded multifunctional photocrosslinker. Angew. Chem. Int. Ed. 2017;56:14521–14525. doi: 10.1002/anie.201708151. [DOI] [PubMed] [Google Scholar]
- Hino N., Okazaki Y., Kobayashi T., Hayashi A., Sakamoto K., Yokoyama S. Protein photo-cross-linking in mammalian cells by site-specific incorporation of a photoreactive amino acid. Nat. Methods. 2005;2:201–206. doi: 10.1038/nmeth739. [DOI] [PubMed] [Google Scholar]
- Hu Y., O’Boyle K., Auer J., Raju S., You F., Wang P., Fikrig E., Sutton R.E. Multiple UBXN family members inhibit retrovirus and lentivirus production and canonical NFκΒ signaling by stabilizing IκBα. PLoS Pathog. 2017;13:e1006187. doi: 10.1371/journal.ppat.1006187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang I.C., Bailey C.C., Weyer J.L., Radoshitzky S.R., Becker M.M., Chiang J.J., Brass A.L., Ahmed A.A., Chi X., Dong L. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7:e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubel P., Urban C., Bergant V., Schneider W.M., Knauer B., Stukalov A., Scaturro P., Mann A., Brunotte L., Hoffmann H.H. A protein-interaction network of interferon-stimulated genes extends the innate immune system landscape. Nat. Immunol. 2019;20:493–502. doi: 10.1038/s41590-019-0323-3. [DOI] [PubMed] [Google Scholar]
- Jia R., Pan Q., Ding S., Rong L., Liu S.-L., Geng Y., Qiao W., Liang C. The N-terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization. J. Virol. 2012;86:13697–13707. doi: 10.1128/JVI.01828-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia R., Xu F., Qian J., Yao Y., Miao C., Zheng Y.-M., Liu S.-L., Guo F., Geng Y., Qiao W. Identification of an endocytic signal essential for the antiviral action of IFITM3. Cell. Microbiol. 2014;16:1080–1093. doi: 10.1111/cmi.12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L.-Q., Xia T., Hu Y.-H., Sun M.-S., Yan S., Lei C.-Q., Shu H.-B., Guo J.-H., Liu Y. IFITM3 inhibits virus-triggered induction of type I interferon by mediating autophagosome-dependent degradation of IRF3. Cell. Mol. Immunol. 2018;15:858–867. doi: 10.1038/cmi.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S.P., Chin C.R., Perreira J.M., Feeley E.M., Aker A.M., Savidis G., Smith S.E., Elia A.E.H., Everitt A.R., Vora M. The CD225 domain of IFITM3 is required for both IFITM protein association and inhibition of influenza A virus and dengue virus replication. J. Virol. 2013;87:7837–7852. doi: 10.1128/JVI.00481-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner P., Bug M., Meyer H. Ubiquitination of the N-terminal region of caveolin-1 regulates endosomal sorting by the VCP/p97 AAA-ATPase. J. Biol. Chem. 2013;288:7363–7372. doi: 10.1074/jbc.M112.429076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner R.E., Hang L.E., Molloy K.R., Chait B.T., Kapoor T.M. A chemical proteomics approach to reveal direct protein-protein interactions in living cells. Cell Chem. Biol. 2018;25:110–120.e3. doi: 10.1016/j.chembiol.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.-Y.J., Fu R.M., Liang C., Sloan R.D. IFITM proteins inhibit HIV-1 protein synthesis. Sci. Rep. 2018;8:14551. doi: 10.1038/s41598-018-32785-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Lin S., Wang J., Jia S., Yang M., Hao Z., Zhang X., Chen P.R. Ligand-free palladium-mediated site-specific protein labeling inside Gram-negative bacterial pathogens. J. Am. Chem. Soc. 2013;135:7330–7338. doi: 10.1021/ja402424j. [DOI] [PubMed] [Google Scholar]
- Li K., Markosyan R.M., Zheng Y.-M., Golfetto O., Bungart B., Li M., Ding S., He Y., Liang C., Lee J.C. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 2013;9:e1003124. doi: 10.1371/journal.ppat.1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Goraya M.U., Yuan X., Zhang B., Chiu S.-H., Chen J.-L. Functional involvement of interferon-inducible transmembrane proteins in antiviral immunity. Front. Microbiol. 2019;10:1097. doi: 10.3389/fmicb.2019.01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T.-Y., Chin C.R., Everitt A.R., Clare S., Perreira J.M., Savidis G., Aker A.M., John S.P., Sarlah D., Carreira E.M. Amphotericin B increases influenza A virus infection by preventing IFITM3-mediated restriction. Cell Rep. 2013;5:895–908. doi: 10.1016/j.celrep.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Chen L., Fan Y., Hong Y., Yang X., Li Y., Lu J., Lv J., Pan X., Qu F. IFITM3 promotes bone metastasis of prostate cancer cells by mediating activation of the TGF-β signaling pathway. Cell Death Dis. 2019;10:517. doi: 10.1038/s41419-019-1750-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Pan Q., Rong L., Liu S.-L., Liang C. The IFITM proteins inhibit HIV-1 infection. J. Virol. 2011;85:2126–2137. doi: 10.1128/JVI.01531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking J.D. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat. Rev. Immunol. 2012;12:367–382. doi: 10.1038/nri3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H., Bug M., Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol. 2012;14:117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- Min J., Feng Q., Liao W., Liang Y., Gong C., Li E., He W., Yuan R., Wu L. IFITM3 promotes hepatocellular carcinoma invasion and metastasis by regulating MMP9 through p38/MAPK signaling. FEBS Open Biol. 2018;8:1299–1311. doi: 10.1002/2211-5463.12479. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Muller U., Steinhoff U., Reis L., Hemmi S., Pavlovic J., Zinkernagel R., Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- Ong S.-E., Blagoev B., Kratchmarova I., Kristensen D.B., Steen H., Pandey A., Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Ong S.-E., Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat. Protoc. 2006;1:2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- Panda D., Rose P.P., Hanna S.L., Gold B., Hopkins K.C., Lyde R.B., Marks M.S., Cherry S. Genome-wide RNAi screen identifies SEC61A and VCP as conserved regulators of sindbis virus entry. Cell Rep. 2013;5:1737–1748. doi: 10.1016/j.celrep.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos C., Kirchner P., Bug M., Grum D., Koerver L., Schulze N., Poehler R., Dressler A., Fengler S., Arhzaouy K. VCP/p97 cooperates with YOD1, UBXD1 and PLAA to drive clearance of ruptured lysosomes by autophagy. EMBO J. 2017;36:135–150. doi: 10.15252/embj.201695148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T., Hang H.C. Bifunctional fatty acid chemical reporter for analyzing S-palmitoylated membrane protein–protein interactions in mammalian cells. J. Am. Chem. Soc. 2015;137:556–559. doi: 10.1021/ja502109n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T., Hang H.C. Site-specific bioorthogonal labeling for fluorescence imaging of intracellular proteins in living cells. J. Am. Chem. Soc. 2016;138:14423–14433. doi: 10.1021/jacs.6b08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percher A., Ramakrishnan S., Thinon E., Yuan X., Yount J.S., Hang H.C. Mass-tag labeling reveals site-specific and endogenous levels of protein S-fatty acylation. Proc. Natl. Acad. Sci. U.S.A. 2016;113:4302–4307. doi: 10.1073/pnas.1602244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreira J.M., Chin C.R., Feeley E.M., Brass A.L. IFITMs restrict the replication of multiple pathogenic viruses. J. Mol. Biol. 2013;425:4937–4955. doi: 10.1016/j.jmb.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham N.D., Parker R.B., Kohler J.J. Photocrosslinking approaches to interactome mapping. Curr. Opin. Chem. Biol. 2013;17:90–101. doi: 10.1016/j.cbpa.2012.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puschnik A.S., Majzoub K., Ooi Y.S., Carette J.E. A CRISPR toolbox to study virus–host interactions. Nat. Rev. Microbiol. 2017;15:351–364. doi: 10.1038/nrmicro.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz D., Vuk M., Kirchner P., Bug M., Schütz S., Hayer A., Bremer S., Lusk C., Baloh R.H., Lee H. Endolysosomal sorting of ubiquitylated caveolin-1 is regulated by VCP and UBXD1 and impaired by VCP disease mutations. Nat. Cell Biol. 2011;13:1116–1123. doi: 10.1038/ncb2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenherr C., Byron A., Sandilands E., Paliashvili K., Baillie G.S., Garcia-Munoz A., Valacca C., Cecconi F., Serrels B., Frame M.C. Ambra1 spatially regulates Src activity and Src/FAK-mediated cancer cell invasion via trafficking networks. eLife. 2017;6:e23172. doi: 10.7554/eLife.23172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W., Rice C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P., Rice C.M. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P.S., Wojcechowskyj J.A., Eckhardt M., Krogan N.J. Comparative mapping of host–pathogen protein–protein interactions. Curr. Opin. Microbiol. 2015;27:62–68. doi: 10.1016/j.mib.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havli J., Olsen J.V., Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- Shi G., Ozog S., Torbett B.E., Compton A.A. mTOR inhibitors lower an intrinsic barrier to virus infection mediated by IFITM3. Proc. Natl. Acad. Sci. U.S.A. 2018;115:E10069–E10078. doi: 10.1073/pnas.1811892115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J.S., He R., Hoffmann H.-H., Das T., Thinon E., Rice C.M., Peng T., Chandran K., Hang H.C. IFITM3 directly engages and shuttles incoming virus particles to lysosomes. Nat. Chem. Biol. 2019;15:259–268. doi: 10.1038/s41589-018-0213-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stach L., Freemont P.S. The AAA+ ATPase p97, a cellular multitool. Biochem. J. 2017;474:2953–2976. doi: 10.1042/BCJ20160783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suddala K.C., Lee C.C., Meraner P., Marin M., Markosyan R.M., Desai T.M., Cohen F.S., Brass A.L., Melikyan G.B. Interferon-induced transmembrane protein 3 blocks fusion of sensitive but not resistant viruses by partitioning into virus-carrying endosomes. PLoS Pathog. 2019;15:e1007532. doi: 10.1371/journal.ppat.1007532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Bond M.R., Kohler J.J. Photocrosslinkers illuminate interactions in living cells. Mol. Biosyst. 2008;4:473–480. doi: 10.1039/b803218a. [DOI] [PubMed] [Google Scholar]
- Tartour K., Appourchaux R., Gaillard J., Nguyen X.-N., Durand S., Turpin J., Beaumont E., Roch E., Berger G., Mahieux R. IFITM proteins are incorporated onto HIV-1 virion particles and negatively imprint their infectivity. Retrovirology. 2014;11:103. doi: 10.1186/s12977-014-0103-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tippmann E.M., Liu W., Summerer D., Mack A.V., Schultz P.G. A genetically encoded diazirine photocrosslinker in Escherichia coli. ChemBioChem. 2007;8:2210–2214. doi: 10.1002/cbic.200700460. [DOI] [PubMed] [Google Scholar]
- Twomey E.C., Ji Z., Wales T.E., Bodnar N.O., Ficarro S.B., Marto J.A., Engen J.R., Rapoport T.A. Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding. Science. 2019;365:502–505. doi: 10.1126/science.aax1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- Van Engelenburg S.B., Palmer A.E. Imaging type-III secretion reveals dynamics and spatial segregation of Salmonella effectors. Nat. Methods. 2010;7:325–330. doi: 10.1038/nmeth.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Xie J., Schultz P.G. Expanding the genetic code. Annu. Rev. Biophys. Biomol. Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- Wang P., Yang L., Cheng G., Yang G., Xu Z., You F., Sun Q., Lin R., Fikrig E., Sutton R.E. UBXN1 interferes with rig-I-like receptor-mediated antiviral immune response by targeting MAVS. Cell Rep. 2013;3:1057–1070. doi: 10.1016/j.celrep.2013.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zhang A., Wan Y., Liu X., Qiu C., Xi X., Ren Y., Wang J., Dong Y., Bao M. Early hypercytokinemia is associated with interferon-induced transmembrane protein-3 dysfunction and predictive of fatal H7N9 infection. Proc. Natl. Acad. Sci. U.S.A. 2014;111:769–774. doi: 10.1073/pnas.1321748111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Kawakami E., Shoemaker J.E., Lopes T.J.S., Matsuoka Y., Tomita Y., Kozuka-Hata H., Gorai T., Kuwahara T., Takeda E. Influenza virus-host interactome screen as a platform for antiviral drug development. Cell Host Microbe. 2014;16:795–805. doi: 10.1016/j.chom.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidner J.M., Jiang D., Pan X.-B., Chang J., Block T.M., Guo J.-T. Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J. Virol. 2010;84:12646–12657. doi: 10.1128/JVI.01328-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston S., Czieso S., White I.J., Smith S.E., Kellam P., Marsh M. A membrane topology model for human interferon inducible transmembrane protein 1. PLoS One. 2014;9:e104341. doi: 10.1371/journal.pone.0104341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Li X.-M., Qin F., Lin J., Zhang G., Zhao J., Bao X., Zhu R., Song H., Li X.D. Genetically encoded photoaffinity histone marks. J. Am. Chem. Soc. 2017;139:6522–6525. doi: 10.1021/jacs.7b01431. [DOI] [PubMed] [Google Scholar]
- Yang Y., Song H., He D., Zhang S., Dai S., Lin S., Meng R., Wang C., Chen P.R. Genetically encoded protein photocrosslinker with a transferable mass spectrometry-identifiable label. Nat. Commun. 2016;7:12299. doi: 10.1038/ncomms12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Tang W.K., Zhang T., Xia D. A mighty “protein extractor” of the cell: structure and function of the p97/CDC48 ATPase. Front. Mol. Biosci. 2017;4:39. doi: 10.3389/fmolb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D.D., Schultz P.G. Playing with the molecules of life. ACS Chem. Biol. 2018;13:854–870. doi: 10.1021/acschembio.7b00974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount J.S., Karssemeijer R.A., Hang H.C. S-Palmitoylation and ubiquitination differentially regulate interferon-induced transmembrane protein 3 (IFITM3)-mediated resistance to influenza virus. J. Biol. Chem. 2012;287:19631–19641. doi: 10.1074/jbc.M112.362095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount J.S., Moltedo B., Yang Y.-Y., Charron G., Moran T.M., López C.B., Hang H.C. Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat. Chem. Biol. 2010;6:610–614. doi: 10.1038/nchembio.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zani A., Yount J.S. Antiviral protection by IFITM3 in vivo. Curr. Clin. Microbiol. Rep. 2018;5:229–237. doi: 10.1007/s40588-018-0103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zani A., Zhang L., McMichael T.M., Kenney A.D., Chemudupati M., Kwiek J.J., Liu S.-L., Yount J.S. Interferon-induced transmembrane proteins inhibit cell fusion mediated by trophoblast syncytins. J. Biol. Chem. 2019;294:19844–19851. doi: 10.1074/jbc.AC119.010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Hu Y., Hau R., Musharrafieh R., Ma C., Zhou X., Chen Y., Wang J. Identification of NMS-873, an allosteric and specific p97 inhibitor, as a broad antiviral against both influenza A and B viruses. Eur. J. Pharm. Sci. 2019;133:86–94. doi: 10.1016/j.ejps.2019.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., He D., Lin Z., Yang Y., Song H., Chen P.R. Conditional chaperone–client interactions revealed by genetically encoded photo-cross-linkers. Acc. Chem. Res. 2017;50:1184–1192. doi: 10.1021/acs.accounts.6b00647. [DOI] [PubMed] [Google Scholar]
- Zhang M., Lin S., Song X., Liu J., Fu Y., Ge X., Fu X., Chang Z., Chen P.R. A genetically incorporated crosslinker reveals chaperone cooperation in acid resistance. Nat. Chem. Biol. 2011;7:671–677. doi: 10.1038/nchembio.644. [DOI] [PubMed] [Google Scholar]
- Zhang Y.-H., Zhao Y., Li N., Peng Y.-C., Giannoulatou E., Jin R.-H., Yan H.-P., Wu H., Liu J.-H., Liu N. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat. Commun. 2013;4:1418. doi: 10.1038/ncomms2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The proteomics database search results have been deposited to the Mendeley database (https://doi.org/10.17632/tds8by96gc.1).