Abstract
CRISPR-Cas adaptive immune systems protect bacteria and archaea against their invading genetic parasites, including bacteriophages/viruses and plasmids. In response to this immunity, many phages have anti-CRISPR (Acr) proteins that inhibit CRISPR-Cas targeting. To date, anti-CRISPR genes have primarily been discovered in phage or prophage genomes. Here, we uncovered acr loci on plasmids and other conjugative elements present in Firmicutes, using the Listeria acrIIA1 gene as a marker. The four identified genes, found in Listeria, Enterococcus, Streptococcus, and Staphylococcus genomes, can inhibit Type II-A SpyCas9 or SauCas9, and are thus named acrIIA16-19. In Enterococcus faecalis, conjugation of a Cas9-targeted plasmid was enhanced by anti-CRISPRs derived from Enterococcus conjugative elements, highlighting a role for Acrs in the dissemination of plasmids. Reciprocal co-immunoprecipitation showed that each Acr protein interacts with Cas9, and Cas9:Acr complexes were unable to cleave DNA. Northern blotting suggests that these anti-CRISPRs manipulate sgRNA loading or stability. Mirroring their activity in bacteria, AcrIIA16 and AcrIIA17 provide robust and highly potent broad-spectrum inhibition of distinct Cas9 proteins in human cells (e.g. SpyCas9, SauCas9, SthCas9, NmeCas9, CjeCas9). This work presents a focused analysis of non-phage Acr proteins, demonstrating a role in horizontal gene transfer bolstered by broad spectrum CRISPR-Cas9 inhibition.
Introduction
Bacteria are constantly exposed to invasive mobile genetic elements (MGEs) that can either benefit or harm the host. Many MGEs encode antibiotic resistance pathogenicity factors that can enhance microbe virulence1,2, however, most are regarded as parasitic entities3. To combat MGE invasions, bacteria possess defense mechanisms, including restriction modification and CRISPR-Cas adaptive immunity4, which can limit the exchange of destructive genetic material5–7. CRISPR-Cas systems are widespread, found in roughly half of bacteria and over 80% of archaea8, and can protect host genomes against phage infection and plasmid conjugation9. Yet, occurrence of horizontal gene transfer (HGT) persists across species, evident by DNA sequence estimates suggesting that, on average 5–6% of genes in bacterial genomes are derived from HGT10, with numbers as high as 10–20% for some microbes11.
Bacteriophages have responded to CRISPR-Cas with anti-CRISPR (Acr) proteins12, that can inhibit CRISPR-Cas complex formation/stability13,14 or target DNA binding or cleavage15–18. To date, 46 distinct Acr protein families inhibiting various CRISPR-Cas subtypes have been discovered, in which type II-A Cas9 inhibitors alone constitute 1119–23. Numerous strategies have been employed for Acr discovery, including bioinformatic19,24, experimental12,20, and metagenomic screening22,23. Many of these approaches have discovered Acrs on phages and prophages, however, it is not clear how other MGEs avoid CRISPR targeting.
Here, we utilize the widespread phage and plasmid-encoded acrIIA1, previously identified in Listeria prophages, as a marker gene to discover four distinct inhibitors of the Type II-A CRISPR-Cas system, named acrIIA16–19. These proteins are predominantly encoded by non-phage elements, including plasmids and integrative and conjugative elements (ICEs). We demonstrate that AcrIIA16–19 inactivate Cas9-mediated cleavage of foreign DNA in vivo, both during phage infection and plasmid conjugation, in vitro, and in human cells. In vitro analyses suggest that these inhibitors interact with SpyCas9 through mechanisms distinct from the DNA mimics AcrIIA225,26 and AcrIIA427,28 and may modulate sgRNA expression/stability/loading. Interestingly, AcrIIA16 displays broad spectrum inhibition of SpyCas9 and SauCas9, similar in potency to previously identified AcrIIA521,29, while AcrIIA17 potently inhibits Type II-C NmeCas9. Together they provide useful off-switches for multiple phylogenetically distinct Cas9s.
Results
Type II-A anti-CRISPRs (AcrIIA16–19) inhibit SpyCas9 upstream of DNA-binding
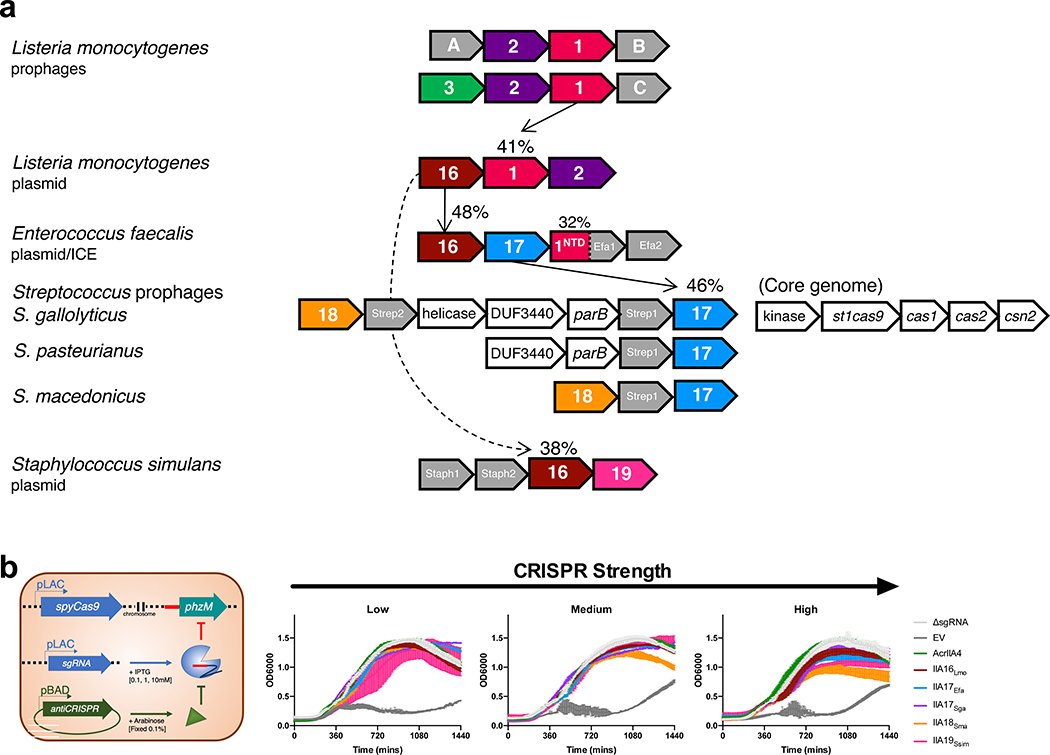
To better understand how MGEs interact with CRISPR-Cas immunity, we sought to identify undiscovered acr genes. We utilized the widespread acrIIA1 gene as an anchor in bioinformatic searches across genomes on NCBI (Fig. 1a). An AcrIIA1 homolog (41% amino acid sequence identity) was previously identified within an L. monocytogenes plasmid, along with an AcrIIA2 homolog that was recently characterized (AcrIIA2b.3, Jiang et al., 2019). Genomic neighbors in this locus were tested against the Type II-A Cas9 system using an established SpyCas9 phage-targeting screening system in Pseudomonas aeruginosa25,30 (Fig. 1b). Gene AWI79_RS12835 (now acrIIA16) inhibited SpyCas9 in this assay. Using acrIIA16 as the anchor gene, testing of its neighbors revealed three more distinct anti-CRISPR genes (acrIIA17-19) identified in Enterococcus, Streptococcus, and Staphylococcus (Fig. 1a). To quantify the strength of SpyCas9 inhibition, Cas9 and the sgRNA were titrated via IPTG induction. At the lowest CRISPR-Cas expression level, all identified acrIIA genes inhibited SpyCas9, restoring phage replication to nearly the same levels as in the strain lacking CRISPR immunity (ΔsgRNA, Fig. 1b). However, at higher CRISPR-Cas expression levels, only AcrIIA16Lmo, AcrIIA17Sga, and control AcrIIA4 maintained inhibition against SpyCas9 (Fig. 1b). In agreement with this result, the AcrIIA proteins also protect against self-genome cleavage assay with similar strength (Extended Data Fig. 1b).
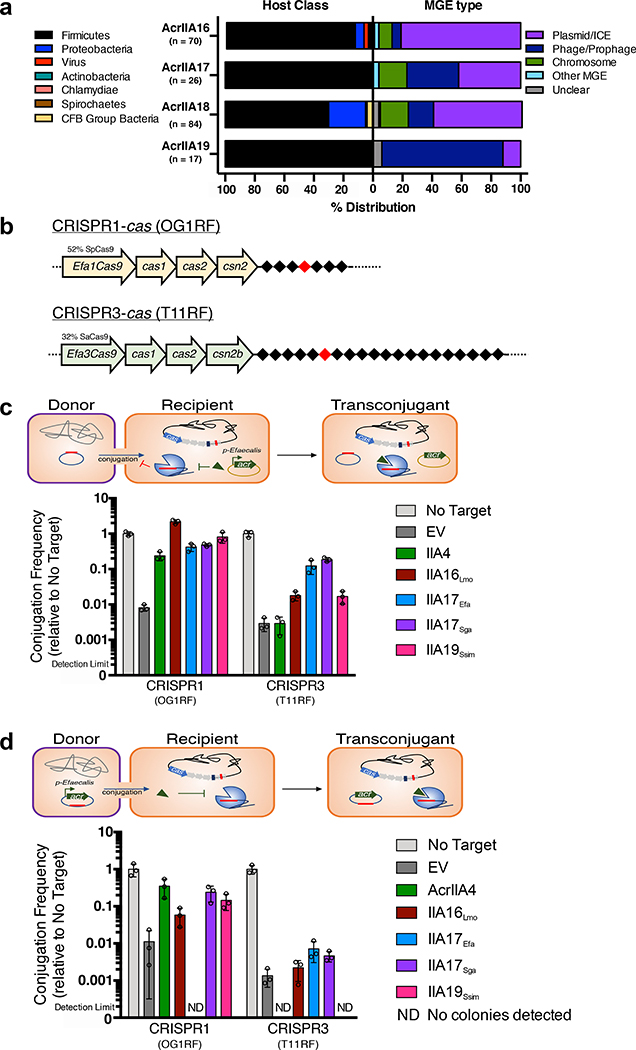
Fig. 1|. Identification of four Type II-A Cas9 inhibitors, AcrIIA16–19.
a, Schematic representation of Type II-A acr genes with vertical arrows indicating relationships between acr loci and percent protein sequence identity. Numbers in genes correspond to AcrIIA number. Grey genes are proteins of unknown function that tested negative for AcrIIA activity. b, Schematic of phage plaque assays to assess CRISPR-SpyCas9 inhibition. 10-fold serial dilutions of targeted phage (black circles) are spotted on a lawn of P. aeruginosa (grey background) expressing the Type II-A CRISPR-Cas system and indicated acr genes. CRISPR strength is determined by expression of sgRNA from the chromosome (low), or from a multicopy plasmid at increasing induction levels [0.1, 1, 10 mM IPTG]. ΔsgRNA lacks a phage-targeting sgRNA. EV, empty vector. Representative picture of at least three biological replicates for each are shown. c, Schematic of experiment to assess CRISPRi inhibition. Chromosomally-integrated dCas9 (yellow asterisks) in P. aeruginosa programmed to bind the phzM gene promoter with sgRNA expressed from a multicopy plasmid at low or medium IPTG induction levels, in the presence of indicated AcrIIA proteins. CRISPRi inhibition was assessed by quantification of pyocyanin levels in response to phzM gene repression, relative to ΔsgRNA. Percent pyocyanin levels at low and medium CRISPR strength are represented as the mean of three ± SD and two biological replicates respectively, and representative picture at medium CRISPR strength are shown (bottom).
To inspect the mechanism of these AcrIIA proteins in vivo, we established a CRISPRi assay, where catalytically dead SpyCas9 (dCas9) is programmed to bind the promoter of the phzM gene. Repression of phzM halts the production of green pigment called pyocyanin, generating a yellow culture15. In the presence of AcrIIA4, DNA-binding by dCas9 is inhibited, generating a green culture. AcrIIA16–19 all presented a similar phenotype to AcrIIA4, at two dCas9 induction levels, suggesting that these AcrIIAs inhibit SpyCas9 at the step of target DNA binding or an upstream stage (Fig. 1c).
acrIIA genes inhibit Cas9 during conjugation
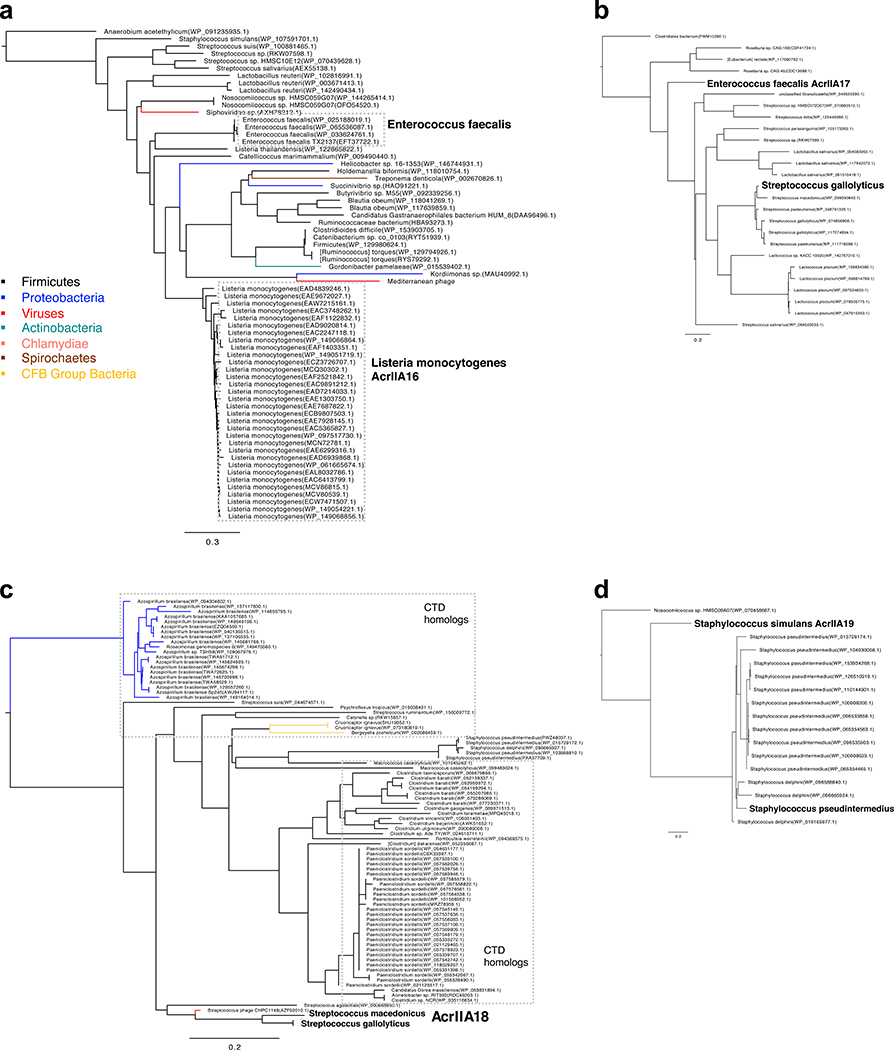
To determine the distribution of the identified acr loci, adjacent genes were examined for presence of signature genes that denote the locus to be phage, plasmid, MGE-like, or chromosome (see methods for details). A comprehensive list of Acr orthologues are listed in Supplementary Table 2. Analysis of AcrIIA16–18 distribution revealed that most orthologs are present in conjugative MGEs of Firmicutes, with some found in phages, bacterial chromosomes, or other mobile elements including transposons or integrons (Fig. 2a). AcrIIA16 is widespread in plasmids or ICE’s of various Firmicutes. AcrIIA17 is equally distributed in plasmids and prophages, predominantly found in Streptococcus and Lactococcus species. Full length AcrIIA18 is commonly found on Streptococcus and Staphylococcus prophages, while it’s C-terminal domain is not only found on a Streptococcus phage, but also on plasmids and core genomes of other Firmicutes (e.g. Clostridium sp. and Paeniclostridium sp.) and Azospirillium sp. (Proteobacteria). AcrIIA19Ssim was initially identified on a plasmid, but its homologs are commonly found in Staphylococcus prophages. Moreover, it possesses a helix-turn-helix domain, reminiscent of AcrIIA1, suggesting a dual regulatory and anti-CRISPR function31. Altogether, these Acr proteins are encoded by a variety of microbes and mobile elements, including phages, plasmids, and conjugative elements.
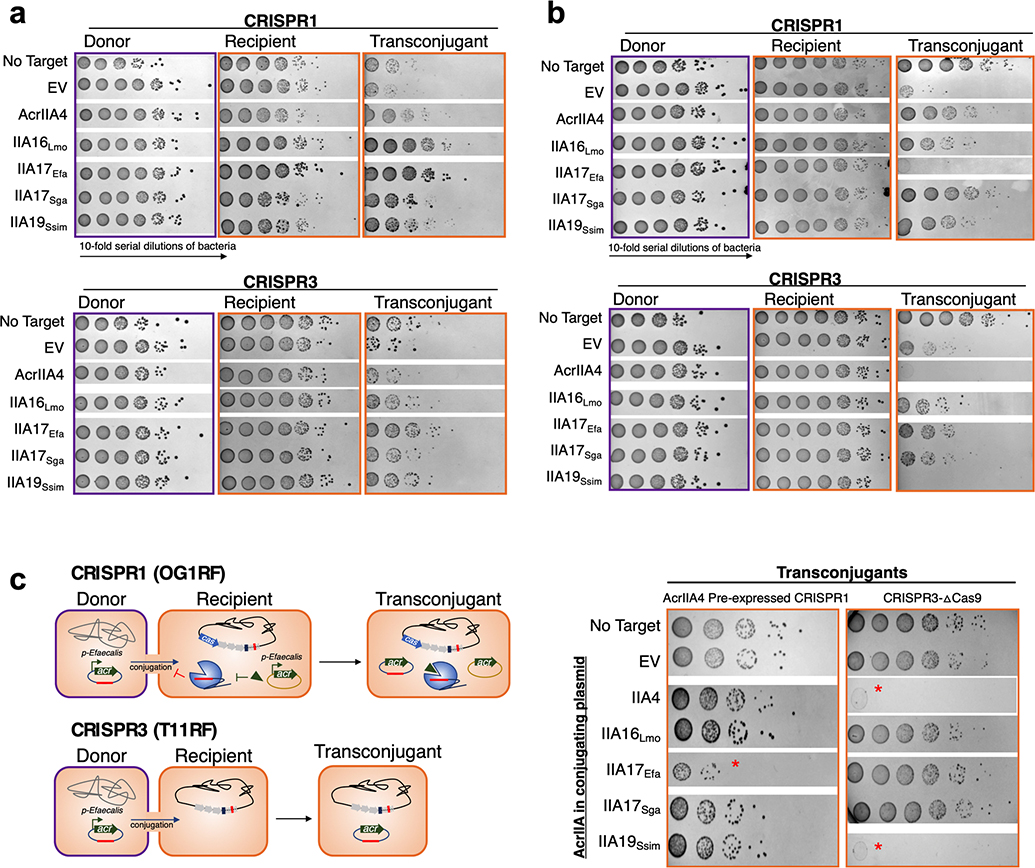
Fig. 2|. Prevalence of acrIIA genes in integrative mobile genetic elements and their effect on CRISPR-targeting during conjugation.
a, Left: Host distribution of acrIIA16–19 based on phylogenetic analysis, see Extended Data Fig 2. Right: Mobile genetic element distribution of acrIIA16–19 based on genomic neighbors characteristic of phage, plasmid, chromosomal or mobile genes including transposons and integrons. “Unclear” denote genomic regions that could not be identified as known elements. For every genomic region, at least one signature gene is identified to characterize the MGE type, see methods and Supplementary Table 2 for details. b, Schematic of the native CRISPR-Cas system in E. faecalis strains OG1RF for CRISPR1 and T11RF for CRISPR3 utilized for all conjugation experiments. Black diamonds denote spacers in the CRISPR array and red indicates spacer that matches the engineered protospacer in the targeted plasmids. c, Schematic of conjugation in E. faecalis encoding a Type II-A CRISPR system that targets the protospacer-bearing plasmid in the presence of indicated acrIIA genes episomally expressed in recipient cells. Conjugation frequency is quantified as transconjugants per donor relative to a non-targeted plasmid, and represented as the mean of three biological replicates ± SD. d, Schematic of plasmid conjugation in E. faecalis from a donor to recipient. The conjugating plasmid carries the indicated acrIIA gene and is targeted by the host’s Type II-A CRISPR-Cas system. Conjugation frequency is quantified as transconjugants per donor relative to a non-targeted plasmid, and represented as the mean of three biological replicates ± SD
Given the prevalence of many of these genes on plasmids, we chose to investigate the plasmid-encoded acrIIA16, 17 and 19 orthologues against CRISPR-targeting during plasmid conjugation. We tested the ability of Cas9 to target a plasmid when an AcrIIA protein is expressed either in the recipient or by the conjugating element. Previously reported E. faecalis strains32 were engineered to express acrIIA genes individually from an E. faecalis promoter native to the acr locus. E. faecalis encodes two distinct endogenous Type II-A CRISPR-Cas variants – CRISPR1, which is 52% identical to SpyCas9 and CRISPR3, which is 32% identical to SauCas9 (Fig. 2b). Two different conjugating plasmids were used, each engineered to contain a protospacer matching a natural spacer found in recipient cells OG1RF (CRISPR1) or T11RF (CRISPR3). Conjugation efficiency was reduced by 100–500-fold due to Cas9 targeting (Fig. 2c). When acrIIA16, 17, or 19 were pre-expressed in recipient cells, all inhibited CRISPR1 robustly, and CRISPR3 to a lesser degree (Fig. 2c, Extended Data Fig. 3a). acrIIA4 only inhibited CRISPR1 activity, which encodes a Cas9 that has a similar PAM-interacting domain to SpyCas9 (Fig. 2c).
To determine whether AcrIIA proteins could function when the conjugating CRISPR-targeted plasmid carries the acrIIA gene, the targeted plasmids were engineered to express acrIIA16–17 or acrIIA19 from the same Enterococcus acr promoter. These acr genes were indeed protective against plasmid targeting by CRISPR1 when produced during conjugation, with acrIIA17 providing modest protection against CRISPR3 (Fig. 2d, Extended Data Fig. 3b). Oddly, plasmids expressing certain acr genes did not produce detectable transconjugants (e.g. acrIIA17Efa when challenged with CRISPR1 and acrIIA4/acrIIA19Ssim against CRISPR3), but this was independent of CRISPR-targeting (Extended Data Fig. 3c), for a reason that is unknown. We conclude that acrIIA genes are able to inhibit both CRISPR-Cas9 systems during plasmid conjugation in E. faecalis and can enhance HGT by >1 order of magnitude when pre-expressed in recipient cells.
AcrIIA16–19 proteins interact with SpyCas9
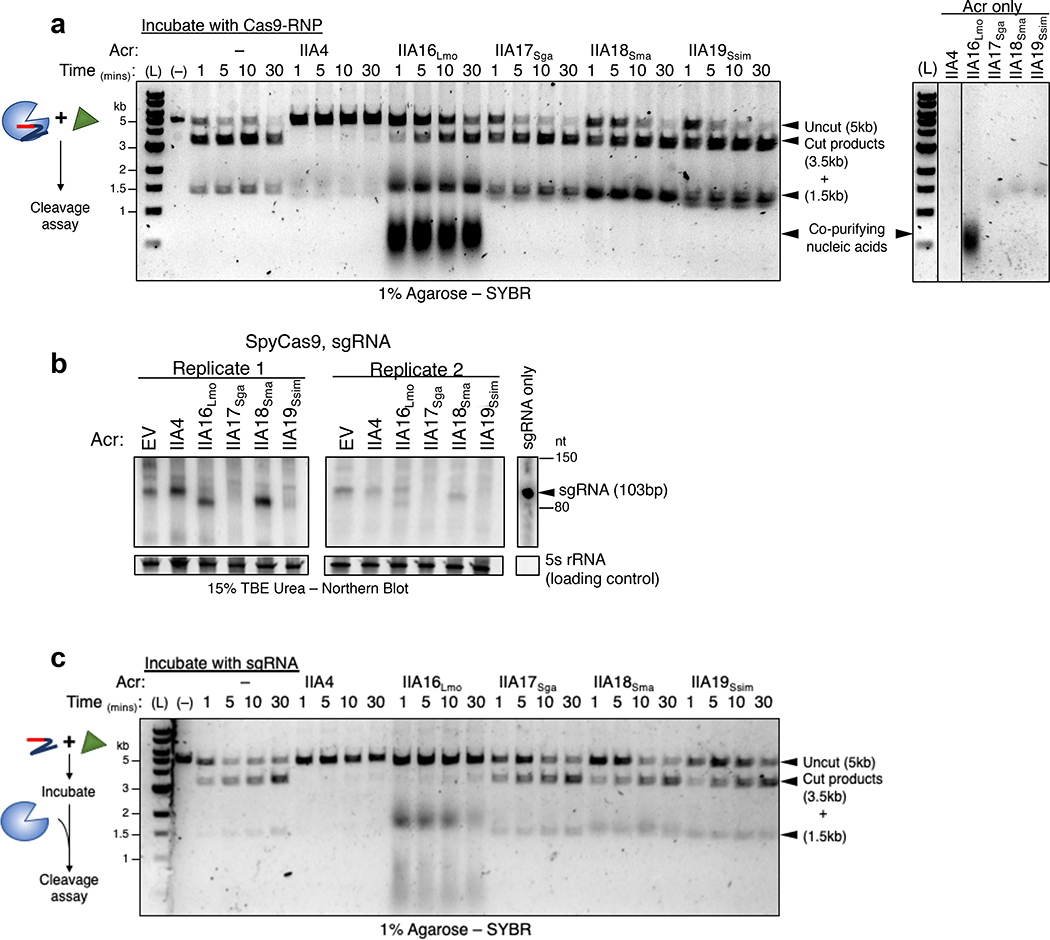
To further investigate the mechanism of Cas9 inhibition by the AcrIIA proteins, we purified one homolog of AcrIIA16–19 to directly test their effect on SpyCas9 activity. Surprisingly, in vitro cleavage experiments using the purified AcrIIA16–19 proteins and guide-loaded SpyCas9 did not directly inhibit DNA binding or cleavage under these conditions, while the positive control AcrIIA4 did (Fig. 3a). Due to the CRISPRi results above, suggesting that a step upstream of DNA-binding could be inhibited, we next considered guide-RNA stability or loading. Total RNA was harvested from the P. aeruginosa strains co-expressing SpyCas9, sgRNA and Acr proteins, followed by probing for sgRNA with a Northern blot (Fig. 3b, two independent biological replicates are shown). Interestingly, sgRNA in cells expressing AcrIIA16 are visible as two distinct bands – full length and a shorter version relative to its wildtype length. The presence of AcrIIA17 and AcrIIA19 lead to undetectable sgRNA in the cell, while AcrIIA18 enrich for a slightly truncated sgRNA. These results suggest inhibition mechanisms for AcrIIA16–19 that may involve manipulation of sgRNA levels or loading. To test whether any of the Acr proteins can directly interfere with sgRNA loading, the in vitro cleavage experiment was repeated, but with AcrIIA proteins first incubated with sgRNA before complexing with ApoSpyCas9. Remarkably, this change enabled inhibition by AcrIIA16, blocking SpyCas9-mediated DNA cleavage (Fig. 3c). AcrIIA17–19 activity were unaffected by this change in protocol. This suggests that AcrIIA16 acts on the sgRNA, ApoCas9 or both, to prevent activity.
Fig. 3|. In vitro binding and inhibition activities of AcrIIA16–19 against SpyCas9.
a, Left: Time courses of prebound SpyCas9-sgRNA cleavage reactions targeting a double stranded linear DNA template in the presence of purified Acr proteins. Right: Purified Acr proteins loaded on agarose gel to visualize presence of copurifying nucleic acids. (L) 1kb dsDNA ladder, (–) DNA template alone. Data shown are representative of two replicates. b, Northern blot analysis of sgRNA in P. aeruginosa expressing SpyCas9-sgRNA and indicated Acr’s. For all blots, 5S rRNA served as loading control. Data are shown for two independent experiments. c, Time courses of target DNA cleavage reactions using sgRNA preincubated with Acr proteins followed by Apo-SpyCas9 to form RNP. Representative time-points are shown at the top of each lane. (L) 1kb dsDNA ladder, (–) DNA template alone. Data shown are representative of two replicates.
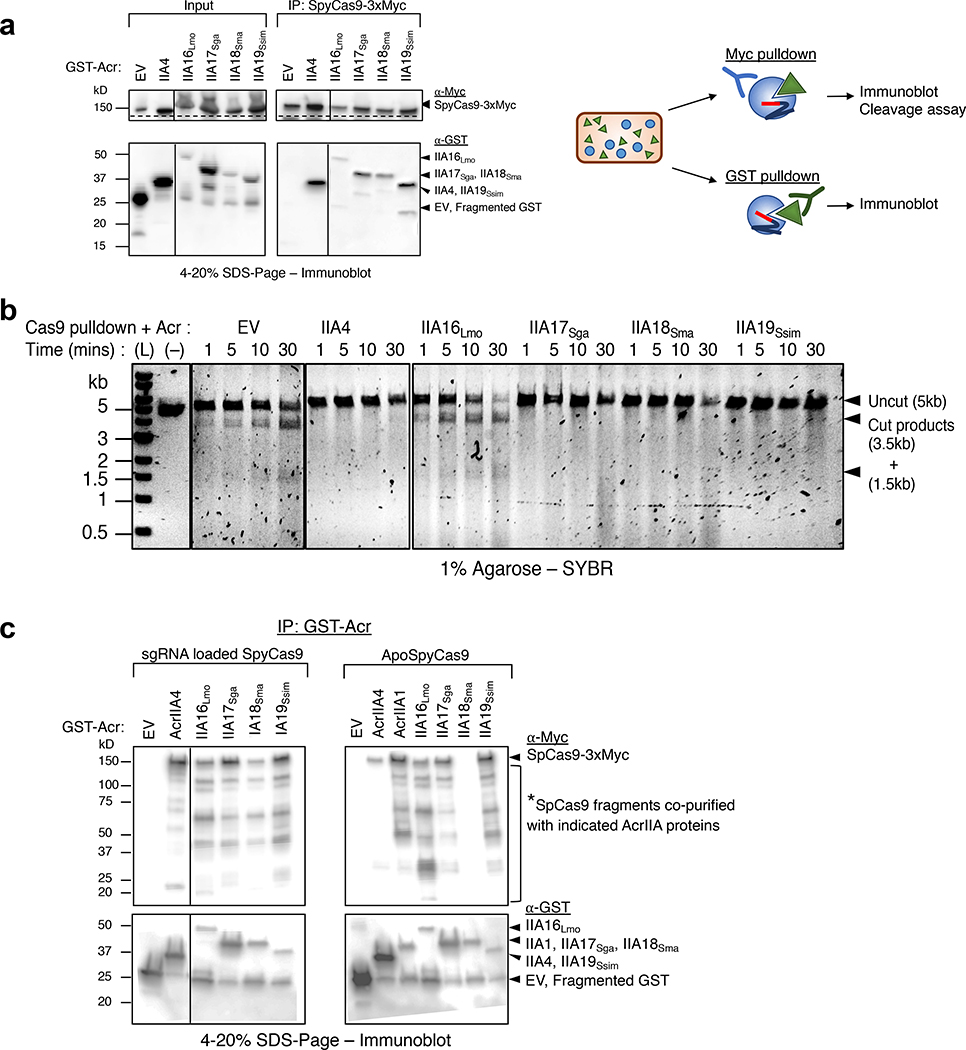
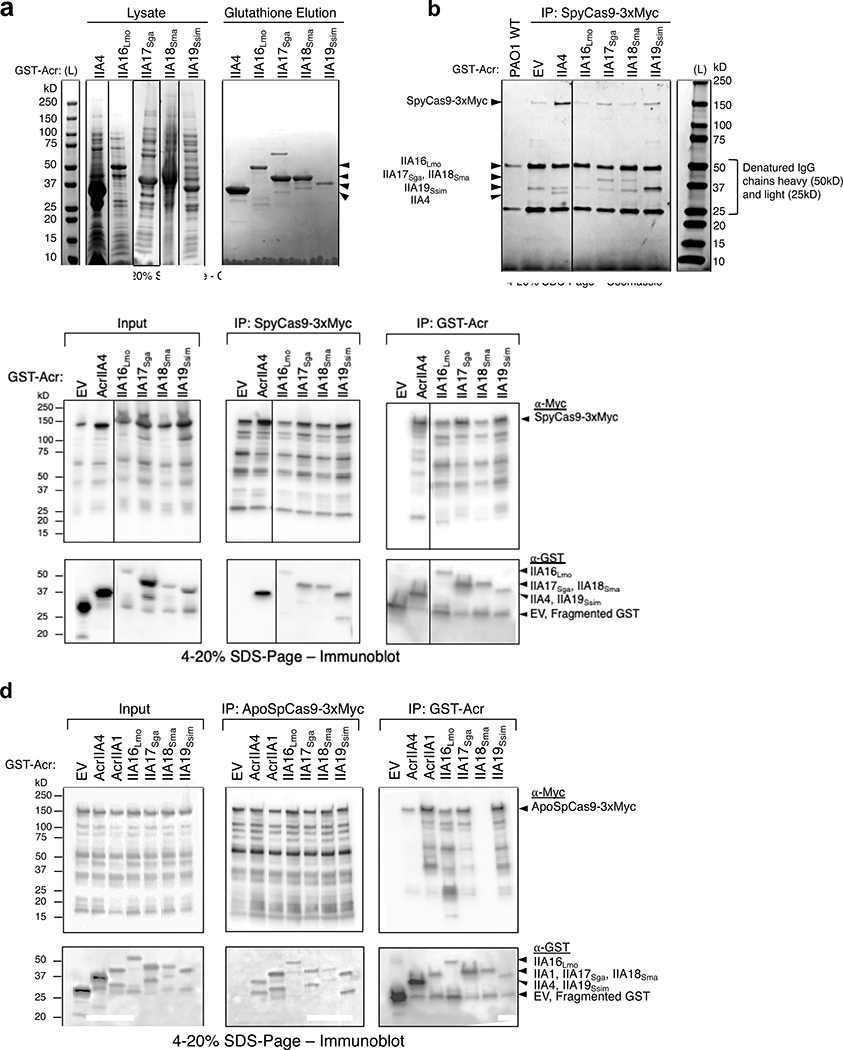
To determine whether AcrIIA16–19 interact with Cas9, myc-tagged SpyCas9 was immunoprecipitated from the P. aeruginosa strains introduced above. This experiment revealed that all four AcrIIA proteins co-purify with SpyCas9 (Fig. 4a). Interestingly, SpyCas9 purified from cells co-expressing AcrIIA17–19 did not perform DNA cleavage. The absence of any obvious stoichiometric, co-purifying proteins suggests a direct interaction between Cas9 and the Acr proteins (Extended Data Fig. 4b). SpyCas9 co-purified with low amounts of AcrIIA16Lmo, however was not inhibited (Fig. 4b). The failure of AcrIIA16Lmo to inhibit immunoprecipitated SpyCas9 in vitro may be due to its low expression level, as visualized in the input western blot (Fig. 4a) and/or the fact that it inhibits when exposed to the sgRNA first. Northern blot also suggested that traces of wildtype length sgRNA are still present in cells expressing AcrIIA16Lmo (Fig. 3b) which could form active RNP.
Fig. 4|. In vivo co-immunoprecipitation of AcrIIA16–19 and SpyCas9.
a, Immunoprecipitation of Myc-tagged SpyCas9-sgRNA (162kD) or GST-tagged Acr proteins (free-GST 27kD, IIA4 37kD, IIA16Lmo 50kD, IIA17Sga 39kD, IIA18Sma 48kD and IIA19Ssim 42kD). Left: Immunoblot probed with α-Myc (top) and α-GST (middle). Dashed lines indicate where image is cropped to show only the bands corresponding to full-length SpyCas9, see Extended Data Fig. 4c for uncropped version. Data shown are representative of two independent experiments. Right: Schematic of immunoprecipitation from P. aeruginosa cells co-expressing SpyCas9 and Acr proteins followed by analysis. b, Time courses of target DNA cleavage reactions using SpyCas9 co-immunoprecipitated with AcrIIA-proteins from Fig. 4b. Representative time-points are shown at the top of each lane. (L) 1kb dsDNA ladder, (–) DNA template alone. Data shown are representative of two independent experiments. c, Immunoprecipitation of GST-tagged Acr proteins (free-GST 27kD, IIA4 37kD, IIA1 44kD, IIA16Lmo 50kD, IIA17Sga 39kD, IIA18Sma 48kD and IIA19Ssim 42kD) from P. aeruginosa co-expressing guide-loaded SpyCas9 or ApoSpyCas9 without sgRNA (162kD). Immunoblot for Myc-Cas9 (top) or GST-Acr (bottom). Data shown are representative of two independent experiments.
We next conducted the reciprocal co-immunoprecipitation experiment, confirming that SpyCas9 co-purifies with each tagged Acr (Fig. 4c). Moreover, we observed that SpyCas9 expressed in P. aeruginosa exhibits a series of degradation products when blotted for the C-terminal Myc tag. The enriched SpyCas9 fragments co-immunoprecipitated with AcrIIA16–19 appeared to be different from those of AcrIIA4, suggesting distinct binding sites (Fig. 4c). This experiment, coupled with observations of sgRNA degradation led us to test whether these Acr proteins bind Apo-SpyCas9, a complex previously reported to be only a weak AcrIIA4 binding partner28 and strong in vitro interaction partner for AcrIIA131. AcrIIA16–17 and AcrIIA19 co-purified with Apo-SpyCas9 similar to expected levels of AcrIIA1. AcrIIA4 showed weak binding (comparing the relative amount of AcrIIA4 to Cas9) and AcrIIA18 showed no interaction with Apo-SpyCas9 (Fig. 4c and Extended Data Fig. 4d). These results suggest that AcrIIA16, 17, and 19 have distinctive SpyCas9 interacting mechanism from AcrIIA4 and AcrIIA18, and may modulate sgRNA stability or loading, via an interaction with ApoCas9.
AcrIIA16Efa and AcrIIA17Efa potently inhibit Cas9 orthologs in human cells
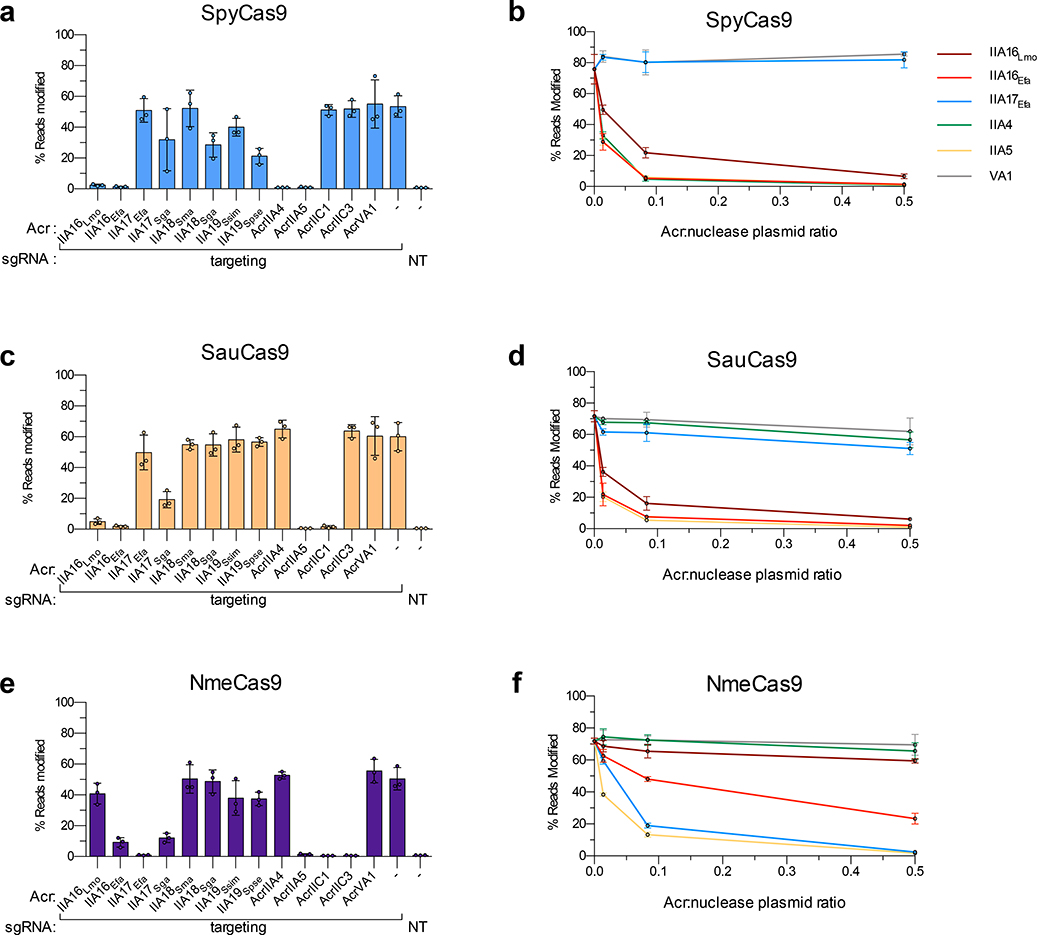
Given the increasing use of various Cas9 orthologs for gene editing applications, we examined the ability of our AcrIIA proteins to prevent SpyCas9 activity in human cells. HEK 293T cells were co-transfected with plasmids expressing Cas9, sgRNAs programmed to target sites located in endogenous genes, and 13 different acrIIA genes – two homologs each for AcrIIA16–19 along with five previously validated control acr genes. By using targeted deep sequencing to evaluate editing activity, we observed near-complete inhibition of SpyCas9 by the two AcrIIA16 orthologs at levels comparable to the well validated AcrIIA427,28 and the broad-spectrum AcrIIA529 (Fig. 5a). Furthermore, by titrating the molar ratio of Acr plasmids transfected (Fig. 5b), inhibition with AcrIIA16 was observed even with very low amounts of Acr plasmid, at levels comparable to other “gold standard” SpyCas9 inhibitors AcrIIA4 and AcrIIA5. The other six AcrIIA17–19 proteins exhibited more modest and inconsistent levels of inhibition, where at least one homolog of each moderately inhibited SpyCas9 (Figs. 5a–b).
Fig. 5|. Acr-mediated inhibition of Cas9 orthologs during gene editing in human cells.
a to f, Reported Acr proteins in this study and from previous work tested for inhibition of genome editing activities of SpyCas9 (a, b), SauCas9 (c, d), and NmeCas9 (e, f). Inhibition is assessed at a fixed Acr:nuclease ratio for all Acr proteins (3:1 for panels a, c, and e), or at various ratios of Acr:nuclease plasmid (0.5:1, 0.083:1, and 0.014:1) for select Acr proteins (b, d and f). Editing efficiencies against endogenous genes in HEK 293T cells were assessed by targeted sequencing and quantified as the percentage of reads containing a nuclease-induced alteration; the no-Acr condition contains an EGFP expression plasmid; the NT control includes an empty U6 expression plasmid. Percent reads modified are represented as the mean of three biological replicates ± SD.
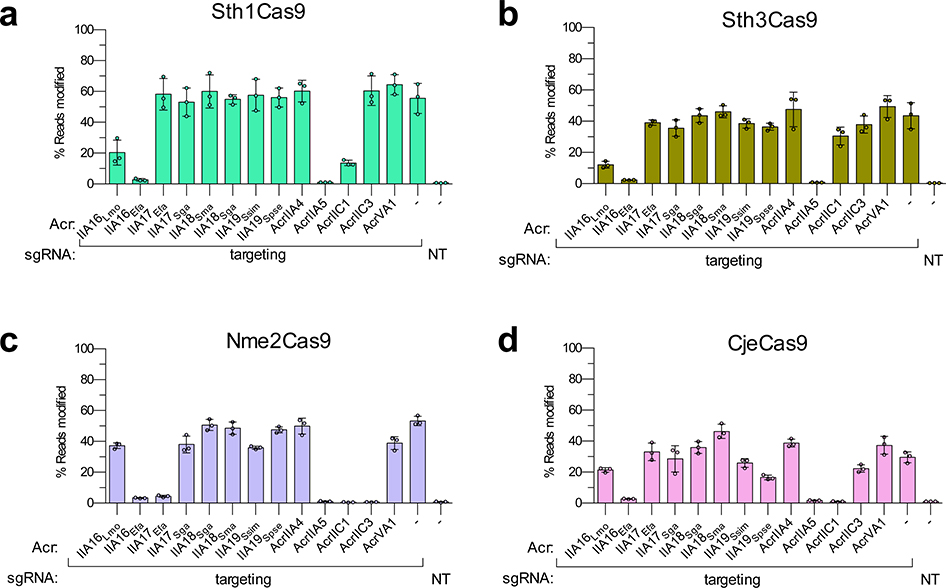
Next, because our Enterococcus experiments suggested the potential for broad spectrum inhibition with the reported Acr proteins, we examined the activities of these same 13 Acr proteins against other commonly used type II-A and II-C Cas9 orthologs: SauCas9, NmeCas9, Sth1Cas9, Sth3Cas9, Nme2Cas9, and CjeCas9 (Figs. 5c–f, and Extended Data Fig. 5). Interestingly, AcrIIA16Efa inhibited gene editing by all six additional Cas9 proteins, to levels comparable with control inhibitors of each specific system (Fig. 5 and Extended Data Fig. 5). Titrations revealed potent inhibition of SauCas9 by AcrIIA16Efa, similar to AcrIIA5 (Figs. 5c–d), and AcrIIA17Efa robustly inhibited NmeCas9 (Figs. 5e–f), confirming the broad-spectrum nature of these Acr proteins. Taken together, we observe that the previously unidentified AcrIIA16–19 are found in many MGEs (phages, plasmids, etc.) and are capable of inhibiting Cas9 orthologues in different cell backgrounds, including native and heterologous bacterial and human cells.
Discussion
Numerous strategies continue to be developed for identification of Acrs, with a remarkably diverse range of disclosed inhibition mechanisms33,34. Here, we discovered unusual acr loci in various MGEs, led initially by gene associations with acrIIA119,31. The acr genes reported here are found in diverse MGEs including plasmids, ICEs, prophages, transposons, integrons and other uncharacterized elements. These Cas9 inhibitors protect phage DNA during infection and plasmid DNA during conjugation. AcrIIA16–19 interact with SpyCas9 via distinct binding mechanisms compared to AcrIIA4 and AcrIIA2, to ultimately inhibit target DNA cleavage. Finally, AcrIIA16 and AcrIIA17 displayed potent inhibition of Type II-A and II-C Cas9 orthologs, respectively.
It is of high clinical relevance to find acrIIA genes in E. faecalis, where the spread of antibiotic resistance genes is frequently promoted through plasmid transfer despite the presence of host-encoded CRISPR-Cas systems. This work opens the door to the identification of more acr genes in this organism and its relatives. Previous work has shown that multidrug resistant E. faecalis strains are more likely to lack CRISPR-Cas9 but can acquire MGEs with protospacer matches due to low levels of Cas9 expression, and tolerate those plasmids transiently32,35,36. Our results suggest that these complex interactions have an additional layer and that a state of plasmid self-targeting could be stabilized for some time prior to potential CRISPR-Cas or spacer loss. We demonstrated that AcrIIA proteins not only could enhance the spread of the antibiotic resistance plasmid that encodes them, but they also impair the host’s ability to limit the acquisition of other MGEs. Future work on the mechanism and diversity of acr genes in E. faecalis will be necessary to understand their prevalence and importance in HGT.
The AcrIIA proteins reported in this work appear to modulate sgRNA levels or lengths when co-expressed with sgRNA and Cas9. Additionally, AcrIIA16 can directly impair Cas9 function when exposed to sgRNA and ApoSpyCas9 separately prior to loading, but not when exposed to loaded RNP in vitro. Further investigation of these Acr proteins in their native host may be required to truly understand their mechanism (i.e. where crRNA and tracrRNA are encoded separately and processing must occur) and direct in vitro interaction mapping coupled with structural analysis is needed.
With the increasing use of CRISPR-Cas systems for various genome editing applications, the discovery and characterization of natural inhibitors that regulate a variety of Cas9 orthologs via different mechanisms remains critical. The broad-spectrum inhibitors AcrIIA16 and IIA17, are attractive as practical regulators of multiple distinct Cas9 proteins. We also observed that AcrIIA5 is a good candidate for broad spectrum Cas9 inhibition, as reported previously21,29. AcrIIC1 also performed well against SauCas9 and NmeCas9, however, was reported in a previous assay to not inhibit SauCas9 in vitro37. The discovery of Acr proteins in organisms with more than one Type II-A CRISPR-Cas9 system (e.g. Streptococcus, Listeria, and Enterococcus) may lead to the identification of other broad-spectrum inhibitors, as there is a selective pressure to inhibit multiple Cas9-based systems. Conjugative elements with a broader host range than phages may face extensive and variable pressure and thereby are promising for the discovery of uncharacterized acr genes and mechanisms.
Methods
Microbes
Escherichia coli (DH5α, XL1Blue, NEB 10-beta, or NEB turbo) were routinely cultured in lysogeny broth (LB) at 37 °C supplemented with antibiotics at the following concentrations: gentamicin (30 μg/mL), carbenicillin (100 μg/mL), kanamycin (25 μg/mL), chloramphenicol (25 μg/mL), erythromycin (300 μg/mL) or tetracycline (10 μg/mL). Pseudomonas aeruginosa (PAO1) was cultured in LB medium at 37°C with supplemented antibiotics for plasmid maintenance: gentamicin (50 μg/mL) or carbenicillin (250 μg/mL). For maintaining multiple plasmids in the same P. aeruginosa strain, antibiotic concentrations were adjusted to 30 μg/mL gentamicin and 100 μg/mL carbenicillin. All Enterococcus faecalis strains (C173, OG1RF, T11RF, T11RFΔCas9) were cultured in brain-heart-infusion (BHI) medium at 37 °C, unless otherwise mentioned. Antibiotics were used in the following concentrations: spectinomycin (500 μg/mL), streptomycin (500 μg/mL), rifampicin (50 μg/mL), fusidic acid (25 μg/mL), chloramphenicol (15 μg/mL) or erythromycin (50 μg/mL).
Cell lines
Human HEK293T cells were obtained from ATCC and authenticated by STR profiling. Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% heat-inactivated FBS (HI-FBS) and 1% penicillin/streptomycin. Media supernatant from cell cultures was analyzed monthly for the absence of mycoplasma using MycoAlert PLUS (Lonza).
Construction of P. aeruginosa and E. faecalis strains
P. aeruginosa heterologous type II-A system was generated as previously described30 under “construction of PAO1::SpyCas9 expression strain,” with sgRNA integrated into the bacterial genome using the mini-CTX2 vector38 or expressed from multi-copy episomal plasmid pMMB67HE-PLac for in vivo assays, and plasmid pHERD30T-PBad for in vitro assays. All acr candidate genes were synthesized as gene fragments (Twist Biosciences) and cloned using Gibson Assembly into plasmids of P. aeruginosa vectors pHERD30T or pMMB67HE, and E. faecalis vectors pKH12 or pMSP3535 (gifts from Kelli L. Palmer and Gary M. Dunny RRID:Addgene_46886 respectively). Plasmids were electroporated into PAO1 for all P. aeruginosa strains39, and E. faecalis strains C173, OG1RF, T11RF and T11RFΔCas9 using previously published protocols40. All strains and plasmids constructed and used in this study are listed in Supplementary Table 3a–b.
Bacteriophage plaque assays in P. aeruginosa
Plaque assays were performed as previously described25,30 with sgRNA designed to target Pseudomonas phage JBD30. The PLac promoter driving chromosomally integrated SpyCas9 and sgRNA, or pMMB67HE-sgRNA was induced with titrating levels of IPTG (0.1, 1, 10mM) and the PBad promoter driving pHERD30T-acr with 0.1% arabinose. One representative plate for each candidate were imaged using Gel Doc EZ Gel Documentation System (Bio-Rad) and Image Lab software.
Self-genome targeting and CRISPRi assay in P. aeruginosa
Strains with chromosomally integrated WT SpyCas9 or dCas9 are programmed with pMMB67HE-sgRNA to target the PAO1 chromosomal phzM gene promoter in the presence of pHERD30T-acr. Cultures were grown overnight in LB supplemented with appropriate antibiotics for plasmid maintenance and 0.1% arabinose to pre-induce anti-CRISPR expression. Overnight cultures are diluted in 1:100 LB supplemented with inducers 0.1% arabinose and IPTG (0.1, 0.25, 1, 10 mM to titrate CRISPR strength) in a 96-well Costar plate (150 μL/well) for self-targeting survival analysis or glass tubes (3 mL) for CRISPRi, in triplicates. Self-genome targeting was assayed by measuring bacterial growth curves for 16–24 hours in Synergy H1 microplate reader (BioTek, using Gen5 software) at 37 °C with continuous shaking, and data displayed as the mean OD600 of at least three biological replicates ± standard deviation (error bars) as a function of time. For CRISPRi, cells were grown for 20–24 hours with continuous shaking. Next, pyocyanin was extracted and quantified as previously described15. Data are displayed as the mean OD520 of at least three biological replicates ± standard deviation (error bars) and representative pictures are shown.
Phylogenetic tree construction
Homologs were identified using 1 run of iterative PSI-BLASTp and e-value cut off < 0.1. Distance tree of results was generated on BLAST using fast minimum evolution tree method, 0.85 maximum sequence difference, and Grishin (protein) distance model. Further labels and annotations were performed on FigTree.
Host and MGE distribution prediction
Genomes were first annotated as plasmids or phages and their host class according to NCBI description. Next, genes adjacent to the specified loci were examined for presence of phage, plasmid or bacteria chromosomal proteins and identified as signature genes as listed in Supplementary Table 2. For draft genomes where signature genes cannot be identified, PlasFlow was used to predict putative plasmid elements with threshold adjusted to 0.5 and Phaster was used to predict putative prophages.
Conjugation assay in E. faecalis
Protospacers perfectly matching to indicated spacers in CRISPR1 or CRISPR3 array (Fig. 2b) were synthesized as complementary oligonucleotides (IDT) and cloned into pKH1232 to generate the targeted conjugative plasmid. The promoter region of the of acr loci in E. faecalis (nucleotide sequence 350 bp upstream) was synthesized (Twist Bioscience) and cloned upstream the acr genes of the targeted pKH12 conjugative plasmid or pMSP3535. The derivatives of pKH12 were introduced into the C173 donor strain as the transferring plasmid, and pMSP3535 into OG1RF, T11RF or T11RFΔCas9 to pre-express the Acr proteins in recipient cells.
Conjugation mating experiments were performed as previously described5, except for the following adjustments. Diluted cultures of plasmid-donor and recipient strains were grown to OD600 0.9–1.0, after which 100 μL of donor strain was mixed with 900 μL of OG1RF recipient strains or 500 μL donor with 500 μL of T11RF recipients. Resuspended pellets were plated on Mixed Cellulose Ester filter membranes (Advantec #A020H047A) on BHI agar plates without selection and incubated overnight at 37 °C. The next day, mated cells were collected by washing the filter membrane with 1.5 mL of 1X PBS and 10-fold serial dilutions were plated or spotted on BHI agar plates supplemented with antibiotics to quantify donor (spectinomycin, streptomycin and chloramphenicol), recipient (rifampicin and fusidic acid, and erythromycin for pMSP353 containing strains) or transconjugant (rifampicin, fusidic acid and chloramphenicol, with erythromycin for pre-expressed Acr strains) populations. Plates were incubated for 48 to 72 hours at 30 °C to allow colonies to develop. Plates with 30 to 300 colonies were used to calculate CFU/mL and conjugation frequency was determined by dividing the number of transconjugants over donors. For plates with spotted dilutions, the fold reductions in transconjugants were qualitatively derived by examining at least three replicates of each experiment. Plate images were acquired as in “bacteriophage plaque assays in P. aeruginosa” and a representative picture is shown.
Expression and purification of anti-CRISPR proteins
N-terminally GST-tagged Acr proteins were purified from E. coli BL21 following a previous protocol31 under “Cas9 and anti-CRISPR protein expression and purification”. Lysates were incubated with Glutathione Sepharose (GE 17–0756-05) followed by dialysis by centrifugation into storage buffer (100 mM Tris-Cl pH 8, 150 mM KCl, 10% Glycerol, 1mM DTT) to remove reduced glutathione used in elution.
Cleavage assays using purified proteins
Lyophilized sgRNA was resuspended in Nuclease-free Duplex Buffer following protocol from IDT, and stored frozen at −80 °C or incubated with SpyCas9 (NEB) at room temperature for 15 mins to form SpyCas9-RNP. All reactions were carried out in 1X MST Buffer (50 mM Tris-Cl pH 7.4, 150 mM NaCl, 20 mM MgCl2, 5 mM DTT, 5% Glycerol, 0.05% Tween-20 [v/v]). 25 nM SpyCas9-RNP was incubated with 2500 nM of Acr protein for 10 mins at room temperature. For sgRNA preincubation experiment, 25 nM sgRNA alone was incubated with 2500 nM Acr protein for 15 mins at room temperature, followed by SpyCas9 for 10 mins at room temperature to form 25 nM RNP. DNA substrate linearized by NheI digestion was added to a final concentration of 2 nM and the reaction was allowed to cut for 1, 5, 10 and 30 mins, at each timepoints the reaction was quenched in warm Quench Buffer (50 mM EDTA, 0.02% SDS) followed by heating at 95 °C for 10mins. Products were analyzed on 1% agarose gel and stained with SYBR Safe.
Co-immunoprecipitation of SpyCas9–3xMyc and GST-Acr
Chromosomally integrated SpyCas9 and pHERD30T-sgRNA for guide-loaded Cas9 or empty vector for apo-Cas9 were expressed off the PBad promoter, and pMMB67HE-GST-AcrIIA expressed of PLac in P. aeruginosa PAO1 strain. Saturated overnight cultures were diluted 1:100 the next morning in a total volume of 50 mL, induced with 0.3% arabinose and 1 mM IPTG at OD600 0.3–0.4, and harvested at OD600 1.8–2.0 by centrifugation at 6,000 × g for 10 mins at 4 °C. Cell pellets were flash frozen on dry ice, resuspended in 1mL lysis buffer (50 mM Tris-Cl pH 7.4, 150 mM NaCl, 20 mM MgCl2, 0.5% NP40, 5% Glycerol [v/v], 5 mM DTT, and 1 mM PMSF), lysed by sonication (20 s pulse for 4 cycles with cooling on ice between cycles, and lysates were clarified by centrifugation at 14,000 × g for 10 mins at 4 °C. For input samples, 10 μLlysates were added in 3X volume of 4X Laemmli Sample Buffer. Using a magnetic stand, Anti-c-Myc Magnetic Beads #88842 or Gluthathione Magnetic Agarose Beads #78601 (Thermo Fisher Scientific) were prewashed with 1mL of cold wash buffer (50 mM Tris-Cl pH 7.4, 150 mM NaCl, 20 mM MgCl2), and remaining lysate were added to bead slurry in a volume ratio of 20:1 for Myc or 40:1 for GST followed by overnight incubation at 4°C with end-over-end rotation. Beads were washed five times using a magnetic stand at room temperature with 1mL of cold wash buffer with addition of 5mM DTT, gradual decreasing concentrations of detergent NP40 (0.5%, 0.05%, 0.01%, 0.005%, 0) and glycerol (5%, 0.5%, 0.05%, 0.005%, 0). Bead-bound proteins were resuspended in 100 μLof final wash buffer without detergent and glycerol. For analysis, 10 μL of beads-bound protein were added to equal volume of 4X Laemmli Sample Buffer. Samples were analyzed on 4–20% SDS-Page gel and stained with Coomassie (Bio-Safe Coomassie Stain, Bio-Rad).
Immunoblotting
Protein samples were separated by SDS-Page using 4–20% gel (Mini-PROTEAN TGX Precast Gels, Bio-Rad) and transferred in 1X Tris/Glycine Buffer (Bio-Rad) with 20 % Methanol onto 0.2 μm Immun-Blot PVDF Membrane (Bio-Rad). Blots were probed with the following antibodies diluted 1:5000 in 1X TBS-T containing 5% nonfat dry milk: mouse anti-Myc (Cell Signaling Technology #2276, RRID:AB_331783), rabbit anti-GST (Cell Signaling Technology #2625, RRID:AB_490796), HRP-conjugated goat anti-mouse IgG (Santa Cruz Biotechnology #sc-2005, RRID:AB_631736) and HRP-conjugated goat anti-rabbit IgG (Bio-Rad #170–6515, RRID:AB_11125142). Blots were developed using Clarity ECL Western Blotting Substrate (Bio-Rad), and chemiluminescence was detected on an Azure c400 Biosystems Imager.
Cleavage assays using SpyCas9–3xMyc tagged pull downs
DNA substrate linearized by NheI digestion was added into beads-bound protein slurry to a final concentration of 1.5 nM and the reaction was allowed to react for 1, 5, 10 and 30 mins in the thermomixer at 25 °C with gentle shaking 1000 rpm. At each timepoint, the reaction was quenched in warm Quench Buffer (50 mM EDTA, 0.02% SDS), followed by heating at 95 °C for 10 mins. Products were analyzed on 1% agarose gels stained with SYBR Safe.
RNA extraction
Chromosomally integrated SpyCas9 and pHERD30T-sgRNA were expressed off the PBad promoter, and pMMB67HE-GST-AcrIIA expressed of PLac in P. aeruginosa PAO1 strain. Saturated overnight cultures were diluted 1:100 the next morning in a total volume of 10 mL LB containing inducers 0.1% arabinose and 1 mM IPTG and harvested at OD600 0.8–0.9 by centrifugation at 6,000 × g for 10 mins at 4 °C. Total RNA was extracted using TRIzol Max Bacterial RNA Isolation Kit (Life Technologies 16096–020), and treated with DNase I (Turbo DNA-free kit AM1907 from Life Technologies) according to manufacturer’s protocol. The concentration of RNA in each sample was further normalized following spectrophotometry measurements using NanoDrop.
Northern Blot analysis
Northern blot was carried out as previously described12, with exceptions described below. Radiolabeled probe was generated by amplifying a fragment containing the sgRNA construct from pHERD30T plasmid with primers CCAAACCGGTAACCCCGCTTA and GATTAAGTTGGGTAACGCCAGGGTTTTC, cleaning the PCR product (DNA Clean and Concentrator Kit D4034 from Zymo Research), labeling 200 ng of the clean product with α−32P dCTP using DNA Polymerase I Klenow Fragment (NEB M0210L) and purification using G25 columns (GE Healthcare) to remove unincorporated nucleotides. 5 ug of total RNA extracts were loaded using 2X RNA Loading Dye (NEB B0363S) onto a 15% denaturing gel (Mini-PROTEAN TBE-Urea Gel from Bio-Rad) and separated by electrophoresis. RNA was transferred onto Hybond-N+ membrane (GE Healthcare RPN303B) via semi-dry apparatus (Trans-Blot Turbo Transfer System from Bio-Rad) at 200 mA for 1 hour, and then crosslinked with 10 mJ UV burst over 30 seconds (Stratagene). The membrane was blocked with pre-hybridization buffer (50% formamide, 5X Denhardts solution and 6X SSC) containing 100 ug/mL salmon sperm DNA at 42 °C for 1.5 hr with rotation, followed by hybridization with radiolabeled probe at 42 °C overnight with rotation. The blot was washed with wash solution 1 (2X SSC and 1% SDS) for two 10 mins at 25 °C, two 30 mins at 65 °C, and wash solution 2 (0.2X SSC and 0.1% SDS) for one 10 min at 25 °C. Blots were developed using a phosphor screen and Typhoon imager.
Plasmid for human cell experiments
Descriptions of all plasmids used for expression of nucleases and Acr proteins in human cells, sgRNA/crRNA entry vectors, and all sgRNA/crRNA target sequences are listed in Supplementary Table 4. U6 promoter sgRNA and crRNA expression plasmids were generated by annealing and ligating oligonucleotide duplexes into BsmBI-digested entry vectors (Supplementary Table 4b). Human codon optimized Acr constructs containing a C-terminal SV40 nuclear localization signal were cloned into NotI/AgeI of Addgene plasmid ID 43861. New human expression plasmids described in this study have been deposited with Addgene (Supplementary Table 4a).
Human cell culture and transfection
HEK293T cells were seeded at 2×104 cells/well in 96-well plates approximately 20 hours prior to transfection. Each transfection reaction consisted of 1.25 μL of TransIT-X2 (Mirus Bio) with 70 ng of nuclease, 30 ng of sgRNA/crRNA, and 110 ng of anti-CRISPR expression plasmids in a final volume of 20 μL otherwise containing Opti-MEM (Thermo Fisher Scientific). For control conditions containing no Acr plasmid, 110 ng of a pCMV-EGFP plasmid was utilized; for non-targeting sgRNA/crRNA conditions, 30 ng of an empty U6 promoter plasmid was used. For titration experiments, cells were transfected with 70 ng of nuclease, 30 ng sgRNA/crRNA, varying amounts of acr expression and DNA stuffer plasmids totaling 96.5 ng (0.5 ng Acr with 96 ng stuffer; 2.75 ng acr with 93.75 ng stuffer; 16 ng acr with 80.5 ng stuffer), and 1.17 μL of TransIT-X2 (Mirus Bio) in 20 μl Opti-MEM. DNA stuffer plasmids were an orthogonal and incompatible pCAG-MbCas12a expression plasmid. Genomic DNA was harvested approximately 72 hours post-transfection by suspending cells in 100 μL of lysis buffer (20 mM Hepes pH 7.5, 100 mM KCl, 5 mM MgCl2, 5% glycerol, 25 mM DTT, 0.1% Triton X-100, and 30 ng/μL Proteinase K (NEB)), followed by incubation at 65 °C for 6 minutes and 98 °C for 2 minutes. All experiments were performed with at least 3 independent biological replicates.
Assessment of genome editing in human cells
Genome modification was measured by next-generation sequencing using a 2-step PCR-based Illumina library construction method. Briefly, genomic regions were initially amplified using Q5 High-Fidelity DNA Polymerase (NEB), human cell lysate containing ~100 ng of genomic DNA, and gene-specific round 1 primers (Supplementary Table 4c). PCR products were purified using paramagnetic beads as previously described41 and diluted 1:100 to serve as template for a second round of PCR using Q5 High-Fidelity Polymerase and primers encoding Illumina barcodes and adapter sequences (Supplementary Table 4c). PCR products were purified prior to quantification (via Qiagen QIAxcel electrophoresis), normalization, and pooling. Final libraries were quantified by qPCR (Illumina Library qPCR Quantification Kit, KAPA Biosystems) and sequenced on a MiSeq sequencer using a 300-cycle v2 kit (Illumina). Genome editing activities were determined from the sequencing data using CRISPResso2 (Clement et al., 2019) with commands --min_reads_to_use_region 100, -w 10, and for certain sequencing data sets --ignore_substitutions.
Data Availability
All data generated or analysed during this study are included in this published article and its Extended Data, Source Data, and Supplementary Information files. All relevant accession codes are available in Table 1.
Table 1.
Summary of Anti-CRISPRs reported and inhibition activity.
| Anti-CRISPR | Strain | Accession Numbera | Inhibits Cas9?b | Inhibits SpyCas9 in P.aeruginosa heterologous system?b | Inhibits EfaCas9 in E. faecalis native system?b | Inhibits Cas9 orthologs in mammalian cells system?b | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CRISPR1 (Spy-like) | CRISPR3 (Sau-like) | Spy | Sau | Sth1 | Nme | ||||||
| IIA16-Lmo | Listeria monocytogenes | WP_061665674.1 | Yes | yes | yes | yes | yes | yes | yes | no | |
| IIA16-Efa | Enterococcus faecalis | WP_025188019.1 | Yes | ND | ND | ND | yes | yes | yes | yes | |
| IIA17-Efa | Enterococcus faecalis | WP_002401839.1 | Yes | yes | yes | yes | no | no | no | yes | |
| IIA17-Sga | Streptococcus gallolyticus | WP_074626943.1 | Yes | yes | yes | yes | yes | yes | no | yes | |
| IIA18-Sma | Streptococcus macedonicus | WP_099390844.1 | Yes | yes | ND | ND | no | no | no | no | |
| IIA18-Sga | Streptococcus gallolyticus | WP_074627086.1 | Yes | ND | ND | ND | yes | no | no | no | |
| IIA19-Ssim | Staphylococcus simulans | WP_107591702.1 | Yes | yes | yes | yes | no | no | no | no | |
| IIA19-Spse | Staphylococcus pseudintermedius | WP_100006909.1 | Yes | ND | ND | ND | yes | no | no | no | |
List of accession numbers for AcrIIA16–19 proteins reported in this study
summary of their inhibition activity against Cas9 orthologs
ND = not determined
Extended Data
Extended Data Fig. 1. Schematic of acr loci and lethal self-genome cleavage assay.
a, Full schematic of acr loci with relevant neighboring genes displayed. b, Schematic of SpyCas9 in P. aeruginosa programmed to cause lethal self-genome cleavage to assess bacterial survival in the presence of AcrIIA proteins. CRISPR strength is determined by titrating levels of IPTG, which induces expression of sgRNA targeting the chromosomal phzM gene from a multicopy plasmid. OD600 measurements are represented as the mean of three biological replicates ± SD.
Extended Data Fig. 2. Anti-CRISPR distribution in integrative mobile genetic elements across bacterial taxa.
Phylogenetic analysis based on acrIIA16–19 homologs (panels a to d, respectively) reconstructed from a midpoint rooted minimum-evolution of full-length protein sequences identified following an iterative PSI-BLASTp search, see methods for details. Number of genomes included to construct each tree for acrIIA16-19 are seventy, twenty-six, eighty-four and seventeen respectively. Branches are labeled with species name and colored according to species class (see legend). Species for which AcrIIA homologs have been tested in this study are shown in bold.
Extended Data Fig. 3. AcrIIA enhance conjugation-mediated horizontal gene transfer in E. faecalis, related to Fig 2. a, b,
Mating outcomes during plasmid conjugation of a targeted plasmid from donor to recipient cells where indicated acrIIA genes are (a) pre-expressed in recipient cells, or (b) encoded on conjugating plasmid. Data displayed as 10-fold colony serial dilution spots of donor, recipient or transconjugant cells on selective antibiotic plates. Mating assays were performed in biological triplicate and produced similar outcomes. c, Schematic of E. faecalis conjugation of protospacer and acrIIA-bearing plasmid transferring into CRISPR-defective recipients. For CRISPR1, the bona fide AcrIIA4 is utilized to suppress CRISPR-targeting, and a ΔCas9 strain from previously reported work is used for CRISPR3 (Price et al., 2016). Red * denotes plasmids that have lost conjugation ability. Mating assays were performed in biological duplicate or triplicate and produced similar outcomes.
Extended Data Fig. 4. AcrIIA16–19 biochemical analysis, related to Figs 3 and 4.
a, Coomassie-stained polyacrylamide gel showing GST-tagged AcrIIA proteins (IIA4 37kD, IIA16Lmo 50kD, IIA17Sga 39kD, IIA18Sma 48kD and IIA19Ssim 42kD) purified from E. coli by elution from Glutathione Sepharose columns. Visible bands at different sizes are co-purifying proteins from E. coli. Data shown are representative of two independent experiments. b, Coomassie-stained polyacrylamide gel showing co-immunoprecipitation of Acr proteins with Myc-tagged sgRNA-bound SpyCas9 pulled down from P. aeruginosa. Data shown are representative of two independent experiments. c, d, Uncropped versions of both Myc and GST pulldowns from Figs 4a and c, displaying all fragments of (c) sgRNA-bound SpyCas9, or (d) Apo- SpyCas9 without sgRNA present. Data shown are representative of two independent experiments.
Extended Data Fig. 5. Acr inhibition activity in human cells tested against different Cas9 orthologs, related to Fig 5.
Reported Acr proteins in this study and from previous works tested for inhibition of genome editing activities of Sth1Cas9, Sth3Cas9, Nme2Cas9 and CjeCas9 (a-d, respectively). Editing efficiencies against endogenous genes in HEK 293T cells were assessed by targeted sequencing and quantified as the percentage of reads containing a nuclease-induced alteration; the no-Acr condition contains an EGFP expression plasmid; the NT control includes an empty U6 expression plasmid. Percent reads modified are represented as the mean of three biological replicates ± SD.
Supplementary Material
Acknowledgements
We thank Kelli L. Palmer (UT Dallas) and Gary M. Dunny (University of Minnesota) for providing all E. faecalis strains, vector and protocols. We thank Matthew Johnson for his assistance in executing PlasFlow, Nicole Marino for her profound wisdom and comedic wit, and all members of the Bondy-Denomy Lab for their continuous support and thoughtful discussions contributing to this study. Bondy-Denomy Lab was supported by the UCSF Program for Breakthrough Biomedical Research funded in part by the Sandler Foundation, the Searle Fellowship, the Vallee Foundation, an NIH Director’s Early Independence Award DP5-OD021344, and R01GM127489. Research in the Kleinstiver Lab is supported by NIH R00-CA218870, an A.S.G.C.T. Career Development Award, and the Margaret Q. Landenberger Research Foundation.
Footnotes
Competing Interests
J.B.-D. is a scientific advisory board member of SNIPR Biome and Excision Biotherapeutics, and a scientific advisory board member and co-founder of Acrigen Biosciences. B.P.K. is an inventor on various patents and patent applications that describe gene editing and epigenetic editing technologies, is a consultant for Avectas Inc., and an advisor for Acrigen Biosciences.
References
- 1.Palmer KL, Kos VN & Gilmore MS Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr. Opin. Microbiol 13, 632–639 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waldor MK & Mekalanos JJ Lysogenic Conversion by a Filamentous Phage Encoding Cholera Toxin. Science (80-. ). 272, 1910–1914 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Koonin EV Viruses and mobile elements as drivers of evolutionary transitions. Philosophical Transactions of the Royal Society B: Biological Sciences vol. 371 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Labrie SJ, Samson JE & Moineau S Bacteriophage resistance mechanisms. Nat. Rev. Microbiol 8, 317–327 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Price VJ, Huo W, Sharifi A & Palmer KL CRISPR-Cas and Restriction-Modification Act Additively against Conjugative Antibiotic Resistance Plasmid Transfer in Enterococcus faecalis. mSphere 1, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edgar R & Qimron U The Escherichia coli CRISPR system protects from λ lysogenization, lysogens, and prophage induction. J. Bacteriol 192, 6291–6294 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y et al. Processing-Independent CRISPR RNAs Limit Natural Transformation in Neisseria meningitidis. Mol. Cell 50, 488–503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makarova KS et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol 13, 722–736 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garneau JE et al. The CRISPR/cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Clark DP & Pazdernik NJ Horizontal Gene Transfer in Molecular Biology (Elsevier Inc., 2013). [Google Scholar]
- 11.Casjens S Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol 49, 277–300 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Bondy-Denomy J, Pawluk A, Maxwell KL & Davidson AR Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429–432 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Y et al. Diverse Mechanisms of CRISPR-Cas9 Inhibition by Type IIC Anti-CRISPR Proteins. Mol. Cell 74, 296–309.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thavalingam A et al. Inhibition of CRISPR-Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2. Nat. Commun 10, 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bondy-Denomy J et al. Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins. Nature 526, 136–139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrington LB et al. A Broad-Spectrum Inhibitor of CRISPR-Cas9. Cell 170, 1224–1233.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong L et al. An anti-CRISPR protein disables type V Cas12a by acetylation. Nat. Struct. Mol. Biol 26, 308–314 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Knott GJ et al. Broad-spectrum enzymatic inhibition of CRISPR-Cas12a. Nat. Struct. Mol. Biol 26, 315–321 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rauch BJ et al. Inhibition of CRISPR-Cas9 with Bacteriophage Proteins. Cell 168, 150–158.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hynes AP et al. An anti-CRISPR from a virulent streptococcal phage inhibits Streptococcus pyogenes Cas9. Nat. Microbiol 2, 1374–1380 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Hynes AP et al. Widespread anti-CRISPR proteins in virulent bacteriophages inhibit a range of Cas9 proteins. Nat. Commun 9, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uribe RV et al. Discovery and Characterization of Cas9 Inhibitors Disseminated across Seven Bacterial Phyla. Cell Host Microbe 25, 233–241.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Forsberg KJ et al. Functional metagenomics-guided discovery of potent cas9 inhibitors in the human microbiome. Elife 8, 1–32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pawluk A et al. Naturally Occurring Off-Switches for CRISPR-Cas9. Cell 167, 1829–1838.e9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang F et al. Temperature-Responsive Competitive Inhibition of CRISPR-Cas9. Mol. Cell 73, 601–610.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L, Yin M, Wang M & Wang Y Phage AcrIIA2 DNA Mimicry: Structural Basis of the CRISPR and Anti-CRISPR Arms Race. Mol. Cell 73, 611–620.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Dong D et al. Structural basis of CRISPR–SpyCas9 inhibition by an anti-CRISPR protein. Nature 546, 436 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Shin J et al. Disabling Cas9 by an anti-CRISPR DNA mimic. Sci. Adv 3, 1–9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia B et al. Anti-CRISPR AcrIIA5 Potently Inhibits All Cas9 Homologs Used for Genome Editing. Cell Rep. 29, 1739–1746.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borges AL et al. Bacteriophage Cooperation Suppresses CRISPR-Cas3 and Cas9 Immunity. Cell 174, 917–925.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osuna BA et al. Listeria phages induce Cas9 degradation to protect lysogenic genomes. bioRxiv (2019) doi: 10.1101/787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hullahalli K, Rodrigues M & Palmer KL Exploiting CRISPR-Cas to manipulate Enterococcus faecalis populations. Elife 6, 1–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trasanidou D et al. Keeping crispr in check: diverse mechanisms of phage-encoded anti-crisprs. FEMS Microbiol. Lett 366, 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang F, Song G & Tian Y Anti‐CRISPRs: The natural inhibitors for CRISPR‐Cas systems. Anim. Model. Exp. Med 1–7 (2019) doi: 10.1002/ame2.12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palmer KL & Gilmore MS Multidrug-resistant enterococci lack CRISPR-cas. MBio 1, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hullahalli K, Rodrigues M, Nguyen UT & Palmer K An attenuated CRISPR-cas system in enterococcus faecalis permits DNA acquisition. MBio 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seamon KJ, Light YK, Saada EA, Schoeniger JS & Harmon B Versatile High-Throughput Fluorescence Assay for Monitoring Cas9 Activity. Anal. Chem 90, 6913–6921 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Hoang TT, Kutchma AJ, Becher A & Schweizer HP Integration-proficient plasmids for Pseudomonas aeruginosa: Site- specific integration and use for engineering of reporter and expression strains. Plasmid 43, 59–72 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Choi KH, Kumar A & Schweizer HPA 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: Application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64, 391–397 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Bhardwaj P, Ziegler E & Palmer KL Chlorhexidine induces VanA-type Vancomycin resistance genes in enterococci. Antimicrob. Agents Chemother 60, 2209–2221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kleinstiver BP et al. Engineered CRISPR–Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nature Biotechnology vol. 37 276–282 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its Extended Data, Source Data, and Supplementary Information files. All relevant accession codes are available in Table 1.
Table 1.
Summary of Anti-CRISPRs reported and inhibition activity.
| Anti-CRISPR | Strain | Accession Numbera | Inhibits Cas9?b | Inhibits SpyCas9 in P.aeruginosa heterologous system?b | Inhibits EfaCas9 in E. faecalis native system?b | Inhibits Cas9 orthologs in mammalian cells system?b | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CRISPR1 (Spy-like) | CRISPR3 (Sau-like) | Spy | Sau | Sth1 | Nme | ||||||
| IIA16-Lmo | Listeria monocytogenes | WP_061665674.1 | Yes | yes | yes | yes | yes | yes | yes | no | |
| IIA16-Efa | Enterococcus faecalis | WP_025188019.1 | Yes | ND | ND | ND | yes | yes | yes | yes | |
| IIA17-Efa | Enterococcus faecalis | WP_002401839.1 | Yes | yes | yes | yes | no | no | no | yes | |
| IIA17-Sga | Streptococcus gallolyticus | WP_074626943.1 | Yes | yes | yes | yes | yes | yes | no | yes | |
| IIA18-Sma | Streptococcus macedonicus | WP_099390844.1 | Yes | yes | ND | ND | no | no | no | no | |
| IIA18-Sga | Streptococcus gallolyticus | WP_074627086.1 | Yes | ND | ND | ND | yes | no | no | no | |
| IIA19-Ssim | Staphylococcus simulans | WP_107591702.1 | Yes | yes | yes | yes | no | no | no | no | |
| IIA19-Spse | Staphylococcus pseudintermedius | WP_100006909.1 | Yes | ND | ND | ND | yes | no | no | no | |
List of accession numbers for AcrIIA16–19 proteins reported in this study
summary of their inhibition activity against Cas9 orthologs
ND = not determined