In late December 2019, a cluster of pneumonia cases of unknown cause was first noted in human patients in the city of Wuhan, Hubei Province, China.1 The patients with this emerging disease presented with clinical signs of viral pneumonia similar to those observed in severe acute respiratory syndrome (SARS).1 Chinese scientists, including those in the Shanghai Public Health Clinical Center, the Chinese Center for Disease Control and Prevention, and the Wuhan Institute of Virology, immediately took actions to probe for the obligated emerging pathogen(s) through next-generation sequencing. A novel Betacoronavirus was soon identified, and its genome sequence was first made accessible to the world on January 12, 2020.2 On February 11, 2020, this novel coronavirus was named SARS-coronavirus 2 (SARS-CoV-2) by the International Committee on Taxonomy of Viruses and the associated disease was named coronavirus disease 2019 (COVID-19) by the World Health Organization.
COVID-19 is highly contagious. It has been reported that SARS-CoV-2 had an estimated basic case reproduction number (R 0) as high as 3.15 before the implementation of the emergency response on January 23, 2020, in mainland China.3 SARS-CoV-2 viral load in the upper respiratory system is highly dynamic along the progression of COVID-19, and it exhibits significant variation in different patients,4 with peaks on the order of 108 viral genome copies/mL in throat swabs and 1011 copies/mL in sputum samples.5 SARS-CoV-2 also showed differential tissue tropism in different infected individuals. Viral load could be higher in fecal samples than in nasopharyngeal swab samples in some cases, likely because of oral transmission.6 The current standard operating procedure in clinical diagnosis uses pharyngeal swabbing to collect samples from the upper respiratory system. Limitations in preparing viral genetic materials for PCR tests include timing of sample collection and anatomic locations of sampling, which can be further complicated by practical variations in technical operations such as those in shipping, storage, and material/reagent preparation. The genetic sequence of SARS-CoV-2 enabled the rapid development of point-of-care real-time PCR diagnostic tests specific for SARS-CoV-2. PCR tests were used as a criterion standard measure to confirm active SARS-CoV-2 infection. There was, however, a false-negative rate close to 40%7 because of various issues, most of which are associated with sample preparation. This 40% false-negative rate is the rate of PCR testing in diagnosing SARS-CoV-2 infection, but not the false-negative rate of PCR test kits. In a recent external quality assessment program directed by the National Center for Clinical Laboratories in China, SARS-CoV-2 PCR test kits approved by China’s National Medical Products Administration were reported with an overall 95.4% positive test result when the input viral genomic load was 3200 copies/mL. The PCR test kit developed by the Beijing Genomics Institution detected 100% of the positive samples when the input was 3200 copies/mL and 97.1% of those with input level at 640 copies/mL; this kit was approved by the US Food and Drug Administration through emergency use authorization, and it became available for COVID-19 diagnosis in the United States on March 26, 2020.
There is a broad spectrum of COVID-19 symptoms, including fever, dry cough, shortness of breath, sputum production, diarrhea, fatigue, and others, most of which are associated with the respiratory and gastrointestinal systems.8 In mainland China, about 80% of the patients with confirmed infection have mild symptoms and 2.3% have fatal symptoms, whereas 1.2% are asymptomatic (calculated on the basis of data reported before February 11).9 The reported asymptomatic patients here were defined by a positive PCR test result accompanied by a lack of symptoms clinically, as described by patients, measured by doctors, and visualized by imaging, and the patients did not develop any symptoms after a fortnight quarantine. Patients who are still in the incubation period may remain asymptomatic in their upper respiratory system for weeks, although signs of pneumonia might have been developing in their lower respiratory system. There is evidence that infected individuals can be contagious even when they show no symptoms,9 and light or mild cases can develop into severe and even critical cases after a long period of time on account of lack of rest, mental stress, and/or malnutrition.10 An early and accurate diagnosis is extremely critical for containment and disease intervention, as well as for epidemiologic investigation.
Some patients in whom the virus was suspected had a clear history of contacts with confirmed and symptomatic patients but tested negative by PCR multiple times, whereas their symptoms varied from signs of viral pneumonia to mild or even no symptoms. Some patients had a documented history of exposure and showed typical signs of ground glass opacity in computed tomography (CT) scans; these patients were immediately admitted and treated as having COVID-19 with clinical diagnosis in China, with most of them testing positive by PCR and/or antibody assays during disease progression and recovery. Because of variations in the viral load of different individuals, and timing and anatomic locations of sampling for PCR, some infected patients were tested more than once but never had a sample with viral load that passed the PCR detection threshold. These patients with COVID-19 could have fully recovered without any positive PCR test results. Their immune systems, however, kept the footprints of the infection, virus-specific antibodies. By testing for SARS-CoV-2–specific antibodies, it was possible to determine whether the patients in whom the virus was suspected had a history of infection, even if the virus had been cleared after recovery. To obtain a more accurate number of COVID-19 cases, the Chinese Center for Disease Control and Prevention, along with scientists and doctors, developed a standard operating procedure for screening patients in whom the virus was suspected and confirming cases involving patients with infection who showed negative results in PCR tests by using SARS-CoV-2–specific antibody assays. A combination of information on exposure history, clinical presentation visualized on CT scans, presence of viral genes, and presence of virus-specific antibodies was used for diagnosing COVID-19 in mainland China. Of note, in an antibody (IgG) test trial conducted with >600 serum samples collected from >300 patients with “clinically diagnosed” COVID-19, more than 95% of the samples collected 21 to 25 days after disease onset showed strong positive results (An et al, data not shown).
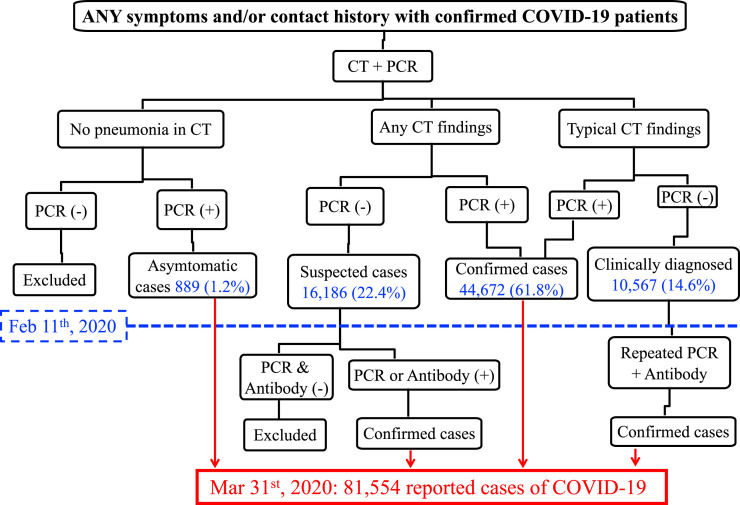
The schematic flowchart delineating the standard operating procedure of the COVID-19 diagnostic process in mainland China is depicted in Fig 1 . In brief, patients in whom COVID-19 is suspected and who have exhibited any COVID-19 symptoms and/or have a confirmed history of exposure are first evaluated by CT scanning and PCR tests for SARS-CoV-2. These patients are then divided into 3 categories: (1) those with no signs of pneumonia on their CT scan; (2) those with atypical signs of lung opacity on their CT scan; and (3) those with typical signs of viral pneumonia ground glass opacity on their CT scan. In these 3 categories, patients who have a positive PCR test result are confirmed as having the infection. In the second category, patients who have atypical signs of lung opacity on their CT scan and a negative test result by PCR will be retested with PCR and antibody assays. In the third category, patients who have a documented history of exposure and have shown signs of typical viral pneumonia on their CT scan are reported as having a “clinically diagnosed case of COVID-19.”7 In February, in Hankou Hospital in the city of Wuhan, an antibody test trial with serum samples collected from more than 300 “clinically diagnosed” patients showed that over 95% of these patients had a history of infection (An et al, data not shown); therefore, it is unlikely that clinical diagnosis would lead to an overestimation of COVID-19 cases in mainland China. On the basis of this scheme of diagnostic process as analyzed and published by the Chinese Center for Disease Control and Prevention, by February 11, 2020, there were 72,314 patients with COVID-19 reported in mainland China, of whom 1.2% were asymptomatic, 61.8% showed signs of pneumonia with a positive PCR result, and 14.6% were clinically diagnosed (Fig 1).10 In early February, mainland China started to report asymptomatic cases of SARS-CoV-2 infection and COVID-19 (infection plus disease symptoms) separately, according to the fourth edition of the Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia. Antibody assays were adopted to retest suspected cases involving patients who tested negative by PCR assay, and by March 31, 2020, a total of 81,554 confirmed cases of COVID-19 in mainland China were reported by the National Health Commission of China.
Fig 1.
Scheme of the COVID-19 diagnostic process in mainland China. In mainland China, patients with any COVID-19 symptoms and/or a documented history of contact with individuals with a confirmed case of COVID-19 were examined with CT scanning and PCR testing. Patients were then divided into 3 categories on the basis of findings in lung imaging. Signs of pneumonia in imaging and a positive PCR test result were used to diagnose and confirm COVID-19. Patients who had a contact history and typical signs of pneumonia in imaging but had a negative PCR test results were clinically diagnosed as having COVID-19 and subsequently retested with PCR and antibody assays. Before February 11, 2020, asymptomatic cases were reported as COVID-19 in mainland China. In early February, the 4th edition of Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia was released in mainland China; it recommended reporting SARS-CoV-2 infection in 2 categories: (1) COVID-19 (cases with confirmed infection and disease symptoms) and (2) asymptomatic infection. Any symptoms, including discomfort reported by patients, disease parameters measured by doctors (eg, high body temperature and low fingertip pulse oximeter reading), and signs of pneumonia in CT images, will define a case as symptomatic. The current reported COVID-19 cases in mainland China include all cases of COVID-19 (cases involving patients with infection plus symptoms) and asymptomatic cases (cases involving patients who never had symptoms) of SARS-CoV-2 infection determined before February 11, 2020.
The goal of this Editorial is to provide a concise and clear flowchart to depict the COVID-19 diagnostic process developed and implemented in mainland China. We hope that this helps our readers better understand how the math was done to determine the number of reported cases of SARS-CoV-2 infection in mainland China. The math that results in the reported number of confirmed cases in mainland China is clearly beyond pneumonia. Despite our general understanding that COVID-19 is an acute respiratory disease caused by a novel coronavirus, clinical diagnosis was often complicated by clinical presentations, individual variations in patients and medical/laboratory operators, and the sensitivity and specificity of the tests. This diagnostic process used in mainland China may not be directly applicable to other areas in the world during the COVID-19 pandemic; however, it exploited current knowledge of epidemiology, disease characteristics, viral genetics, and immunology about COVID-19, and it provides an example of a comprehensive diagnostic approach.
Acknowledgments
We thank Dr Avery August at Cornell University for critical reading and Dr Pengcheng Zhang at the University of California, San Francisco, and Dr Taixue An at Southern Medical University for helpful discussions. Research related to viral pneumonia and lung inflammation in the authors’ laboratories is supported in part by grants from the National Institutes of Health (R56AI146226, R21AI137822, and P20GM130555-6610).
Footnotes
Disclosure of potential conflict of interest: W. Huang received research support from MegaRobo Technologies Corporation. F. Wu declares that she has no relevant conflicts of interest.
References
- 1.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tian H, Liu Y, Li Y, Wu CH, Chen B, Kraemer MUG, et al. An investigation of transmission control measures during the first 50 days of the COVID-19 epidemic in China [e-pub ahead of print]. Science https://doi.org/10.1126/science.abb6105. Accessed March 31, 2020. [DOI] [PMC free article] [PubMed]
- 4.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA https://doi.org/10.1001/jama.2020. Accessed March 11, 2020. [DOI] [PMC free article] [PubMed]
- 7.Novel Coronavirus Pneumonia Emergency Response Epidemiology Team The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41:145–151. [Google Scholar]
- 8.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Z., Song C., Xu C., Jin G., Chen Y., Xu X. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci China Life Sci. 2020;63:706–711. doi: 10.1007/s11427-020-1661-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen S, Zhang Z, Yang J, Wang J, Zhai X, Barnighausen T, et al. Fangcang shelter hospitals: a novel concept for responding to public health emergencies. Lancet https://doi.org/10.1016/S0140-6736(20)30744-3. Accessed April 8, 2020. [DOI] [PMC free article] [PubMed]