Abstract
A20 is an anti-inflammatory protein that is strongly linked to human disease. Here we find that mice expressing three distinct targeted mutations of A20’s ZF7 ubiquitin binding motif uniformly developed digit arthritis that shares features with psoriatic arthritis, while mice expressing point mutations in A20’s OTU or ZF4 motifs did not exhibit this phenotype. Arthritis in A20ZF7 mice required T cells and MyD88, was exquisitely sensitive to tumor necrosis factor (TNF) and interleukin 17A, and persisted in germ-free conditions. A20ZF7 cells exhibited prolonged IKK kinase activity that drove exaggerated transcription of late-phase NF-κB-response genes in vitro and in pre-diseased mouse paws in vivo. In addition, mice expressing double-mutant A20 proteins in A20’s ZF4 and ZF7 motifs died perinatally with multi-organ inflammation. Therefore, A20’s ZF4 and ZF7 motifs synergistically prevent inflammatory disease in a non-catalytic manner.
Introduction
A20, also known as TNFAIP3, is a potent anti-inflammatory protein that is prominently associated with human disease 1. Genome-wide association studies have linked single nucleotide polymorphisms (SNPs) at the TNFAIP3 locus with reduced A20 expression and susceptibility to psoriatic arthritis, psoriasis, rheumatoid arthritis, systemic lupus erythematosus, Crohn’s disease, and other diseases 2. Reduced A20 expression has been described in tissues from diseased patients independently of germline SNPs 3, 4. Finally, emerging studies reveal that individuals born with one mutant allele of TNFAIP3 rapidly develop inflammatory diseases resembling Behcet’s disease, juvenile onset arthritis, early onset inflammatory bowel disease, hepatitis, and other diseases 5, 6, 7, 8. Hence, reduced A20 expression is strongly linked to inflammatory and autoimmune human diseases.
Causal relationships between A20 and disease have been defined in murine models of A20 deficiency. Global deficiency of A20 (i.e., in Tnfaip3−/− mice, herein referred to as A20−/− mice) causes perinatal lethality and widespread tissue inflammation, establishing the profound physiologic importance of this protein 9. A20 is pleiotropically expressed, and deletions of A20 from a number of hematopoietic and non-hematopoietic cell lineages leads to distinct autoimmune and autoinflammatory phenotypes 1,2. Therefore, A20 directly preserves immune homeostasis and prevents disease.
A20’s ability to regulate cellular responses and prevent disease has led to efforts to understand how this protein regulates intracellular signals. A20 contains several ubiquitin (Ub)-dependent biochemical motifs and regulates Ub-dependent signaling. Studies with recombinant A20 proteins reveal that a catalytic cysteine at C103 cleaves unanchored K48-linked Ub chains and anchored K63-linked chains 10, 11. A20’s ZF4 motif binds to K63-linked Ub chains and supports E3 Ub ligase activity 12, 13. A20’s ZF7 motif binds M1-linked Ub chains 14,15. These motifs have been elegantly linked to several signaling functions in vitro 12–20. However, a major conundrum has arisen from the observations that A20−/− mice die perinatally with multi-organ inflammation, while mice bearing strategic point mutations abrogating either A20’s C103 (A20OTU mice) or A20’s ZF4 motif (A20ZF4 mice) survive for up to one year without overt disease 9, 18, 21. In this study, we have used six distinct gene-targeted lines of Tnfaip3 knock-in mice to dissect how A20 prevents disease. We find that non-catalytic Ub binding by A20 plays dominant roles in preventing disease in vivo.
Results
A20’s ZF7 motif prevents distal digit arthritis
To begin to test the role of A20’s ZF7 motif in preserving immune and tissue homeostasis, we used TALEN-directed gene targeting in C57BL/6 derived embryonic stem cells to generate mice expressing dual cysteine to alanine mutations in zinc-coordinating cysteines within ZF7, which we call A20ZF7-CC mice (Fig. 1a). A20ZF7-CC mice were born in Mendelian ratios and survived for at least three months (Extended Data Fig. 1A). We studied cohorts of these mice alongside wild-type mice, as well as mice bearing a cysteine to alanine mutation of A20’s catalytic cysteine, C103 (A20OTU mice), and mice bearing dual cysteine to alanine mutations in A20’s ZF4 motif (A20ZF4 mice)18. All these strains of mice were generated and maintained on inbred C57BL/6 backgrounds in the same mouse room, ensuring genetic and environmental consistency. While wild-type, A20OTU/OTU, and A20ZF4/ZF4 mice were all grossly normal for the first three months of life, A20ZF7-CC/ZF7-CC mice uniformly and spontaneously developed distal digit swelling followed by nail loss (Fig. 1b,c). While 3-week-old A20ZF7-CC/ZF7-CC mice exhibited no disease, most A20ZF7-CC/ZF7-CC mice possessed visibly swollen digits by 6 weeks of age, and disease progressed in all mice until all 20 digits were afflicted by 12 weeks of age (Fig. 1c). Male and female mice developed disease at similar ages and with similar severity (Extended Data Fig. 1b). Distal interphalangeal (DIP) joints were uniformly affected, while more proximate paw joints and other organs were grossly normal through 12 weeks of age (Fig. 1b,c). In addition, the marked difference between arthritic A20ZF7-CC/ZF7-CC mice on one hand and non-arthritic A20OTU/OTU, A20ZF4/ZF4, and wild-type mice on the other indicates that A20’s ZF7 motif prevents disease in a manner that requires neither A20’s C103-mediated deubiquitinase (DUB) function nor its ZF4-mediated Ub binding/E3 ligase functions.
Figure 1: A20’s ZF7 motif prevents arthritis of distal interphalangeal joints.
(a) Schematic of A20 protein (above) and diagram of A20’s ZF7 residues targeted in A20ZF7-CC, A20ZF7-ΔFGN, and A20ZF7-FG knock-in mice (highlighted in red) (below).
(b) Representative rear paws from 12 week old mice of the indicated genotypes. WT=wild-type mice; ZF7=A20ZF7/ZF7 mice; ZF4= A20ZF4/ZF4 mice; and OTU=A20OTU/OTU mice throughout this and subsequent figures. Representative of at least six independent mice.
(c) Longitudinal quantitation of digital disease by numbers of digits with dactylitis (distal digit swelling) and nail loss in mice at the indicated ages (from 4–12 weeks of age). WT mice (black squares); ZF7 mice (red circles); ZF4 mice (blue upright triangles; and OTU mice (inverted green triangles) are shown. Note that only ZF7 mice develop any signs of arthritis.
(d) Histology (H&E) of distal digits from 3-week-old WT and ZF7 mice. Scale bar = 1mm. Representative of four independent mice.
(e) Histology (H&E) of distal digits from indicated genotypes of mice at 3 months of age. Distal digit from ZF7 mouse expanded to reveal epidermal neutrophilic microabscesses (marked with *), cutaneous epidermal hyperplasia and hyperkeratosis (closed triangle), nail loss, as well as periosteal inflammation with bony erosion (open arrow) and distal digit resorption. Scale bar = 1mm. Representative of four independent mice.
(f) Histology of paws from WT and ZF7 mice (left) with boxed area expanded to reveal enthesitis and bone formation (right). Scale bar = 1 mm (left) and 100μM (right). Representative of three independent mice.
(g) Micro-CT of paws from WT and ZF7 mice. Note destructive arthritis in ZF7 mice with distal erosions and resorption (marked with *) and juxta-articular bony hypertrophy of ZF7 mice (marked with arrow). Scale bar = 1 mm. Representative of two independent mice.
The dual cysteine to alanine substitutions in A20’s ZF7 motif should destabilize the structure of ZF7. This ZF7 motif directly interacts with ubiquitin via conserved phenylalanine (Phe) and glycine (Gly) residues 14. To more directly interrogate the importance of ZF7’s ubiquitin binding function versus other potential ZF7 dependent functions, we generated a second mouse line lacking these Phe and Gly residues along with the neighboring Asn, A20ZF7-ΔFGN mice (Fig. 1a). Similarly to A20ZF7-CC/ZF7-CC mice, A20ZF7-ΔFGN/ZF7-ΔFGN mice uniformly developed distal arthritis (Extended Data Fig. 1c). As the deletion of the FGN residues in A20ZF7-ΔFGN proteins would alter spacing of the four cysteines that coordinate Zn++ binding and hence potentially perturb ZF7’s stability, we generated a third line of mutant mice expressing point mutations substituting two alanines for the same Phe and Gly residues of A20’s ZF7 motif (A20ZF7-FG mice) (Fig. 1a). These A20ZF7-FG/ZF7-FG mice spontaneously developed arthritis similarly to both A20ZF7-CC/ZF7-CC and A20ZF7-ΔFGN/ZF7-ΔFGN mice (Extended Data Fig. 1c). The similar phenotypes of these three lines of mice suggest that ubiquitin binding is the major physiological function performed by A20’s ZF7 motif and that this shared function prevents arthritis in vivo. Given the similarities between these three distinct A20 ZF7 knock-in mouse lines, A20ZF7-CC/ZF7-CC mice were used for subsequent studies, and will be referred to as A20ZF7/ZF7 mice unless otherwise noted.
Histological analyses of 3-week-old pre-diseased A20ZF7/ZF7 mice revealed that the distal phalanges of these mice were formed normally, ruling out developmental defects in bone formation, and exhibited no significant inflammation (Fig. 1d). By contrast, histological studies of distal digits from 12-week-old A20ZF7/ZF7 mice revealed epidermal hyperplasia, hyperkeratosis, and neutrophilic microabscesses, all hallmarks of psoriasis. These paws also contained subjacent dermal inflammation and bone destruction, together representing onychoperiostitis (Fig. 1e). Histological examination of tendon attachment sites of the distal phalanx revealed enthesitis and bone formation (Fig. 1f). Micro-CT analyses of paws from A20ZF7/ZF7 mice confirmed aberrant bone formation (Fig. 1g). These analyses also confirmed the selective erosive arthritis of distal phalanges and DIP joints with relative sparing of more proximate paw joints (Fig. 1g). Histological surveys of other organs in 8–12-week-old A20ZF7/ZF7 mice revealed mild inflammation in kidneys, livers, colons, and lungs, although no gross disease was observed (Extended Data Fig. 2 and data not shown). Taken together with the clinical pattern of DIP involvement, the histological skin and joint features are characteristic of psoriasis and psoriatic arthritis.
Arthritis requires T cells but not B cells
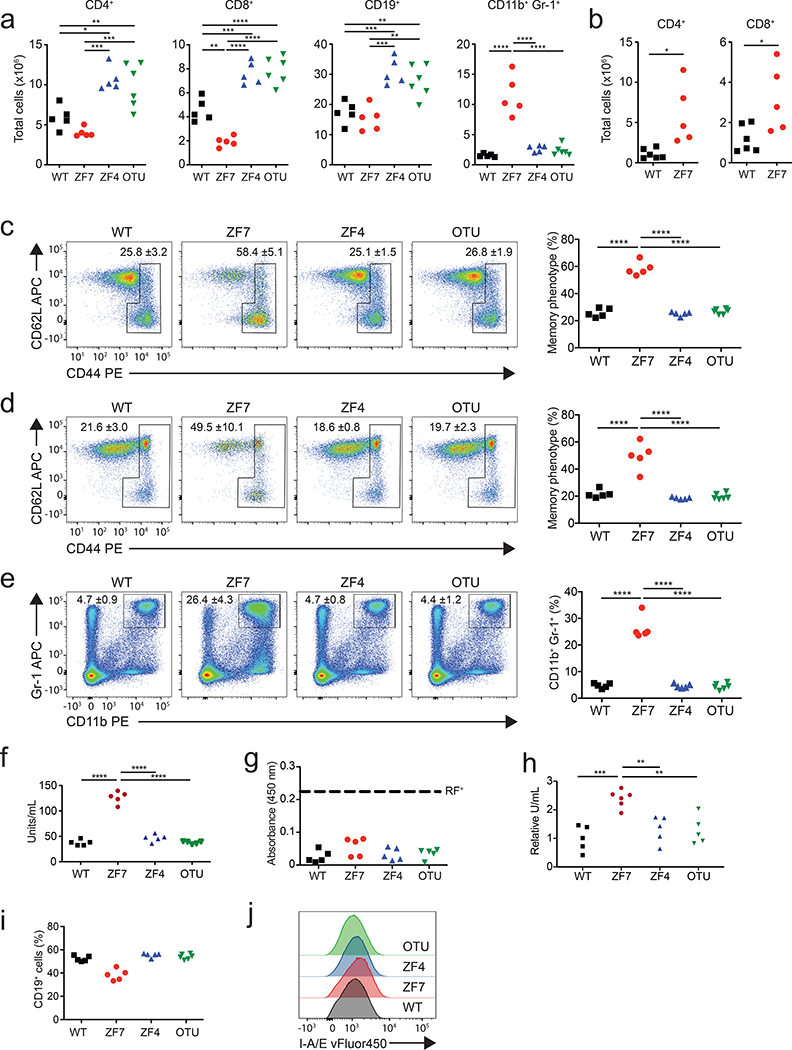
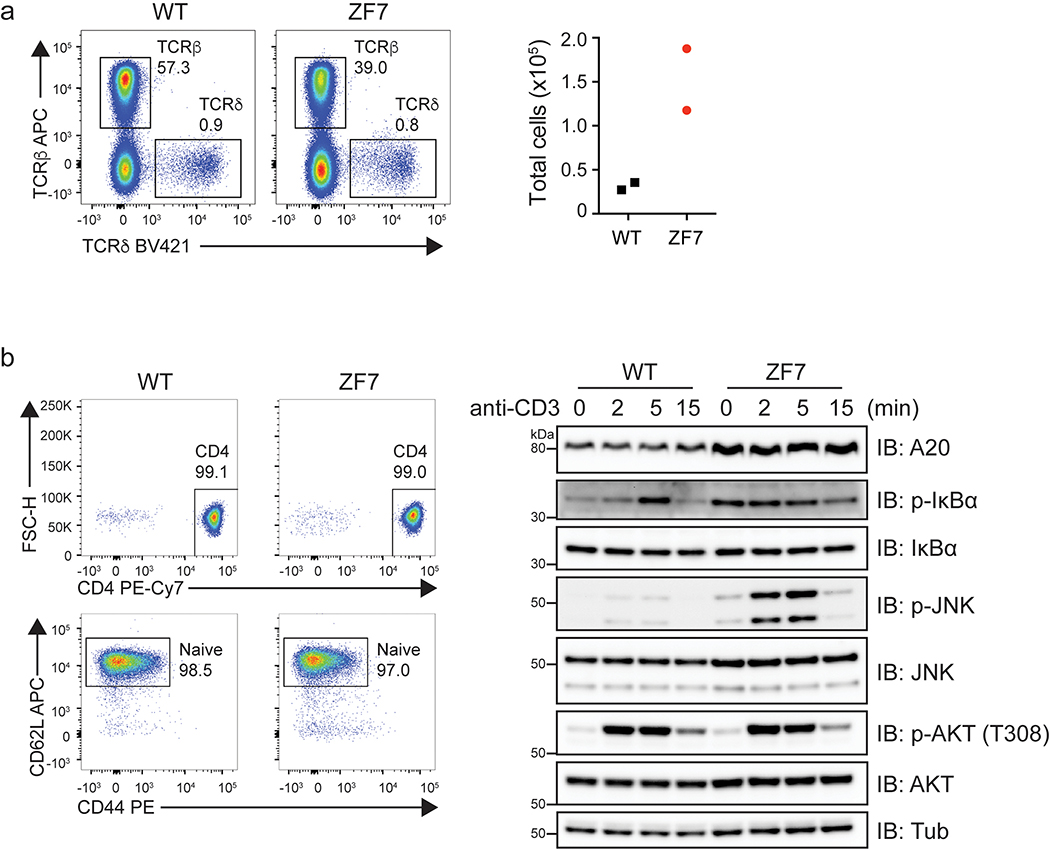
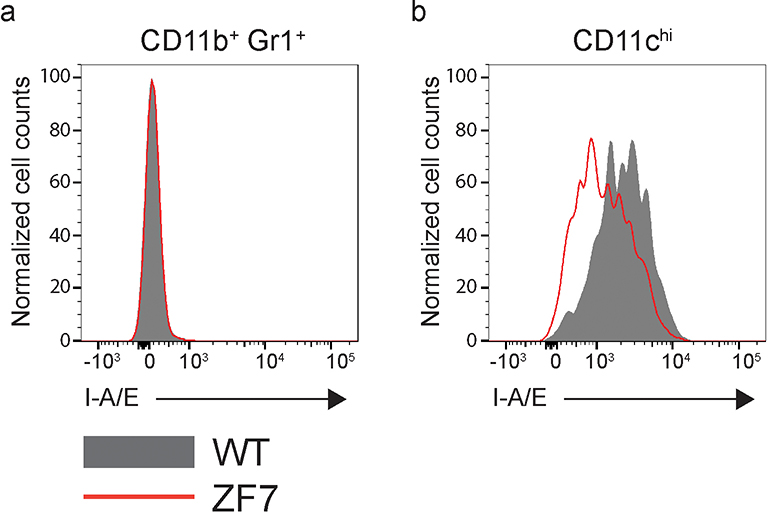
Aberrant lymphocyte functions can cause arthritis in experimental models and have been implicated in human disease. We thus analyzed lymphocyte populations from 3-month-old mice by flow cytometry. A20ZF7/ZF7 mice contained increased numbers of CD4+ and CD8+ T cells, when compared with A20OTU/OTU, A20ZF4/ZF4, and wild-type mice (Fig. 2a,b). T cell expansion was largely comprised of TCRαβ+ cells, although TCRγδ+ cells were also proportionally increased (Extended Data Fig. 3a). The percentages of memory phenotype T cells were markedly increased in A20ZF7/ZF7 mice (Fig. 2c,d), and naïve A20ZF7/ZF7 T cells exhibited increased NF-κB and JNK signaling after TCR stimulation when compared with wild-type T cells (Extended Data Fig. 3b). Hence, A20’s ZF7 motif restricts T cell activation and expansion. The numbers of TCR−CD19−CD11b+Gr-1+ myeloid cells were also expanded in A20ZF7/ZF7 mice, relative to A20OTU/OTU, A20ZF4/ZF4, and wild-type mice (Fig. 2a,e). Neither these CD11b+Gr-1+ cells nor TCR−CD19−CD11chi dendritic cells from A20ZF7/ZF7 mice exhibited elevated expression of MHC II (Extended Data Fig. 4). Inflammatory arthritides in humans can be associated with anti-rheumatoid factor (anti-RF) and/or anti-cyclic citrullinated peptide (anti-CCP) antibodies. We thus assayed serum from the various A20 mutant and control mice, and found that A20ZF7/ZF7 mice expressed increased amounts of anti-CCP and ANA, but not anti-RF antibodies (Fig. 2f–h). These antibodies were present without B cell expansion (Fig. 2a,i), although mild increases in MHCII expression on A20ZF7/ZF7 B cells suggested increased B cell activation (Fig. 2j). Hence, A20ZF7/ZF7 mice exhibit spontaneous immune activation.
Figure 2. Spontaneous immune activation in A20ZF7/ZF7 (ZF7) knock-in mice.
(a) Flow cytometric (FACS) quantitation of absolute numbers of splenic immune subsets from WT (black squares), ZF7 (red circles), ZF4 (blue triangles), and OTU (inverted green triangles) mice. Biologically independent samples (n=5 for WT, ZF7 and ZF4 mice, n=6 for OTU mice). One-way ANOVA with Tukey’s multiple comparison test.
(b) FACS quantitation of absolute numbers of LN T cells from WT (black squares) and ZF7 (red circles) mice. Biologically independent samples compared by unpaired T test, two-tailed (n=6 for WT mice; n=5 for ZF7 mice).
(c-e) FACS quantitation of percentages of memory phenotype CD4+ T cells (c), memory phenotype CD8+ T cells (d), and myeloid cells (e) from WT (black squares), ZF7 (red circles), ZF4 (blue triangles), and OTU (inverted green triangles) mice. Representative FACS plots shown on left; quantitation from multiple mice shown in graphs on right. Mean ± SD of biologically independent samples (n=5 for WT, ZF7 and ZF4 mice, n=6 for OTU mice). One-way ANOVA with Tukey’s multiple comparison test.
(f-h) ELISA of serum anti-CCP antibodies (f), rheumatoid factor (RF) (g), and anti-nuclear antibodies (h) from 12 week old mice of the indicated genotypes (n=6 mice for each genotype). Dashed line shows threshold of positive RF assay. One-way ANOVA with Tukey’s multiple comparison test.
(i) FACS quantitation of percentages of splenic B cells. Independent mice indicated as in panels (f-h) (n=5 for WT, ZF7 and ZF4 mice, n=6 for OTU mice).
(j) FACS (histogram) analysis of MHCII expression of splenic B cells.
In all assays, statistical significance indicated: * p < 0.05, ** p < 0.01, *** p < 0.005; **** p< 0.0001.
To test the potential roles of adaptive lymphocytes and/or autoantibodies in causing arthritis in A20ZF7/ZF7 mice, we interbred these mice with lymphocyte deficient Rag1–/– mice. The uniform incidence of arthritis in A20ZF7/ZF7 mice, with gross digit swelling leading to loss of nails, allowed robust longitudinal comparisons of these compound mutant mice along with control mice. No A20ZF7/ZF7 Rag1–/– mice developed arthritis through 12 weeks of age, and histological studies of paws from these mice revealed normal joints (Fig. 3a–c). Thus, adaptive lymphocytes are essential for the arthritis in A20ZF7/ZF7 mice. To test the potential roles of B cells and/or autoantibodies in causing arthritis, we interbred A20ZF7/ZF7 mice with μMT mice. B cell-deficient A20ZF7/ZF7 μMT mice uniformly developed digital arthritis with similar kinetics to B cell-competent A20ZF7/ZF7 mice (Fig. 3b,d). Histological studies of paws from A20ZF7/ZF7 μMT mice revealed that the enthesitis and onychoperiostitis observed in A20ZF7/ZF7 mice were preserved in A20ZF7/ZF7 μMT mice (Fig. 3b,d). Hence, while B cells and autoantibodies do not influence arthritis incidence or severity in A20ZF7/ZF7 mice, T cells are absolutely required.
Figure 3. Arthritis in A20ZF7/ZF7 mice requires T cells but not B cells, and requires MyD88 but not TRIF dependent signals.
Functional interrogation of lymphocytes and TLR signaling in arthritis of A20ZF7/ZF7 (ZF7) mice. (a-d) Longitudinal quantitation of digital disease by numbers of digits with dactylitis (distal digit swelling) and nail loss (graphs in left panels), representative gross (middle panels) and histology (H&E) (right panels) of the following genotypes of mice at 12 weeks of age: (a) WT (n=6), (b) ZF7 (n=6), (c) ZF7 RAG–/– (n=7), (d) ZF7 μMT–/– (n=6). Scale bar = 1mm.
(e, f) ELISA of TNF (e) and IL-6 (f) secretion from LPS stimulated BMDMs from WT (black squares), ZF7 (red circles), ZF4 (blue triangles), and OTU (green inverted triangles) mice. Biologically independent samples (n=3). Results are representative of 3 independent experiments. One-way ANOVA with Tukey’s multiple comparison test. * p < 0.05, ** p < 0.01, *** p < 0.005; **** p< 0.0001.
(g, h) Longitudinal quantitation of digital disease by numbers of digits with dactylitis and nail loss (graphs in left panels) followed by representative gross (middle panels) and histology (H&E) (right panels) of the indicated genotypes of mice at 12 weeks of age: (g) ZF7 Myd88–/– (n=5), (h) ZF7 Triflps/lps2 (n=7). Scale bar = 1mm.
Arthritis requires innate immune signals
Innate signals in myeloid cells trigger both inflammatory myeloid responses as well as T cell activation, and A20 restricts both MyD88- and TRIF-dependent Toll-like receptor (TLR) responses 10, 22, 23, 24. Accordingly, we tested whether A20’s ZF7 motif is required for A20’s ability to restrict TLR responses. Bone marrow-derived macrophages (BMDMs) from A20ZF7/ZF7 mice secreted greater amounts of TNF and interleukin 6 (IL-6) than BMDMs from A20OTU/OTU, A20ZF4/ZF4, and wild-type mice after stimulation with lipopolysaccharide (LPS) (Fig. 3e,f). LPS-induced signals engage both MyD88 and TRIF adaptors. We thus tested whether arthritis in A20ZF7/ZF7 mice requires MyD88 or TRIF by interbreeding A20ZF7/ZF7 mice with Myd88–/– and Triflps2/lps2 mice. Compound mutant A20ZF7/ZF7Myd88–/– mice failed to develop any signs of arthritis through 12 weeks of age, while A20ZF7/ZF7 Triflps2/lps2 mice developed arthritis similarly to A20ZF7/ZF7 mice (Fig. 3g,h). Hence, MyD88-dependent signals are completely required for the development of arthritis in A20ZF7/ZF7 mice, while TRIF is dispensable.
In addition to stimulating secretion of various cytokines, LPS primes NLRP3 inflammasomes, and inflammasomes have been implicated in the pathogenesis of inflammatory arthritides. NLRP3 inflammasome activation in murine macrophages normally requires a priming signal, e.g., LPS, that induces transcription of Il1b and Nlrp3, followed by a second signal, e.g., extracellular ATP, that activates the inflammasome complex. A20-deficient macrophages secrete mature IL-1β after stimulation with LPS alone, and this aberrant “spontaneous” inflammasome activity requires TRIF engagement and RIP3 25. In addition, mice lacking A20 expression in LysM-Cre expressing myeloid cells develop arthritis that requires NLRP3 26. We first tested whether A20’s ZF7 motif is required for preventing “spontaneous” NLRP3 inflammasome activation. Despite increased LPS induced responses, A20ZF7/ZF7 macrophages failed to secrete IL-1β after stimulation with LPS alone (Fig. 4a). This result contrasted with the IL-1β secretion observed in A20–/– macrophages stimulated in the same conditions (Fig. 4a). Thus, A20ZF7/ZF7 macrophages do not phenocopy A20–/– macrophages, and A20’s ZF7 motif is not required for A20’s ability to prevent spontaneous NLRP3 inflammasome activation. We next tested whether arthritis in A20ZF7/ZF7 mice requires NLRP3-dependent signals by interbreeding A20ZF7/ZF7 mice with Nlrp3–/– mice. Compound mutant A20ZF7/ZF7 Nlrp3–/– mice developed arthritis similarly to A20ZF7/ZF7 mice (Fig. 4b). Hence, NLRP3 inflammasomes are dispensable for arthritis in A20ZF7/ZF7 mice.
Figure 4. A20’s ZF7 motif prevents IL-17 and TNF dependent arthritis.
Functional contributions of NLRP3 inflammasomes, IL-17, commensal flora, and TNF in arthritis pathophysiology of A20ZF7/ZF7 mice. (a) ELISA of IL-1β secretion from BMDMs of the indicated genotypes (“KO” = A20–/– cells, grey diamonds) after stimulation with LPS alone (without ATP). ND = none detected. Biologically independent samples (n=3). Results are representative of 3 independent experiments. One-way ANOVA with Tukey’s multiple comparison test.
(b) Longitudinal quantitation of digital disease by numbers of digits with dactylitis and nail loss followed by representative gross and histology (H&E) of 12 week old ZF7 Nlrp3–/– mice (n=8).
(c) FACS quantitation of IL-17+ CD4+ cells in lymph nodes of 12 week old WT, ZF7, ZF4, and OTU mice. Biologically independent samples (n=3), Sidak’s multiple comparison test, one-way ANOVA. ** = p < 0.01.
(d) Longitudinal quantitation of digital arthritis in ZF7 mice at the indicated ages when treated with isotype control antibody (red circles, n=5) or anti-IL-17A antibody (black triangles, n=4) between 3–11 weeks of age.
(e) FACS quantitation of lymph node IL-17+ CD4+ cells in 12 week old WT and ZF7 mice housed in germ-free conditions. Biologically independent samples (n=3 for WT mice; n=6 for ZF7 mice). unpaired T test, two-tailed. * = p < 0.05.
(f) Representative gross and histological images of 12 week old germ-free WT and ZF7 mice. Scale bar = 1mm. Representative of 4 independent mice.
(g) Longitudinal quantitation of digital disease by numbers of digits with dactylitis (distal digit swelling) and nail loss in ZF7 (red circles, n=10) and WT (black triangles, n=9) mice housed in germ free conditions.
(h) Longitudinal quantitation of digital disease by numbers of digits with dactylitis and nail loss in heterozygous ZF7 Tnf+/– mice at the indicated ages in SPF conditions (n=4).
(i) Longitudinal quantitation of digital disease by numbers of digits with dactylitis and nail loss followed by representative gross and histology (H&E) of 12 week old homozygous ZF7 Tnf–/– (n=7) in SPF conditions. Scale bar = 1mm.
Arthritis requires IL-17A but not commensal flora
IL-17-targeted therapies are effective in many psoriatic arthritis patients, and CD4+ helper type-17 (TH17) cells can elaborate IL-17 and drive synoviocyte proliferation and joint inflammation 27, 28. As our findings with A20ZF7/ZF7 Rag1–/– and A20ZF7/ZF7 μMT compound mutant mice indicate that T cells are required for arthritis in A20ZF7/ZF7 mice, we quantitated the numbers of IL-17 expressing T cells in A20ZF7/ZF7 and control mice. These studies revealed that A20ZF7/ZF7 mice possess markedly increased numbers of CD4+ IL-17 expressing cells when compared with A20OTU/OTU, A20ZF4/ZF4, and wild-type mice (Fig. 4c). CD4+ IFN-γ+ TH1 cells and CD25+ FoxP3+ regulatory T were also expanded in A20ZF7/ZF7 mice (Extended Data Fig. 5). To interrogate the potential functional importance of IL-17 to arthritis development in A20ZF7/ZF7 mice, we injected either antibodies specific for IL-17A or isotype control antibodies into A20ZF7/ZF7 mice. We injected antibodies weekly between 3 and 11 weeks of age, and monitored the A20ZF7/ZF7 mice for development of gross arthritis until 12 weeks of age. These experiments revealed that neutralization of IL-17A completely prevented arthritis in A20ZF7/ZF7 mice (Fig. 4d).
Commensal microbial organisms such as segmented filamentous bacterium (SFB) drive the expansion of CD4+ TH17 cells and are associated with arthritis 29, 30, 31. We thus tested whether the spontaneous development of arthritis in A20ZF7/ZF7 mice requires commensal bacteria by re-deriving these mice into germ-free conditions. Notably, germ-free A20ZF7/ZF7 mice contained elevated numbers of TH17 cells when compared to germ-free wild-type mice (Fig. 4e). Hence, A20’s ZF7 motif limits TH17 cell expansion in both the presence and absence of commensal flora. We next monitored the development of arthritis in germ-free A20ZF7/ZF7 mice. Germ-free wild-type mice remained free of arthritis. Remarkably, germ-free A20ZF7/ZF7 mice all developed arthritis between 5–12 weeks of age (Fig. 4f,g). Paws from germ-free A20ZF7/ZF7 mice also exhibited histological pathology resembling SPF A20ZF7/ZF7 mice (Fig. 4f). These findings suggest that A20’s ZF7 motif restricts TH17 cell expansion and arthritis in a commensal-independent manner.
Arthritis is exquisitely sensitive to TNF
Anti-TNF therapies are effective in many patients with inflammatory arthritides 32, 33. While A20 restricts TNF signaling in vitro, elimination of TNF does not prevent spontaneous disease in A20–/– mice or A20Flox LysM-Cre mice 10, 34. We thus tested the potential role of TNF in the arthritis observed in A20ZF7/ZF7 mice by interbreeding these mice with Tnf–/– mice. Heterozygous deficiency of TNF abrogated the incidence and severity of arthritis in A20ZF7/ZF7 mice, as most A20ZF7/ZF7 Tnf+/– mice were disease-free while a small fraction developed disease in 1–2 digits by 12 weeks of age (Fig. 4h). Moreover, homozygous TNF deficiency completely eliminated arthritis in A20ZF7/ZF7 Tnf–/– mice (Fig. 4i). Hence, arthritis in A20ZF7/ZF7 mice is exquisitely TNF dependent. More broadly, these results provide the first evidence that TNF drives A20-dependent disease in vivo.
A20’s ZF7 motif restricts late phase NF-κB response genes
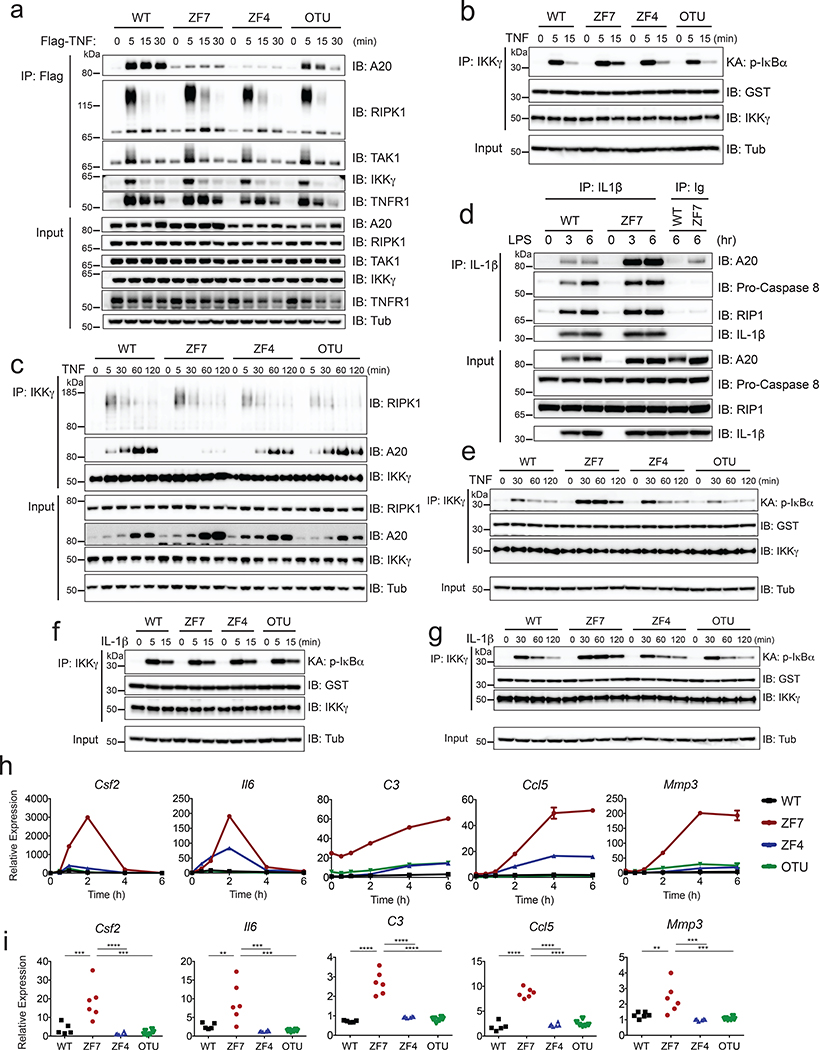
The exquisite sensitivity of arthritis in A20ZF7/ZF7 mice to TNF dosage could reflect aberrant cellular responses to TNF. We thus generated murine embryonic fibroblasts (MEFs) from the various A20 knock-in mice and compared TNF responses of these cells. We performed signaling studies using cells that were pre-incubated with TNF at 4 οC, washed away from soluble TNF, and then warmed to 37 οC to trigger a synchronous and transient TNF stimulus. This approach affords more precise comparison of signaling kinetics 35. We analyzed ligand-engaged TNF receptor complexes by stimulating cells with FLAG-TNF and immunoprecipitating with anti-FLAG antibodies. These experiments revealed that recruitment of A20ZF7 proteins to FLAG-TNF was decreased compared to wild-type and A20OTU proteins (Fig. 5a). TNF-associated RIP1 ubiquitination was modestly increased in A20ZF7/ZF7 cells compared to wild-type cells (Fig. 5a). IKK kinase activity was similarly increased in A20ZF7/ZF7 cells at 15 min (Fig. 5b). Thus, consistent with recent studies conducted with heterologous ZF7 mutant constructs 19, 20, A20’s ZF7 motif supports A20’s recruitment to and regulation of acute TNFR signaling complexes.
Figure 5. A20’s ZF7 motif (versus C103 or ZF4 motifs) selectively restricts IKK activity.
(a) TNFR complex formation assayed after synchronous pulse stimulation of MEFs of the indicated genotypes with FLAG-TNF. Cells were pre-incubated with FLAG-TNF at 4 °C, washed, and then warmed to 37 °C for the indicated times as described in METHODS. Lysates from cells were immunoprecipitated with anti-FLAG and immunoblotted for the indicated proteins. Input levels of indicated proteins in whole cell lysates are shown below.
(b) Acute IKK kinase activity of the indicated genotypes of cells after synchronized TNF stimulation as in (a). Lysates from cells were immunoprecipitated with anti-IKKγ antibody, incubated with GST-IκBα, and immunoblotted for phospho-IκBα at the indicated times after warming cells to 37 °C. GST and IKKγ expression in IPs are shown as IP control. Whole cell lysate expression of tubulin (input) are shown as loading control.
(c) IKK complex analyses of the indicated genotypes of cells between 5 min and 2 h after synchronized TNF stimulation. Lysates from cells were immunoprecipitated with anti-IKKγ antibody and immunoblotted for the indicated proteins at the indicated times after warming cells to 37 °C. Whole cell lysate expression of proteins (input) are shown below as controls.
(d) pro-IL-1β complex analyses of LPS stimulated BMDMs from WT and ZF7 mice. Lysates from cells were immunoprecipitated with anti-IL-1β antibody and immunoblotted for the indicated proteins at 3 and 6 h after LPS stimulation. Whole cell lysate expression of proteins (input) shown below as controls.
(e) IKK kinase assay of TNF stimulated MEFs of the indicated genotypes as in (B) assessed at the indicated (longer) time points.
(f and g) IKK kinase assays of IL-1β stimulated MEFs. Cells of the indicated genotypes were pre-incubated with IL-1β at 4 °C, washed, and warmed for the indicated time periods as described in METHODS. IKK kinase assays performed as in (b, e) above at the indicated short (f) and long (g) time points.
Figures in panels (a-g) are representative of at least 2 independent experiments.
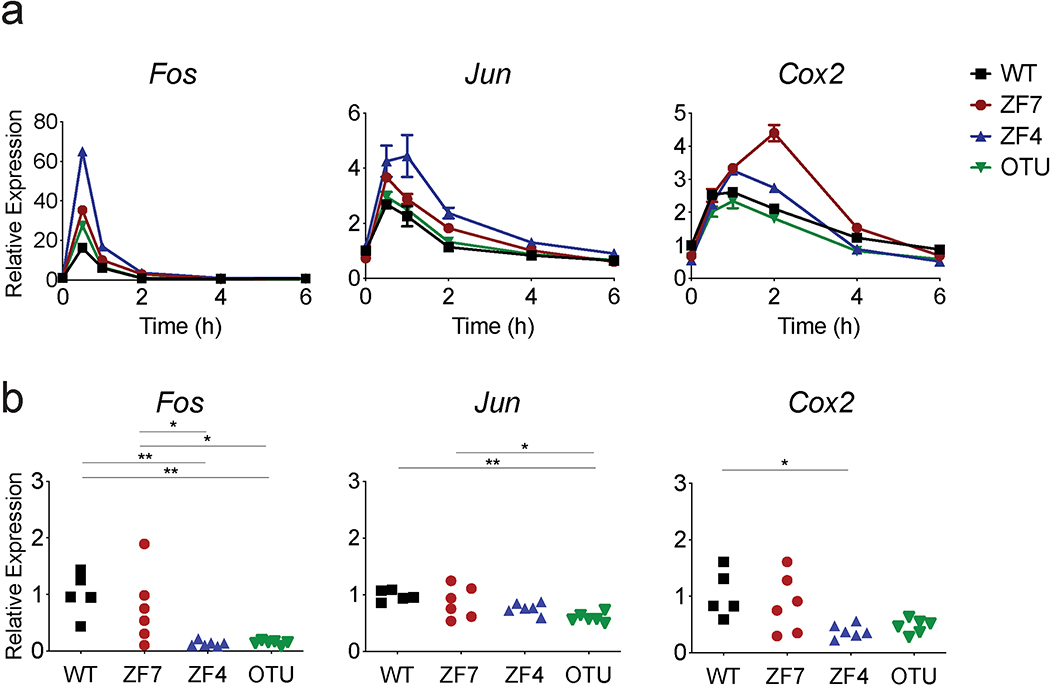
(h) Quantitative PCR analyses of the indicated genes in MEFs of the indicated genotypes transiently stimulated with TNF for 15 min. WT (black), ZF7 (red), ZF4 (blue), and OTU (green). Mean ± SD for technical replicates. Results are representative from 3 independent experiments.
(i) Quantitative PCR analyses of indicated genes in paws from pre-diseased 3-week-old mice of the indicated genotypes. (n=5 independent mice for WT mice [black squares]; n=6 for ZF7 [red circles], ZF4 [blue triangles], and OTU [inverted breen triangles] mice). * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. One-way ANOVA with Tukey’s multiple comparison test.
Our synchronized signaling studies in knock-in cells revealed that TNFR-associated RIP1 ubiquitination fell to negligible levels in all four genotypes of cells by 30 min (Fig. 5a). Recruitment of the kinase TAK1 and IKKγ to FLAG-TNF also peaked at 5 min and returned to baseline by 30 min, irrespective of Tnfaip3 genotype (Fig. 5a). As the IKKγ complex phosphorylates IκBα, and as this complex normally dissociates from TNFR within 15–30 min, we examined IKKγ complexes in A20 knock-in cells. Co-immunoprecipitation analyses of IKKγ complexes confirmed that ubiquitinated RIPK1 dissociated from IKKγ after 30 min in all genotypes (Fig. 5c). Importantly, wild-type A20 protein was recruited to IKKγ between 5 and 30 min after TNF stimulation and remained associated with IKKγ for at least 2 h (Fig. 5c). Thus, although IKKγ disengages from the TNFR and proximate signaling factors such as RIP1 and TAK1 within 30 min, A20 remains bound to the IKK complex for 2 h or more after synchronous TNF stimulation. By contrast, A20ZF7 proteins were poorly recruited to IKKγ throughout this 2 h window despite being superinduced in A20ZF7/ZF7 cells (Fig. 5c). These results raised the possibility that A20’s ZF7 might regulate IKK complexes in addition to acute TNFR complexes. The failure of A20ZF7 mutant proteins to co-precipitate with IKKγ after TNF stimulation contrasts with their robust recruitment to pro-IL-1β inflammasome complexes in LPS stimulated BMDMs, suggesting that A20ZF7 proteins fold normally and retain the ability to regulate other A20-dependent ubiquitinated signaling complexes (Fig. 5d). These results are also consistent with our findings that A20ZF7/ZF7 cells retain competency to prevent spontaneous NLRP3 inflammasome activation and that arthritis in A20ZF7/ZF7 mice does not require NLRP3. Therefore, A20’s ZF7 motif is required for A20 binding to IKKγ kinase complexes but not for A20 binding to pro-IL-1β inflammasome complexes.
The failure of A20ZF7 proteins to bind IKKγ over several hours in A20ZF7/ZF7 cells suggests that A20 may use ZF7 to directly restrict IKKγ kinase activity at longer time points after ligand engagement. Indeed, we observed exaggerated IKKγ kinase activity in these cells relative to wild-type, A20OTU/OTU, and A20ZF4/ZF4 cells between 30 min and 2 h after TNF stimulation (Fig. 5e). Consistent with prior studies, RIPK1, TRAF2, and TAK1 were not co-precipitated with IKKγ at these latter time points (Fig. 5c and data not shown). Hence, A20 utilizes ZF7 to bind to IKKγ complexes and restrict their activity long after initial TNFR signaling complexes dissociate from IKKγ. These results align with prior studies suggesting that A20 can inhibit IKKγ activity without affecting RIP1 ubiquitination 17. We further tested the idea that physiologically induced A20 utilizes ZF7 to regulate IKKγ activity separately from its regulation of RIP1 ubiquitination by measuring IL-1β induced NF-κB signaling, a pathway that requires IKKγ but not RIP1. Similar to the TNF stimulation experiments, we pre-incubated cells with IL-1β at 4 οC for 60 min, washed the cells, and then added warm, ligand-free media to initiate cell signaling. These experiments revealed that IL-1R induced IKKγ kinase activity was induced similarly in wild-type, A20OTU/OTU, A20ZF4/ZF4, and A20ZF7/ZF7 cells between 5 and 15 min after stimulation (Fig. 5f). By contrast, IL-1R-induced IKKγ kinase activity was increased in A20ZF7/ZF7 cells compared to wild-type, A20OTU/OTU and A20ZF4/ZF4 cells between 30 min and 2 h (Fig. 5g). As IL-1R-induced signals require MyD88, exaggerated responses of A20ZF7/ZF7 cells to IL-1β may also help explain the sensitivity of arthritis in A20ZF7/ZF7 mice to MyD88 but not commensal organisms. Taken together, restricting IKK complex activity can explain the selective role of A20’s ZF7 motif (versus A20’s OTU or ZF4 motifs) in limiting NF-κB signaling downstream of TNF and IL-1 receptors.
Prolonged IKK activity in A20ZF7/ZF7 cells could drive increased transcription of inflammatory genes. To reflect physiological cytokine dynamics, we transiently stimulated cells with TNF for 15 min. Quantitative PCR (qPCR) analyses of the various A20 knock-in MEFs revealed that TNF stimulated similar amounts of Fos, Jun, and Cox2 mRNAs, genes that have been characterized as early NF-κB response genes based on their rapid transcriptional induction (Extended Data Fig. 6a) 36. In response to this transient stimulus, wild-type and A20OTU/OTU cells expressed negligible amounts of Csf2, Il6, C3, Ccl5, and Mmp3 mRNAs, genes that are induced over longer time periods (Fig. 5h). These “late phase” NF-κB response genes are associated with both experimental and clinical arthritis 37, 38. A20ZF4/ZF4 cells expressed modestly elevated levels of Il6, and Ccl5, consistent with partially compromised recruitment of A20ZF4 proteins to IKKγ (Figs. 5c, h). Importantly, A20ZF7/ZF7 cells elaborated markedly more of these late phase transcripts over several hours (Fig. 5h). As in vitro studies may or may not reflect pathophysiologically relevant processes in vivo, we performed transcriptomic analyses on paws from the various A20 knock-in mice. We analyzed 3-week-old pre-diseased mice that exhibited no digital swelling and had histologically normal paws to avoid biases caused by infiltrating immune cells. Remarkably, in parallel with our results with TNF stimulated MEFs, paws from pre-diseased A20ZF7/ZF7 mice expressed elevated levels of the same Csf2, Il6, C3, Ccl5, and Mmp3 mRNAs when compared with paws from wild-type, A20OTU/OTU and A20ZF4/ZF4 mice (Fig. 5i). By contrast, paws from pre-diseased A20ZF7/ZF7 mice expressed normal amounts of early NF-κB response genes that were also controlled properly in TNF stimulated MEFs (Extended Data Fig. 6b). These findings provide in vivo validation of the importance of exaggerated NF-κB signaling activity to arthritis pathogenesis in A20ZF7/ZF7 mice. They also suggest that aberrant TNF induced signals may be an early pathophysiological trigger of arthritis in these mice. Taken together, our findings reveal that A20’s ZF7 motif — but not its OTU and ZF4 motifs — prevents prolonged IKK activity, expression of late phase NF-κB-dependent genes, and arthritis.
A20’s ZF4 motif synergizes with A20’s ZF7 motif
While A20ZF7/ZF7 mice uniformly develop arthritis within 8 weeks of age, these mice typically live for more than 6 months, far longer than A20–/– mice that die perinatally 9. Hence, the loss of A20’s ZF7-based Ub binding does not abrogate all of A20’s functions, and other motifs of A20 must also perform important homeostatic functions in vivo. As proteins with multiple Ub-binding motifs can utilize these motifs to coordinate binding to polyubiquitin chains 39, 40, we hypothesized that A20’s ZF7 and ZF4 Ub-binding motifs might collaborate to optimize A20 binding to ubiquitinated signaling complexes and to regulate cell signaling. In this scenario, simultaneously mutating both ZF4 and ZF7 motifs could compromise A20 function to a greater degree than mutating either motif alone.
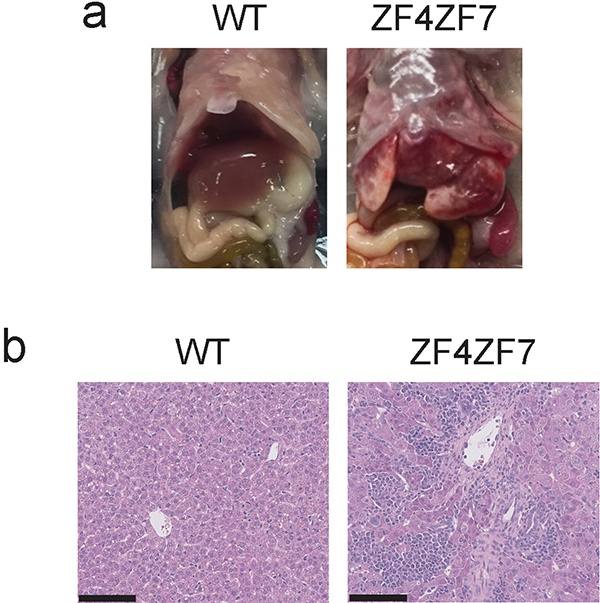
To test this hypothesis, we generated mice expressing double mutant A20 proteins containing point mutations in both ZF4 and ZF7 motifs. We harvested fertilized oocytes from timed matings of A20ZF4 mice with C57BL/6 mice, and used CRISPR-directed gene targeting to generate FG to AA substitutions in A20’s ZF7 ubiquitin binding surface in these A20ZF4/+ oocytes (Fig. 6a). After outbreeding and sequencing, we identified mice in which the desired ZF7 mutations were introduced on the ZF4 mutant allele. We call these mice A20ZF4ZF7 mice. Remarkably, homozygous A20ZF4ZF7/ZF4ZF7 mice were born in roughly Mendelian ratios but died within three weeks of birth (Fig. 6b). Two-week-old A20ZF4ZF7/ZF4ZF7 mice were markedly runted, weighing less than half of their wild-type littermates (Fig. 6c). Livers from A20ZF4ZF7/ZF4ZF7 mice showed signs of gross disease, and histological studies confirmed widespread inflammation and tissue damage (Extended Data Fig. 7a,b). These severe phenotypes resemble those observed in A20–/– mice 9, and were never observed in A20OTU/OTU, A20ZF4/ZF4, or A20ZF7/ZF7 mice. Hence, abrogating both A20’s ZF4 and ZF7 motifs causes profoundly greater disease than mutating either of these ZF motifs (or A20’s OTU motif) alone.
Figure 6. A20’s ZF7 motif synergizes with A20’s ZF4 motif to restrict inflammation in vitro and in vivo.
(a) Scheme of targeting ZF7FG residues in A20ZF4 embryos. Generation of A20ZF4ZF7 mice described in METHODS.
(b) Quantitation of live A20ZF4ZF7 pups at 14 days of age born from intercrossed A20ZF4ZF7/+ heterozygote parents.
(c) Weights of 14 day old A20ZF4ZF7/ZF4ZF7 and WT littermate mice. **** p < 0.0001. Unpaired two-tailed t-test.
(d) Quantitative PCR analyses of indicated genes in MEFs transiently stimulated with TNF for 15 min and measured at the indicated times. WT (black), ZF7 (red), ZF4ZF7 (purple) MEFs are shown. Mean ± SD for technical replicates. Results are representative from three independent experiments.
(e) IKK kinase assays of TNF-stimulated MEFs of the indicated genotypes. Lysates were immunoprecipitated with anti-IKKγ, incubated with GST-IκBα and assayed for phospho-IκBα at the indicated times after warming cells to 37 °C. GST and IKKγ levels in IPs shown as IP control. Immunoblot analyses of phospho-IKKα/β, phospho-IκBα and control signaling proteins in whole cell lysates shown in input samples.
(f) Immunoblot analyses of A20 expression in MEFs of indicated genotypes before and 60 min after TNF stimulation. Representative of 3 independent experiments.
(g, h) ELISA of IL-6 (g) and IL-1β (h) secretion from indicated genotypes of BMDMs after LPS stimulation. WT (black), ZF4 (blue), ZF7 (red), ZF4ZF7 (purple) mice. Biologically independent culture samples. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. One-way ANOVA, Tukey’s Multiple Comparison Test.
To assess the cell autonomous impact of dual ZF4 and ZF7 mutations upon A20 dependent signaling, we generated MEFs from A20ZF4ZF7/ZF4ZF7 mice and tested their responses to TNF. Importantly, A20ZF4ZF7/ZF4ZF7 MEFs expressed markedly more NF-κB dependent mRNAs than either A20ZF7/ZF7 or wild-type cells after transient TNF stimulation (Fig. 6d). Consistent with these transcriptional responses, acute signaling studies with these cells revealed that A20ZF4ZF7/ZF4ZF7 cells exhibited exaggerated phosphorylated IKKβ beyond 30 min when compared to A20ZF7/ZF7 or wild-type cells (Fig. 6e). In parallel, A20ZF4ZF7/ZF4ZF7 cells displayed exaggerated phosphorylated IκBα expression relative to IκBα (Fig. 6e). These cells also possessed increased IKK kinase activity at these longer time points (Fig. 6e). Hence, A20’s ZF4 and ZF7 ubiquitin binding motifs cooperatively restrict TNF induced NF-κB signaling. Notably, A20ZF4ZF7 double mutant proteins were induced to higher levels than wild-type A20 proteins by TNF stimulation (Fig. 6f). Hence, A20ZF4ZF7 mutant proteins are stable, and are superinduced by exaggerated NF-κB signaling. Despite these high levels of expression, A20ZF4ZF7 mutant proteins are unable to control NF-κB signaling. Therefore, the dramatic in vivo synergy between A20’s ZF4 and ZF7 based functions corresponds with cooperative, cell autonomous restriction of NF-κB signaling.
As A20 expression in myeloid cells is important for immune homeostasis, we tested whether A20’s ZF4 and ZF7 motifs synergize in regulating TLR signaling in macrophages. These studies revealed that A20ZF4ZF7/ZF4ZF7 macrophages elaborate more IL-6 than A20ZF4/ZF4, A20ZF7/ZF7, and wild-type cells after LPS stimulation (Fig. 6g). We next tested the roles of these ubiquitin binding motifs in regulating NLRP3 inflammasomes, a process that involves NF-κB signaling as well as IL-1β inflammasome complex ubiquitination. Interestingly, although WT, A20ZF4/ZF4, and A20ZF7/ZF7 BMDMs all restrained IL-1β secretion normally after treatment with LPS alone, A20ZF4ZF7/ZF4ZF7 macrophages displayed spontaneous inflammasome activity (Fig. 6h). Thus, A20’s ZF4 and ZF7 motifs synergistically restrict NLRP3 inflammasome activity. Cell autonomous regulation of NF-κB and NLRP3 inflammasome signaling functions hence aligns with the potent synergy between these Ub binding motifs in vivo.
Discussion
Our findings with A20ZF7/ZF7 mice unveil a biochemical function of A20 that is essential for preventing spontaneous disease in vivo. A20ZF7/ZF7 mice uniformly and rapidly develop arthritis of distal digits. In this regard, they may resemble recently described mice targeting the ZF7 motif 41. Features of the inflammatory arthritis seen in A20ZF7/ZF7 mice that strongly resemble psoriatic arthritis include dactylitis, enthesitis, nail destruction, and significant erosive disease adjacent to areas of bone formation. These features, along with the critical roles for TNF and IL-17 align with the pathophysiology of human psoriatic arthritis 42. The unexpected role of A20’s ZF7 motif in restraining TH17 cells in the absence of commensal organisms also provides intriguing clues linking A20 with the regulation of IL-17 expressing cells and other IL-17 dependent diseases. In contrast to animal models driven solely by overexpression of TNF, arthritis in A20ZF7/ZF7 mice is also dependent on T cells 43,44. A20ZF7/ZF7 mice also differ significantly from recently described A20Flox LysM-Cre mice that require NLRP3 but do not require T cells or TNF for the spontaneous development of arthritis 26. In contrast to most human psoriatic arthritis, A20ZF7/ZF7 mice produce autoantibodies including anti-CCP antibodies more commonly seen in human rheumatoid arthritis. These findings could provide pathophysiological insight into the rare subset of psoriatic arthritis patients that express anti-CCP antibodies. As A20 is also genetically linked to rheumatoid arthritis, A20 ZF7 regulated pathways may be pathogenetic in both psoriatic and rheumatoid arthritis patients 45. Overall, A20ZF7/ZF7 mice constitute a compelling spontaneous model of inflammatory arthritis.
We have found that spontaneous arthritis in A20ZF7/ZF7 mice is exquisitely sensitive to TNF, demonstrating for the first time that A20 prevents TNF dependent disease in vivo. The critical pathophysiological importance of TNF gene dosage to this A20 dependent disease highlights the intricate cross-talk between A20 and TNF functions. Furthermore, the dichotomy between TNF dependent disease in A20ZF7/ZF7 mice and TNF independent pathologies in A20–/– mice provides unique opportunities to dissect how A20’s distinct biochemical activities regulate diverse disease pathways. Understanding these pathways is clinically paramount, given the extensive experience that anti-TNF therapy is effective in some but not all patients with arthritis.
While our results with A20ZF7/ZF7 knock-in cells are consistent with prior studies suggesting that A20’s ZF7 motif helps recruit A20 to TNFR complexes to deubiquitinate RIP1 20, A20’s ZF7 motif likely also directly regulates NF-κB signaling in a manner that does not involve A20’s C103 based deubiquitinating activity. This non-catalytic function may involve a RIP1 independent role in regulating IKKγ, as defective A20-IKKγ association corresponds with prolonged IKK kinase activity and as A20’s ZF7 restricts IL-1β induced IKK kinase activity, a cascade that does not involve RIP1. In this regard, our results reinforce prior studies showing that A20 utilizes its ZF7 motif to bind Ub and IKKγ, and restrict IKK kinase activity without affecting RIP1 ubiquitination 17. The physiological relevance of our results is strengthened by our exclusive use of knock-in cells expressing A20 mutant proteins under endogenous physiological regulation. Furthermore, the importance of A20 binding to IKKγ aligns with a subset of patients bearing IKKγ mutations that fail to bind A20, as these patients exhibit inflammatory phenotypes that are not observed in patients bearing other IKKγ mutations 46.
Our studies reveal profound cooperativity between A20’s ZF4 and ZF7 Ub binding motifs. By performing serial gene targeting of these motifs, we discovered that A20ZF4ZF7/ZF4ZF7 mice die within two to three weeks of birth while A20ZF4/ZF4 and A20ZF7/ZF7 mice both survive for more than six months. In parallel with this cooperativity in vivo, cell autonomous synergy between A20’s ZF4 and ZF7 motifs was evident in responses of A20ZF4ZF7/ZF4ZF7 MEFs and BMDMs. This synergy may reflect cooperative Ub binding by A20’s ZF4 and ZF7 motifs. While A20’s ZF7 motif exhibits distinct binding affinities and Ub chain preferences from ZF4 14, 15, 13, 20, K63 Ub chains can be a substrate for M1 Ub chains in cells, and a mixture of K63 and M1 linked Ub chains exist in IKK complexes 47. These mixed Ub chain complexes could thus be cooperatively recognized by A20’s ZF4 and ZF7 motifs. A20’s ZF4 has also been shown to exert E3 Ub ligase activity upon RIP1, so Ub binding by ZF7 might also support Ub ligase activity. Future studies can explore this possibility as well as the ability of A20 to perform RIP1 independent functions. Our studies reveal a stepwise hierarchy of these motifs as WT, A20ZF4/ZF4, A20ZF7/ZF7, and A20ZF4ZF7/ZF4ZF7 proteins exhibit progressively severe defects in binding to IKKγ, leading to progressively increased IKKγ activity and more severe in vivo disease. As these defects were not observed in syngeneic A20OTU/OTU cells or mice, cooperative Ub binding by A20’s ZF4 and ZF7 motifs likely restricts IKKγ activity independently of A20’s DUB activity.
In summary, we have identified a critical biochemical function of A20 required for preventing spontaneous disease. A20ZF7/ZF7 mice are a robust model of arthritis that shares clinical, histological, and pathophysiological features with psoriatic arthritis. More broadly, our studies reveal how non-catalytic Ub binding by A20 is critical for preserving immune and tissue homeostasis, and that A20’s ZF4 and ZF7 Ub binding motifs cooperate to regulate Ub dependent signaling and prevent disease. Our genetic dissection of A20’s biochemical functions has unveiled distinct molecular disease pathways, and shed insight into why this multifaceted protein is linked to multiple human diseases. These insights should help re-focus future efforts to diagnose and treat the wide array of A20 dependent diseases.
Methods
Generation of A20 mutant mice
Tnfaip3–/– and Tnfaip3FL/FL mice were previously described 9, 51, 52. A20OTU and A20ZF4 mice were previously described 18. To generate A20C764A, C767A (A20ZF7-CC) mice, nucleotide substitutions were generated in PRXB6T (C57BL/6) embryonic stem (ES) cells using transcription activator-like effector nuclease (TALEN) homology-directed repair. TAL effector DNA binding domain (Cellectis) sequences were: TGATCACTTTGGCAATGCC (left) and TACTGCAATGAGTGCTA (right). The codons encoding cysteine (C) 764 and C767 in ZF7 are underlined. The homology directed repair matrix was 180 bp single-stranded oligonucleotide (ssODN, IDT) DNA with the following sequence: left homology arm (42bp)-TGCGATCACTTTGGCAATGCCAAGTGTAATGGTTACGCCAATGAGGCCTACCAGTTTAAA-right homology arm (78bp). Nucleotide substitutions are underlined. The TGC–>GCC and TGC–>GCC substitutions change C764 and C767 to alanines. The T–>C substitution in the repair exchange matrix is a silent mutation to prevent further TALEN binding and cleavage of mutated alleles. The TTC–>TTT and AAG–>AAA are silent mutations that generate a novel PmeI site. Plasmid DNA encoding the TALENs and the ssODN were co-transfected with a neomycin-resistance vector into ES cells using the Neon Transfection System (Thermo). Properly targeted ES cell clones were identified by PCR amplification and digestion of the amplicon with PmeI, and subsequently confirmed by cloning and sequencing. ES cells were injected into C57BL/6 blastocysts by the Gladstone Transgenic Gene-Targeting Core Laboratory, and chimeric mice bred to C57BL/6 mice. Two ES clones were successfully transmitted to the germline, generating two independently derived lines of A20ZF7-CC mice. Both mouse lines displayed similar phenotypes. The data in this manuscript are derived from one clone.
A20 ZF7FG mice were generated by CRISPR-Cas9 genome editing in zygotes. AltR crRNA (GCCCCTGCTTGTGATCACTT) — AltR trcrRNA — high HiFi Cas9 nuclease ribonucleoprotein (IDT) and a 113 base single-stranded oligonucleotide (ACGCCTGAAGAGCCCCCTAAACAGCGCTGCCGGGCCCCTGCTTGTGACCACGCTGCCAATGCCAAGTGTAATGGTTACTGCAATGAGTGCTACCAGTTCAAGCAGATGTATGG) were injected into heterozygous A20ZF4/+ fertilized oocytes. Nucleotides that differed from wild-type C57BL/6 DNA sequence are underlined. Nucleotides that encode F755 G756 (TTT, GGC) were changed to encode alanines (GCT, GCC). A silent mutation (GAT–>GAC) that destroyed a Bcl I restriction enzyme site was included for initial screening by PCR. Offspring that were confirmed by sequencing to have successfully inserted A20ZF7-FG mutations were bred to C57BL/6 mice. These offspring were screened for the A20ZF4 mutation. A mouse that did not carry the A20ZF4 mutation founded the A20ZF7-FG line; a mouse that also screened positive for the A20ZF4 mutation established the A20ZF4ZF7 line.
A20 ZF7ΔFGN were generated using CRISPR sgRNA (GCCCCTGCTTGTGATCACTTTGG) and Cas9 (PNA Bio), along with the ssODN repair DNA (IDT) injected into C57BL/6 zygotes. One had an in-frame deletion of nine base pairs that led to the deletion of the three amino acids F755 G756 N757. This mouse was bred to C57BL/6 mice to derive the A20 ZF7ΔFGN mouse line. All mutations were confirmed by cloning and sequencing.
The following mouse strains were purchased from Jackson Laboratories: B6.129S6- Nlrp3tm1Bhk/J,(021302), B6.129S2-Ighmtm1Cgn/J (μMT, 002288), B6.129S7-Rag1tm1Mom/J (Rag1 knockout, 002216), C57BL/6J-Ticam1Lps2/J (Trif deficient), B6;129S-TNF<tm1Gkl>/J (TNF-deficient mice), backcrossed in our lab to C57BL/6J) (Boone et al., 2004). Myd88–/– mice were previously obtained from S. Akira via R. Medzhitov (Yale University) 22.
In vivo mouse studies
Germ-free A20ZF7 mice were derived by Cesarean delivery into the UCSF Gnotobiotic Core Facility and maintained in germ-free isolators during monitoring and scoring for spontaneous arthritis. Stool pellets from isolators in the Gnotobiotic facility were screened monthly by PCR for 16S bacterial and 18S (ITS) fungal ribosomal sequences. Mice were randomly housed based on sex. All mice were maintained on an inbred C57BL/6 background.
For longitudinal analyses of clinical arthritis, mice were scored in a blinded fashion on a weekly basis beginning at the indicated ages. Digits demonstrating visible erythema, swelling, and loss of the nail plate were counted as positive for arthritis. For histological analyses, paws of euthanized mice were collected in 10% phosphate buffered formalin, with further processing for H&E staining performed by the UCSF Mouse Histology Core Facility. Livers of A20ZF4ZF7 mice were collected in 4% paraformaldehyde and samples were processed and H&E stained by HistoWiz. Micro-CT assays were performed as described 24. For antibody inhibition experiments, mice were injected intraperitoneally on a weekly basis with 1.0 mg of anti-IL17A antibody (Bioxcell, Clone: 17F3) or control isotype antibody (Bioxcell, Clone: MOPC-21). For all graphs presenting in vivo experiments, each dot represents a single mouse. All mice were housed and bred in accordance with UCSF’s institutional guidelines under approved protocols.
Cellular Immune Assays
Bone marrow cells from WT, A20OTU/OTU, A20ZF4/ZF4, A20ZF7/ZF7, and A20FL/FL Rosa26/ER-Cre+ mice were cultured for 5 days in media containing macrophage colony stimulating factor to generate BMDMs. Tnfaip3 deletion was induced in bone marrow cultures from Tnfaip3FL/FL Rosa26/ER-Cre+ mice with 20 nM 4-OHT on day 4. For analysis of macrophage responses to LPS, BMDMs were stimulated for 24 h with 100 ng/ml ultrapure LPS-EB (InVivogen). Cytokines secreted into the supernatant were quantified by ELISA according to manufacturer’s instructions (BD Biosciences). Three separate wells were measured in each experiment. ELISAs for quantitation of murine rheumatoid factor (RF), anti-cyclic citrullinated peptide (CCP), and anti-nuclear antigen (ANA) antibodies were performed with respective kits from Alpha Diagnostics International. The threshold of RF+ positive assays was determined by manufacturer’s instructions (method 1).
For FACS analyses, cells were stained with the following antibodies: anti-CD19 PE (1D3; Tonbo), anti-TCRβ PerCP-Cy5.5 (H57–597; Tonbo), anti-CD19 PerCP-Cy5.5 (1D3; Tonbo), anti-Ly-6G/C APC (RB6–8C5; Tonbo), anti-I-A/I-E violetFluor 450 (M5/114.15.2; Tonbo), anti-CD44 PE (IM7; Tonbo) and anti-CD62L APC (MEL-14; Tonbo), anti-Ly-6G/C PerCP-Cy5.5 (RB6–8C5; BioLegend), anti-CD138 APC (281- 2; BioLegend), anti-GL7 FITC (GL7; BioLegend), anti-CD95 PE-Cy7 (Jo2; BD), anti-CD11b PE (M1/70; BioLegend), anti-CD8 PB (53–6.7; BioLegend), anti-CD4 PE-Cy7 (RM4–5; Tonbo), anti-IL-17A (BD 559502). For analysis of intracellular IL-17A expression, cells from peripheral lymph nodes (axillary, brachial and inguinal) were pooled and stimulated with PMA (50 ng/ml) and ionomycin (500 ng/ml) in the presence of brefeldin A for 3 h at 37 °C and assayed by flow cytometry using BD Cytofix/Cytoperm plus Fixation/Permeabilization kit (555028) according to manufacturer’s instructions. Acquisition of all flow cytometry samples were performed on a LSR II flow cytometer (BD) and analyzed with FlowJo software (Tree Star). Each assay was done on a separate mouse. Example gating strategy can be found in Extended Data Fig. 8.
Signaling Assays
TNF and IL-1β signaling
Murine embryonic fibroblasts (MEFs) were incubated at 4 °C with the respective recombinant murine cytokines (Peptrotech) for 1 h, washed three times with phosphate-buffered saline (PBS) at 4 °C and then brought to 37 °C temperature with pre-warmed, cytokine-free media for the duration of the indicated time points. For gene expression experiments, MEFs were stimulated for 15 min with TNF then washed three times with PBS and were incubated for the remainder of the experiment in TNF-free media.
Immunoblots and immunoprecipitations (IPs)
MEFs were lysed in 1% Triton-X100 lysis buffer [20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton-X100, 1.25 mM NaF, 0.2 mM NaVO3, 2 mM β-glycerophosphate, 10 mM N-ethylmaleimide and complete TM protease inhibitor cocktail (Roche)], each lysate was normalized by BCA protein assay (Thermo). IKKγ immunoprecipitations were performed using a sheep polyclonal anti-IKKγ antibody at 4 °C for 4 h. Protein G Dynabeads (Thermo) were added for the final 1 h of incubation. Active TNFR complex IPs were performed with magnetic anti-Flag (M2) beads (Sigma) by incubation at 4 °C for 2 h. IL-1β IPs were performed by incubating lysates with either biotinylated anti-IL-1β (BioLegend B122) or biotinylated IgG1 isotype control for 16 h followed by adding Neutravidin Agarose (Pierce) for 1 h. Beads were then washed three times and samples were eluted from the beads using 2X LDS buffer (Thermo) containing 715 mM β-mercaptoethanol.
IKK kinase assays
MEFs were lysed in RIPA buffer [20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton-X100, 0.5% Na deoxycholate, 0.1% SDS, 1.25 mM NaF, 0.2 mM NaVO3, 2 mM β-glycerophosphate and complete TM protease inhibitor cocktail (Roche)], each lysate normalized to 500 g total protein, and IKK complex was immunoprecipitated using 1 μg of a sheep polyclonal anti-IKKγ antibody and 15 μl Protein G Dynabeads (Thermo) by incubation at 4 °C for 1 h. Beads were then washed 3 times in RIPA buffer and 1 time in kinase assay buffer (Cell Signaling #9802), and kinase assays were performed in kinase assay buffer plus 1 mM ATP and 2 μg of GST-IκBα 1–54 peptide in a total volume of 20 μl for 30 min at 30 °C. The reaction was terminated with 7.5 μl 4X LDS buffer (Thermo NP0007) plus 1 μl 14.3 M β-mercaptoethanol. Kinase activity was determined by immunoblot analysis using a p-IκBα specific antibody. Antibodies used were: TNFR (Cell Signaling C25C1), NEMO (R&D AF4365, LSBio LS-C346449, or SCBT sc-166398), tubulin (Millipore 05–829), RIPK1 (Cell Signaling 3493), p-IKKα/β (CST 2078S), total IKK (CST: 8943S), (p-IκBα: CST: CS9246), total IκBα (CST: 9242L), Pro-IL-1β (BioLegend B122), GST (BioLegend 640802), A20 (Cell Signaling 5630S), Pro-Caspase8 (Cell Signaling D35G2).
Transcriptomic Analyses
RNA Extraction for gene expression analysis was performed with Qiagen RNeasy kit. Mouse paws were homogenized with MP Bio bead mill. MEFs were collected in RLT buffer. Generation of cDNA was performed with Thermo High-Capacity cDNA Reverse Transcription Kit. Quantitative PCR was performed with Taqman probes and FastAdvanced Master Mix on a Thermofisher (ABI) QuantStudio™ 6 Flex Real-Time PCR System. In figures showing gene expression of mouse paws, each dot represents a separate mouse. In figures showing gene expression of MEFs, technical replicates are shown.
Micro-CT Tomography
After serial fixation in 4% phosphate-buffered formaldehyde and 70% ethanol, mouse bones and paws were scanned ex vivo using a μCT 40 (Scanco Medical), structural and 3D parameters were calculated using Scanco Medical software. Images of arthritis paws were reconstructed from scans performed with a voxel size of 20 μm, filter AL0.5mm, x-ray energy/intensity of 55KvP, 109 mA, 6 W and calibration: 55 kVP, 0.5 mm Al, BH: 1200 mg HA/ccm, scaling 4096.
Statistical Analysis
Statistical analyses of data are provided in figure legends.
Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request. Source data for Fig. 1–3 are provided with this paper.
Extended Data
Extended Data Fig. 1. Arthritis in A20ZF7/ZF7 mice.
(a) Quantitation of live born pups at 12 days of age. (b) Longitudinal quantitation of digital disease of A20ZF7/ZF7 (ZF7) mice by numbers of digits with dactylitis (distal digit swelling) and nail loss in male and female mice at the indicated ages (from 4–12 weeks of age, Mean ± SD for independent mice, n=10 for each genotype). (c) Gross photos (left) and H&E histology (right) of forepaws of A20ZF7-ΔFGN/ZF7-ΔFGN and A20ZF7-FG/ZF7-FG mice at 3 months of age. Scale bars = 1mm. Representative of three independent mice
Extended Data Fig. 2. Mild systemic inflammation in A20ZF7/ZF7 mice.
Representative histology (H&E) from Kidney (a), Liver (b), Colon (c), and Lung (d) of wild-type and ZF7 mice at 3months of age. Scale bars are 100μm for Kidney and Liver, 200μm for Colon, and 1mm for Lung. Representative of three independent mice.
Extended Data Fig. 3. WT and ZF7 T cell signaling and T cell profiling.
(a) Flow cytometry analysis of TCRβ+ and TCRδ+ T cells in lymph nodes (axillary, brachial and inguinal combined, left) and total quantitation of TCRδ+ cells in lymph nodes (right). (b) (left) Representative flow cytometry analysis of naive CD4+ T cells after separation from spleen and lymph nodes (EasySep Mouse Naive CD4+ T cell Isolation Kit). (right) Immunoblot analysis of TCR signaling from purified naive CD4+ T cells shown at left. Purified naive CD4+ T cells were stimulated with anti-CD3 mAb (145–2C11; Tonbo) followed by crosslinking with goat anti-hamster IgG (Jackson Immuno) for the indicated times at 37 °C with gentle shaking. Data are representative of three independent experiments.
Extended Data Fig. 4. Myeloid activation status in wild-type and A20ZF7/ZF7 mice.
Representative flow cytometry analysis of I-A/E cell surface expression on CD11b+ Gr1+ (a) and CD11chi (b) cells in the spleen of 3-month old WT and ZF7 mice. Cells were gated on lineage negative population (TCRβ, CD19). Data are representative of 5 biological replicates per genotype.
Extended Data Fig. 5. Quantitation of T cell subsets in 3-month old wild-type and A20ZF7/ZF7 mice.
(a) Flow cytometric analysis of CD25 vs FoxP3 expression on CD4+ T cells. (b) Flow cytometric analysis of intracellular IFNγ on ex vivo CD4+ T cells stimulated with PMA and ionomycin. Data are representative of two independent experiments.
Extended Data Fig. 6. Expression of early NF-κB -induced genes.
(a) Quantitative qPCR analyses of indicated early NF-κB-induced genes in mouse embryonic fibroblasts (MEFs) transiently stimulated with TNF for 15 minutes. Mean ± SD for technical replicates. Results are representative from three independent experiments. (b) Quantitative PCR analyses of indicated genes in paws from pre-diseased 3-week-old mice of the indicated genotypes. Data are representative of paws from multiple independent mice (WT n = 5, ZF7, ZF4, OTU n=6). * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. One-way ANOVA with Tukey’s multiple comparison test.
Extended Data Fig. 7. Liver inflammation in A20ZF4ZF7/ZF4ZF7 mice.
Gross (a) and hepatic H&E histology (b) of A20ZF4ZF7/ZF4ZF7 and WT littermate mice. Scale bar = 100 μM. Representative of three independent mice.
Extended Data Fig. 8. Gating strategy for flow cytometric analysis.
Representative gating strategy for flow cytometric analysis of intracellular IL-17A staining is shown.
Supplementary Material
Acknowledgements
We thank J. Ashouri Sinha, J. Zikherman, H.-E. Liang, and J. Zhang for helpful discussions and advice, and N. Szeto for micro-CT reconstructions. This work was supported by NIH R01 AI135198 and R01 AI117908 (A.M.), AR070155 (M.C.N.), and a Dermatology Foundation Fellowship (B.R.). MCN is also supported by the Russell/Engleman Arthritis Center at UCSF and the Department of Veteran’s Affairs Health Care System. B.R. is also supported by the Department of Veteran’s Affairs Health Care System. Micro-CT analyses were performed by the UCSF Core Center for Musculoskeletal Biology and Medicine (CCMBM) supported by P30AR066262 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS). FACS analyses were supported by the UCSF Liver Center P30DK026743 from the National Institute of Digestive Diseases and Kidney (NIDDK).
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- 1.Malynn BA & Ma A A20: A multifunctional tool for regulating immunity and preventing disease. Cell. Immunol 340, 103914 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Catrysse L, Vereecke L, Beyaert R & van Loo G A20 in inflammation and autoimmunity. Trends Immunol. 35, 22–31 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Schuijs MJ et al. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science 349, 1106–1110 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Aki A, Nagasaki M, Malynn BA, Ma A & Kagari T Hypomorphic A20 expression confers susceptibility to psoriasis. PloS One 12, e0180481 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Q et al. Loss-of-function mutations in TNFAIP3 leading to A20 haploinsufficiency cause an early-onset autoinflammatory disease. Nat. Genet. 48, 67–73 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aeschlimann FA et al. A20 haploinsufficiency (HA20): clinical phenotypes and disease course of patients with a newly recognised NF-kB-mediated autoinflammatory disease. Ann. Rheum. Dis 77, 728–735 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Berteau F et al. Autosomic dominant familial Behçet disease and haploinsufficiency A20: A review of the literature. Autoimmun. Rev 17, 809–815 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Kadowaki T et al. Haploinsufficiency of A20 causes autoinflammatory and autoimmune disorders. J. Allergy Clin. Immunol 141, 1485–1488.e11 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Lee EG et al. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science 289, 2350–2354 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boone DL et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat. Immunol 5, 1052–1060 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Lin S-C et al. Molecular basis for the unique deubiquitinating activity of the NF-kappaB inhibitor A20. J. Mol. Biol 376, 526–540 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wertz IE et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430, 694–699 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Bosanac I et al. Ubiquitin binding to A20 ZnF4 is required for modulation of NF-κB signaling. Mol. Cell 40, 548–557 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Tokunaga F et al. Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-κB regulation. EMBO J. 31, 3856–3870 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verhelst K et al. A20 inhibits LUBAC-mediated NF-κB activation by binding linear polyubiquitin chains via its zinc finger 7. EMBO J. 31, 3845–3855 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shembade N, Ma A & Harhaj EW Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science 327, 1135–1139 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skaug B et al. Direct, noncatalytic mechanism of IKK inhibition by A20. Mol. Cell 44, 559–571 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu TT et al. Dimerization and ubiquitin mediated recruitment of A20, a complex deubiquitinating enzyme. Immunity 38, 896–905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Draber P et al. LUBAC-Recruited CYLD and A20 Regulate Gene Activation and Cell Death by Exerting Opposing Effects on Linear Ubiquitin in Signaling Complexes. Cell Rep. 13, 2258–2272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wertz IE et al. Phosphorylation and linear ubiquitin direct A20 inhibition of inflammation. Nature 528, 370–375 (2015). [DOI] [PubMed] [Google Scholar]
- 21.De A, Dainichi T, Rathinam CV & Ghosh S The deubiquitinase activity of A20 is dispensable for NF-κB signaling. EMBO Rep. 15, 775–783 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turer EE et al. Homeostatic MyD88-dependent signals cause lethal inflamMation in the absence of A20. J. Exp. Med 205, 451–464 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kool M et al. The ubiquitin-editing protein A20 prevents dendritic cell activation, recognition of apoptotic cells, and systemic autoimmunity. Immunity 35, 82–96 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Hammer GE et al. Expression of A20 by dendritic cells preserves immune homeostasis and prevents colitis and spondyloarthritis. Nat. Immunol 12, 1184–1193 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duong BH et al. A20 restricts ubiquitination of pro-interleukin-1β protein complexes and suppresses NLRP3 inflammasome activity. Immunity 42, 55–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vande Walle L et al. Negative regulation of the NLRP3 inflammasome by A20 protects against arthritis. Nature 512, 69–73 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blauvelt A & Chiricozzi A The Immunologic Role of IL-17 in Psoriasis and Psoriatic Arthritis Pathogenesis. Clin. Rev. Allergy Immunol 55, 379–390 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robert M & Miossec P IL-17 in Rheumatoid Arthritis and Precision Medicine: From Synovitis Expression to Circulating Bioactive Levels. Front. Med 5, 364 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivanov II et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu H-J et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815–827 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scher JU et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2, e01202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elliott MJ et al. Randomised double-blind comparison of chimeric monoclonal antibody to tumour necrosis factor alpha (cA2) versus placebo in rheumatoid arthritis. Lancet Lond. Engl 344, 1105–1110 (1994). [DOI] [PubMed] [Google Scholar]
- 33.Ritchlin CT, Colbert RA & Gladman DD Psoriatic Arthritis. http://dx.doi.org/10.1056/NEJMra1505557https://www.nejm.org/doi/10.1056/nejmra1505557https://www.nejm.org/doi/10.1056/nejmra1505557 (2017) doi: 10.1056/NEJMra1505557. [DOI]
- 34.Matmati M et al. A20 (TNFAIP3) deficiency in myeloid cells triggers erosive polyarthritis resembling rheumatoid arthritis. Nat. Genet 43, 908–912 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Hoffmann A, Levchenko A, Scott ML & Baltimore D The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science 298, 1241–1245 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Hao S & Baltimore D The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat. Immunol 10, 281–288 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McInnes IB & Schett G Cytokines in the pathogenesis of rheumatoid arthritis. Nat. Rev. Immunol 7, 429–442 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Zhang F et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol 20, 928–942 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sims JJ & Cohen RE Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol. Cell 33, 775–783 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sato Y et al. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J. 28, 2461–2468 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polykratis A et al. A20 prevents inflammasome-dependent arthritis by inhibiting macrophage necroptosis through its ZnF7 ubiquitin-binding domain. Nat. Cell Biol 21, 731–742 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Veale DJ & Fearon U The pathogenesis of psoriatic arthritis. Lancet Lond. Engl 391, 2273–2284 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Jacques P et al. Proof of concept: enthesitis and new bone formation in spondyloarthritis are driven by mechanical strain and stromal cells. Ann. Rheum. Dis. 73, 437–445 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Armaka M et al. Mesenchymal cell targeting by TNF as a common pathogenic principle in chronic inflammatory joint and intestinal diseases. J. Exp. Med. 205, 331–337 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plenge RM et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat. Genet 39, 1477–1482 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zilberman-Ru J et al. Recruitment of A20 by the C-terminal domain of NEMO suppresses NF-κB activation and autoinflammatory disease. Proc. Natl. Acad. Sci. U. S. A 113, 1612–1617 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emmerich CH et al. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc. Natl. Acad. Sci. U. S. A 110, 15247–15252 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tavares RM et al. The ubiquitin modifying enzyme A20 restricts B cell survival and prevents autoimmunity. Immunity 33, 181–191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Onizawa M et al. The ubiquitin-modifying enzyme A20 restricts ubiquitination of the kinase RIPK3 and protects cells from necroptosis. Nature Immunol 16, 618–627 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request. Source data for Fig. 1–3 are provided with this paper.