Abstract
Immunohistochemistry for mismatch repair protein expression is widely used as a surrogate for microsatellite instability status- an important signature for immunotherapy and germline testing. There are no systematic analyses examining the sensitivity of immunohistochemistry for microsatellite instability-high status. Mismatch repair immunohistochemistry and microsatellite instability testing were performed routinely as clinically validated assays. We classified germline/somatic mutation types as truncating (nonsense, frameshift, in/del) versus missense and predicted pathogenicity of the latter. Discordant cases were compared to concordant groups: microsatellite instability-high/ mismatch repair-deficient for mutation comparison and microsatellite stable/ mismatch repair-proficient for immunohistochemical comparison. 32 of 443 (7%) microsatellite instability-high cases had immunohistochemistry. Four additional microsatellite instability-high research cases had discordant immunohistochemistry. Of 36 microsatellite instability-high cases with discordant immnohistochemistry, 30 were mismatch repair-proficient while 6 (5 MLH1 and 1 MSH2) retained expression of the defective mismatch repair protein and lost its partner. In microsatellite instability-high tumors with discordant immunohistochemistry, we observed an enrichment in deleterious missense mutations over truncating mutations, with nearly 70% (25/36) of cases having pathogenic germline or somatic missense mutations, as opposed to only 17% (6/36) in a matched microsatellite instability-high group with concordant immunohistochemistry (p=0.0007). In microsatellite instability-high cases with discordant immunohistochemistry and MLH1 or PMS2 abnormalities, less cells showed expression (p=0.015 and p=0.00095 respectively) compared to microsatellite stable/ mismatch repair-proficient cases. Tumor mutation burden, MSIsensor score, and truncating mismatch repair gene mutations were similar between microsatellite instability-high cases with concordant versus discordant immunohistochemical expression. Approximately 6% of microsatellite instability-high cases have retained mismatch repair protein expression and would be missed by immunohistochemistry-based testing, hindering patient access to immunotherapy. Another 1% of microsatellite instability-high cases show isolated loss of the defective gene’s dimerization partner, which may lead to germline testing of the wrong gene. These cases are enriched for pathogenic mismatch repair missense mutations.
Keywords: mismatch repair, immunohistochemistry, microsatellite instability, next generation sequencing
INTRODUCTION
Mismatch repair protein expression via immunohistochemistry and microsatellite instability status are integral parts of the management of many patients with solid cancers. Microsatellite instability testing is performed using DNA, either by polymerase chain reaction or next generation sequencing. Microsatellite instability polymerase chain reaction measures the degree of indel mutations involving microsatellite loci via a set of 5 mononucleotide microsatellites in the tumor vs normal DNA (1). Microsatellite instability assessment via next generation sequencing with MSIsensor is performed via assessment of all available microsatellites covered by a given panel (2). We have previously validated use of MSIsensor with our institutional next generation sequencing assay, MSK-IMPACT (3). Immunohistochemistry is used to assess the presence or absence of mismatch repair protein expression, and complete absence of MLH1, PMS2, MSH2, and/or MSH6 usually correlates with the presence of microsatellite instability-high status in the DNA. The National Comprehensive Cancer Network (NCCN) universally recommends microsatellite instability testing/ mismatch repair protein immunohistochemistry screening for all patients with colorectal carcinoma as well as various other types of solid cancer (4). In addition, patients with stage II-III microsatellite instability-high/ mismatch repair-deficient colorectal carcinoma are managed differently than mismatch repair-proficient/ microsatellite stable colorectal carcinoma as the former do not benefit from 5-fluorouracil adjuvant therapy (5). More broadly, mismatch repair protein loss or microsatellite instability-high status signifies eligibility for immune checkpoint inhibitor treatment (6) as well as the possibility of Lynch syndrome (7) in all types of solid malignancies. Patients with microsatellite instability-high status and advanced malignancies are often eligible for immune checkpoint inhibitor therapy based solely on microsatellite instability status whereas microsatellite instability-high or mismatch repair-deficient status in itself is not diagnostic of Lynch Syndrome but is generally a pre-requisite before further work-up with clinical, personal, and family history as well as germline sequencing to rule in or out the possibility of Lynch Syndrome. It is important that mismatch repair-deificent/ microsatellite instability-high status is detected when present so that patients do not miss an opportunity to be treated with immune checkpoint inhibitors such as pembrolizumab.
Mismatch repair protein expression status corresponds with DNA-based microsatellite instability (microsatellite instability) testing results over 90% of the time (7). Due to the high concordance rate and certain advantages that immunohistochemistry has over microsatellite instability testing, including turnaround time, knowledge of which mismatch repair gene is abnormal, use of only 4 slides, and sensitivity in low tumor purity cases; many institutions perform mismatch repair protein immunohistochemistry rather than microsatellite instability testing.
However, in certain circumstances, mismatch repair protein expression status and microsatellite instability status are discordant. Low tumor purity affects microsatellite instability results, usually by resulting in an underestimation of the degree of microsatellite instability. Neoadjuvant therapy can induce loss of MSH6 expression in cases with microsatellite stable status (8). Even rarer, authors have described isolated microsatellite instability-high cases with proficient (some degree of nuclear expression) mismatch repair protein expression (9–11) as well as MLH1 germline mutant Lynch syndrome cancers with retained MLH1 expression (12–13). Here, we systematically review 443 microsatellite instability-high cases also assessed by mismatch repair protein immunohistochemistry and report the incidence, molecular, and clinicopathologic features of mismatch repair-proficient yet microsatellite instability-high cases.
METHODS
Selection criteria
Selection criteria for this study included formalin-fixed paraffin embedded tumor samples with MSK-IMPACT results from January 2014 to December 2018, microsatellite instability-high status by MSIsensor, and mismatch repair protein immunohistochemistry results available.
Microsatellite instability testing
The microsatellite instability status of all cancers sequenced with MSK-IMPACT from January 2014 to December 2018 was reviewed. MSK-IMPACT is a hybridization capture based assay that assesses somatic mutations, copy number, structural variants in 468 genes in its current iteration (410 in v5 and 341 in v3) against a patient’s matched blood sample (14). Microsatellite instability status is also routinely assessed using a modified and clinically validated version of MSIsensor v0.2 (github: https://github.com/ding-lab/MSIsensor) (3). MSIsensor is a bioinformatic program that interrogates all available microsatellite loci with coverage of at least 20x against a matched normal. The median number of loci assessed by each version of MSK-IMPACT was 1152 in v3, 1241 in v5, and 1581 in v6. Each available locus is evaluated with a goodness of fit test to determine whether the locus is stable or unstable loci. The percentage of unstable loci are then expressed as a score (2).
Tumor mutation burden determination
We used all non-silent exonic and splicing (within the +/− 2bp of intron/exon boundary) mutations (single nucleotide and insertions/deletions) that were reported to the patients and divided the total number of bases where we report mutations in each version of the panel: v3: 896,665 ; v5: 1,016,478 and v6: 1,139,322.
Mismatch repair protein immunohistochemistry
Immunohistochemical results for mismatch repair proteins (MLH1, PMS2, MSH2, and MSH6) were reviewed for tumors that were microsatellite instability-high on MSK-IMPACT; and slides for any immunohistochemistry results signed out as ‘retained,’ ‘equivocal,’ ‘weak,’ or ‘faint’ were further reviewed. Mismatch repair-deficient status was defined as complete loss of nuclear expression of mismatch repair protein(s) within the tumor as per prior studies (11). Mismatch repair-deficient cases with discordant patterns of germline or somatic mutations (ex: isolated PMS2 deficiency with MLH1 hypermethylation and negative germline results) were also reviewed. When available, immunohistochemistry slides were reviewed for cases with discordant mismatch repair statuses and the percent of tumor cells expressing each mismatch repair protein was estimated. A 3:1 control set of immunohistochemistry slides from mismatch repair-P/ microsatellite stable cases was also assessed, and the percent of tumor cells expressing each mismatch repair protein was visually estimated by JFH for all tumor cells available and recorded from microsatellite instability-high cases with discordant mismatch repair protein expression statuses as well as the control set of mismatch repair-proficient /microsatellite stable cases.
MLH1 promoter hypermethylation status analysis
MLH1 promoter hypermethylation testing was performed via bisulfite conversion followed by pyrosequencing as previously described (15) on samples that were microsatellite instability-high with no known germline or somatic mismatch repair gene mutations.
All immunohistochemistry and molecular assays were clinically validated and performed in CLIA accredited laboratories.
Analysis of mismatch repair gene missense mutations
All coding regions of EPCAM, MLH1, PMS2, MSH2, and MSH6 are covered in all versions of MSK-IMPACT. Because truncating mutations (frameshift, stop gain, and splicing) should result in a premature stop codon, nonsense-mediated decay of the transcript, and subsequent lack of mismatch repair protein expression, we theorized that deleterious mismatch repair gene missense mutations are a potential cause for retained mismatch repair protein expression in microsatellite instability-high cases. We therefore assessed whether deleterious mismatch repair gene missense and in-frame indel mutations were enriched in this subset, as compared to truncating mutations (frameshift, stop gain, and splicing). The pathogenicity of somatic missense mutations was predicted using Rare Exome Variant Ensemble Learner (REVEL) (16). The number of cases with a pathogenic mismatch repair gene missense mutation was compared against a 3:1 control set of microsatellite instability-high cases with concordant immunohistochemistry to investigate whether microsatellite instability-high cases with discordant mismatch repair protein expression statuses are enriched in mismatch repair gene missense mutations which lead to expressed but potentially dysfunctional proteins.
Statistics
A Mann-Whitney Test was used for comparison of percentage of tumor cells expressing mismatch repair immunohistochemistry, and the frequencies of mismatch repair gene mutations were compared with two-sided Fisher’s exact test.
RESULTS
Incidence
Examination of 29,530 clinical cases sequenced with MSK-IMPACT assay revealed 582 (2%) tumors were microsatellite instability-high. We reviewed 443 microsatellite instability-high cases with available mismatch repair protein immunohistochemistry, which identified 32 cases (7.2%) with discrepant immunohistochemistry results. These 32 tumors included 17 colorectal carcinomas, 9 endometrial carcinomas, and 1 each of various other cancer types (Table 1). Discordant mismatch repair immunohistochemistry occurred in 6.4% of microsatellite instability-high colorectal carcinomas and 4.9% of microsatellite instability-high endometrial carcinomas.
Table 1.
Characteristic of microsatellite instability-high tumor with discordant immunohistochemical mismatch repair protein expression results.
| CbioPortal ID | Tumor type | MMR IHC | Germline MMR Mutation | Somatic MMR Mutation | REVEL | VAF | MLH1 Promoter Hypermethylation | Dysfunctional MMR Gene |
|---|---|---|---|---|---|---|---|---|
| P-0020886 | Cervical Squamous | Retained | None | MLH1 p.E605Q (c.1813G>C) | 0.74 | 0.54 | Absent | MLH1 |
| MLH1 p.E620K (c.1858G>A) | 0.65 | 0.52 | ||||||
| MLH1 p.E669K (c.2005G>A) | 0.78 | 0.55 | ||||||
| CMO42 | CRC | Retained | MLH1 p.E102K (c. 304G>A) | MSH6 p.F1088Sfs*2 (c.3261delC) | 0.17 | Not Performed | MLH1 | |
| CMO43 | CRC | Retained | MLH1 p.E102K (c. 304G>A) | MLH1 p.X196_splice (c.588+1G>A) | 0.29 | Not Performed | MLH1 | |
| P-0005455 | CRC | PMS2 loss only | None | MLH1 p.M35del (c.105_107delGAT) | 0.14 | Absent | MLH1 | |
| MLH1 p.E414Rfs*77 (c.1240delG) | 0.18 | |||||||
| MSH6 p.F1088Lfs*5 (c.3261dupC) | 0.28 | |||||||
| P-0013462 | CRC | dot-like MLH1 | not performed | None | - | - | Present | MLH1 |
| P-0012115 | CRC | dot-like MLH1 | None | MLH1 p.P300Hfs*40 (c.899_963delCCCAGAATGTGGATGTTAATGTGCACCCCACAAAGCATGAAGTTCACTTCCTGCACGAGGAGAGC) | 0.25 | Not Performed | MLH1 | |
| P-0014258 | CRC | Retained | MLH1 exons 1–19 deletion | MLH1 p.D41N (c.121G>A) | 0.90 | 0.25 | Not Performed | MLH1 |
| MSH2 p.C697Y (c.2090G>A) | 0.85 | 0.12 | ||||||
| P-0016938 | CRC | Retained | MLH1 intron 7 34 bp insertion | MLH1 p.Y157Tfs*3 (c.469delT) | 0.17 | Not Performed | MLH1 | |
| P-0025715 | CRC | Retained | MLH1 p.N306K (c. 918T>A) | None | - | - | Not Performed | MLH1 |
| P-0018118 | CRC | PMS2 loss only | None | MSH6 p.R1076H (c.3227G>A) | 0.84 | 0.22 | Present | MLH1 |
| P-0036383 | CRC | PMS2 loss only | MLH1 p.E102A (c. 305G>C) | None | - | - | Not Performed | MLH1 |
| P-0024289 | UEC | PMS2 loss only | None | MSH6 p.N897Ifs*9 (c.2690delA) | 0.22 | Present | MLH1 | |
| P-0019360 | UEC | Retained | None | MLH1 p.R425Gfs*66 (c.1272delT) | 0.16 | Absent | MLH1 | |
| MLH1 p.G98D (c.293G>A) | 0.98 | 0.17 | ||||||
| MLH1 p.Y750* (c.2250C>G) | 0.32 | |||||||
| MLH1 p.F261Sfs*6 (c.782_785delTCAT) | 0.22 | |||||||
| P-0034967 | Small bowel carcinoma | Retained | MLH1 p.I19F (c. 55A>T) | None | - | - | Not Performed | MLH1 |
| P-0006482 | Breast carcinoma | Retained | None | MSH2 p.F474_L481del (c.1420_1443delTTTGATCCTAATCTCAGTGAATTA) | 0.39 | Absent | MSH2 | |
| CMO40 | CRC | Retained | MSH2 p.A636P (c. 1906G>C) | MSH2 p.V34Pfs*2 (c.2690delA) | 0.33 | Not Performed | MSH2 | |
| CMO41 | CRC | MSH6 loss only | MSH2 p.R524H (c. 1571G>A) | MSH6 p.F1088Sfs*2 (c.3261delC) | 0.38 | Not Performed | MSH2 | |
| P-0010504 | CRC | Retained | MSH2 p.A636P (c. 1906G>C) | MSH2 p.E572del (c.1715_1717delAAG) | 0.20 | Not Performed | MSH2 | |
| MSH6 p.F1088Lfs*5 (c.3261dupC) | 0.17 | |||||||
| P-0017697 | CRC | Retained | None | MSH2 p.D603N (c.1807G>A) | 0.92 | 0.14 | Absent | MSH2 |
| PMS2 p.M136I (c.408G>A) | 0.20 | 0.14 | ||||||
| P-0027324 | CRC | Retained | MSH2 p.R389* (c. 1165C>T) | MSH2 p.E749K (c.2245G>A) | 0.98 | 0.24 | Not Performed | MSH2 |
| P-0020757 | UEC | Retained | MSH2 c. 942A+3>T | MSH2 p.P385Qfs*27 (c.1152_1153delinsT) | 0.05 | Not Performed | MSH2 | |
| P-0027238 | Anaplastic thyroid | Retained | not performed | MSH2 p.S494* (c.1481C>G) | 0.61 | Not Performed | MSH2 | |
| PMS2 p.R151H (c.452G>A) | 0.56 | 0.33 | ||||||
| P-0004379 | UEC | Retained | None | MSH2 p.R621Q (c.1862G>A) | 1.00 | 0.28 | Not Performed | MSH2 or MSH6 (POLE p.V411L, ultramutated) |
| MSH6 p.E547D (c.1641A>C) | 0.88 | 0.32 | ||||||
| MSH6 p.K70N (c.210G>T) | 0.04 | 0.34 | ||||||
| MSH6 p.E877* (c.2629G>T) | 0.29 | |||||||
| MSH6 p.R178H (c.533G>A) | 0.80 | 0.32 | ||||||
| P-0030372 | UEC | Retained | None | MSH2 p.E580* (c.1738G>T) | 0.16 | Absent | MSH2 or MSH6 (POLE p.P286R, ultramutated) | |
| MSH2 p.A272V (c.815C>T) | 0.89 | 0.15 | ||||||
| MSH2 p.D386Y (c.1156G>T) | 0.98 | 0.12 | ||||||
| MSH2 p.R524C (c.1570C>T) | 0.85 | 0.30 | ||||||
| MSH2 p.E859* (c.2575G>T) | 0.23 | |||||||
| MSH6 p.R121H (c.362G>A) | 0.54 | 0.14 | ||||||
| MSH6 p.E699K (c.2095G>A) | 0.78 | 0.18 | ||||||
| MSH6 p.R922Q (c.2765G>A) | 0.64 | 0.39 | ||||||
| P-0013010 | CRC | Retained | MSH6 p.Y1249Lfs*26 (c. 3743_3744insT) | None | - | - | Not Performed | MSH6 |
| P-0030908 | CRC | Retained | MSH6 p.N911* (c. 2731C>T) | MSH6 p.I871Lfs*2 (c.2611delA) | 0.31 | Not Performed | MSH6 | |
| MSH6 p.F1088Sfs*2 (c.3261delC) | 0.33 | |||||||
| MSH6 p.R1331Q (c.3992G>A) | 0.44 | 0.31 | ||||||
| P-0036500 | CRC | Retained | None | MSH6 p.T1219I (c.3656C>T) | 0.93 | 0.36 | Not Performed | MSH6 |
| MSH6 p.F1088Pfs*3 (c.3260_3261dupCC) | 0.21 | |||||||
| P-0028580 | CRC | Retained | None | MSH6 p.T1219I (c.3656C>T) | 0.93 | 0.33 | Absent | MSH6 |
| MSH6 p.F1088Lfs*5 (c.3261dupC) | 0.28 | |||||||
| P-0021117 | UEC | Retained | None | MSH6 p.T1219I (c.3656C>T) | 0.93 | 0.24 | Absent | MSH6 |
| MSH6 p.F1088Lfs*5 (c.3261dupC) | 0.20 | |||||||
| P-0021505 | UEC | Retained | None | MSH6 p.T1219I (c.3656C>T) | 0.93 | 0.20 | Not Performed | MSH6 |
| MSH6 p.G1105Wfs*3 (c.3312dupT) | 0.20 | |||||||
| P-0019598 | UEC | Retained | None | MSH6 p.F1088Lfs*5 (c.3261dupC) | 0.16 | Not Performed | MSH6 | |
| P-0001821 | uterine leiomyosarcoma | Retained | None | MSH2 p.R214I (c.641G>T) | 0.80 | 0.49 | Not Performed | MSH6 |
| MSH6 p.A1162P (c.3484G>C) | 0.93 | 0.81 | ||||||
| P-0030265 | CRC | Retained | PMS2 p.R563* (c. 1687C>T) | MSH2 p.X359_splice (c.1076+1G>A) | 0.22 | Not Performed | PMS2 | |
| MSH6 p.C869Y (c.2606G>A) | 0.33 | 0.20 | ||||||
| PMS2 p.R151Pfs*22 (c.448_451dupCCCC) | 0.10 | |||||||
| P-0031050 | CRC | Retained | not performed | PMS2 p.R107W (c.319C>T) | 0.76 | 0.33 | Absent | PMS2 |
| P-0000449 | Prostate | Retained | PMS2 p.S46I (c. 137G>T) | None | - | - | Not Performed | PMS2 |
MMR= Mismatch repair, IHC= Immunohistochemistry, REVEL: Rare Exome Variant Ensemble Learner, VAF= Variant Allele Frequency, CRC= Colorectal C=carcinoma, UEC= Uterine endometrioid carcinoma
Four additional microsatellite instability-high colon cancers from Lynch syndrome patients with discordant mismatch repair protein expression statuses were also identified via MSK-IMPACT testing performed in the research setting, for a total of 36 discordant cases identified for further analysis. These cases were compared against mutation data from a 1:1 matched cohort of 36 microsatellite instability-high, mismatch repair immunohistochemistry concordant cases and mismatch repair immunohistochemistry data from a 1:1 matched cohort of 36 microsatellite stable, mismatch repair-proficient cases.
Mismatch repair protein immunohistochemistry patterns and expression
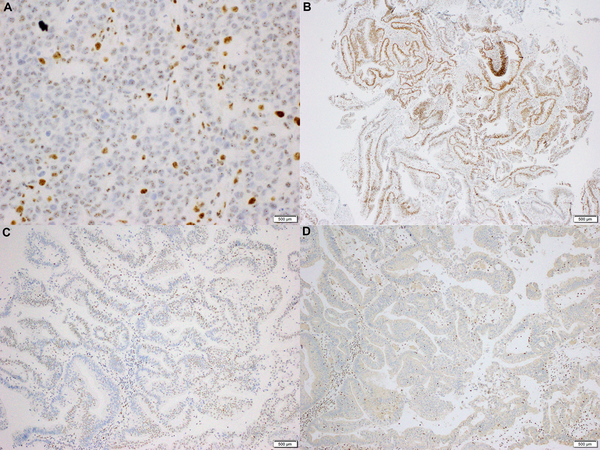
Nuclear dot-like MLH1 staining has been reported in the literature in cases with abnormal mismatch repair expression and MLH1 germline mutations (17, 18). In the current study, 2 microsatellite instability-high/ mismatch repair immunohistochemistry discordant case showed nuclear dot-like MLH1 expression (Figure 1A): 1 of these cases had MLH1 promoter hypermethylation while the other had a truncating somatic MLH1 mutation (p. P300Hfs*40). Interestingly, 1 of 108 microsatellite stable/ mismatch repair-proficient cases also showed nuclear dot-like MLH1 expression, demonstrating that while nuclear dot-like (also called nucleolar or granular) MLH1 expression is rare, it is not specific for mismatch repair-deficient/ microsatellite instability-high status.
Figure 1.
A) A microsatellite instability-high colorectal carcinoma with a nuclear dotlike or ‘nucleolar’ MLH1 expression pattern. No germline or somatic mismatch repair gene mutations were present, yet MLH1 promoter hypermethylation was present (MLH1 immunohistochemistry, Ventana, 400x original magnification). B) A microsatellite instability-high colorectal carcinoma with retain MSH6 expression in approximately 75% of tumor cells. Germline mutation analysis was negative for mismatch repair gene mutations, yet 2 somatic mutations in MSH6 were present: p. F1088Pfs*3 (c. 3260_3261dupCC) at 20.6% variant allele fraction (VAF), which is truncating, and p.T1219I (c.3656C>T) at 36.4% variant allele frequency, which is a missense mutation that is predicted to be pathogenic (MSH6 immunohistochemistry, Cell Marque, 40x original magnification).
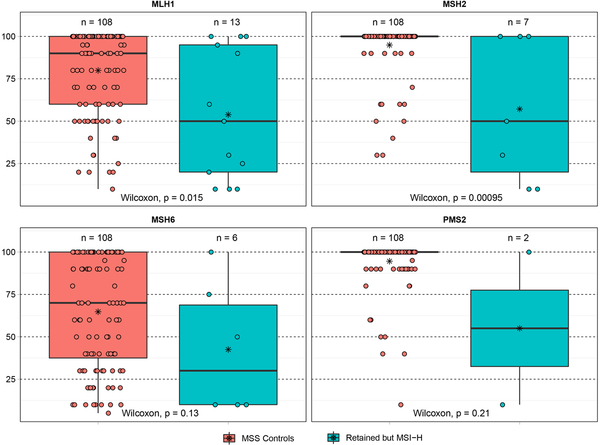
While the majority of cases displayed a degree of expression of all 4 mismatch repair proteins (Figure 1B), 5 cases with MLH1 abnormalities displayed isolated loss of PMS2 (Figure 1C, 1D) while 1 case with MSH2 abnormalities (both somatic and germline) displayed isolated loss of MSH6. The DNA level abnormalities in these 6 cases are specified in table 1. The remaining 30 cases displayed varying degrees of expression of all 4 mismatch repair proteins, with the abnormal mismatch repair gene often showing a decreased percent of tumor cells with mismatch repair protein expression in comparison to a mismatch repair-proficient/ microsatellite stable cohort (Figure 2): the decreased percentage of tumor cells expressing mismatch repair proteins was statistically significant for cases with abnormal MLH1 (p=0.015) and PMS2 (p=0.00095), yet cases with abnormal MSH2 and MSH6 did not show statistical significant decreased expression despite trends.
Figure 2.
Box plot of percentage of tumor cells expressing mismatch repair proteins by immunohistochemistry. The percentage of tumor cells expressing A) MLH1, B) MSH2, C) MSH6, and D) PMS2 was in general lower for microsatellite instability-high cases with retained mismatch repair protein expression (right, green) in comparison to microsatellite stable cases (left, red).
Tumor mutation burden and MSIsensor score comparison
To assess whether microsatellite instability-high cases with discordant immunohistochemistry have a lesser degree of microsatellite instability or lower mutation burden, we compared them against microsatellite instability-high cases with concordant immunohistochemical results. The median tumor mutation burden and MSIsensor score for microsatellite instability-high cases with concordant mismatch repair immunohistochemistry was 44.6 mutations/ megabase and 29.96, respectively. The median tumor mutation burden and MSIsensor score for microsatellite instability-high cases with discordant mismatch repair immunohistochemistry were similar: 54.1 mutations/ megabase and 27.08, respectively.
Mismatch repair gene mutations
Deleterious somatic mismatch repair gene missense mutations were more common in microsatellite instability-high cases with discordant immunohistochemistry:16 (44%) discordant cases harbored deleterious missense mutations in comparison to only 6 (17%) microsatellite instability-high cases in a matched cohort of concordant mismatch repair-deficient/ microsatellite instability-high cases (p=0.02). Nine (25%) microsatellite instability-high cases with discordant mismatch repair protein expression harbored pathogenic germline alterations in an mismatch repair gene (Lynch syndrome), while 1 (3%) microsatellite instability-high/ mismatch repair-deficient cases harbored a pathogenic germline missense mutation in an mismatch repair gene. Together, 25 (69%) microsatellite instability-high/ mismatch repair immunohistochemistry discordant cases harbored either a pathogenic germline or somatic mismatch repair gene missense mutation while only 6 (16%) microsatellite instability-high/ mismatch repair-deficient cases harbored a pathogenic germline or somatic mismatch repair gene missense mutation (p=0.0001). The incidence of truncating mismatch repair gene mutations in microsatellite instability-high cases with discordant mismatch repair immunohistochemistry was not significantly different from that in microsatellite instability-high cases with concordant immunohistochemistry (Table 2).
Table 2.
Comparison of mutation profiles in microsatellite instability-high cases with discordant versus concordant immunohistochemical mismatch repair protein expression results.
| Number of Cases with MMR Gene Mutations | Discordant MMR IHC | Concordant MMR IHC | P value |
|---|---|---|---|
| Pathogenic Somatic Missense Mutations | 16 | 6 | 0.02 |
| Pathogenic Germline Missense Mutations | 9 | 1 | 0.01 |
| Total Cases with Pathogenic Missense Mutations | 25 | 7 | 0.0001 |
| Truncating Germline Mutations | 7 | 4 | 0.51 |
| Truncating Somatic Mutations | 20 | 18 | 0.64 |
| Total Cases with Truncating Mutations | 27 | 22 | 0.2 |
MMR= mismatch repair, IHC= immunohistochemistry
Several mismatch repair gene missense mutations occurred repeatedly in multiple microsatellite instability-high cases with discordant immunohistochemistry. These recurrent missense mutations included 2 patients with germline MLH1 p. E102K (c. 304G>A) mutations as well as a germline MLH1 p. E102A (c. 305 G>A), 2 cases of germline MSH2 p. A636P (c. 1906G>C), and 4 cases with the somatic MSH6 mutation p. T1219I (c. 3656C>T). Each case mentioned above had retained expression of the mutated mismatch repair gene.
Response to Immune Checkpoint Blockade
Three patients with microsatellite instability-high/ mismatch repair immunohistochemistry discordant cancers were treated with the anti-PD1 antibody pembrolizumab as monotherapy. All 3 patients benefited from treatment with best response, one each, of complete response, partial response, and stable disease. All three patients remained on pembrolizumab for over 12 months without progression of disease. Response data are summarized in table 3.
Table 3.
Pembrolizumab response in patients with microsatellite instability-high cancers and discordant immunohistochemical mismatch repair protein expression results.
| CBioPortal ID | Time on pembrolizumab | Best Response | Reason for discontinuation |
|---|---|---|---|
| P-0013462 | 35 months | Complete Response | Stopped after 3 years/course completed |
| P-0012115 | 12 months | Partial Response | Stopped after 1 year/course completed |
| P-0019598 | 24 months | Stable Disease | Treatment is ongoing |
DISCUSSION
We found that approximately 7% of microsatellite instability-high cancers in our study population have discordant (either retained expression or different pattern of protein loss) mismatch repair immunohistochemistry staining patterns, so the presence of microsatellite instability-high status would be missed with immunohistochemistry testing. These tumors are enriched in both germline and somatic mismatch repair gene missense mutations and often have a decrease in the percentage of tumor cells expressing the abnormal mismatch repair protein. Treatment with immune checkpoint inhibitors in three of the patients with discordant immunohistochemistry indicated prolonged clinical benefit, validating the identification of these cases as microsatellite instability-high by next generation sequencing and highlighting the importance to properly classify these cases.
Mismatch repair protein expression is often interpreted as ‘retained’ or ‘lost’ with ‘lost’ meaning complete loss of expression (11), and the literature to date has only scarcely dealt with mismatch repair gene missense mutations associated with patchy mismatch repair protein expression (11–13). Our findings warrant caution in the interpretation of mismatch repair immunohistochemistry, especially when focal or when dot-like nuclear MLH1 expression is present. Potential DNA-based reflex testing for microsatellite instability (next generation sequencing, polymerase chain reaction) may be warranted and may uncover microsatellite instability-high status that would otherwise have been missed, along with opportunities for immune checkpoint inhibition therapy. Particularly, colorectal carcinomas undergoing next generation sequencing testing benefit from having microsatellite instability testing integrated into the next generation sequencing test, even if mismatch repair immunohistochemistry shows proficient expression by clinical standards. Additionally, when mismatch repair expression status and microsatellite instability polymerase chain reaction results are discordant, deciphering which test result is accurate is challenging. In these scenarios, certain findings including germline or somatic missense mutations in mismatch repair genes or patchy staining may help with clinical decision making (whether to manage the case as microsatellite instability-high/ mismatch repair-deficient or mismatch repair-proficient/ microsatellite stable). For example, rare next generation sequencing cases may show falsely elevated MSIsensor scores, resulting in a false microsatellite instability-high status. This may be due to low or borderline coverage. Other factors that elevate the MSIsensor score with the version we used include unmatched analysis (no matched blood sample, history of bone marrow transplant). Further scrutiny may reveal a problem with the next generation sequencing/ microsatellite instability result in some cases. Yet in other cases, mismatch repair immunohistochemical expression is retained and microsatellite instability-high status is called confidently. In the latter scenario, presence of a mismatch repair gene missense mutation helps to verify the microsatellite instability-high result and explain the discordance.
Mismatch repair protein immunohistochemistry is often performed without microsatellite instability polymerase chain reaction/ next generation sequencing in patients with early stage malignancies such as colorectal carcinoma as a screen for Lynch Syndrome. The sensitivity of immunohistochemistry remains high in general, yet we have shown that for some patients with Lynch Syndrome (including several with a pathogenic germline missense mutation), immunohistochemistry may still show retained mismatch repair protein expression. We recommend further work-up with a DNA-based microsatellite instability assessment method if tumor purity is sufficient when any of the 5 Revised Bethesda Guidelines criteria are met or if staining is focal and/or weak. If tumor purity and/or coverage is low, the degree of microsatellite instability might be underestimated, and germline sequencing should be offered. Patients with advanced (metastatic or unresectable) malignancies should be offered comprehensive molecular testing that covers targets for currently approved tumor agnostic therapies including NTRK1–3 fusions and microsatellite instability status, as well as targets for their specific malignancy (for example, extended RAS mutation testing in colorectal carcinoma). If tumor purity or coverage is lower than a specific DNA based assay’s limit of detection for microsatellite instability-high status, mismatch repair immunohistochemistry should be considered based on the type of cancer (colorectal, endometrial, and esophagogastric all having relatively high rates of microsatellite instability-high) and the discretion of the oncologist and pathologist.
Rare studies have demonstrated the phenomenon of MLH1 gene missense mutations (particularly germline mutations) resulting in retained MLH1 expression (12) and occasionally isolated loss of PMS2 expression (13). This study confirms that association on a larger scale, yet the mechanism behind the isolated loss of PMS2 expression in MLH1 germline mutant Lynch patients has yet to be elucidated.
Abnormal dot-like patterns of MLH1 expression may sometimes be associated with mismatch repair-deficient status, specifically deficient MLH1 (17,18). However, we also identified this pattern of expression in the mismatch repair-proficient/ microsatellite stable control group, indicating that dot-like ‘nucleolar’ MLH1 expression is not specific for abnormal MLH1 function or microsatellite instability-high status. Since this phenomenon is relatively rare (occurring in only 1 of 108 mismatch repair-proficient/ microsatellite stable cases in the current study), further testing for microsatellite instability is warranted.
The main limitation of this study is a relatively low number of discordant mismatch repair immunohistochemistry cases due to rarity, which decreases the power for statistical analyses and makes a retrospective approach necessary.
In conclusion, retained mismatch repair protein expression occurs in approximately 6% of microsatellite instability-high cases while another 1% of microsatellite instability-high cases harbor isolated loss of the defective mismatch repair gene’s dimerization partner. Microsatellite instability-high cases with retained mismatch repair protein expression do not have a lower mutation burden or degree of microsatellite instability and benefit from immune checkpoint blockade. The majority of these cases harbor germline or somatic mismatch repair gene missense mutations and often express mismatch repair proteins in a lower percentage of tumor cells.
Acknowledgments
This study was funded in part by the National Cancer Institute (NCI) under the MSK Cancer Center Support Grant/Core Grant (P30 CA008748), and by the Rome Milio Lynch Syndrome Foundation.
Footnotes
This study was approved by the institutional review board at Memorial Sloan Kettering Cancer Center.
REFERENCES
- 1.Bacher JW, Flanagan LA, Smalley RL, et al. Development of a fluorescent multiplex assay for detection of microsatellite instability-highigh tumors. Dis Markers 2004;20:237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niu B, Ye K, Zhang Q, Lu C, Xie M, McLellan MD, Wendl MC, Ding L. microsatellite instabilitysensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 2014. ;30:1015–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Middha S, Zhang L, Nafa K et al. Reliable pan-cancer microsatellite instability assessment by using targeted next-generation sequencing data. JCO Precis Oncol 2017;1:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hechtman JF, Middha S, Stadler ZK et al. Universal screening for microsatellite instability in colorectal cancer in the clinical genomics era: new recommendations, methods, and considerations. Fam Cancer 2017;16:525–529. [DOI] [PubMed] [Google Scholar]
- 5.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol 2005;23:609–618. [DOI] [PubMed] [Google Scholar]
- 6.Overman MJ, McDermott R, Leach JL et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol 2017;18:1182–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Latham A, Srinivasan P, Kemel Y et al. Microsatellite instability is associated with the presence of Lynch syndrome pan-cancer. J Clin Oncol 2019; 37:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao F, Panarelli NC, Rennert H et al. Neoadjuvant therapy induces loss of MSH6 expression in colorectal carcinoma. Am J Surg Pathol 2010;34:1798–1804. [DOI] [PubMed] [Google Scholar]
- 9.Richman S Deficient mismatch repair: read all about it. Int J Oncol 2015;47:1189–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shia J Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome: part I. The utility of immunohistochemistry. J Mol Diagn 2008;10:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCarthy AJ, Capo-Chichi JM, Spence T, et al. Heterogenous loss of mismatch repair (mismatch repair) protein expression: a challenge for immunohistochemical interpretation and microsatellite instability (microsatellite instability) evaluation. J Pathol Clin Res 2019;5:115–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Riel E, Ausems MG, Hogervorst FB, et al. A novel pathogenic MLH1 missense mutation, c.112A > C, p. Asn38His, in six families with Lynch syndrome. Hered Cancer Clin Pract 2010;8:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Jong AE, van Puijenbroek M, Hendriks Y, et al. Microsatellite instability, immunohistochemistry, and additional PMS2 staining in suspected hereditary nonpolyposis colorectal cancer. Clin Cancer Res 2004;10:972–980. [DOI] [PubMed] [Google Scholar]
- 14.Cheng DT, Mitchell TN, Zehir A et al. Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): A hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 2015;17:251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cocco E, Benhamida J, Middha S et al. Colorectal carcinomas containing hypermethylated MLH1 promoter and Wild-Type BRAF/KRAS are enriched for targetable kinase fusions. Cancer Res 2019;79:1047–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ioannidis NM, Rothstein JH, Pejaver V et al. REVEL: An ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet 2016;99:877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarode VR, Robinson L. Screening for lynch syndrome by immunohistochemistry of mismatch repair proteins: significance of indeterminate result and correlation with mutational studies. Arch Pathol Lab Med 2019;143:1225–1233. [DOI] [PubMed] [Google Scholar]
- 18.Tarancón-Diez M, Büttner R, Friedrichs N. Enhanced Tumoral MLH1 expression in MLH1-/PMS2-deficient colon cancer is indicative of sporadic colon cancer and cot HNPCC. Pathol Oncol Res 2019. Epub ahead of print. [DOI] [PubMed] [Google Scholar]