Abstract
The oocyte is a complex cell that executes many crucial and unique functions at the start of each life. These functions are fulfilled by a unique collection of macromolecules and other factors, all of which collectively support meiosis, oocyte activation, and embryo development. This review focuses on the effects of oocyte components on developmental processes that occur after the initial stages of embryogenesis. These include long-term effects on genome function, metabolism, lineage allocation, post-natal progeny health, and even subsequent generations. Factors that regulate chromatin structure, genome programming, and mitochondrial function are elements that contribute to these oocyte functions.
Keywords: oocyte, maternal effect, chromatin, embryo gene regulation
Introduction
In addition to the essential maternal genetic contribution to each offspring, the oocyte has the unique function of providing a vast reservoir of macromolecules that sustain early embryogenesis, control essential metabolic functions, and mediate initial programming of the embryonic genome so that it can execute the appropriate sequence of events to create a new individual. Many maternal effects are readily apparent through early, direct negative consequences during embryo life when they are disrupted. Across diverse species such effects encompass mutations that compromise essential early oocyte functions such as meiosis or yolk deposition, localized ooplasmic determinants in some species, and chromatin regulators and other factors that are required for genome programming and activation (Condic, 2016).
But oocyte factors can also exert long-term effects in development. Early events occurring soon after fertilization may restrict or dictate how later events will unfold, impacting the development or health of the progeny much later, possibly even during post-natal and adult life or even transgenerationally (Figure 1). The scope and impacts of these less immediate and less direct maternal effects continue to be discovered. Such maternal effects are also taking on new and expanding importance as researchers endeavor to understand the oocyte’s role in developmental origins of health and disease (DOHaD), particularly in the context of maternal health and nutrition effects on oocyte quality (Andreas, Reid, Zhang, & Moley, 2019; Matorras et al., 2020) and, for example, adult cardiovascular disease (Ferey et al., 2019; Velazquez, Fleming, & Watkins, 2019). Not only might the environment impact the oocyte, and thereby lead to short- and long-term effects on embryogenesis, but genetic variation in the inherent features of oocytes may also determine the responses of oocytes and embryos to different environmental factors and stressors. This lends new relevance to achieving a more complete understanding of maternal effect genes of the oocyte, and how they are connected to long-term health and viability in progeny.
Figure 1.
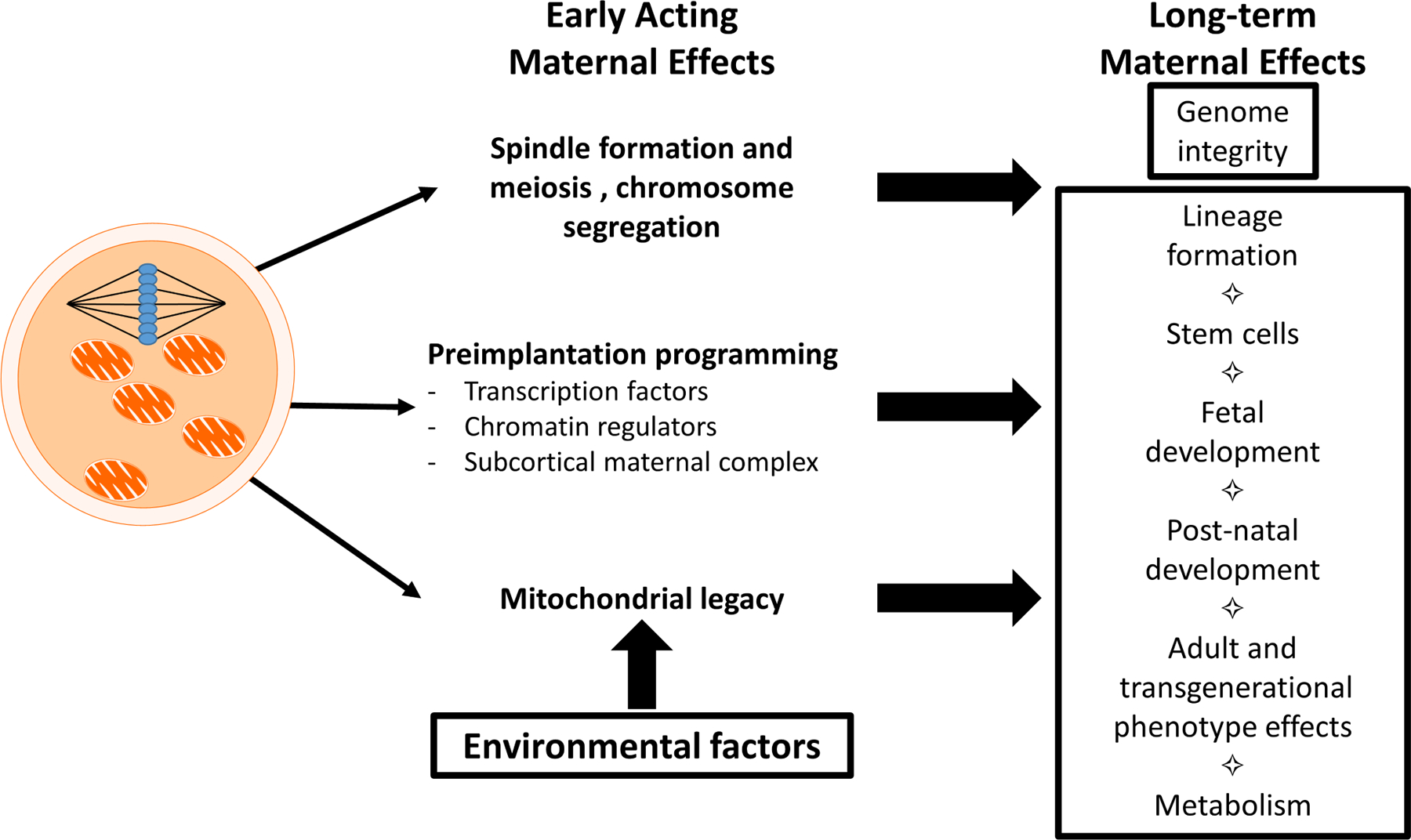
Short-term and long-term maternal effects mediated by oocyte components and processes. Short-term effects include immediate effects of oocyte components on processes in the oocyte and early embryo before the embryonic genome assumes control of development. These include spindle formation, meiosis and chromosome segregation, early aspects of genome programming that determine initial transcriptional activity, and the precise control of oocyte mitochondrial activity. Long-term effects are those that are observed after the embryonic genome assumes control of development, with effects on later preimplantation stages, fetal development, adult phenotype, and even transgenerational impacts. Such long-term effects emerge through persistent epigenetic programming of the early embryonic genome, impacts of these programming events on lineage formation and stem cell formation and later developmental processes, consequences of early programming of mitochondrial function and dynamics. Such long-term effects can also impact later generations. Environmental factors acting upon the oocyte an also impact long-term phenotype via effects on mitochondrial function.
Many other maternal effect genes in mammals have been described that impact key early processes, which, when disrupted, are associated with early embryo demise (Lu, Gao, Qin, & Li, 2017) (Condic, 2016). While the study of such mutations is valuable for understanding essential early developmental processes, its relevance to understanding long-term phenotypic effects is limited. This review focuses on components of mammalian oocytes that have more long-term, less immediate and broader-acting maternal effects on embryo and progeny phenotype. These examples include some specific maternal effect genes, but also include organelle and genome-wide factors in oocytes that have long-term, even life-long impacts on progeny phenotype. The lessons taught to us by recognizing such long-term maternal effects in mammals should provide insight into mechanisms by which early processes that occur in the oocyte set the stage for later developmental events that determine progeny viability and health.
Oocyte histones and histone modifiers
One of the main tasks of oocytes is to progress through meiosis and segregate a haploid complement of chromosomes to the embryo. This task is classified as one of the early acting maternal effects and is essential for normal meiosis and quality of the oocyte and prevent of aneuploidy in the embryo (Figure 1). Recent studies revealed epigenetic changes in the genome that enable meiosis (Huang et al., 2012; Ma & Schultz, 2013). These changes also control gene expression, at least during initial stages of embryogenesis. Additional epigenetic changes in the zygote are mediated by oocyte-expressed histone modifying proteins or mRNAs encoding them. Because such epigenetic changes can exert long-term effects on gene expression, epigenetic actions of oocyte components may dictate more long-term events.
One of the changes that appears to be key for meiosis is histone deacetylation. The meiotic spindle assembly is driven in part by the binding of the chromosome passenger complex to the chromosomes via deacetylated histone H3 and H4 (Akiyama, Kim, Nagata, & Aoki, 2004; Akiyama, Nagata, & Aoki, 2006; Balboula, Stein, Schultz, & Schindler, 2014; Bui et al., 2007; Q. Wang et al., 2006). Histone deacetylation also contributes to transcriptional silencing and formation of the surrounded nucleolus nuclear state in oocytes (De La Fuente, Viveiros, Burns, et al., 2004; De La Fuente, Viveiros, Wigglesworth, & Eppig, 2004; Lee, Wildt, & Comizzoli, 2015). Inadequate histone deacetylation is associated with increased aneuploidy, aberrant oocyte meiosis (Huang et al., 2012; Ma & Schultz, 2013, 2016) and with reduced oocyte quality accompanying aging (van den Berg et al., 2011). Histone deacetylases also regulate the activities of other proteins, and inhibition of this process can also disrupt spindle formation in mitotic and meiotic cells (Chuang, Pan, Hawke, Lin, & Yu-Lee, 2013; Gabrielli & Brown, 2012; X. Li et al., 2017; Shin et al., 2003). Excess histone acetylation may account in part for increased rates of aneuploidy seen in cloned constructs made by transferring somatic cell nuclei to oocytes (Mizutani et al., 2012; Nolen et al., 2005). Although histone deacetylase inhibitors (HDACis) may help program some genes for expression in clones to improve early development (Laguna-Barraza, Sanchez-Calabuig, Gutierrez-Adan, Rizos, & Perez-Cerezales, 2018; Mizutani, Wakayama, & Wakayama, 2015), a potential downside could be disruption of spindle formation and possibly aneuploidy. Histone deacetylation may be sensitive to exogenous factors such as excessive follicle stimulating hormone or stress, which can also induce oocyte aneuploidy (Roberts et al., 2005; Salvador et al., 2001; Xu et al., 2011). The histone deacetylases: HDAC1, HDAC2, HDAC3, RBBP4, RBBP7, and Haspin are implicated in histone deacetylation in oocytes (Balboula et al., 2014; Balboula, Stein, Schultz, & Schindler, 2015; De La Fuente, 2014; Eot-Houllier, Fulcrand, Watanabe, Magnaghi-Jaulin, & Jaulin, 2008; Ma & Schultz, 2013; Q. Wang et al., 2006). One study in porcine oocytes found that class I HDACs in the germinal vesicle were not responsible for global deacetylation, and that instead other cytoplasmic HDACs were responsible (Endo, Kano, & Naito, 2008). Histone methylation may also contribute to meiosis (X. Wang et al., 2014). The correct modulation of histone acetylation and methylation in the oocyte, which regulates not only meiosis but also potentially other transcription factors, may set the stage for long-term effects on development and embryo phenotype.
Histone demethylation driven by oocyte-derived histone demethylases has emerged recently as a key process in remodeling paternal genomes following fertilization (Endo et al., 2008; Hatanaka et al., 2017; Jenkins & Carrell, 2012; Yamagata & Okada, 2011). Additionally, cloned embryos display enhanced developmental potential if the somatic cell nuclei are subjected to increased activities of a range of histone demethylases (Chung et al., 2015; X. Liu et al., 2018; Z. Liu et al., 2018; Matoba et al., 2014; Wei et al., 2017; Y. Zhang et al., 2018). The ability of histone demethylases to positively influence cloned embryo developmental potential implies an important role in the correct programming of normal embryonic nuclei for long-term function and embryo phenotype. Histone demethylation also contributes to embryonic genome activation (Dahl et al., 2016).
Another important maternally derived histone is the histone variant H3.3. Although present in both sperm and egg, paternally-derived H3.3 is lost after fertilization (Kong et al., 2018). Maternally-derived H3.3 associates with the paternal chromatin, remains detectable in the nuclei until the morula stage, and is essential for correct transcription from the paternally derived chromosomes, the correct expression of the key pluripotency gene Oct4/Pou5f1, and embryo pre-implantation viability (Kong et al., 2018). This latter effect on Oct4/Pou5f1 suggests a possible long-term maternal effect of oocyte-derived H3.3 level on progeny phenotype.
The histone demethylase, KDM1A, also exerts-long-term maternal effects, as well as contributing to embryonic genome activation. Conditional knockouts achieving complete ablation of oocyte-expressed KDM1A using the Gdf9 or Zp3 promoters in mice prevents embryonic genome activation and arrests embryos by the two-cell stage (Wasson et al., 2016). However, an alternate conditional knockout allele using the Vasa promoter that reduces KDM1A by about 67% allows approximately 5% blastocyst formation and a low rate of development to birth. But this is followed by a significant rate of perinatal mortality and abnormal behavior (excessive scratching, digging and food grinding, and anxiety in survivors as well as imprinting defects (Wasson et al., 2016), demonstrating a long term maternal effect of reduced oocyte expression.
The euchromatic histone lysine N-methyl transferase (EHMT2, a.k.a., G9A), may contribute to essential lineage-regulation during preimplantation development. Ablation of both maternal and embryonic sources of this protein leads to a failure to repress certain genes at the four-cell stage, and subsequent dysregulation of lineage segregation at the blastocyst stage (Zylicz et al., 2018). This suggests that the maternally expressed EHMT2 could contribute to the correct programming of later gene expression patterns that contribute to optimum lineage formation, but long-term studies would be needed to confirm this.
These observations highlight the dramatic remodeling of histones associated with meiosis and early embryogenesis, and mediated by oocyte-expressed histones and histone modifiers. They also provide a powerful demonstration of how the histone modifications driven in oocytes and by oocyte-expressed factors contribute to long-term gene regulation and developmental phenotype.
Other oocyte-expressed chromatin remodeling factors impacting later events
Besides the histone variants and histone modifiers described above, other ooplasmic factors drive early events that regulate the genome in the embryo, with long-term impacts. In mammals, for example, many studies have been published describing roles for oocyte-expressed factors in mediating initial embryonic transcriptional genome activation, such as double homeobox (DUX) (De Iaco et al., 2017; Hendrickson et al., 2017), a homolog of yeast Sin3 (SIN3A), Yes-associated protein (YAP), tripartite motif-containing 24 (TRIM24, a.k.a. TIF1A), mediator complex-13 (MED13), histone demethylases, components of the Polycomb Repressor Complex 1 (e.g., Ring finger proteins 1 and 2) and many others (Ancelin et al., 2016; Dahl et al., 2016; Jimenez et al., 2015; Miao et al., 2018; Posfai et al., 2012; Torres-Padilla & Zernicka-Goetz, 2006; Winata et al., 2018; Yu et al., 2016). Here, however, we focus on a special class of maternal effect genes comprised of oocyte-expressed chromatin and transcription regulators that exert long-term effects well beyond the initial stages of early embryonic gene expression (Figure 1).
One such factor is TET3, which oxidizes 5-methylcytosine in DNA as a step in removing cytosine methylation, and plays a key role in embryonic genome reprogramming at the two-cell stage in mice. Ablation of oocyte expressed TET3 does not impede oogenesis, fertilization, or development to birth, but increases embryonic gene transcription at the two-cell stage and results in post-natal growth deficiency and death (Tsukada, Akiyama, & Nakayama, 2015), a striking example of the impact of oocyte-expressed chromatin regulators on long-term embryo development.
Structural maintenance of chromosome flexible domain containing 1 (Smchd1) is a maternally expressed chromatin regulator that provides early actions that contribute to long-term viability. siRNA Smchd1 knockdown inhibits the termination of the first wave of embryonic gene transcription, revealing an important early maternal effect role for SMCHD1 (Ruebel, Vincent, Schall, Wang, & Latham, 2019; Schall, Ruebel, & Latham, 2019). But in addition, transient reduction of maternal plus early embryonic sources of this protein through the morula stage by siRNA knockdown leads to reduced blastocyst formation and hatching, reduced total cell number, reduced inner cell mass allocation, and disruptions in gene regulation, with increased expression of genes related to the trophectoderm lineage (Midic et al., 2018). Importantly, these modest preimplantation developmental effects are accompanied by a nearly 40% reduction in term development from the two-cell stage (Midic et al., 2018). Smchd1 knockdown also reduced expression of the mRNA encoding the essential cell cycle driver, S-phase kinase associated protein 2, (SKP2). These results contrast with previous studies of Smchd1 mutations, in which homozygous mutant embryos from heterozygous mothers displayed mid-gestation lethality with no apparent early effect. Because those studies did not limit maternally expressed SMCHD1, the viability through the mid-gestation stage indicates that the oocyte-derived Smchd1 expression remaining in these early mutant embryos contributed to long-term viability, possibly through support of correct lineage segregation and stem cell proliferation.
Subcortical maternal complex (SCMC) effects
In addition to the above mentioned oocyte-expressed chromatin and transcription regulators, the subcortical maternal complex (SCMC) can exert long term effects on future progeny. The SCMC in mammalian oocytes and embryos includes a number of maternal effect proteins, including NLRP5, OOEP, TLE6, and KHDC3L and proteins NLRP5 (Mater), Fila, OOEP (Floped), and KHDC3L (TLE6) (Amoushahi, Sunde, & Lykke-Hartmann, 2019). This complex is required for development of the mouse embryos past the 2-cell stage and controls formation of cytoplasmic lattices, F-actin dynamic, distribution of ER and mitochondria, and mRNA degradation during the oocyte to embryo transition (Amoushahi et al., 2019). A recent clinical study showed that mutations in SCMC genes lead to embryonic arrest, with embryos failing to form blastocysts, including arrest well after the earliest SCMC actions and indicates more long-term impacts of early SCMC functions (Figure 1) (X. Wang et al., 2018).
Maternal antigen that embryos require (MATER), also known as Pyrin domain (PYD) containing proteins 5 (NLRP5), was one of the first maternal effect genes to be identified in mammals and a component of the SCMC, but it is still incompletely understood how NLRP5 controls embryo development (Fernandes et al., 2012; Kim & Lee, 2014). Nlrp5 mRNA is highly expressed in germinal vesicle stage oocytes but not detected in early embryos, although NLRP5 protein is detected throughout preimplantation development (Amoushahi et al., 2019). NLRP5 protein is localized in nucleoli and mitochondria of the oocyte, demonstrating that it has both nuclear and cytoplasmic functions (Amoushahi et al., 2019). MATER has also been reported to play a role in forming cytoplasmic lattices (B. Kim, Kan, Anguish, Nelson, & Coonrod, 2010), mRNA translation, spindle positioning, and epigenetic genome modification (Bebbere, Masala, Albertini, & Ledda, 2016) (Bebbere et al., 2016). The first Nlrp5 knockout mice demonstrated an essential role in early embryo development. Deficient embryos arrested at the 2-cell stage, despite there being no difference in follicle distribution or number of ovulated oocytes compared to control mice (Kim & Lee, 2014). This observation was expanded in non-human primates, where depletion of NLRP5 caused a disruption in embryo development with embryos arresting before the 16-cell stage (X. Wu, 2009). Importantly, while some studies demonstrate such short-term maternal effects of SCMC component deficiency, other studies reveal that oocyte deficiency can also have longer-term effects, impairing blastocyst formation and later development (Amoushahi et al., 2019).
In clinical studies, mutations of NLRP5 are associated with spontaneous abortions, reproductive wastage, and multi-locus imprinting disorders, through aberrant methylation of imprinted loci (Amoushahi et al., 2019), which can have significant long term consequences. Additionally, NLRP5 deficiency in mice alters mitochondrial localization and increases ATP levels compared to controls (Fernandes et al., 2012). This leads to increased cellular stress through increased reactive oxygen species (ROS) production and changes to mitochondrial membrane potential, which caused depletion of mitochondria in the oocyte (Fernandes et al., 2012), possibly impacting long-term phenotype. NLRP5 deficiency in the oocyte is thought to cause mitochondrial damage by triggering premature activation of the mitochondrial pool (Amoushahi et al., 2019; Fernandes et al., 2012). However, the exact involvement of NLRP5 and the SCMC in redistribution or translocation of mitochondria within the embryo is still not fully understood. Regardless of mechanism, these observations reveal both short-term and long-term effects of NLRP5 and SCMC. The extent to which other SCMC components may contribute to long-term effects in the embryo (beyond initial cleavage divisions) remains to be determined.
Mitochondrial legacy
The foregoing discussion makes repeated reference to mitochondrial function in the oocyte. The health and development of the mitochondria, which are exclusively matrilineally transmitted, are critical for oocyte quality and long-term embryo viability, as well as diverse aspects of progeny phenotype. The oocyte mitochondrial legacy can have a large impact on long-term embryo development, adult phenotype, and even transgenerational effects in progeny health (Andreas et al., 2019; Ferey et al., 2019; St John et al., 2019; Udagawa & Ishihara, 2019). Through ATP production, oocyte mitochondria not only support essential metabolic and homeostatic needs of the oocyte and early embryo, but also provide the energy required for meiosis, the vast amount of genome reprogramming that occurs, DNA replication and repair, and the production of the glutathione that is needed for mitigating effects of ROS. Because disruptions in these broader processes can impact long-term phenotype, oocyte mitochondrial function constitutes a significant maternal effect factor relevant to understanding long-term effects of oocyte factors (Figure 1).
A key point to bear in mind regarding mitochondrial activity in the oocyte is that the precise level of mitochondrial activity appears to be critical (Leese et al., 2016). Mitochondrial insufficiency is highly detrimental to short-term embryonic phenotype (Al-Zubaidi et al., 2019), but, conversely too much mitochondrial activity can be detrimental, as it can be associated with excess production of ROS (Leese et al., 2016). The production of excess ROS is especially problematic for oocytes and zygotes because, compared to somatic cells, they are deficient in some of the enzymes that are needed to eliminate ROS, acquiring these ROS scavenging molecules instead from the follicular or oviductal fluid (Guerin, El Mouatassim, & Menezo, 2001; Ufer & Wang, 2011). Indeed, oocyte mitochondria appear to be adapted during oogenesis to minimize the risk of excess ROS production. An “under-developed” mitochondrial state emerges during oogenesis, where in mitochondria are small, spherical, contain under-developed cristae, and have lower ATP production (Babayev & Seli, 2015; Van Blerkom, 2004, 2011). Correct regulation of oocyte ROS production is key for oocyte quality and early embryo survival (Johnson & Nasr-Esfahani, 1994; Mihalas, Redgrove, McLaughlin, & Nixon, 2017). Interestingly, recent studies revealed roles for DNA methyltransferases in the epigenetic modulation of nuclear encoded mitochondrial protein genes (Ruebel et al., 2018). Additional evidence that this regulation contributes to reducing ROS production was obtained from oocytes treated with antioxidants (Cao et al., 2019). That the oocyte genome is subject to such epigenetic control of mitochondrial protein genes raises the possibility of long-term effects of such changes on later development. Factors that impact this regulation could thus have long-term effects. These results indicate that correct regulation of oocyte mitochondrial gene activity, mitochondrial function, and cellular reactive oxygen species production may be key for generating high-quality oocytes, which in turn support long-term development of healthy progeny. A recent study also provided evidence for an epigenetic effect of maternal diet on progeny cardiac mitochondrial function in mice (Ferey et al., 2019).
Another mechanism for long-term impact of the oocyte mitochondrial legacy on progeny phenotype relates to the control of mitochondrial dynamics, which ultimately determine mitochondrial number in the early embryo and beyond. Normal somatic cells undergo mitophagy to eliminate damaged mitochondria, but there appears to be little to no activation of mitophagy in the oocyte, leading to an increase in the number of mitochondria susceptible to damage (Boudoures et al., 2017; Igosheva et al., 2010; Luzzo et al., 2012). How the oocyte reduces the transmission of impaired mitochondria to the early embryo is unclear (Tworzydlo, Sekula, & Bilinski, 2020). Recent studies revealed roles for GAS6 and PINK1 in this process (Boudoures et al., 2017; Kim, Kim, Ko, & Lee, 2019; Niu, Nie, Shin, Zhou, & Cui, 2019). Wu et al. also demonstrated a connection between mitochondrial damage, mitochondrial dynamics, and endoplasmic reticulum (ER) stress (L. L. Wu, Russell, Norman, & Robker, 2012). The uncoupled protein response (UPR) can play a role in the regulation and function of the mitochondrial dynamics (Senft & Ronai, 2015) by inducing mitophagy, regulating mitochondrial biogenetics, and reducing mitochondrial membrane potential in somatic cells and increasing ROS production (Senft & Ronai, 2015). Dysregulation of mitochondrial dynamics in oocytes can also have long-term consequences. Imbalanced mitochondrial dynamics in oocytes and other somatic cells is associated with increased ATP levels, increased mtDNA levels, ROS production, and oxidative phosphorylation (Liesa & Shirihai, 2013; Udagawa & Ishihara, 2019; Wai & Langer, 2016). Overexpression of mitofusion proteins in oocytes negatively affects both mitochondrial morphology and dynamics, and is associated with chromosome misalignment when compared to control oocytes (Wakai, Harada, Miyado, & Kono, 2014). Other observations reveal the sensitivity of mitochondrial dynamics to stress and metabolic alterations, and the attendant sensitivity of oocyte developmental potential to mitochondrial regulation (Babayev et al., 2016; Igosheva et al., 2010; Saben et al., 2016). Bu the changes in oocyte mitochondrial dynamics have long-term effects as well as immediate effects on the oocyte and early embryo energetics. One striking series of studies found that a high fat/high sucrose maternal diet can damage oocyte mitochondria (Boudoures et al., 2017; Grindler & Moley, 2013; Saben et al., 2016), and that this can lead to changes in cardiac mitochondrial structure and function for multiple generations (Ferey et al., 2019), as well as oocyte and skeletal muscle (Andreas et al., 2019; Saben et al., 2016). Thus, correct regulation of a healthy oocyte mitochondrial legacy contributes to long-term healthy progeny phenotype.
Understanding the large impact of abnormal mitochondrial function on oocyte and embryo development has also led to new research looking at the effects of mitochondria replacement therapy. The thought behind these techniques, which can include pronuclear transfer, mitochondria spindle transfer, ooplasm transfer, or mitochondrial microinjection, is that the mutant or defective mitochondria can be replaced or augmented with healthy donor mitochondria (Mobarak et al., 2019). However, these techniques are subject to both ethical and safety concerns (Adashi & Cohen, 2018). One study reported a healthy baby boy born after oocyte spindle transfer to prevent transfer of a mitochondrial disease, Leigh syndrome, in a woman undergoing IVF treatment (J. Zhang et al., 2017), and other reports of using oocyte spindle transfer to treat infertility have surfaced since. In non-human primates, mitochondria spindle transfer experiments led to viable progeny that showed growth and development comparable to control offspring (Adashi & Cohen, 2018; Tachibana et al., 2009). In addition, mouse studies yielded progeny from mitochondria replacement procedures designed to overcome mitochondrial genetic defects (Adashi & Cohen, 2018; Sato et al., 2005). These methods generally had low mtDNA carryover, but heteroplasmy can be seen in progeny (Mobarak et al., 2019). However, studies to date from mitochondrial microinjection indicate that benefits may be limited in some contexts and may be affected by the cellular source of the mitochondria (Igarashi et al., 2016; Mobarak et al., 2019). The possible transgenerational effects of any heteroplasmy that would result from these methods have not been well studied. Potential epigenetic changes in the progeny genomes have also not been rigorously studied. Further studies are needed to understand the safety and efficacy of this method.
Environmental factors impacting long-term maternal effects
A growing body of evidence suggests that environmental factors that create stress in the oocytes as well as the peri-conception stage embryos can have substantial long-term effects on progeny phenotype. Stressors can include oocyte in vitro maturation medium, embryo culture, medium, maternal low-protein diet, maternal obesity, diabetes, environmental toxins and chemicals, ethanol consumption, and excess hormonal stimulation (Andreas et al., 2019; Ecker et al., 2004; Eichenlaub-Ritter & Pacchierotti, 2015; Fleming, Eckert, & Denisenko, 2017; Hart, 2016; L. Li et al., 2020; Mann et al., 2004; Ng, Lau, Yeung, & Ho, 2003; Snider & Wood, 2019; VandeVoort, Grimsrud, Midic, Mtango, & Latham, 2014; Velazquez, 2015). Of particular interest here, is the impact of such factors on the oocyte. In rhesus macaques, administration of ethanol to simulate binge drinking can compromise oocyte quality, even after the treatment is terminated (VandeVoort et al., 2014). Exposure to environmental endocrine disruptors can impact meiosis and oocyte quality and have multi-generational and transgenerational effects in the ovary (Rattan & Flaws, 2019). Maternal diets deemed unhealthy can lead to mitochondrial dysfunction, inflammation, changes in ER stress and UPR related gene expression (Atf4, Xbp1s, Hspa1a, Hapa1b) and reduced developmental competence in ovulated oocytes compared to control litter mates (Boudoures et al., 2017; Ruebel et al., 2017; Snider & Wood, 2019; L. L. Wu et al., 2015). Effects of unhealthy maternal diets have also been observed transgenerationally. Abnormal mitochondrial function, morphology, and dynamics of oocytes from F1 and F2 generation offspring were demonstrated in offspring that were exposed to maternal high fat/high sucrose diet (Boudoures et al., 2017; Jaeger, Saben, & Moley, 2017; Saben et al., 2016). These observations indicate that the oocyte is a sensitive target for effects of diverse exogenous and endogenous factors that exert multi-and transgenerational effects. The mechanism responsible for such effects remain incompletely understood, but these studies highlight the importance of oocyte quality in the next and even subsequent generations, highlighting the crucial role played by maternal factors in the oocyte.
Perspectives
The oocyte is richly supplied with a multitude of factors that support not only early embryogenesis, but also long-term progeny health. Many of these factors play crucial roles in early programming of the embryonic genome for correct transcription, correct regulation of mitochondria, essential production of ATP, metabolism, repairing DNA damage, preventing ongoing damage to DNA and other macromolecules from ROS, and many other vital processes. One impressive observation to emerge in the last decade is the finding that early insults to the mother can negatively impact the oocyte and lead to transgenerational effects. Studies in cattle revealed that even mild nutrient restriction during a narrow peri-conceptional window of time can lead to reduced ovarian reserve, aorta malformation, and hypertension in female progeny (Mossa, Latham, Ireland, & Veiga-Lopez, 2019), demonstrating the high potential of maternal factors in impacting progeny health. Maternal exposure to low levels of environmental toxins (Hunt et al., 2003; Hunt, Susiarjo, Rubio, & Hassold, 2009; Lawson et al., 2011; Muhlhauser et al., 2009; Susiarjo, Hassold, Freeman, & Hunt, 2007), maternal obesity (Boudoures et al., 2017; Grindler & Moley, 2013), and potential effects of ovarian stimulation and oocyte/embryo in vitro culture and manipulations likewise can disrupt the fragile biology of the oocyte, impacting progeny health. These observations are important because they suggest that a significant fraction of adult health disorders and disease, including the number one killer in our society—cardiovascular disease—may emerge in part through the impact of comparatively mild and seemingly innocuous factors affecting the oocyte, by compromising these long-term functions of the oocyte in supporting progeny development and health. Genetic factors that increase or decrease susceptibility to such exogenous influences warrant further study, as do potential approaches to mitigate the effects of these factors following exposure, as well as approaches to reducing exposure. Additionally, recognition that oocyte functions that are related to progeny health may be compromised well in advance of conception provides an incentive for further research and education related to identifying and minimizing such detrimental exposures throughout reproductive life.
Acknowledgements and Funding
The authors thank Mr. Peter Schall for assistance with the manuscript. The work was supported in part by grants from the National Institutes of Health, Eunice Kennedy Shriver National Institute of Child Health and Human Development (RO1HD075903 and T32HD087166), MSU AgBioResearch, and Michigan State University. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Declaration of Interests: Authors have no conflicting interests to declare.
References
- Adashi EY, & Cohen IG (2018). Preventing mitochondrial diseases: embryo-sparing donor-independent options. Trends Mol Med, 24(5), 449–457. doi: 10.1016/j.molmed.2018.03.002 [DOI] [PubMed] [Google Scholar]
- Akiyama T, Kim JM, Nagata M, & Aoki F (2004). Regulation of histone acetylation during meiotic maturation in mouse oocytes. Mol Reprod Dev, 69(2), 222–227. doi: 10.1002/mrd.20121 [DOI] [PubMed] [Google Scholar]
- Akiyama T, Nagata M, & Aoki F (2006). Inadequate histone deacetylation during oocyte meiosis causes aneuploidy and embryo death in mice. Proc Natl Acad Sci U S A, 103(19), 7339–7344. doi: 10.1073/pnas.0510946103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Zubaidi U, Liu J, Cinar O, Robker RL, Adhikari D, & Carroll J (2019). The spatio-temporal dynamics of mitochondrial membrane potential during oocyte maturation. Mol Hum Reprod. doi: 10.1093/molehr/gaz055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoushahi M, Sunde L, & Lykke-Hartmann K (2019). The pivotal roles of the NOD-like receptors with a PYD domain, NLRPs, in oocytes and early embryo development. Biol Reprod. doi: 10.1093/biolre/ioz098 [DOI] [PubMed] [Google Scholar]
- Ancelin K, Syx L, Borensztein M, Ranisavljevic N, Vassilev I, Briseno-Roa L, … Heard E (2016). Maternal LSD1/KDM1A is an essential regulator of chromatin and transcription landscapes during zygotic genome activation. Elife, 5. doi: 10.7554/eLife.08851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreas E, Reid M, Zhang W, & Moley KH (2019). The effect of maternal high-fat/high-sugar diet on offspring oocytes and early embryo development. Mol Hum Reprod, 25(11), 717–728. doi: 10.1093/molehr/gaz049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babayev E, & Seli E (2015). Oocyte mitochondrial function and reproduction. Curr Opin Obstet Gynecol, 27(3), 175–181. doi: 10.1097/GCO.0000000000000164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babayev E, Wang T, Szigeti-Buck K, Lowther K, Taylor HS, Horvath T, & Seli E (2016). Reproductive aging is associated with changes in oocyte mitochondrial dynamics, function, and mtDNA quantity. Maturitas, 93, 121–130. doi: 10.1016/j.maturitas.2016.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboula AZ, Stein P, Schultz RM, & Schindler K (2014). Knockdown of RBBP7 unveils a requirement of histone deacetylation for CPC function in mouse oocytes. Cell Cycle, 13(4), 600–611. doi: 10.4161/cc.27410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboula AZ, Stein P, Schultz RM, & Schindler K (2015). RBBP4 regulates histone deacetylation and bipolar spindle assembly during oocyte maturation in the mouse. Biol Reprod, 92(4), 105. doi: 10.1095/biolreprod.115.128298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebbere D, Masala L, Albertini DF, & Ledda S (2016). The subcortical maternal complex: multiple functions for one biological structure? J Assist Reprod Genet, 33(11), 1431–1438. doi: 10.1007/s10815-016-0788-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudoures AL, Saben J, Drury A, Scheaffer S, Modi Z, Zhang W, & Moley KH (2017). Obesity-exposed oocytes accumulate and transmit damaged mitochondria due to an inability to activate mitophagy. Dev Biol, 426(1), 126–138. doi: 10.1016/j.ydbio.2017.04.005 [DOI] [PubMed] [Google Scholar]
- Bui HT, Van Thuan N, Kishigami S, Wakayama S, Hikichi T, Ohta H, … Miyano T (2007). Regulation of chromatin and chromosome morphology by histone H3 modifications in pig oocytes. Reproduction, 133(2), 371–382. doi:133/2/371 [pii] 10.1530/REP-06-0099 [DOI] [PubMed] [Google Scholar]
- Cao Z, Gao D, Tong X, Xu T, Zhang D, Wang Y, … Pu Y (2019). Melatonin improves developmental competence of oocyte-granulosa cell complexes from porcine preantral follicles. Theriogenology, 133, 149–158. doi: 10.1016/j.theriogenology.2019.05.003 [DOI] [PubMed] [Google Scholar]
- Chuang C, Pan J, Hawke DH, Lin SH, & Yu-Lee LY (2013). NudC deacetylation regulates mitotic progression. PLoS One, 8(9), e73841. doi: 10.1371/journal.pone.0073841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung YG, Matoba S, Liu Y, Eum JH, Lu F, Jiang W, … Zhang Y (2015). Histone demethylase expression enhances human somatic cell nuclear transfer efficiency and promotes derivation of pluripotent stem cells. Cell Stem Cell, 17(6), 758–766. doi: 10.1016/j.stem.2015.10.001 [DOI] [PubMed] [Google Scholar]
- Condic ML (2016). The role of maternal-effect genes in mammalian development: are mammalian embryos really an exception? Stem Cell Rev Rep, 12(3), 276–284. doi: 10.1007/s12015-016-9648-6 [DOI] [PubMed] [Google Scholar]
- Dahl JA, Jung I, Aanes H, Greggains GD, Manaf A, Lerdrup M, … Klungland A (2016). Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature, 537(7621), 548–552. doi: 10.1038/nature19360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A, Planet E, Coluccio A, Verp S, Duc J, & Trono D (2017). DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet, 49(6), 941–945. doi: 10.1038/ng.3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Fuente R (2014). Histone deacetylation: establishing a meiotic histone code. Cell Cycle, 13(6), 879–880. doi: 10.4161/cc.28214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Fuente R, Viveiros MM, Burns KH, Adashi EY, Matzuk MM, & Eppig JJ (2004). Major chromatin remodeling in the germinal vesicle (GV) of mammalian oocytes is dispensable for global transcriptional silencing but required for centromeric heterochromatin function. Dev Biol, 275(2), 447–458. doi:S0012–1606(04)00584–6 [pii] 10.1016/j.ydbio.2004.08.028 [DOI] [PubMed] [Google Scholar]
- De La Fuente R, Viveiros MM, Wigglesworth K, & Eppig JJ (2004). ATRX, a member of the SNF2 family of helicase/ATPases, is required for chromosome alignment and meiotic spindle organization in metaphase II stage mouse oocytes. Dev Biol, 272(1), 1–14. doi: 10.1016/j.ydbio.2003.12.012 S0012160603007930 [pii] [DOI] [PubMed] [Google Scholar]
- Ecker DJ, Stein P, Xu Z, Williams CJ, Kopf GS, Bilker WB, … Schultz RM (2004). Long-term effects of culture of preimplantation mouse embryos on behavior. Proc Natl Acad Sci U S A, 101(6), 1595–1600. doi: 10.1073/pnas.0306846101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenlaub-Ritter U, & Pacchierotti F (2015). Bisphenol A effects on mammalian oogenesis and epigenetic integrity of oocytes: A case study exploring risks of endocrine disrupting chemicals. Biomed Res Int, 2015, 698795. doi: 10.1155/2015/698795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Kano K, & Naito K (2008). Nuclear histone deacetylases are not required for global histone deacetylation during meiotic maturation in porcine oocytes. Biol Reprod, 78(6), 1073–1080. doi: 10.1095/biolreprod.107.067397 [DOI] [PubMed] [Google Scholar]
- Eot-Houllier G, Fulcrand G, Watanabe Y, Magnaghi-Jaulin L, & Jaulin C (2008). Histone deacetylase 3 is required for centromeric H3K4 deacetylation and sister chromatid cohesion. Genes Dev, 22(19), 2639–2644. doi: 10.1101/gad.484108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferey JLA, Boudoures AL, Reid M, Drury A, Scheaffer S, Modi Z, … Moley KH (2019). A maternal high-fat, high-sucrose diet induces transgenerational cardiac mitochondrial dysfunction independently of maternal mitochondrial inheritance. Am J Physiol Heart Circ Physiol, 316(5), H1202–H1210. doi: 10.1152/ajpheart.00013.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes R, Tsuda C, Perumalsamy AL, Naranian T, Chong J, Acton BM, … Jurisicova A (2012). NLRP5 mediates mitochondrial function in mouse oocytes and embryos. Biol Reprod, 86(5), 138, 131–110. doi: 10.1095/biolreprod.111.093583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming TP, Eckert JJ, & Denisenko O (2017). The role of maternal nutrition during the periconceptional period and its effect on offspring phenotype. Adv Exp Med Biol, 1014, 87–105. doi: 10.1007/978-3-319-62414-3_5 [DOI] [PubMed] [Google Scholar]
- Gabrielli B, & Brown M (2012). Histone deacetylase inhibitors disrupt the mitotic spindle assembly checkpoint by targeting histone and nonhistone proteins. Adv Cancer Res, 116, 1–37. doi: 10.1016/B978-0-12-394387-3.00001-X [DOI] [PubMed] [Google Scholar]
- Grindler NM, & Moley KH (2013). Maternal obesity, infertility and mitochondrial dysfunction: potential mechanisms emerging from mouse model systems. Mol Hum Reprod, 19(8), 486–494. doi: 10.1093/molehr/gat026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerin P, El Mouatassim S, & Menezo Y (2001). Oxidative stress and protection against reactive oxygen species in the pre-implantation embryo and its surroundings. Hum Reprod Update, 7(2), 175–189. doi: 10.1093/humupd/7.2.175 [DOI] [PubMed] [Google Scholar]
- Hart RJ (2016). Physiological aspects of female fertility: Role of the environment, modern lifestyle, and genetics. Physiol Rev, 96(3), 873–909. doi: 10.1152/physrev.00023.2015 [DOI] [PubMed] [Google Scholar]
- Hatanaka Y, Tsusaka T, Shimizu N, Morita K, Suzuki T, Machida S, … Ogura A (2017). Histone H3 methylated at arginine 17 is essential for reprogramming the paternal genome in zygotes. Cell Rep, 20(12), 2756–2765. doi: 10.1016/j.celrep.2017.08.088 [DOI] [PubMed] [Google Scholar]
- Hendrickson PG, Dorais JA, Grow EJ, Whiddon JL, Lim JW, Wike CL, … Cairns BR (2017). Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet, 49(6), 925–934. doi: 10.1038/ng.3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Li T, Ding CH, Brosens J, Zhou CQ, Wang HH, & Xu YW (2012). Insufficient histone-3 lysine-9 deacetylation in human oocytes matured in vitro is associated with aberrant meiosis. Fertil Steril, 97(1), 178–184 e173. doi: 10.1016/j.fertnstert.2011.10.023 [DOI] [PubMed] [Google Scholar]
- Hunt PA, Koehler KE, Susiarjo M, Hodges CA, Ilagan A, Voigt RC, … Hassold TJ (2003). Bisphenol a exposure causes meiotic aneuploidy in the female mouse. Curr Biol, 13(7), 546–553. doi:S0960982203001891 [pii] [DOI] [PubMed] [Google Scholar]
- Hunt PA, Susiarjo M, Rubio C, & Hassold TJ (2009). The bisphenol A experience: a primer for the analysis of environmental effects on mammalian reproduction. Biol Reprod, 81(5), 807–813. doi: 10.1095/biolreprod.109.077008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi H, Takahashi T, Abe H, Nakano H, Nakajima O, & Nagase S (2016). Poor embryo development in post-ovulatory in vivo-aged mouse oocytes is associated with mitochondrial dysfunction, but mitochondrial transfer from somatic cells is not sufficient for rejuvenation. Hum Reprod, 31(10), 2331–2338. doi: 10.1093/humrep/dew203 [DOI] [PubMed] [Google Scholar]
- Igosheva N, Abramov AY, Poston L, Eckert JJ, Fleming TP, Duchen MR, & McConnell J (2010). Maternal diet-induced obesity alters mitochondrial activity and redox status in mouse oocytes and zygotes. PLoS One, 5(4), e10074. doi: 10.1371/journal.pone.0010074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger K, Saben JL, & Moley KH (2017). Transmission of metabolic dysfunction across generations. Physiology (Bethesda), 32(1), 51–59. doi: 10.1152/physiol.00017.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TG, & Carrell DT (2012). Dynamic alterations in the paternal epigenetic landscape following fertilization. Front Genet, 3, 143. doi: 10.3389/fgene.2012.00143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez R, Melo EO, Davydenko O, Ma J, Mainigi M, Franke V, & Schultz RM (2015). Maternal SIN3A regulates reprogramming of gene expression during mouse preimplantation development. Biol Reprod, 93(4), 89. doi: 10.1095/biolreprod.115.133504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MH, & Nasr-Esfahani MH (1994). Radical solutions and cultural problems: could free oxygen radicals be responsible for the impaired development of preimplantation mammalian embryos in vitro? BioEssays, 16(1), 31–38. doi: 10.1002/bies.950160105 [DOI] [PubMed] [Google Scholar]
- Kim KH, Kim EY, Ko JJ, & Lee KA (2019). Gas6 is a reciprocal regulator of mitophagy during mammalian oocyte maturation. Sci Rep, 9(1), 10343. doi: 10.1038/s41598-019-46459-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, & Lee KA (2014). Maternal effect genes: Findings and effects on mouse embryo development. Clin Exp Reprod Med, 41(2), 47–61. doi: 10.5653/cerm.2014.41.2.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q, Banaszynski LA, Geng F, Zhang X, Zhang J, Zhang H, … Wen D (2018). Histone variant H3.3-mediated chromatin remodeling is essential for paternal genome activation in mouse preimplantation embryos. J Biol Chem, 293(10), 3829–3838. doi: 10.1074/jbc.RA117.001150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguna-Barraza R, Sanchez-Calabuig MJ, Gutierrez-Adan A, Rizos D, & Perez-Cerezales S (2018). Effects of the HDAC inhibitor scriptaid on the in vitro development of bovine embryos and on imprinting gene expression levels. Theriogenology, 110, 79–85. doi: 10.1016/j.theriogenology.2017.12.043 [DOI] [PubMed] [Google Scholar]
- Lawson C, Gieske M, Murdoch B, Ye P, Li Y, Hassold T, & Hunt PA (2011). Gene expression in the fetal mouse ovary is altered by exposure to low doses of bisphenol A. Biol Reprod, 84(1), 79–86. doi: 10.1095/biolreprod.110.084814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PC, Wildt DE, & Comizzoli P (2015). Nucleolar translocation of histone deacetylase 2 is involved in regulation of transcriptional silencing in the cat germinal vesicle. Biol Reprod, 93(2), 33. doi: 10.1095/biolreprod.115.129106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leese HJ, Guerif F, Allgar V, Brison DR, Lundin K, & Sturmey RG (2016). Biological optimization, the Goldilocks principle, and how much is lagom in the preimplantation embryo. Mol Reprod Dev, 83(9), 748–754. doi: 10.1002/mrd.22684 [DOI] [PubMed] [Google Scholar]
- Li L, Jing Y, Dong MZ, Fan LH, Li QN, Wang ZB, … Sun QY (2020). Type 1 diabetes affects zona pellucida and genome methylation in oocytes and granulosa cells. Mol Cell Endocrinol, 500, 110627. doi: 10.1016/j.mce.2019.110627 [DOI] [PubMed] [Google Scholar]
- Li X, Liu X, Gao M, Han L, Qiu D, Wang H, … Gu L (2017). HDAC3 promotes meiotic apparatus assembly in mouse oocytes by modulating tubulin acetylation. Development, 144(20), 3789–3797. doi: 10.1242/dev.153353 [DOI] [PubMed] [Google Scholar]
- Liesa M, & Shirihai OS (2013). Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab, 17(4), 491–506. doi: 10.1016/j.cmet.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang Y, Gao Y, Su J, Zhang J, Xing X, … Zhang (2018). H3K9 demethylase KDM4E is an epigenetic regulator for bovine embryonic development and a defective factor for nuclear reprogramming. Development, 145(4). doi: 10.1242/dev.158261 [DOI] [PubMed] [Google Scholar]
- Liu Z, Cai Y, Wang Y, Nie Y, Zhang C, Xu Y, … Sun Q (2018). Cloning of macaque monkeys by somatic cell nuclear transfer. Cell, 172(4), 881–887 e887. doi: 10.1016/j.cell.2018.01.020 [DOI] [PubMed] [Google Scholar]
- Lu X, Gao Z, Qin D, & Li L (2017). A maternal functional module in the mammalian oocyte-to-embryo transition. Trends Mol Med, 23(11), 1014–1023. doi: 10.1016/j.molmed.2017.09.004 [DOI] [PubMed] [Google Scholar]
- Luzzo KM, Wang Q, Purcell SH, Chi M, Jimenez PT, Grindler N, … Moley KH (2012). High fat diet induced developmental defects in the mouse: oocyte meiotic aneuploidy and fetal growth retardation/brain defects. PLoS One, 7(11), e49217. doi: 10.1371/journal.pone.0049217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P, & Schultz RM (2013). Histone deacetylase 2 (HDAC2) regulates chromosome segregation and kinetochore function via H4K16 deacetylation during oocyte maturation in mouse. PLoS Genet, 9(3), e1003377. doi: 10.1371/journal.pgen.1003377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P, & Schultz RM (2016). HDAC1 and HDAC2 in mouse oocytes and preimplantation embryos: Specificity versus compensation. Cell Death Differ, 23(7), 1119–1127. doi: 10.1038/cdd.2016.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann MR, Lee SS, Doherty AS, Verona RI, Nolen LD, Schultz RM, & Bartolomei MS (2004). Selective loss of imprinting in the placenta following preimplantation development in culture. Development, 131(15), 3727–3735. doi: 10.1242/dev.01241 [DOI] [PubMed] [Google Scholar]
- Matoba S, Liu Y, Lu F, Iwabuchi KA, Shen L, Inoue A, & Zhang Y (2014). Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell, 159(4), 884–895. doi: 10.1016/j.cell.2014.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matorras R, Exposito A, Ferrando M, Mendoza R, Larreategui Z, Lainz L, … Ruiz-Sanz JI (2020). Oocytes of women who are obese or overweight have lower levels of n-3 polyunsaturated fatty acids compared with oocytes of women with normal weight. Fertil Steril, 113(1), 53–61. doi: 10.1016/j.fertnstert.2019.08.059 [DOI] [PubMed] [Google Scholar]
- Miao YL, Gambini A, Zhang Y, Padilla-Banks E, Jefferson WN, Bernhardt ML, … Williams CJ (2018). Mediator complex component MED13 regulates zygotic genome activation and is required for postimplantation development in the mouse. Biol Reprod. doi: 10.1093/biolre/ioy004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midic U, Vincent KA, Wang K, Lokken A, Severance AL, Ralston A, … Latham KE (2018). Novel key roles for structural maintenance of chromosome flexible domain containing 1 (Smchd1) during preimplantation mouse development. Mol Reprod Dev, 85(7), 635–648. doi: 10.1002/mrd.23001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihalas BP, Redgrove KA, McLaughlin EA, & Nixon B (2017). Molecular Mechanisms Responsible for Increased Vulnerability of the Ageing Oocyte to Oxidative Damage. Oxid Med Cell Longev, 2017, 4015874. doi: 10.1155/2017/4015874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani E, Wakayama S, & Wakayama T (2015). Treatment of donor cell/embryo with different approaches to improve development after nuclear transfer. Methods Mol Biol, 1222, 101–111. doi: 10.1007/978-1-4939-1594-1_8 [DOI] [PubMed] [Google Scholar]
- Mizutani E, Yamagata K, Ono T, Akagi S, Geshi M, & Wakayama T (2012). Abnormal chromosome segregation at early cleavage is a major cause of the full-term developmental failure of mouse clones. Dev Biol, 364(1), 56–65. doi: 10.1016/j.ydbio.2012.01.001 [DOI] [PubMed] [Google Scholar]
- Mobarak H, Heidarpour M, Tsai PJ, Rezabakhsh A, Rahbarghazi R, Nouri M, & Mahdipour M (2019). Autologous mitochondrial microinjection; a strategy to improve the oocyte quality and subsequent reproductive outcome during aging. Cell Biosci, 9, 95. doi: 10.1186/s13578-019-0360-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossa F, Latham KE, Ireland JJ, & Veiga-Lopez A (2019). Undernutrition and hyperandrogenism during pregnancy: Role in programming of cardiovascular disease and infertility. Mol Reprod Dev, (in press). [DOI] [PubMed] [Google Scholar]
- Muhlhauser A, Susiarjo M, Rubio C, Griswold J, Gorence G, Hassold T, & Hunt PA (2009). Bisphenol A effects on the growing mouse oocyte are influenced by diet. Biol Reprod, 80(5), 1066–1071. doi: 10.1095/biolreprod.108.074815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng EH, Lau EY, Yeung WS, & Ho PC (2003). Oocyte and embryo quality in patients with excessive ovarian response during in vitro fertilization treatment. J Assist Reprod Genet, 20(5), 186–191. doi: 10.1023/a:1023670010031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu YJ, Nie ZW, Shin KT, Zhou W, & Cui XS (2019). PINK1 regulates mitochondrial morphology via promoting mitochondrial fission in porcine preimplantation embryos. FASEB J, 33(7), 7882–7895. doi: 10.1096/fj.201802473R [DOI] [PubMed] [Google Scholar]
- Nolen LD, Gao S, Han Z, Mann MR, Gie Chung Y, Otte AP, … Latham KE (2005). X chromosome reactivation and regulation in cloned embryos. Dev Biol, 279(2), 525–540. doi:S0012–1606(05)00043–6 [pii] 10.1016/j.ydbio.2005.01.016 [DOI] [PubMed] [Google Scholar]
- Posfai E, Kunzmann R, Brochard V, Salvaing J, Cabuy E, Roloff TC, … Peters AH (2012). Polycomb function during oogenesis is required for mouse embryonic development. Genes Dev, 26(9), 920–932. doi: 10.1101/gad.188094.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattan S, & Flaws JA (2019). The epigenetic impacts of endocrine disruptors on female reproduction across generationsdagger. Biol Reprod. doi: 10.1093/biolre/ioz081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R, Iatropoulou A, Ciantar D, Stark J, Becker DL, Franks S, & Hardy K (2005). Follicle-stimulating hormone affects metaphase I chromosome alignment and increases aneuploidy in mouse oocytes matured in vitro. Biol Reprod, 72(1), 107–118. doi: 10.1095/biolreprod.104.032003 [DOI] [PubMed] [Google Scholar]
- Ruebel ML, Cotter M, Sims CR, Moutos DM, Badger TM, Cleves MA, … Andres A (2017). Obesity modulates inflammation and lipid metabolism oocyte gene expression: a single-cell transcriptome perspective. J Clin Endocrinol Metab, 102(6), 2029–2038. doi: 10.1210/jc.2016-3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruebel ML, Schall PZ, Midic U, Vincent KA, Goheen B, VandeVoort CA, & Latham KE (2018). Transcriptome analysis of rhesus monkey failed-to-mature oocytes: deficiencies in transcriptional regulation and cytoplasmic maturation of the oocyte mRNA population. Mol Hum Reprod, 24(10), 478–494. doi: 10.1093/molehr/gay032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruebel ML, Vincent KA, Schall PZ, Wang K, & Latham KE (2019). SMCHD1 terminates the first embryonic genome activation event in mouse two-cell embryos and contributes to a transcriptionally repressive state. Am J Physiol Cell Physiol, 317(4), C655–C664. doi: 10.1152/ajpcell.00116.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saben JL, Boudoures AL, Asghar Z, Thompson A, Drury A, Zhang W, … Moley KH (2016). Maternal metabolic syndrome programs mitochondrial dysfunction via germline changes across three generations. Cell Rep, 16(1), 1–8. doi: 10.1016/j.celrep.2016.05.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador LM, Park Y, Cottom J, Maizels ET, Jones JC, Schillace RV, … Hunzicker-Dunn M (2001). Follicle-stimulating hormone stimulates protein kinase A-mediated histone H3 phosphorylation and acetylation leading to select gene activation in ovarian granulosa cells. J Biol Chem, 276(43), 40146–40155. doi: 10.1074/jbc.M106710200 [DOI] [PubMed] [Google Scholar]
- Sato A, Kono T, Nakada K, Ishikawa K, Inoue S, Yonekawa H, & Hayashi J (2005). Gene therapy for progeny of mito-mice carrying pathogenic mtDNA by nuclear transplantation. Proc Natl Acad Sci U S A, 102(46), 16765–16770. doi: 10.1073/pnas.0506197102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schall PZ, Ruebel ML, & Latham KE (2019). A new role for SMCHD1 in life’s master switch and beyond. Trends Genet, 35(12), 948–955. doi: 10.1016/j.tig.2019.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senft D, & Ronai ZA (2015). UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem Sci, 40(3), 141–148. doi: 10.1016/j.tibs.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin HJ, Baek KH, Jeon AH, Kim SJ, Jang KL, Sung YC, … Lee CW (2003). Inhibition of histone deacetylase activity increases chromosomal instability by the aberrant regulation of mitotic checkpoint activation. Oncogene, 22(25), 3853–3858. doi: 10.1038/sj.onc.1206502 [DOI] [PubMed] [Google Scholar]
- Snider AP, & Wood JR (2019). Obesity induces ovarian inflammation and reduces oocyte quality. Reproduction, 158(3), R79–R90. doi: 10.1530/REP-18-0583 [DOI] [PubMed] [Google Scholar]
- St John JC, Makanji Y, Johnson JL, Tsai TS, Lagondar S, Rodda F, … Temple-Smith P (2019). The transgenerational effects of oocyte mitochondrial supplementation. Sci Rep, 9(1), 6694. doi: 10.1038/s41598-019-43135-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susiarjo M, Hassold TJ, Freeman E, & Hunt PA (2007). Bisphenol A exposure in utero disrupts early oogenesis in the mouse. PLoS Genet, 3(1), e5. doi: 10.1371/journal.pgen.0030005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sparman M, Sritanaudomchai H, Ma H, Clepper L, Woodward J, … Mitalipov S (2009). Mitochondrial gene replacement in primate offspring and embryonic stem cells. Nature, 461(7262), 367–372. doi: 10.1038/nature08368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Padilla ME, & Zernicka-Goetz M (2006). Role of TIF1alpha as a modulator of embryonic transcription in the mouse zygote. J Cell Biol, 174(3), 329–338. doi: 10.1083/jcb.200603146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y, Akiyama T, & Nakayama KI (2015). Maternal TET3 is dispensable for embryonic development but is required for neonatal growth. Sci Rep, 5, 15876. doi: 10.1038/srep15876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tworzydlo W, Sekula M, & Bilinski SM (2020). Transmission of functional, wild-type mitochondria and the fittest mtDNA to the next generation: bottleneck phenomenon, balbiani body, and mitophagy. Genes (Basel), 11(1). doi: 10.3390/genes11010104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udagawa O, & Ishihara N (2019). Mitochondrial dynamics and interorganellar communication in the development and dysmorphism of mammalian oocytes. J Biochem. doi: 10.1093/jb/mvz093 [DOI] [PubMed] [Google Scholar]
- Ufer C, & Wang CC (2011). The Roles of Glutathione Peroxidases during Embryo Development. Front Mol Neurosci, 4, 12. doi: 10.3389/fnmol.2011.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Blerkom J (2004). Mitochondria in human oogenesis and preimplantation embryogenesis: engines of metabolism, ionic regulation and developmental competence. Reproduction, 128(3), 269–280. doi: 10.1530/rep.1.00240 [DOI] [PubMed] [Google Scholar]
- Van Blerkom J (2011). Mitochondrial function in the human oocyte and embryo and their role in developmental competence. Mitochondrion, 11(5), 797–813. doi: 10.1016/j.mito.2010.09.012 [DOI] [PubMed] [Google Scholar]
- van den Berg IM, Eleveld C, van der Hoeven M, Birnie E, Steegers EA, Galjaard RJ, … van Doorninck JH (2011). Defective deacetylation of histone 4 K12 in human oocytes is associated with advanced maternal age and chromosome misalignment. Hum Reprod, 26(5), 1181–1190. doi: 10.1093/humrep/der030 [DOI] [PubMed] [Google Scholar]
- VandeVoort CA, Grimsrud KN, Midic U, Mtango N, & Latham KE (2014). Transgenerational effects of binge drinking in a primate model: implications for human health. Fertil Steril, 103, 560–569. doi: 10.1016/j.fertnstert.2014.10.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez MA (2015). Impact of maternal malnutrition during the periconceptional period on mammalian preimplantation embryo development. Domest Anim Endocrinol, 51, 27–45. doi: 10.1016/j.domaniend.2014.10.003 [DOI] [PubMed] [Google Scholar]
- Velazquez MA, Fleming TP, & Watkins AJ (2019). Periconceptional environment and the developmental origins of disease. J Endocrinol, 242(1), T33–T49. doi: 10.1530/JOE-18-0676 [DOI] [PubMed] [Google Scholar]
- Wai T, & Langer T (2016). Mitochondrial dynamics and metabolic regulation. Trends Endocrinol Metab, 27(2), 105–117. doi: 10.1016/j.tem.2015.12.001 [DOI] [PubMed] [Google Scholar]
- Wakai T, Harada Y, Miyado K, & Kono T (2014). Mitochondrial dynamics controlled by mitofusins define organelle positioning and movement during mouse oocyte maturation. Mol Hum Reprod, 20(11), 1090–1100. doi: 10.1093/molehr/gau064 [DOI] [PubMed] [Google Scholar]
- Wang Q, Yin S, Ai JS, Liang CG, Hou Y, Chen DY, … Sun QY (2006). Histone deacetylation is required for orderly meiosis. Cell Cycle, 5(7), 766–774. doi: 10.4161/cc.5.7.2627 [DOI] [PubMed] [Google Scholar]
- Wang X, Gao W, Ma X, Wang X, Song C, Huang X, & Liu H (2014). Dot1L mediated histone H3 lysine79 methylation is essential to meiosis progression in mouse oocytes. Neuro Endocrinol Lett, 35(6), 523–530. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/25433842 [PubMed] [Google Scholar]
- Wang X, Song D, Mykytenko D, Kuang Y, Lv Q, Li B, … Wang L (2018). Novel mutations in genes encoding subcortical maternal complex proteins may cause human embryonic developmental arrest. Reprod Biomed Online, 36(6), 698–704. doi: 10.1016/j.rbmo.2018.03.009 [DOI] [PubMed] [Google Scholar]
- Wasson JA, Simon AK, Myrick DA, Wolf G, Driscoll S, Pfaff SL, … Katz DJ (2016). Maternally provided LSD1/KDM1A enables the maternal-to-zygotic transition and prevents defects that manifest postnatally. Elife, 5. doi: 10.7554/eLife.08848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Antony J, Meng F, MacLean P, Rhind R, Laible G, & Oback B (2017). KDM4B-mediated reduction of H3K9me3 and H3K36me3 levels improves somatic cell reprogramming into pluripotency. Sci Rep, 7(1), 7514. doi: 10.1038/s41598-017-06569-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winata CL, Lapinski M, Pryszcz L, Vaz C, Bin Ismail MH, Nama S, … Mathavan S (2018). Cytoplasmic polyadenylation-mediated translational control of maternal mRNAs directs maternal-to-zygotic transition. Development, 145(1). doi: 10.1242/dev.159566 [DOI] [PubMed] [Google Scholar]
- Wu LL, Russell DL, Norman RJ, & Robker RL (2012). Endoplasmic reticulum (ER) stress in cumulus-oocyte complexes impairs pentraxin-3 secretion, mitochondrial membrane potential (DeltaPsi m), and embryo development. Mol Endocrinol, 26(4), 562–573. doi: 10.1210/me.2011-1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LL, Russell DL, Wong SL, Chen M, Tsai TS, St John JC, … Robker RL (2015). Mitochondrial dysfunction in oocytes of obese mothers: transmission to offspring and reversal by pharmacological endoplasmic reticulum stress inhibitors. Development, 142(4), 681–691. doi: 10.1242/dev.114850 [DOI] [PubMed] [Google Scholar]
- Wu X (2009). Maternal depletion of NLRP5 blocks early embryogenesis in rhesus macaque monkeys (Macaca mulatta). Hum Reprod, 24(2), 415–424. doi: 10.1093/humrep/den403 [DOI] [PubMed] [Google Scholar]
- Xu YW, Peng YT, Wang B, Zeng YH, Zhuang GL, & Zhou CQ (2011). High follicle-stimulating hormone increases aneuploidy in human oocytes matured in vitro. Fertil Steril, 95, 99–104. doi:S0015–0282(10)00658–8 [pii] 10.1016/j.fertnstert.2010.04.037 [DOI] [PubMed] [Google Scholar]
- Yamagata K, & Okada Y (2011). Understanding paternal genome demethylation through live-cell imaging and siRNA. Cell Mol Life Sci, 68(10), 1669–1679. doi: 10.1007/s00018-010-0623-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Ji SY, Dang YJ, Sha QQ, Yuan YF, Zhou JJ, … Fan HY(2016). Oocyte-expressed yes-associated protein is a key activator of the early zygotic genome in mouse. Cell Res, 26(3), 275–287. doi: 10.1038/cr.2016.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu H, Luo S, Lu Z, Chavez-Badiola A, Liu Z, … Huang T (2017). Live birth derived from oocyte spindle transfer to prevent mitochondrial disease. Reprod Biomed Online, 34(4), 361–368. doi: 10.1016/j.rbmo.2017.01.013 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang Q, Liu K, Gao E, Guan H, & Hou J (2018). Treatment of donor cells with recombinant KDM4D protein improves preimplantation development of cloned ovine embryos. Cytotechnology, 70(5), 1469–1477. doi: 10.1007/s10616-018-0224-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylicz JJ, Borensztein M, Wong FC, Huang Y, Lee C, Dietmann S, & Surani MA (2018). G9a regulates temporal preimplantation developmental program and lineage segregation in blastocyst. Elife, 7. doi: 10.7554/eLife.33361 [DOI] [PMC free article] [PubMed] [Google Scholar]


