Abstract
Background
SARS-CoV-2 is a new coronavirus that has spread globally, infecting more than 150000 people, and being declared pandemic by the WHO. We provide here bio-informatic, evolutionary analysis of 351 available sequences of its genome with the aim of mapping genome structural variations and the patterns of selection.
Methods
A Maximum likelihood tree has been built and selective pressure has been investigated in order to find any mutation developed during the SARS-CoV-2 epidemic that could potentially affect clinical evolution of the infection.
Finding
We have found in more recent isolates the presence of two mutations affecting the Non-Structural Protein 6 (NSP6) and the Open Reding Frame10 (ORF 10) adjacent regions. Amino acidic change stability analysis suggests both mutations could confer lower stability of the protein structures.
Interpretation
One of the two mutations, likely developed within the genome during virus spread, could affect virus intracellular survival. Genome follow-up of SARS-CoV-2 spread is urgently needed in order to identify mutations that could significantly modify virus pathogenicity.
Keywords: SARS-CoV-2, COVID-19, Coronavirus, Bio-informatic, Autophagy, Molecular evolution
Introduction
SARS-CoV-2 is the agent of Covid-19, a new coronavirus infection, recently declared pandemic by the WHO, which causes severe pneumonia and acute respiratory distress syndrome (ARDS).1 As of March 16th, more than 150,000 cases of Covid-19 have been notified. While most cases have occurred in mainland China and other Asiatic countries, the virus has also spread to Europe, particularly Italy, where it has caused more than thousand deaths and is overstressing the national health system.
The SARS-CoV-2 genome has been intensely investigated for diagnostics and pathogenicity insights into this virus, as well as to trace its evolution. Presently, more than 350 sequences of virus isolated from several countries are shared in GISAID database. Studies have highlighted the basic structure of the RNA genome, its probable source from a bat coronavirus at Wuhan food and wild animal market (with or without a still unidentified secondary animal host) and the rather close similarity in viral sequences of isolates from different patients2. However, interpretation of genome-driven virus evolution has remained difficult because the published data do still refer to a relatively low number of viral isolates, most of which from China, and few ones from other countries. In particular, there is little information about the evolutionary impact of the few mutations that have been reported by various Authors.
We have here examined all available SARS-CoV-2 sequences with the aim of mapping structural variations of this new coronavirus genome and the patterns of selection, if any, of viral protein genes. We describe the presence of two mutations affecting the Non-Structural Protein 6 (NSP6) and the Open Reading Frame10 (ORF 10) adjacent aminoacidic regions of SARS-CoV-2 and discuss their potential relevance for virus-host interaction, particularly virus-induced cellular autophagy.
Material and Methods
All the 351 sequences available of COVID-19 isolated from humans have been downloaded from GISAID (https://www.gisaid.org/) databank. A dataset has been built including sequences from human and excluding sequences from animals (like bat or pangolin). The Dataset has been aligned using multiple sequence alignment (MAFFT) online tool3 and manually edited using Bioedit program v7.0.5.4 The complete dataset was assessed for presence of phylogenetic signal by applying the likelihood mapping analysis implemented in the IQ-TREE 1.6.8 software (http://www.iqtree.org).5 A maximum likelihood (ML) phylogeny was reconstructed using IQ-TREE 1.6.8 software under the HKY nucleotide substitution model with four gamma categories (HKY+G4), which was inferred in jModelTest (https://github.com/ddarriba/jmodeltest2) as the best fitting model.6
Adaptive Evolution Server (http://www.datamonkey.org/) was used to find possible sites of positive or negative selection. To this purpose the following tests has been used: Fixed Effects Likelihood (FEL)7, Mixed Effects Model of Evolution (MEME)8 and Bayesian Graphical Models for co-evolving sites (BGM).9 These tests allowed to infer the site-specific pervasive selection, the episodic diversifying selection across the region of interest, to identify episodic selection at individual sites and to verify the presence of some co-evolving sites.10 Statistically significant positive or negative selection was based on p value < 0.05 .11
Protein homology modelling has been attempted using the websites SwissModel12 and HHPred.13 I-Tasser has also been used as an alternative source of SARS-CoV-2 protein structure models. I-Mutant2.014 online server has been used to predict the effect of the mutations found under selective pressure on protein stability. Secondary structure and trans-membrane predictions have been carried out with Jpred15, TMHMM16 and Protter17 services. Three-dimensional structures have been analyzed and displayed using PyMOL.18
Role of the funding source
No specific funding source has been received
Results
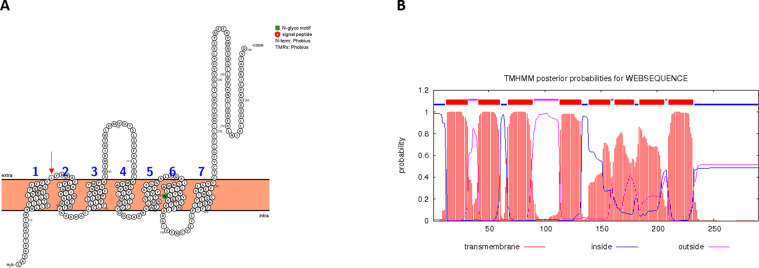
A Maximum Likelihood tree using HKY+G4 model has been built and results have been compared with epidemiological information. Sequences from several different countries have been found in the same clusters while sequences from the same countries have not been found in the same cluster. No separated clade is evident, but all the sequences are part of the same clade. The mutation on the amino acid position 3691 does not appear to be associated within the same cluster with sequences with a leucine on the residue position 9659. Moreover 3 sequences have been found to have a mutation on both the 3691 and the 9659 amino acidic positions. Sequences with a histidine on the position 9659 have been found to belong to distinct clusters. At any rate, clustering of sequence presenting amino acidic mutations did not indicate geographical/epidemiological link with the patients from whom SARS-CoV-2 was isolated (Supplementary Fig. 1). No reliable homology model could be built using SwissModel and HHpred servers. For this reason, the three-dimensional model of NSP6 has been downloaded from I-Tasser website. (https://zhanglab.ccmb.med.umich.edu/C-I-TASSER/2019-nCov/QHD43415_6.pdb.gz). The structural analysis performed using TMHMM and Protter servers have shown that NSP6 protein has 7 putative trans-membrane helices like in other coronaviruses.19
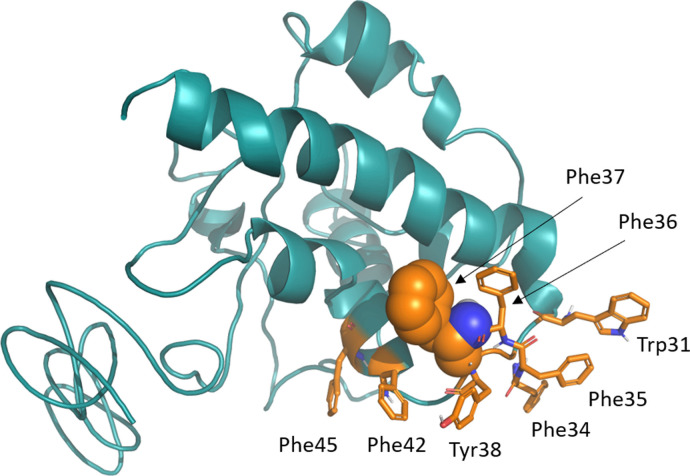
The MEME analysis has shown evidence for episodic synonymous mutations mostly concerning the 3rd codon and not impacting on the overall proteomic asset of the virus. Regarding the FEL analysis, the presence of potential sites under positive selective pressure have been found on 2 sites, on the amino acidic positions 3691 and 9659. These mutations fall on NSP6 and on a region near the Open Reading Frame 10 (ORF 10), respectively. The amino acidic change stability (ACS) analysis has shown that both mutations lead to a lower stability of the protein structures. Namely, at amino acid position 3691 (corresponding to NSP6 position 37), most of the SARS-CoV-2 sequences have a leucine residue while some more recent sequences from Asia, America, Oceania and Europe isolates show phenylalanine (Table 1 ). Both amino acids are non-polar, but phenylalanine has a benzoic ring in the side chain which may stiffen the secondary structure by means of aromatic-aromatic, hydrophobic or stacking interactions. The ACS analysis has shown that this mutation lead to a lower stability of the protein structure (Fig. 1). The mutant position is predicted to be at the C-terminal side of the first transmembrane helix corresponding to the first outer membrane site, close to a sequence region rich of phenylalanine residues (from NSP6 residue position 32 to 40: SLFFFFYENA) of SARS-CoV-2 (Fig. 1 b). According to the structural model, the mutant position is part of a constellation of aromatic residues which includes, in addition to the sequentially contiguous residues, Trp31, Phe42 and Phe45 (Fig. 2). Jpred attributes a helical conformation also to the cytosolic portion of the segment connecting the first to the second transmembrane helix which may facilitate hydrophobic interactions among these aromatic residues.
Table 1.
Table reporting GISAID accession number and country of isolation of the sequences with a mutation on the 3691 aminoacidic position.
| GISAID Accession Number | Country of isolation |
|---|---|
| 408480 | Yunnan |
| 408481 | Chongqing |
| 407988 | Singapore |
| 406223 | USA - Arizona |
| 410984 | France |
| 412974 | Italy |
| 413016 | Brazil |
| 411218 | France |
| 413490 | New Zeland |
| 412975 | Australia |
| 408430 | France |
| 410546 | Italy |
| 413214 | Australia |
| 413213 | Australia |
| 413597 | Australia |
| 413598 | Australia |
| 413597 | Australia |
| 413600 | Australia |
| 410546 | Italy |
| 413595 | Australia |
| 412030 | Hong Kong |
| 412968 | Japan |
| 412969 | Japan |
| 413459 | Japan |
| 408482 | Shandong |
| 412981 | Hubei |
| 413019 | Switzerland |
| 413025 | USA - Washington |
| 413603 | Finland |
| 413605 | Finland |
| 413588 | Netherlands |
| 413589 | Netherlands |
| 413585 | Netherlands |
Fig. 1.
I-TASSER model of NSP6. Residue under positive selective pressure with a p< 0.05 is shown as a sphere. Residues found in the structure proximity are shown in sticks. All residues are marked by the corresponding labels.
Fig. 2.
Results obtained with Protter and TMHMM are shown in panel A and B, respectively.
In the panel A, the residue under positive pressure with p< 0.05 is marked by the red arrow (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
At the amino acidic position 9659 (corresponding to ORF 10 position 3 o 4), most of the SARS-CoV-2 sequences have an arginine residue while some sequences from Australia and America isolates have a histidine residue. The BGM analysis has highlighted the presence of co-evolution between the amino acidic position 9375 and the position 9659 (Table 2 ). Both amino acids are polar, but histidine has an imidazole side chain that suggests a more rigid secondary structure. In fact, ACS analysis has shown that this mutation leads to a lower stability of the protein structure. On the same position, other sequences of SARS-CoV-2 isolates from Australia and New Zealand have shown the presence of a non-polar (leucine) aminoacidic residue (Table 3 ). Also, in this case, the mutation leads to a lower stability of the protein structure, as indicated by ACS analysis.
Table 2.
Table reporting GISAID accession number and country of isolation of the sequences with a histidine on the 9659 aminoacidic position.
| GISAID Accession Number | Country of isolation |
|---|---|
| 412965 | Canada |
| 413490 | New Zeland |
| 412975 | Australia |
| 413214 | Australia |
| 413213 | Australia |
Table 3.
Table reporting GISAID accession number and country of isolation of the sequences with a Leucine on the 9659 aminoacidic position.
| GISAID Accession Number | Country of isolation |
|---|---|
| 411954 | USA - California |
| 410717 | Australia |
| 410718 | Australia |
| 407896 | Australia |
| 407894 | Australia |
Discussion
In this paper, we have examined all available genome sequences (352) of the recently emerged, new coronavirus SARS-CoV-2 which causes a dreadful pneumonia pandemic termed Covid-19 (19). This virus has infected so far more than 120,000 subjects worldwide, with several thousand casualties. Almost all countries have been affected and some of them are now experiencing a rampant rise of disease cases with severe consequences on the stability of health systems. Since disease probable emergence in a wet market of Wuhan, city in the Hubei region of China and recognition of its causative agent, a number of studies on SARS-CoV-2 genome have been published and showed its close similarities (and differences) with the genomes of other coronaviruses isolated from bat, snake, pangolin and SARS CoV.20, 21, 22
Now the attention of most investigators is focused on the potential capability of the viral genome to evolve through mutation, recombination and gene gain and losses, as verified in other human coronaviruses.23 Despite contrary expectations, the selective pressure analysis reported here points out that the genome of SARS-CoV-2 has so far undergone very few mutations, which mostly affect the 3rd codon, and are synonymous, meaning are not going to influence the general molecular structure of this new virus. In addition, it remains difficult to prove the biological relevance of these mutations by pure bioinformatic approach, in the absence of experimental correlates. To somewhat overcome these difficulties, we have here joined bioinformatic and phylogenetic with structural analysis of SARS-CoV-2 protein encoded by mutated genes, in an attempt to obtain some insights into the biological significance and plausibility of the noted mutations. We posit that some of these mutations can provide the virus with useful adaptations in its fight to persist and multiply within humans.
We have particularly assessed two SARS-CoV-2 mutations of non-structural viral proteins, NSP6 and an aminoacidic region near ORF 10, with particular interest into the former protein. NSP6, a common component of both α and β-coronaviruses, locates to the endoplasmic reticulum (ER) and generates autophagosomes.24 We notice that the presence of multiple phenylalanine residues in the outer membrane region of NSP6 should favor the affinity between this region and the ER membrane inducing a more stable binding of the protein to ER. It has been shown that this binding may favor coronavirus infection by compromising the ability of autophagosomes to deliver viral components to lysosomes for degradation.25 Thus, its role would be to limit autophagosome expansion, directly or indirectly by starvation or chemical inhibition of mTOR signaling.26 Nonetheless, the role of autophagy in viral infection is a double-edge sword and we don't have direct evidence that NSP6 mutation does in fact favor viral replication and evasion from cellular immunity or the opposite. In this context, it should be noted that mutational protein analysis speaks for a lower stability of NSP6 upon changing phenylalanine from leucine, but it should be considered that ACS analysis doesn't consider trans-membrane position and other protein interactions. Regarding the aminoacidic region near the ORF 10, previous studies performed on the SARS-CoV reported a 29 nucleotides deletion segment disrupting ORF 9 and, simultaneously, eliminating ORFs 10 and 11. The clinical significance of this deletion is unclear also because it has been found to co-exist with the non-deleted variant in the same host and same clinical specimen (25). A comparison of data from evolutionary and phylogenetic analysis leads us to hypothesize that the mutations are probably unrelated to a strain or a sub-family of the COVID-19 but are due to independent converging evolution of the virus that promote these changes in the viral genome.
In conclusion, the analysis of a relatively wide database of SARS-CoV-2 genomes of worldwide isolates representative of Covid-19, from the start of epidemic in China up to the recent virus spread to European countries, has revealed only two synonymous mutations. Nonetheless, we here speculate that one of these two mutations, i.e the NSP6, could bring to some appreciable change in the expression of SARS-CoV-2 relationship with its host, particularly concerning a critical host anti-viral defense, such as the autophagic lysosomal machinery. Changes in these viral regions should be constantly monitored as they could significantly modify SARS-CoV-2 pathogenicity.
Funding
No specific funding source has been received
Contributors
DB and AC designed the study. DB, MB and MG did the experiments. DB, MB, SP and MC analysed data and DB, AC, SA and RC wrote the article.
Data sharing
Data are available on different websites
Declaration of Competing Interest
We declare no competing interests.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2020.03.058.
Appendix. Supplementary materials
References
- 1.Long‐quan L., Tian H., Yong‐qing W. 2019 novel coronavirus patients’ clinical characteristics, discharge rate and fatality rate of meta‐analysis. J Med Virol. 2020 doi: 10.1002/jmv.25757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Na Z., Dingyu Z., Wenling W. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020 doi: 10.1056/nejmoa2001017. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinf. 2019;20(4):1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall T.A. BioEdit A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 5.Nguyen L.T., Schmidt H.A., von Haeseler A. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darriba D., Taboada G.L., Doallo R. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kosakovsky S.L.P., Frost S.D.W. Not so different after all: a comparison of methods for detecting amino acid sites under selection molecular. Biol Evolut. 2005;22(5):1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 8.Murrell B., Wertheim J.O., Moola S., Weighill T., Scheffler K., Kosakovsky Pond S.L. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012;8(7) doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poon A.F., Lewis F.I., Frost S.D., Kosakovsky Pond S.L. Spidermonkey: rapid detection of co-evolving sites using Bayesian graphical models. Bioinformatics. 2008;24(17):1949–1950. doi: 10.1093/bioinformatics/btn313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weaver S., Shank S.D., Spielman S.J, Li M., Muse S.V. and Kosakovsky Pond S.LDatamonkey 2.0: a modern web application for characterizing selective and other evolutionary processes2018;35(3):773-777. doi: 10.1093/molbev/msx335. [DOI] [PMC free article] [PubMed]
- 11.Waterhouse A., Bertoni M., Bienert S. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zimmermann L., Stephens A., Nam S.Z. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J Mol Biol. 2018;430(15):2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER Suite: Protein structure and function prediction. Nat Methods. 2015;12(1):7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capriotti E., Fariselli P., Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33(Web Server issue):W306–W310. doi: 10.1093/nar/gki375. http://gpcr.biocomp.unibo.it/cgi/predictors/I-Mutant2.0/I-Mutant2.0.cgi [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drozdetskiy A, Cole C, Procter J, Barton GJ. JPred4: a protein secondary structure prediction server. Nucleic Acids Res July;43(W1):W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed]
- 16.Moller S., Croning M.D.R., Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17(7):646‐653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- 17.Omasits U., Ahrens C.H., Müller S., Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2014;30(6):884–886. doi: 10.1093/bioinformatics/btt607. https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 18.Schrödinger LLC (2015) The {PyMOL} Molecular Graphics System, Version∼1.8.
- 19.Oostra M., Hagemeijer M.C., van Gent M. Topology and membrane anchoring of the coronavirus replication complex: Not All Hydroph Doma NsP3 and NsP6 Are Membrane Spanning. J Virol. 2008;82(24):12392–12405. doi: 10.1128/JVI.01219-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200313-sitrep-53-covid-19.pdf?sfvrsn=adb3f72_2
- 21.Kandeel M., Ibrahim A.A., Fayez M., Al‐Nazawi M. From SARS and MERS CoVs to SARS‐CoV‐2: moving toward more biased codon usage in viral structural and non‐structural genes. J Med Virol. 2020 doi: 10.1002/jmv.25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X., Zai J., Zhao Q. Evolutionary history, potential intermediate animal host, and cross‐species analyses of SARS‐CoV‐2. J Med Virol. 2020:1–10. doi: 10.1002/jmv.25731. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benvenuto D., Giovanetti M., Salemi M., Prosperi M., De Flora C., Junior Alcantara L.C., Angeletti S., Ciccozzi M.The global spread of 2019-nCoV: a molecular evolutionary analysis, Pathogens and Global Health, doi: 10.1080/20477724.2020.1725339. [DOI] [PMC free article] [PubMed]
- 24.Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25(1):35–48. doi: 10.1016/j.tim.2016.09.001. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou A., Li S., Khan F.A., Zhang S.. Autophagy postpones apoptotic cell death in PRRSV infection through Bad-Beclin1 interaction. Virulence. 2016;7(2):98–109. doi: 10.1080/21505594.2015.1131381. pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang J.W., Cheung J.L., Chu I.M., Sung J.J., Peiris M., Chan P.K. The Large 386-nt Deletion in SARS-associated coronavirus: evidence for quasispecies? J Infect Diseas. 2006;194(6):808–813. doi: 10.1086/507044. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.