Highlights
-
•
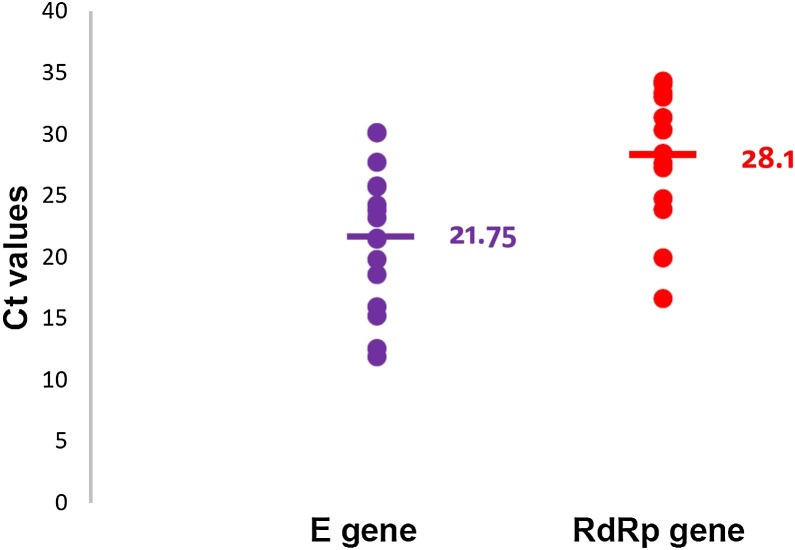
Ct values of E gene were significantly lower than RdRp gene target.
-
•
COVID-19 case definition not specific, other respiratory viruses in 42 % of samples.
-
•
AusDiagnostics assay sensitive but not specific for the detection of SARS-CoV-2.
Keywords: SARS-CoV-2, NAT, Covid-19
Abstract
Introduction
There is limited data on the analytical performance of commercial nucleic acid tests (NATs) for laboratory confirmation of COVID-19 infection.
Methods
Nasopharyngeal, combined nose and throat swabs, nasopharyngeal aspirates and sputum was collected from persons with suspected SARS-CoV-2 infection, serial dilutions of SARS-CoV-2 viral cultures and synthetic positive controls (gBlocks, Integrated DNA Technologies) were tested using i) AusDiagnostics assay (AusDiagnostics Pty Ltd); ii) in-house developed assays targeting the E and RdRp genes; iii) multiplex PCR assay targeting endemic respiratory viruses. Discrepant SARS-CoV-2 results were resolved by testing the N, ORF1b, ORF1ab and M genes.
Results
Of 52 clinical samples collected from 50 persons tested, respiratory viruses were detected in 22 samples (42 %), including SARS CoV-2 (n = 5), rhinovirus (n = 7), enterovirus (n = 5), influenza B (n = 4), hMPV (n = 5), influenza A (n = 2), PIV-2 (n = 1), RSV (n = 2), CoV-NL63 (n = 1) and CoV-229E (n = 1). SARS-CoV-2 was detected in four additional samples by the AusDiagnostics assay. Using the in-house assays as the "gold standard", the sensitivity, specificity, positive and negative predictive values of the AusDiagnostics assay was 100 %, 92.16 %, 55.56 % and 100 % respectively.
The Ct values of the real-time in-house-developed PCR assay targeting the E gene was significantly lower than the corresponding RdRp gene assay when applied to clinical samples, viral culture and positive controls (mean 21.75 vs 28.1, p = 0.0031).
Conclusions
The AusDiagnostics assay is not specific for the detection SARS-CoV-2. Any positive results should be confirmed using another NAT or sequencing. The case definition used to investigate persons with suspected COVID-19 infection is not specific.
1. Introduction
On 31 December 2019, the Chinese Center for Disease Control and Prevention reported to the World Health Organization (WHO) a series of patients with pneumonia of uncertain aetiology in Wuhan city, Hubei province, China [1]. The pathogen responsible for this outbreak was subsequently identified as a novel group 2B betacoronavirus, designated as Severe acute respiratory syndrome coronavirus-2 (SARS CoV-2). It shares approximately79 % homology to the Severe Acute Respiratory Syndrome coronavirus (SARS-CoV) and 50 % with Middle East respiratory syndrome coronavirus (MERS-CoV) and is most closely related to two bat-derived SARS-like coronaviruses [2].
Rapid escalation of case numbers has ensued; as of 7 April 2020, 1, 279,722 cases of COVID-19 infections have been confirmed worldwide. COVID-19 was declared a pandemic on 11 March 2020 by the WHO [WHO, COVID-19 Situation Report-78]. The first case of SARS-CoV-2 in Australia was confirmed on 24 January 2020 in Victoria, and as of 7 April 2020, there have been a total of 5844 cases confirmed in Australia with New South Wales accounting for 2734 infections. (Department of Health, Australia).
Expeditious and accurate laboratory diagnosis of persons with COVID-19 infection followed by appropriate infection control measures is key to preventing further spread of infection, particularly in the absence of effective antiviral therapy. The genetic sequence of SARS-CoV-2 was released on 10 January 2020, and the WHO subsequently recommended several assays for the detection of SARS-CoV-2 [3].
Within NSW, the Centre for Infectious Diseases and Microbiology Laboratory Services, in the NSW Health Pathology-Institute of Clinical Pathology and Medical Research, was designated as the public health laboratory responsible for specific testing of SARS-CoV-2 during the earliest phase of the outbreak. During this initial phase, commercial assays were not available in Australia. However, on 31 January 2020, a commercial assay for the specific detection of SARS-CoV-2 (AusDiagnostics Pty Ltd, Mascot, NSW, Australia) was announced [4]. We evaluated this assay’s performance against real-time PCR (RT-PCR) assays using SARS-CoV-2 gene targets recommended by the WHO [5].
2. Methods
2.1. Clinical samples
Samples were collected from persons with suspected SARS-CoV-2 infection according to the case definition outlined in the Communicable Diseases Network Australia (CDNA) National Guidelines for Public Health Units for managing COVID-19. A suspect case was defined as meeting both clinical criteria, fever ≥ 38 °C or history of fever or acute respiratory infection (e.g. cough, shortness of breath, sore throat) and epidemiology criteria which includes international travel in the 14 days prior to symptom onset to a country known to have cases of COVID-19 or close contact with a confirmed or probable case. This case definition continues to be updated and is (available at: https://www1.health.gov.au/internet/main/publishing.nsf/Content/cdna-song-novel-coronavirus.htm).
Upper respiratory tract (URT) samples [including nasopharyngeal swabs (NPS), combined nose and throat swabs (NTS) and nasopharyngeal aspirates (NPA)] were collected under appropriate infection control measures, placed in 1−3 mL of viral or universal transport media, transferred to the laboratory and tested within 12 h of specimen receipt. As aerosol generating procedures (collection of induced sputum, tracheal aspirates, or bronchoalveolar lavage) were not performed in patients not requiring invasive ventilatory support, sputum was the only lower respiratory tract (LRT) sample available for testing. Serial sampling of the URT and/or LRT was performed in patients with confirmed SARS-CoV-2 infection. The specimen types, collection, and processing used in this study were in accordance with those recommended by the AusDiagnostics Assay package insert.
2.2. SARS-CoV-2 culture and positive controls
Viral cultures using Vero E6 cells were performed on samples where SARS-CoV-2 nucleic acid was detected. Cells were inspected daily for cytopathic effects, and when observed, this was confirmed by nucleic acid testing (NAT) of cell culture supernatant, using the ORF1b target. Serial dilutions of a patient’s SARS-CoV-2 cell culture supernatant (neat to 10−7) and synthetic positive control (neat and 10-9, gBlocks gene fragments covering E, RdRp and N genes, Integrated DNA Technologies, Coralville, Iowa, USA) were also tested by NAT as outlined below.
2.3. Nucleic acid extraction, amplification and detection
RNA extraction was performed using the BioRobot EZ1 and EZ1 Virus Mini Kit v2.0 (Qiagen, Valencia, CA, USA) according to the manufacturers’ instructions, using 200 u L of specimen. Nucleic acid amplification and detection was then performed using the methods outlined below.
From January 22 to January 31, 2020, SARS-CoV-2 was detected using an in-house developed gel-based NAT targeting the E gene as described by Corman et al. [6] as primers but not probes were available. From 1 February 2020 onwards, SARS-CoV-2 detection was performed using RT-PCR assays targeting both the E and RdRp genes. The RdRp gene P2 probe is specific for SARS-CoV-2 and does not detect SARS-CoV [7]. Specimens collected prior to 31 January 2020 previously tested using the gel-based NAT were retrospectively tested using the same RT-PCR assays.
Samples were also tested using the AusDiagnostics assay (Coronavirus Typing version 01) a research use only assay, which is a RT-PCR assay that distinguishes between SARS-CoV-2 and other endemic coronaviruses (HKU-1, OC43, 229E and NL63), SARS-CoV and MERS-CoV. Data analysis is performed using proprietary software (RealTime_PCR v7.7) with positive, negative or inhibited results provided following interpretation. Typically, a positive result satisfies pre-defined criterion for cycle threshold (Ct) and melting temperature (Tm) for the amplified gene target. Where discrepant AusDiagnostics assay results were encountered (relative to the in-house assays targeting the E and/or RdRp genes), the cycling and melt curves of each sample were manually examined, followed by further testing of four other WHO recommended targets (the N, ORF1b, ORF1ab and M genes) [5] for discrepant analysis, with the definitive result determined by the consensus of the assays.
In addition to SARS-CoV-2-specific testing, all samples were also tested using an in-house multiplex respiratory virus panel, which detects influenza A, influenza B, respiratory syncytial virus [RSV], parainfluenza viruses [PIV] 1−3, human adenovirus and human metapneumovirus [hMPV], as previously described [8].The primers and probes, mastermix composition and cycling conditions used in the in-house assays specifically detecting SARS-CoV-2 are outlined in Table 4.
Table 4.
Primers and probes used in the in-house assays specifically detecting SARS-CoV-2, mastermix composition and cycling conditions.
| E_ Sarbeco_F1forward primer | ACAGGTACGTTAATAGTTAATAGCGT |
| E_Sarbeco_E2 reverse primer | ATATTGCAGCAGTACGCACACA |
| E_Sarbeco_P1 probe | FAM-ACACTAGCCATCCTTACTGCGCTTCG-BHQ1 |
| RdRp_ SARSr-F forward primer | GTGARATGGTCATGTGTGGCGG |
| RdRp_SARSr_reverse primer | CARATGTTAAASACACTATTAGCATA |
| RdRp_SARS-P2 probe* | FAM-CAGGTGGAACCTCATCAGGAGATGC-BHQ1 |
| N gene forward | CACATTGGCACCCGCAATC |
| N gene reverse | GAGGAACGAGAAGAGGCTTG |
| N gene probe | FAM-ACTTCCTCAAGGAACAACATTGCCA-BHQ1 |
| ORF1ab forward | CCCTGTGGGTTTTACACTTAA |
| ORF1ab reverse | ACGATTGTGCATCAGCTGA |
| ORF1ab probe | FAM-CCGTCTGCGGTATGTGGAAAGGTTATGG-BHQ1 |
| ORF1b forward | CATGGTGGACAGCCTTTGTTAC |
| ORF1b reverse | TCGCGTGGTTTGCCAAGAT |
| ORF1b probe | FAM- AATGTGAATGCGTCATCATCTGAAGCA-BHQ1 |
| M gene forward | CAAGGACCTGCCTAAAGAAATCAC |
| M gene reverse | ACGCTGCGAAGCTCCCAAT |
| M gene probe | FAM- TGTTGCTACATCACGAACGCTTTC-BHQ1 |
| Master Mix | ||
|---|---|---|
| AgPath-ID™ One-Step RT-PCR Kit (Applied Biosystems™ Catalog number: 4387424 m) | ||
| 1x (20 + 5) reaction uL | ||
| AgPath RT mix 2x | 2x | 10 |
| Forward primer | 20 μM | 0.625 |
| Reverse primer | 20 μM | 0.625 |
| Probe (FAM/BHQ1) | 20uM | 0.312 |
| BGL PCO3 | 100uM | 0.0625 |
| BGL PCO4 | 100uM | 0.0625 |
| BGL probe BGL Quasar 670 | 100uM | 0.0312 |
| Water | 7.4828 | |
| AgPath 25x RT enzyme | 20 μM | 0.8 |
| Total 20 | ||
| Sample extract | 5 | |
| Cycling conditions LC480 II | |||
|---|---|---|---|
| Cycles | Temperature | Time | Ramp rate (°C/s) |
| 1x | 45 °C | 15 min | 4.4 |
| 1x | 95 °C | 15 min | 4.4 |
| 45x | 95 °C | 15 s | 4.4 |
| 60 °C | 45 s | 2.2 | |
| 1x | 40 °C | 30 s | 2.2 |
*Specific for SARS CoV-2 that does not detect SARS-CoV.
Reaction volume 20 μL+5 μL template. We run at 16 μL+ 4 u L volumes (economy).
2.4. Genome sequencing
Three full-length sequences of the SARS-CoV-2 genome from clinical samples and/or viral cultures were obtained by amplicon-based Illumina sequencing (with and without enrichment using the Illumina Pan Viral Kit), and submitted to GISAID, accession IDs: EPI_ISL_407893, 408976 and 408977.
3. Results
The median age of the persons under investigation was 31.5 years of age (range 0–84) and 30 were males (58 %). Samples were collected from the URT and LRT (sputa) from adults. Nine of 50 (18 %) persons were children (<16 years old), with six providing NTS or NPS, and three providing NPA.
Of the 52 specimens tested, respiratory viruses were detected in 22 (42 %) samples, including SARS-CoV-2 (n = 5), rhinovirus (n = 7), enterovirus (n = 5), influenza B (n = 4), hMPV (n = 5), influenza A (n = 2), PIV-2 (n = 1), RSV (n = 2), CoV-NL63 (n = 1) and CoV-229E (n = 1). Coinfections were detected in four samples (one sample each of SARS-CoV-2 plus enterovirus, rhinovirus plus PIV-2, RSV plus rhinovirus, and rhinovirus plus enterovirus).
3.1. AusDiagnostics versus in-house developed assays
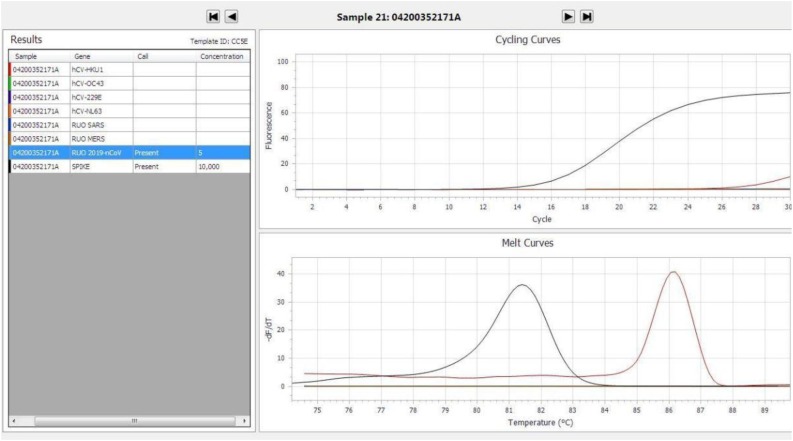
SARS−COV-2 was detected in nine samples by the AusDiagnostics assay, but this was only confirmed by in-house assays targeting E, RdRp genes in five samples. In the remaining four samples, the E, RdRp, N, ORF1b, ORF1ab and M gene results were negative, and these were deemed as false positive results. Using the in-house assays as the “gold standard”, the sensitivity, specificity, positive and negative predictive values of the AusDiagnostics assay 100 %, 92.16 %, 55.56 % and 100 % respectively. Further interrogation of the cycling and melt curves of the false positive samples revealed a relatively flat, non-sigmoidal amplification curve (Fig. 1 ). By manual interpretation, the results of such amplification curves would have been called negative, despite the melt curves suggesting a positive test result.
Fig. 1.
Example of a false positive AusDiagnostics assay result showing a flat, non-sigmoidal amplification curve.
Of note, no other coronaviruses (HKU-1, OC43, 229E, NL63, SARS-CoV and MERS-CoV) were detected by the AusDiagnostics assay in these four samples. However, the AusDiagnostics assay detected other coronaviruses (NL63 and 229E) in two other samples. The results of testing by the in-house and AusDiagnostics assays are shown in Table 1, Table 2, Table 3 .
Table 1.
Results of samples testing positive for SARS-CoV-2 on AusDiagnostics assay.
| Age (yrs) | Sex | Days since symptom onset | Sample | Other respiratory virus | AusDiagnostics RUO 2019-nCoV | E-gene | RdRp-gene | N-gene | ORF1b-gene | ORF1ab gene | M-gene |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Take-off cycle | Ct | Ct | Ct | Ct | Ct | Ct | |||||
| 53 | M | 9 | NPS | ND | 20.76 | 30.14 | 34.11 | ND | 40 | 31.46 | ND |
| 44 | M | 3 | NTS | ND | 8.42 | 21.48 | 24.76 | 27.21 | 24.19 | 21.85 | 27.47 |
| 21 | F | 2 | NTS | ND | 13.99 | 25.72 | 28.46 | 32.09 | 28.58 | 25.79 | 32.19 |
| 35 | M | 7 | NTS | ND | 19.68 | 30.14 | 31.38 | 37.81 | 34.53 | 40 | 40 |
| 44 | M | 8 | Sputum | ND | 15.39 | 24.26 | 27.28 | 34.68 | 29.36 | 26.65 | 30.02 |
| 40 | M | 6 | NTS | ND | 25.16 | ND | ND | ND | ND | ND | ND |
| 43 | F | 4 | NTS | ND | 24.3 | ND | ND | ND | ND | ND | ND |
| 11m | M | 2 | NPA | ND | 14.66 | ND | ND | ND | ND | ND | ND |
| 44 | M | 13 | NPS | ND | 24.83 | ND | ND | ND | ND | ND | ND |
ND – not detected, NTS – combined nose and throat swab, NPS – nasopharyngeal swab, NPA – nasopharyngeal aspirate.
Table 2.
Results of cell culture titres supernatant and Synthetic positive Control (gBlock (E, RdRp, & N gene) titres.
| AusDiagnostics |
In-house |
|||||
|---|---|---|---|---|---|---|
| Take-off Cycle |
Temp |
E-gene | RdRp-gene | |||
| RUO 2019-nCoV | SPIKE | RUO 2019-nCoV | SPIKE | Ct | Ct | |
| Cell Culture NEAT | ND | ND | ND | ND | ND | ND |
| Cell Culture -1 | ND | ND | ND | ND | 15.98 | 19.95 |
| Cell Culture -2 | ND | ND | ND | ND | 19.83 | 23.89 |
| Cell Culture -3 | ND | ND | ND | ND | 23.21 | 27.66 |
| Cell Culture -4 | 13.89 | 12.57 | 85.94 | 81.8 | 25.81 | 30.36 |
| Cell Culture -5 | 18.19 | 12.63 | 85.91 | 81.8 | 27.75 | 33.38 |
| Cell Culture -6 | 21.71 | 12.79 | 85.94 | 81.8 | ND | 35.21 |
| Cell Culture -7 | ND | 14.3 | ND | 81.8 | ND | ND |
| gBlock -3 | ND | 13.36 | ND | 81.8 | 11.9 | 34.35 |
| gBlock -4 | ND | 13.06 | ND | 81.8 | 15.23 | 33.03 |
| gBlock -5 | ND | 13.02 | ND | 81.8 | 18.6 | ND |
| gBlock -6 | ND | 13.01 | ND | 81.8 | 21.57 | ND |
| gBlock -7 | ND | 13.18 | ND | 81.8 | 23.84 | ND |
| gBlock -8 | ND | 13.16 | ND | 81.8 | ND | ND |
| gBlock -9 | ND | 13.01 | ND | 81.8 | ND | ND |
| H2O | ND | 12.85 | ND | 81.8 | ND | ND |
| AUSD RESP CONTROL | ND | 13.43 | ND | 81.8 | ND | ND |
Table 3.
Summary of other results for clinical samples.
| Age (yrs) | Sex | Days since symptom onset | Sample | Other respiratory virus | Aus Diagnostics RUO 2019-nCoV Take-off cycle | E-gene Ct | RdRp-gene Ct |
|---|---|---|---|---|---|---|---|
| 36 | F | 0 | NTS | ND | ND | ND | ND |
| 11 | F | 0 | NTS | ND | ND | ND | ND |
| 61 | F | 0 | NPS | Influenza B | ND | ND | ND |
| 30 | M | 5 | NPA | ND | ND | ND | ND |
| 4 | M | 2 | NPA | ND | ND | ND | ND |
| 32 | F | 2 | NPS | ND | ND | ND | ND |
| 33 | M | 5 | NPA | Influenza B | ND | ND | ND |
| 28 | M | 11 | NTS | ND | ND | ND | ND |
| 21 | F | 8 | NPS | ND | ND | ND | ND |
| 24 | M | 4 | NTS | Rhinovirus & Enterovirus | ND | ND | ND |
| 2 | M | 3 | NPS | Influenza A & Rhinovirus | ND | ND | ND |
| 21 | M | 1 | NTS | ND | ND | ND | ND |
| 52 | M | 7 | NPS | ND | ND | ND | ND |
| 29 | F | 4 | NPS | ND | ND | ND | ND |
| 29 | F | 3 | NPS | ND | ND | ND | ND |
| 23 | F | 2 | NPS | ND | ND | ND | ND |
| 19 | M | NA | NTS | ND | ND | ND | ND |
| 2 | M | 3 | NPA | hMPV | ND | ND | ND |
| 84 | M | 0 | NPS | Enterovirus | ND | ND | ND |
| 36 | F | 5 | NTS | ND | ND | ND | ND |
| 22 | M | 5 | NPS | Enterovirus | ND | ND | ND |
| 74 | F | 0 | NTS | ND | ND | ND | ND |
| 1 | M | 0 | NTS | Influenza B | ND | ND | ND |
| 8 | M | 0 | NTS | hMPV | ND | ND | ND |
| 62 | M | ?? | NTS | ND | ND | ND | ND |
| 50 | 0 | NTS | Rhinovirus & PIV-2 | ND | ND | ND | |
| 22 | M | 1 | NTS | ND | ND | ND | ND |
| 22 | M | 5 | NTS | Rhinovirus | ND | ND | ND |
| 33 | M | 0 | NTS | Rhinovirus | ND | ND | ND |
| 9 | F | 5 | NTS | hMPV | ND | ND | ND |
| 25 | F | 5 | NTS | ND | ND | ND | ND |
| 43 | M | 1 | NTS | ND | ND | ND | ND |
| 39 | M | 7 | NTS | RSV and Enterovirus | ND | ND | ND |
| 32 | F | 3 | NPS | Influenza B & Enterovirus | ND | ND | ND |
| 25 | F | 1 | NTS | ND | ND | ND | ND |
| 25 | F | 2 | NTS | Rhinovirus | ND | ND | ND |
| 33 | M | NA | NTS | Rhinovirus | ND | ND | ND |
| 50 | M | 3 | NTS | Influenza A | ND | ND | ND |
| 5 | F | 7 | NTS | ND | ND | ND | ND |
| 38 | M | 1 | NPS | RSV | ND | ND | ND |
| 36 | M | 2 | NPS | hMPV | ND | ND | ND |
| 40 | M | 5 | NTS | CoV-229E | ND | ND | ND |
| 44 | M | 4 | NPS | hMPV & CoV-NL63 | ND | ND | ND |
ND – not detected, NTS – combined nose and throat swab, NPS – nasopharyngeal swab, NPA – nasopharyngeal aspirate, hMPV – human metapneumovirus, CoV – corona virus, PIV-2 – parainfluenza virus 2, RSV – respiratory syncytial virus.
3.2. Analytical sensitivity of E versus RdRp gene assays
In all clinical samples and serial dilutions of a SARS-CoV-2 culture isolate and gBlock positive synthetic controls tested, the Ct values of the in-house RT-PCR assay targeting the E Gene was significantly lower than the corresponding RdRp gene assay (mean 21.75 vs 28.10, p = 0.0031 by Mann Whitney test) (Fig. 2 ).
Fig. 2.
Comparison of cycle threshold values of the E gene versus RdRp gene for the detection of SARS-CoV-2..
E gene mean Ct value = 21.75, RdRp mean Ct value = 28.1, p = 0.0031.
4. Discussion
Our experience highlights important considerations for laboratory preparedness in the early phase of the COVID-19 epidemic. Laboratory confirmation of SARS CoV-2 infection is important for individual patient care and public health management to limit the spread of infection by quarantine (which may include their close contacts). In this regard, the clinical laboratory is vital in providing accurate and timely diagnostics to confirm or exclude infection.
As SARS-CoV-2 was rapidly identified as the causative pathogen of the cases of pneumonia of unknown aetiology in Wuhan, laboratories had to quickly develop and evaluate the analytical performance of diagnostic NATs. Several assays targeting different regions of the SARS-CoV-2 genome has been proposed [3], but there are limited data on the analytical performance of these assays. In the early phases of the outbreak, no commercial assays were available for testing within Australia. Assays detecting SARS-CoV-2, whether in-house developed or commercial, should be thoroughly evaluated to ensure they are fit for purpose.
The initial NAT in each laboratory may also be limited by the availability of testing reagents, as evidenced in our laboratory initially using a gel-based assay targeting the E gene in the absence of probes. Following probe procurement, the NAT was changed to RT-PCR assays and initial samples tested using gel-based assay were re-tested using RT-PCR. This is the preferred method as viral loads (and hence viral kinetics) may be determined in a semi-quantitative fashion by correlating the Ct values of positive tests. In the present study, the Ct values of the E gene assay were consistently lower than the corresponding values in the RdRp assay in SARS-CoV-2 positive clinical samples, serial dilutions of SARS-CoV-2 cultures and synthetic positive controls, highlighting the superior sensitivity of the E gene assay as a screening test.
In the early phases of the outbreak, it was suggested that laboratories use a pan-coronavirus NAT followed by sequencing of amplicons from non-conserved regions for characterization and confirmation of SARS-CoV-2, or amplification and detection of SARS-CoV-2 specific sequences without further sequencing [3]. The use of sequencing (although valuable for confirmation of NAT) in the diagnostic algorithm increases laboratory turnaround times and may not be ideal when there are substantial numbers of specimens that require testing. Furthermore, although sequencing may be used to confirm the diagnostic accuracy of newly developed tests for novel pathogens, this technology may not be readily available in all diagnostic laboratories. The use of sequencing can assist with investigation of cases with no clear links at a local level and sharing of sequencing data to platforms such as GISAID can contribute to the understanding of the viral evolution.
The optimal type of respiratory sample that should be collected to confirm or exclude SARS-CoV-2 needs to be determined. In our study, four patients were diagnosed with SARS-CoV-2 infection. One patient had SARS-CoV-2 detectable from his LRT (sputum) but not URT sample on day eight of infection following symptom onset. This discrepant result may reflect differences in sampling quality or progression of infection from the URT to the LRT, and supports the recommendations to test LRT samples where available [PHLN], similar to our previous experience with A(H1N1)pdm09 infection [9]. The detection of SARS-CoV-2 in LRT samples is suggestive of active viral replication within the LRT, particularly in patients with deteriorating respiratory function and abnormal radiological imaging. Together with Ct values, this may be useful in guiding the clinical management of such patients. The significance of viral co-infection in COVID-19 disease on clinical outcomes is unknown. Other samples where SARS-CoV-2 have been detected by NATs include saliva, stool, rectal swabs and urine [CCDC, To].
This study has several limitations. Non-respiratory tract samples were unavailable for testing. There is emerging evidence that there may be prolonged viral shedding in stools from persons with COVID-19 infection, similar to those infected with SARS-CoV [10,11]. The specific gene target(s) of the AusDiagnostics assay are not known, so we were not able to determine the exact reason for the reduced analytical specificity. The respiratory multiplex assay used in this study is an in-house assay that does not include other endemic coronaviruses such as NL-63, OC43, 229E and HKU-1 so we were unable to verify the results of non-SARS-CoV-2 coronaviruses tested using the AusDiagnostics assay.
In conclusion, we determined that the commercial AusDiagnostics assay was less reliable than an in-house RT-PCR using WHO recommended gene targets for the detection of SARS-CoV-2. Assay validation and verification are required to ensure that commercial assays used to detect SARS-CoV-2 are fit for purpose. Further data are awaited regarding the use of assays with different SARS-CoV-2 targets, viral loads and kinetics to better guide individual patient management and infection control measures. The optimal testing algorithm to detect SARS-CoV-2 for diagnostic pathology providers and public health reference laboratories will depend on their healthcare systems, laboratory capacity and capability, particularly with the anticipated substantial increase in testing demands as the epidemic progresses and revised case definitions broaden the indications for testing.
Disclosure
The kits for the AusDiagnostics assay were supplied to the laboratory by AusDiagnostics Pty Ltd free of charge, no funding was provided for this study. All work including study design, execution, analysis, write up and editing was undertaken independently by the authors listed.
CRediT authorship contribution statement
H. Rahman: Conceptualization, Methodology, Validation, Formal analysis, Data curation, Writing - original draft, Writing - review & editing, Visualization. I. Carter: Conceptualization, Methodology, Validation, Formal analysis, Resources, Data curation, Project administration. K. Basile: Validation, Formal analysis, Data curation, Writing - original draft, Writing - review & editing, Visualization. L. Donovan: Investigation. S. Kumar: Investigation. T. Tran: Investigation. D. Ko: Investigation. S. Alderson: Investigation. T. Sivaruban: Investigation. J.-S. Eden: Investigation. R. Rockett: Investigation. M.V. O’Sullivan: Writing - review & editing, Supervision. V. Sintchenko: Writing - review & editing, Supervision. S.C-A. Chen: Writing - review & editing, Supervision. S. Maddocks: Writing - review & editing, Supervision. D.E. Dwyer: Writing - review & editing, Supervision. J. Kok: Conceptualization, Methodology, Validation, Formal analysis, Writing - original draft, Writing - review & editing, Supervision, Project administration.
Acknowledgements
This study was supported by the Prevention Research Support Program funded by the New South Wales Ministry of Health and the NHMRC Centre for Research Excellence in Emerging Infectious Diseases and the Australian Partnership for Preparedness Research on Infectious Disease Emergencies.
References
- 1.Zhu N., Zhang D., Wang W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu R., Zhao X., Li J. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020 doi: 10.1016/S0140-6736(20)30251-8). (available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Novel coronavirus (SARS-CoV-2) technical guidance: Laboratory testing for SARS-CoV-2 in humans. (available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance).
- 4.AusDiagnostics Press Release. 3 February 2020 (available at: https://www.ausdiagnostics.com/press-releases.html).
- 5.World Health Organization. Coronavirus disease 2019 (COVID-19) Situation Report – 78 (available at: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200407-sitrep-78-covid-19.pdf?sfvrsn=bc43e1b_2).
- 6.Corman V.M., Landt O., Kaiser M. Detection of 2019 novel coronavirus (SARS-CoV-2) by real time RT-PCR. Euro Surveill. 2020;25:3. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. 23 Jan 2020 (available at: https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2020.25.3.2000045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corman V., Bleicker T., Brunink S., Drosten D., Landt O., Koopmans M., Zambon M. 2020. Diagnostic Detection of SARS-CoV-2 by Real-time RT-PCR. Available at: https://www.who.int/docs/default-source/coronaviruse/protocol-v2-.pdf?sfvrsn=a9ef618c_2) [Google Scholar]
- 8.Ratnamohan V.M., Taylor J., Zeng F. Pandemic clinical case definitions are non-specific: multiple respiratory viruses circulating in the early phases of the 2009 influenza pandemic in New South Wales, Australia. Virol. J. 2014;11:113. doi: 10.1186/17430422X-11-113. (Available at: https://virologyj.biomedcentral.com/articles/10.1186/1743-422X-11-113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blyth C.C., Iredell J.R., Dwyer D.E. Rapid-test sensitivity for novel swine-origin influenza A H1N1 virus in humans. N. Engl. J. Med. 2009;361:2493. doi: 10.1056/NEJMc0909049. [DOI] [PubMed] [Google Scholar]
- 10.Zhang W., Du R., Li B. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 2020;9 doi: 10.1080/22221751.2020.1729071. doi:10/1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu D., Zhang Z., Jin L. Persistent shedding of viable SARS-CoV in urine and stool of SARS patients during the convalescent phase. Eur. J. Clin. Microbiol. Infect. Dis. 2005;24:165–171. doi: 10.1007/s10096-005-1299-5. [DOI] [PMC free article] [PubMed] [Google Scholar]