Abstract
During a survey of xylarialean fungi in Northern Iran, several specimens that showed affinities to the Hypoxylon rubiginosum complex were collected and cultured. A comparison of their morphological characters, combined with a chemotaxonomic study based on high performance liquid chromatography, coupled with diode array detection and mass spectrometry (HPLC-DAD/MS) and a multi-locus phylogeny based on ITS, LSU, rbp2 and tub2 DNA sequences, revealed a new species here described as Hypoxylon guilanense. In addition, Hypoxylon rubiginosumsensu stricto was also encountered. Concurrently, an endophytic isolate of the latter species showed strong antagonistic activities against the Ash Dieback pathogen, Hymenoscyphus fraxineus, in a dual culture assay in our laboratory. Therefore, we decided to test the new Iranian fungi for antagonistic activities against the pathogen, along with several cultures of other Hypoxylon species that are related to H. rubiginosum. Our results suggest that the antagonistic effects of Hypoxylon spp. against Hym. fraxineus are widespread and that they are due to the production of antifungal phomopsidin derivatives in the presence of the pathogen.
Keywords: Ascomycota , Chemotaxonomy, Chemical ecology, Hypoxylaceae , Natural Products, Taxonomy, one new species
Introduction
Hypoxylon Bull., 1791 is one of the largest genera of the Xylariales and comprises more than 200 species, which are mainly associated with angiosperm trees as saprotrophs and endophytes and are predominant in all forest ecosystems of the world (Daranagama et al. 2018; Helaly et al. 2018).
It traditionally belonged to the Xylariaceae until a recent phylogenetic study has resulted in a re-arrangement of the genera of stromatic Xylariales and the resurrection of the family Hypoxylaceae (Wendt et al. 2018). In this study and during the follow-up work by Lambert et al. (2019), genera like Hypomontagnella and Pyrenopolyporus were segregated from Hypoxylon, but the genus remained paraphyletic, indicating that further taxonomic segregation will eventually become necessary.
While the type species of Hypoxylon, H. fragiforme, belongs to a relatively small clade in the phylogeny of Wendt et al. (2018), the largest clades were comprised of the species of the “Hypoxylon rubiginosum complex” sensu Ju and Rogers (1996). Of these species, many had been lumped in H. rubiginosum, according to the broad concept established in the first monograph of the genus by Miller (1961). Miller’s concept was mainly based on teleomorph morphology. In their revision of Hypoxylon, Ju and Rogers (1996) later recognised that anamorph characters, stromatal pigments and the micromorphology of ascospores and asci (in particular the apical apparatus) constitute valuable diagnostic characters. Modern concepts of the genus combine holomorph morphology with molecular phylogenetic data (Hsieh et al. 2005; Kuhnert et al. 2014). Moreover, secondary metabolite profiles generated by high performance liquid chromatography coupled to diode array detection and mass spectrometry (HPLC-DAD/MS) not only proved highly useful for segregation of species, but even led to the discovery of numerous novel natural products with prominent biological activities (see overviews by Stadler and Hellwig 2005a and Helaly et al. 2018).
Hypoxylon rubiginosum and related taxa have been studied rather well on their stromatal secondary metabolites and, in many cases, morphologically similar species may contain entirely different pigments and other compounds (Stadler et al. 2004, 2008; Fournier et al. 2010). Interestingly, several species of the H. rubiginosum complex are known to frequently colonise Fraxinus species in the temperate Northern hemisphere. In some cases (e.g. H. cercidicola, H. fraxinophilum and H. petriniae), stromata are even almost exclusively found on dead wood of ash trees. They have also been frequently reported as endophytes of the same trees where they produce their stromata (Petrini and Petrini 1985) and are widespread endophytes of other host plants on which their stromata do not even occur (U’Ren et al. 2016). Therefore, the modern concepts of the taxonomy of the Hypoxylaceae take this fact into account and are based on the One Fungus-One Name concept. Some species have even been recognised on the basis of their anamorphic traits (Pažoutová et al. 2013) or their life cycle has been elucidated, based on a polythetic approach, i.e., by comparison of morphological, chemotaxonomic and molecular data of ascospore-derived cultures with endophytic isolates (see Bills et al. 2012 for H. pulicicidum and Kuhnert et al. 2014 for H. griseobrunneum).
The Ash Dieback disease caused by the introduced apothecial ascomycete Hym. fraxineus (Leotiomycetes) has become one of the greatest problems in European forestry and the majority of common ash trees have succumbed to the fungal pathogen. We have recently studied the secondary metabolism of Hym. fraxineus (previously also known under the synonyms, Hym. pseudoalbidus or Chalara fraxinea) and its non-pathogenic domestic relative, Hym. albidus, for secondary metabolite production (Halecker et al. 2014, 2018; Surup et al. 2018a). In parallel, we have also isolated endophytic fungi from apparently resistant ash trees in order to find natural antagonists that may be able to combat the devastating disease. One of the best candidates was identified as H. rubiginosum and, as reported recently (Halecker et al. 2020), it was found to produce the anti-fungal beta-tubulin inhibitor phomopsidin in dual culture with virulent strains of the pathogen. This compound was first reported from a marine-derived fungus that was originally assigned to the genus Phomopsis (Kobayashi et al. 2003). However, it has since then been found in other, terrestrial strains of the same genus, which should now be referred to as Diaporthe (Chepkirui and Stadler 2017), a large genus of the order Diaporthales. Interestingly, phomopsidin derivatives have never been reported from cultures of Xylariales before Halecker et al. (2020) found the compound in dual antagonist assays in agar cultures as described above. Moreover, they do not constitute major detectable metabolites of H. rubiginosum in the culture media that were used to study the chemotaxonomy of the genus before (cf. Bitzer et al. 2008).
Concurrently, we were about to study the taxonomy of new collections of Hypoxylon species originating from Iran that also belong to the Hypoxylon rubiginosum complex. Since mycelial cultures of these fungi had just become available, it appeared practical to combine the description of their taxonomy with an evaluation of their antagonistic potential to combat Hym. fraxineus. We have also included a number of other Hypoxylon species that colonise Fraxinus in Europe. The current study therefore provides new evidence on both, the taxonomy and chemical ecology of Hypoxylon.
Materials and methods
Sample sources
Samples were collected from Guilan and Mazandaran provinces (Northern Iran) during 2015–2017. Parts of corticated branches and trunks bearing Hypoxylaceae stromata were transferred to the laboratory. Details of the specimens used for morphological investigations are listed in the Taxonomy section under the respective descriptions. Specimens have been deposited in the fungarium of the Department of Plant Protection, Faculty of Agricultural Science, University of Guilan, Guilan, Iran (GUM). Living cultures have been deposited in MUCL (Louvain, Belgium).
Morphological characterisation
Microscopic characters of the teleomorph were observed in distilled water and 10% potassium hydroxide (KOH). Melzer’s reagent was used for staining of the apical ascus apparatus. The numbers of perithecia, ascospores, asci, conidia and conidiophores that were measured for size in the descriptions are 10, 30, 10, 30 and 5, respectively. Specimens were cultured from single ascospore isolates, using 2 % malt extract agar (MEA). For examination of culture macro-morphology, the strains were grown on Difco Oatmeal Agar (OA), following the protocols by Ju and Rogers (1996). Pigment colours were determined as described in the latter monograph, with colour codes following Rayner (1970). Macrophotographs were obtained with a Keyence VHX-6000 microscope. Light microscopy with Nomarski differential interference contrast (DIC) was done using a Zeiss Axio Imager A1 compound microscope, equipped with a Zeiss Axiocam 506 colour digital camera. SEM of ascospores were recorded using a field-emission scanning electron microscope (FE-SEM Merlin, Zeiss, Germany), in a similar fashion as reported previously (Kuhnert et al. 2017).
DNA extraction, PCR and sequencing
DNA extraction of fresh cultures and amplification of the ITS (nuc rDNA internal transcribed spacer region containing ITS1-5.8S-ITS2), LSU (5' 1200 bp of the large subunit nuc 28S rDNA), rpb2 (partial second largest subunit of the DNA-directed RNA polymerase II) and tub2 (partial β-tubulin) loci were performed as described by Wendt et al. (2018). Sequences were generated by an in-house Sanger capillary sequencing solution on campus. Sequences were processed with Geneious 7.1.9 (http://www.geneious.com).
Molecular phylogenetic analyses
The newly generated sequences were aligned with selected sequences from Wendt et al. (2018) and a combined matrix of the four loci (ITS, LSU, rpb2 and tub2) was concatenated for phylogenetic analyses, with four species (Biscogniauxia nummularia, Graphostroma platystomum, Xylaria arbuscula and Xylaria hypoxylon) added as the outgroup. The GenBank accession numbers of sequences are listed in Table 1. Sequences were aligned with the server version of MAFFT (http://mafft.cbrc.jp/alignment/server/, Katoh et al. 2019), checked and refined using BioEdit v. 7.2.6 (Hall 1999). After exclusion of ambiguously aligned regions and long insertions, the final combined data matrix contained 4369 characters, i.e. 578 nucleotides of ITS, 1301 nucleotides of LSU, 1017 nucleotides of rpb2, and 1473 nucleotides of tub2.
Table 1.
Isolates and accession numbers of sequences used in the phylogenetic analyses. Type specimens are labelled with HT (holotype) ET (epitype) and PT (paratype). Isolates/sequences in bold were isolated/sequenced in the present study.
Maximum Parsimony (MP) analyses were performed with PAUP v. 4.0a165 (Swofford 2002). All molecular characters were unordered and given equal weight; analyses were performed with gaps treated as missing data; the COLLAPSE command was set to MINBRLEN. MP analysis of the combined multilocus matrix was done using 1000 replicates of heuristic search with random addition of sequences and subsequent TBR branch swapping (MULTREES option in effect, steepest descent option not in effect). Bootstrap analyses with 1000 replicates were performed in the same way but using 10 rounds of random sequence addition and subsequent branch swapping during each bootstrap replicate.
Maximum Likelihood (ML) analyses were performed with RAxML (Stamatakis 2006) as implemented in raxmlGUI 1.3 (Silvestro and Michalak 2012), using the ML + rapid bootstrap setting and the GTRGAMMA substitution model with 1000 bootstrap replicates. The matrix was partitioned for the different gene regions. In the Results and Discussion, bootstrap values ≤ 70% are considered low, between 70–90% intermediate and ≥ 90% high.
HPLC profiling
Stromata of Hypoxylon specimens were extracted as described by Kuhnert et al. (2017) and subsequently analysed by high performance liquid chromatography, coupled with diode array and electrospray mass spectrometric detection (HPLC/DAD-ESIMS) instrument settings as described by Halecker et al. (2020). The resulting UV/Vis and mass spectra were compared with an internal database (cf. Bitzer et al. 2008), comprising standards of known Hypoxylaceae.
Dual culture experiments
Dual cultures of Hypoxylon spp. and Hym. fraxineus (STMA 18166) were co-incubated on barley-malt agar by inoculation at opposite sites on 9 cm Petri dishes (cf. Halecker et al. 2020) with Hym. fraxineus being inoculated one week prior the beginning of the dual culturing due to its slow growth. Axenic cultures, containing only one fungus, were inoculated in parallel as a control group. Growth was documented and observed weekly after incubation in the dark for a maximum of four weeks. Thereafter, the agar plates were extracted with acetone following the method described by Halecker et al. (2020), except that the entire agar plate was extracted instead of the fungal interaction zone.
Results
Phylogenetic analyses
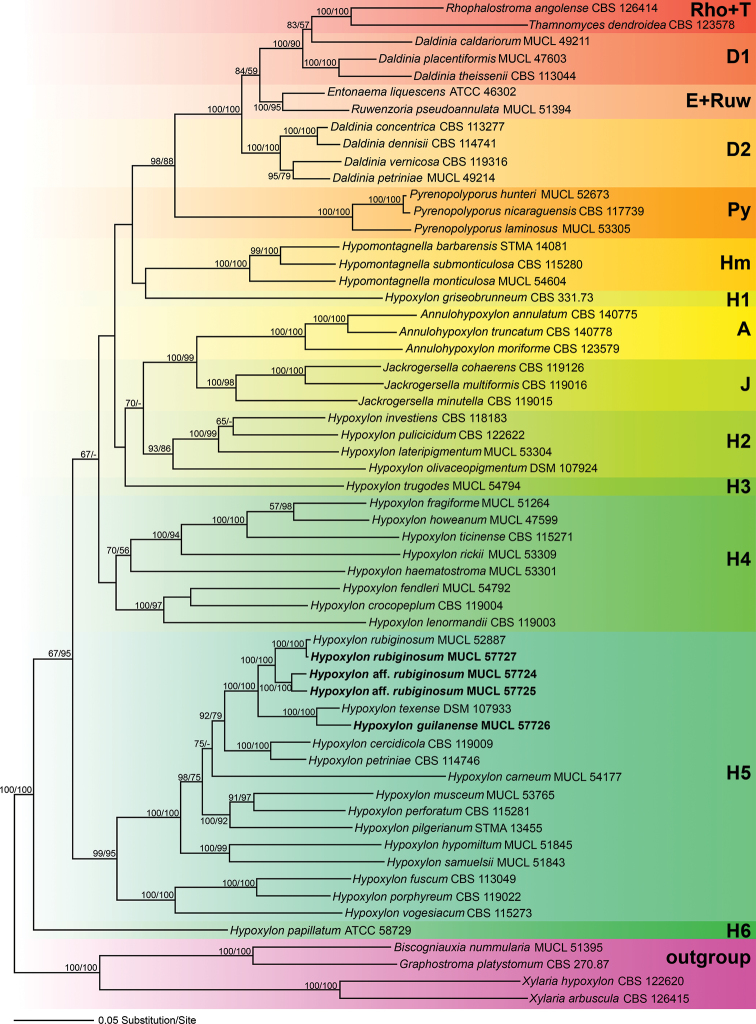
Of the 4369 nucleotide characters of the combined matrix, 1618 are parsimony informative (298 of ITS, 156 of LSU, 487 of rpb2 and 677 of tub2). Fig. 1 shows a phylogram of the best ML tree (lnL = −63870.651550) obtained by RAxML. Maximum parsimony analyses revealed one MP tree comprising 14,014 steps (data not shown). All major groups and deeper, highly supported nodes were consistent between the ML and MP analyses, but topologies of deeper unsupported nodes differed in the MP tree; as these differences are not relevant within the context of our new species, they are not further considered here. The phylogenies reveal a paraphyly of Hypoxylon, with the genera Annulohypoxylon, Daldinia, Entonaema, Jackrogersella, Hypomontagnella, Pyrenopolyporus, Rhopalostroma, Ruwenzoria and Thamnomyces embedded within the former. All of the latter genera appeared monophyletic except for Daldinia (Fig. 1). All of our new Iranian species and records described below are contained within the highly supported Hypoxylon clade H5. The new species H. guilanense clustered together with H. texense with 100% BS support, while sequences of two additional strains (Hypoxylon aff. rubiginosumMUCL 57724) and (Hypoxylon aff. rubiginosumMUCL 57725) formed a highly supported (100% BS support) clade that is the sister group of H. rubiginosum (Fig. 1). The sequences of the Iranian collection of H. rubiginosum (MUCL 57727) are almost identical to those of the ex-epitype culture (MUCL 52887) and they clustered together with maximum support. As in previous studies, the position of H. griseobrunneum and H. trugodes could not be resolved within the family. The remaining clades are in accordance with previous results of Wendt et al. (2018).
Figure 1.
Phylogram of the best ML trees (lnL = −63870.651550) revealed by RAxML from an analysis of the combined ITS–LSU–rpb2–tub2 matrix of selected Xylariales. Strains in bold were sequenced in the current study. ML and MP bootstrap support above 50% are given at the first and second positions, respectively, above or below the branches.
Taxonomy
Hypoxylon guilanense
Pourmoghaddam & C. Lambert sp. nov.
5346E2F6-0AB3-5004-8BE0-A23F251504B6
834521
Figure 2.
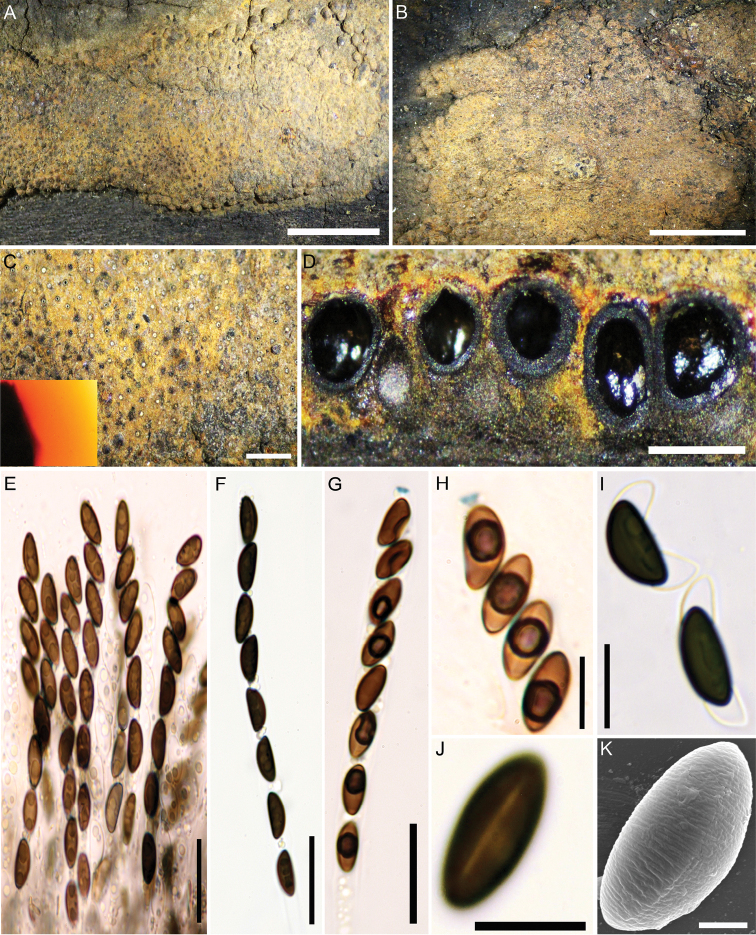
Hypoxylon guilanense (Holotype GUM 989) A stromatal habit B close-up view of stromatal surface, with stromatal pigments in 10% KOHC, H, I ascospores in water, with germ-slits D, E ascospores in 10% KOH with dehiscent perispore F, G ascospore under SEMJ, K culture on 9 cm OA plates after 1 and 3 wk of incubation (left to right). Scale bars: 2.5 mm (A), 1 mm (B); 10 µm (C–E); 2 µm (F, G); 10 µm (H, I).
Holotype.
Iran, Guilan Province, Rasht County, Saravan forest, 37°04'26"N, 49°38'13"E, 183 m elev., on fallen branch of Quercus castaneifolia, 9 Apr 2015, M.J. Pourmoghaddam. (GUM 989; ex-holotype culture MUCL 57726).
Etymology.
Guilanense, refers to its origin in Guilan province, Iran.
Teleomorph.
Stromata superficial, hemispherical to pulvinate, up to 2 cm long × 0.1–0.7 cm wide, with conspicuous perithecial mounds, surface Sienna (8), Umber (9) to Buff (45); Scarlet (5) to Orange (7) granules beneath the surface and between the perithecia, with Orange (7) KOH-extractable pigments. Perithecia spherical to obovoid, 0.33–0.66 high × 0.3–0.55 mm wide. Ostioles umbilicate, inconspicuous. Asci not seen. Ascospores smooth, unicellular, brown to dark brown, ellipsoid, inequilateral with narrowly rounded ends, 12–15 × 5–6 µm, with straight germ slit spore-length on convex side; perispore dehiscent in 10% KOH, conspicuous coil-like ornamentation in SEM; epispore smooth.
Cultures and anamorph.
Colonies on OA covering a 9 cm Petri dish in 4 wk, at first white, becoming Buff (45), cottony, slightly zonate with diffuse margins; finally, becoming Honey (64). Anamorph not produced in culture.
Secondary metabolites.
Orsellinic acid, rubiginosin A and an unknown isomer thereof, as well as mitorubrinol acetate as prevailing stromatal components; cultures produce yet unidentified compounds on barley-malt agar.
Notes.
The description of this taxon is based on a single specimen, which shows the salient features of the teleomorph and can be discriminated easily from all previously described species of the H. rubiginosum complex. The stromata of the holotype specimen differ from H. texense (i.e. the closest relative in the phylogeny), in having stromata with hemispherical to pulvinate shape, Orange (7) KOH-extractable pigments and larger ascospores [12–15 × 5–6 vs. 9.1–10.8 (–11.5) × (4.0–) 4.5–5.4 (–5.7) μm with straight germ slit.
Hypoxylon guilanense can also be easily differentiated from H. rubiginosumsensu stricto and H. petriniae in the peculiar stromatal shape and it also has larger ascospores. H. cercidicola differs from H. guilanense in having erumpent stromata with discoid shape and smaller ascospores [(9–) 9.5–12 × 5–6 μm)] with straight to slightly sigmoid germ slit. Table 2 compares morphological characters of some other taxa that may be confused with H. guilanense.
Table 2.
Diagnostic characters of Hypoxylon rubiginosumsensu stricto and closely related species.
| Taxon | Stromatal shape | Stromatal surface | KOH-extractable pigments | Ascospores (µm) | Germ slit | Host | Known distribution | Anamorph | Secondary metabolites* |
|---|---|---|---|---|---|---|---|---|---|
| Hypoxylon canariense | effused to effused-pulvinate | Fulvous, Dark Brick, Dark Vinaceous | Orange to Sienna | 9.5–11.5 × 4.5–5 | straight | Erica, Ocotea, Laurus, Persea | Spain (Canary Islands) | virgariella-like | Rubiginosins A–C, mitorubrinol acetate |
| Hypoxylon carneum | Effused-pulvinate | Dark purple, Dark vinaceous | Livid violet, absent in old stromata | (7.5–)8–11.5 × 4.5–5 | straight | Various angiosperm hosts including Fraxinus | probably cosmopolitan but rare | sporothrix-like | Carneic acids A and B, BNT |
| Hypoxylon cercidicola | discoid | Dark brick to Sepia | Orange | (9–)9.5–12 × 5–6 | straight to slightly sigmoid | Fraxinus | Europe and North America | unknown | Mitorubrin, rubiginosin A and C |
| Hypoxylon guilanense | hemispherical to pulvinate | Sienna, Umber to Buff | Orange | 12–15 × 5–6 | straight | Quercus | Iran | unknown | Rubiginosin A, mitorubrinol acetate |
| Hypoxylon lusitanicum | effused | Brown Vinaceous | Sienna | 11–13.5 × 5–7 | straight | Rhamnus | Portugal | unknown | Rubiginosins A and C, rutilin A |
| Hypoxylon petriniae | irregularly effused | Lilac, Vinaceous to Brown Vinaceous | Orange to Rust | 8–11.5(–13) × 4.8–6 | straight | Fraxinus (mostly); Acer, Salicaceae | Western and Central Europe | virgariella-like | Rubiginosin A, BNT |
| Hypoxylon retpela | effused-pulvinate | Livid Vinaceous, Brown Vinaceous, | Orange or Scarlet | (9–)9.5–12 × 4.5–5 | straight or slightly sigmoid | unknown | Southeast and East Asia, New Guinea | nodulisporium-like | Mitorubrinol acetate, unknown rubiginosins |
| Hypoxylon rubiginosum | effused-pulvinate | Dark Brick, Brown Vinaceous | Orange | 9–13 × 4–5.5 | straight | Various angiosperm hosts including Fraxinus | Europe, North America | nodulisporium-like | Mitorubrin, rubiginosin A–C, rubiginosic acid, daldinin C |
| Hypoxylon aff. rubiginosum (GUM 1587) | pulvinate to effused-pulvinate | Luteous, Orange to Ochraceous | Orange | 8–10 (–11) × (3–) 4–4.5 (–5) | straight to slightly sigmoid | Quercus | Iran | virgariella-like | like H. rubiginosum |
| Hypoxylon aff. rubiginosum (GUM 1588) | pulvinate | Orange to Apricot | Orange | 10–15 × 5–6.5 | straight to slightly sigmoid | unknown | Iran | not observed- | like H. rubiginosum |
| Hypoxylon salicicola | effused | Dark rust to Sepia, Brown Vinaceous | Fulvous to Rust | 7.2–9.6 × 3–4.2 | straight | Salix, rarely on Fraxinus and Prunus | Northern Europe, USA | nodulisporium-like | Mitorubrinol acetate |
| Hypoxylon texense | effused to effused-pulvinate | Livid Vinaceous to Brown Vinaceous | Rust to Dark Brick | 9.1–10.8(–11.5) × (4.0–)4.5–5.4(–5.7) | straight or slightly sigmoid | unknown | USA | nodulisporium to virgariella-like | Rubiginosin A, mitorubrinol acetate, unknown rubiginosins |
| Hypoxylon urriesii | effused | Dark Brick | Orange | 11–14.5 × 5–6 | straight or slightly sigmoid | unknown | Spain (Canary Islands) | unknown | Mitorubrinol acetate, rubiginosin A |
Hypoxylon rubiginosum
(Pers.) Fr., Summa Veg. Scand. II, p. 384. (1849).
8EE961E2-BCEC-52EA-82A7-BCD87578A61F
Figure 3.
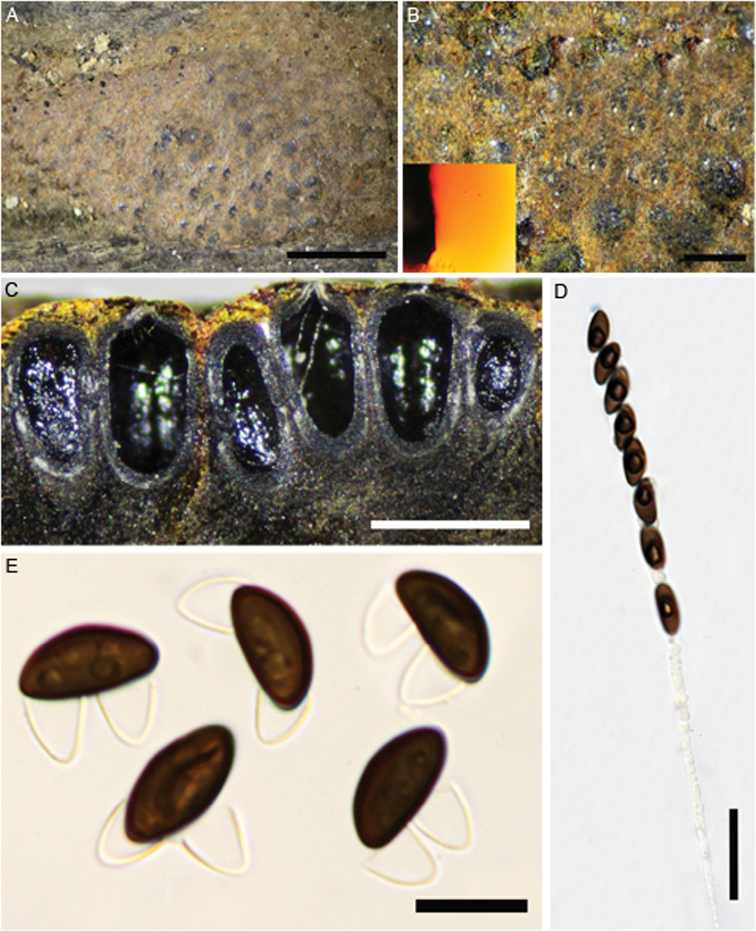
Hypoxylon rubiginosum (GUM 1586) A, B stromatal habit C close-up view of stromatal surface D close-up view of stromatal surface, with stromatal pigments in 10% KOHE ascospores in 10% KOH with dehiscent perispore F mature and immature asci in water G immature ascus in water H mature ascus in water I ascus in Melzer’s reagent J ascospores in water K ascus tip in Melzer’s reagent. Scale bars: 2 cm (A); 1 cm (B); 4 mm (C); 2 mm (D); 10 µm (E); 20 µm (F–I), 10 µm (J, K).
Teleomorph.
Stromata superficial, effused-pulvinate, up to 8 cm long × 0.3–0.2 cm wide; with inconspicuous to conspicuous perithecial mounds, surface Red (2) to Brick (59); Scarlet (5) to Orange (7) granules beneath the surface and between the perithecia, with Orange (7) to Scarlet (5) KOH-extractable pigments. Perithecia spherical to obovoid, 0.2–0.5 high × 0.15–0.45 mm wide. Ostioles umbilicate, inconspicuous. Asci 8-spored, cylindrical, with amyloid, discoid apical apparatus, 0.5–1 µm high × 1.5–2.5 µm wide, stipe up to 180 µm long and spore-bearing portion 40–80 × 6.5–10 µm. Ascospores smooth, unicellular, brown to dark brown, ellipsoid, inequilateral with narrowly rounded ends, 9–12 (–13) × 4–6 µm, with straight germ slit spore-length on convex side; perispore dehiscent in 10% KOH; epispore smooth.
Cultures and anamorph.
Colonies on OA covering a 9 cm Petri dish in 3 wk, at first white, becoming Smoke Grey (105), felty, azonate with diffuse margins; finally becoming Pale Luteous (11) to Straw (46). Asexual morph not produced in culture.
Secondary metabolites.
Rubiginosin A and an unknown compound of the mitorubrin / rubiginosin azaphilone family prevalent; cultures produce phomopsidin and unidentified compounds on barley-malt agar.
Specimens examined.
Iran, Guilan Province, Siahkal County, Deilaman forest, 36°57'25"N, 49°51'54"E, 1100 m elev., on fallen branch of Quercus castaneifolia, 6 Oct 2017 (GUM 1586; culture MUCL 57727); Guilan Province, Shaft County, 36°59'08"N, 49°18'43"E, 594 m elev., on fallen trunk of Pterocarya fraxinifolia, 15 Sep 2015 (GUM 1583); Guilan Province, Langaroud County, Liseroud forest, 37°7'44"N, 50°8'41"E, 28 m elev., on fallen branch of Quercus castaneifolia, 10 Sep 2016 (GUM 1584); Guilan Province, Talesh County, Gisoum forest, 37°37'30"N, 48°58'15"E, 477 m elev., on fallen branch of Populus sp., 20 Oct 2016 (GUM 1585). All specimens collected by M.J. Pourmoghaddam.
Notes.
H. rubiginosumsensu stricto is a very common fungus in the temperate Northern hemisphere (Stadler et al. 2008) and may occur in subtropical areas, such as Florida, USA (Ju and Rogers 1996). Most of the characters of the Iranian specimens are in accordance with previous descriptions (Stadler et al. 2004), aside from insignificant variations in the size of ascospores.
Additional potentially new species of the H. rubiginosum complex
Below, we describe two collections that may eventually be recognised to represent new species. They appear phylogenetically different from the type specimen, as well as from Iranian records of H. rubiginosum, but share salient features with the latter species. It is explained in the Notes why we hesitate to describe them as new taxa in this complicated species complex.
Hypoxylon sp. aff. rubiginosum
GUM 1587
FEA46B16-752F-56D6-9F35-8E509D4414E2
Figure 4.
Hypoxylon aff. rubiginosum (GUM 1587) A, B stromatal habit C close-up view of stromatal surface, with stromatal pigments in 10% KOHD stroma in section showing perithecia and ostioles E mature and immature asci in water F ascus in water G ascus in Melzer’s reagent H ascus tip in Melzer’s reagent I ascospores in 10% KOH with dehiscent perispore J ascospore in water, with germ-slit K ascospore under SEM. Scale bars: 5 mm (A, B); 1 mm (C); 0.5 mm (D); 20 µm (E–G); 10 µm (H–J); 2 µm (K).
Figure 5.
Culture and anamorphic structures of Hypoxylon aff. rubiginosum (GUM 1587) on OAA, B surface of colony after 1 and 8 wk of incubation (respectively, left to right) C–G general view of anamorph structure with virgariella-like branching patterns H, I conidiogenous cells and immature conidia J mature conidia. Scale bars: 20 µm (C–G); 10 µm (H–J).
Teleomorph.
Stromata superficial, pulvinate to effused-pulvinate, up to 5 cm long × 0.6–2 cm wide, with inconspicuous to conspicuous perithecial mounds; surface Luteous (12), Orange (7) to Ochreous (44); Orange (7) granules beneath the surface, Orange (7) and Leaden Black (126) granules between the perithecia, with KOH-extractable pigments Orange (7). Perithecia obovoid, compressed-obovoid to spherical, 0.27–0.50 high × 0.23–0.35 mm wide. Ostioles umbilicate, inconspicuous, usually overlain with conspicuous white substance. Asci 8-spored, cylindrical, with amyloid, discoid apical apparatus, 0.5–1 µm high × 2–2.5 µm wide, stipe up to 180 µm long; spore-bearing portion 80–100 × 5.5–7 µm. Ascospores smooth, unicellular, brown to dark brown, ellipsoid, inequilateral with narrowly rounded ends, 8–10 (–11) × (3–) 4–4.5 (–5) µm, with straight to slightly sigmoid germ slit spore-length on convex side; perispore dehiscent in 10% KOH, conspicuous coil-like ornamentation in SEM; epispore smooth.
Cultures and anamorph.
Colonies on OA covering a 9 cm Petri dish in 3 wk, at first white, becoming Luteous (12) from outwards, cottony, slightly zonate with diffuse margins; finally, attaining a variety of different colours. Conidiogenous structure branching virgariella-like as defined by Ju and Rogers (1996), (Fig. 5C–G). Conidiophores hyaline, smooth to finely roughened. Conidiogenous cells hyaline, smooth to finely roughened, 15–30 × 2–3 μm. Conidia hyaline, smooth to ellipsoid, 4–6 × 2–3 μm.
Specimen examined.
Iran, Guilan Province, Astaneh-Ashrafieh County, Safra-Basteh forest, 37°20'19"N, 49°58'26"E, 14 m elev., on fallen branch of Quercus castaneifolia, 4 Oct 2016, M.J. Pourmoghaddam (GUM 1587; culture MUCL 57724).
Notes.
This specimen resembles H. rubiginosum in many respects. However, it has slightly smaller ascospores [8–10 (–11) × (3–) 4–4.5 (–5) vs. 9–13 × 4–5.5 µm] and the germ slit of the ascospores is often slightly sigmoid. The most significant differences were noted in the anamorphic structures with virgariella-like branching patterns. This anamorph actually resembles that of H. petriniae. However, this species is normally associated with Fraxinus and differs from Hypoxylon aff. rubiginosumGUM 1587 in having Lilac (54), Vinaceous (57) to Brown Vinaceous (84) stromatal surface colours (owing to the presence of BNT, which was not found in the Iranian specimen). It also differs in having more elongate to irregularly effused stromata with black margins and its ascospores are larger (8–11.5 (–13) × 4.8–6 μm) and have a straight germ slit.
Hypoxylon sp. aff. rubiginosum
GUM 1588
69BA22CB-5B18-534E-91A0-A4637BC8E989
Figure 6.
Hypoxylon aff. rubiginosum (GUM 1588) A stromatal habit B close-up view of stromatal surface, with stromatal pigments in 10% KOHC section of stroma showing perithecia and ostioles D ascus in Melzer’s reagent E ascospores in 10% KOH with dehiscent perispore. Scale bars: 2.5 mm (A); 0.5 mm (B, C); 20 µm (D); 10 µm (E).
Teleomorph.
Stromata superficial, pulvinate, up to 1 cm long × 0.2–0.5 cm wide, with inconspicuous to conspicuous perithecial mounds; surface Orange (7) to Apricot (42); Orange (7) granules beneath the surface and Laeden Black (126) granules between the perithecia, with Orange (7) KOH-extractable pigments. Perithecia obovoid to compressed-obovoid, 0.35–0.65 high × 0.3–0.45 mm wide. Ostioles umbilicate, inconspicuous. Asci with amyloid, discoid apical apparatus, 1–1.5 µm high × 2–3 µm wide, stipe up to 160 µm and spore-bearing portion 70–100 × 6–8 µm long. Ascospores smooth, unicellular, brown to dark brown, ellipsoid, inequilateral with narrowly-rounded ends, 10–15 × 5–6.5 µm, with straight to slightly sigmoid germ slit spore-length on convex side; perispore dehiscent in 10% KOH; epispore smooth.
Cultures and anamorph.
Colonies on OA covering a 9 cm Petri dish in 3 wk, at first white, becoming whitish, cottony, azonate with entire margins; remaining mainly uncoloured with Pale Luteous tinges. Anamorph not produced in culture.
Specimen examined.
Iran, Mazandaran Province, Tonekabon County, Do-hezar forest, 36°42'30"N, 50°49'43"E, 456 m elev., on dead branches (host unknown), 28 Oct 2016, M.J. Pourmoghaddam (GUM 1588; culture MUCL 57725).
Notes.
This specimen is morphologically similar to Hypoxylon aff. rubiginosumGUM 1587, but it can be distinguished by its larger ascospores [10–15 × 5–6.5 vs. 8–10 (–11) × (3–) 4–4.5 (–5) μm]. H. rubiginosumsensu stricto differs from this specimen in having smaller ascospores [(8–) 9–12 × 4–5.5 vs. 10–15 × 5–6.5 μm]. In addition, the stromatal secondary metabolite profile is similar to that of H. rubiginosum with two unknown azaphilone compounds of the mitorubrin / rubiginosin family (UC 2, retention time = 8.7 min, 442 Dalton and UC 3, RT = 10.6 min, 884 Da) and rubiginosin A. H. guilanense differs from Hypoxylon aff. rubiginosumGUM 1588 in having stromata with hemispherical to pulvinate shape and difference in average ascospores sizes (12–15 × 5–6 vs. 10–15 × 5–6.5 μm) with straight germ slit. H. texense differs from Hypoxylon aff. rubiginosumGUM 1588 in having Rust (39) to Dark Brick (86) KOH-extractable pigments and much smaller ascospores [9.1–10.8 (–11.5) × (4.0–) 4.5–5.4 (–5.7) vs. 10–15 × 5–6.5 μm].
HPLC profiling of stromata
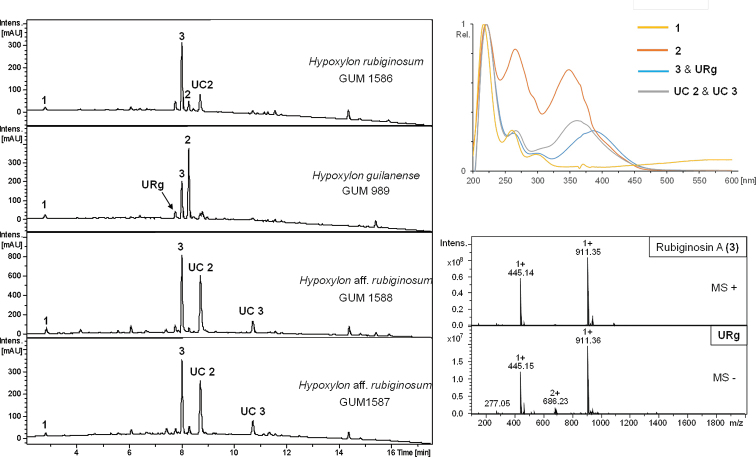
Amongst the four studied Iranian Hypoxylon spp., five major metabolites could be identified. Beneath common secondary metabolites of the H. rubiginosum complex like orsellinic acid (1, Stadler et al. 2008), mitorubrin acetate (2, Steglich et al. 1974; Stadler et al. 2001) and rubiginosin A (3, Quang et al. 2004), three more non-assignable compounds were detected. UV/Vis data of these metabolites tentatively suggested affinities to the rubiginosin azaphilone family (Fig. 9, UC 2 and 3) with one unknown compound sharing the same mass and UV/Vis maxima of mitorubrinol (4), which could possibly constitute a yet undescribed isomer (URg). Compounds URg and UC 2 have been reported from H. texense, which was recently discovered in Texas, USA, as another species of the H. rubiginosum complex (Sir et al. 2019). These findings are further reflected in the taxonomic part of this paper.
Figure 9.
HPLC-UV profiles at 210 nm derived from stromal extracts of strains H. rubiginosum (GUM 1586), H. guilanense (from holotype) and Hypoxylon aff. rubiginosumGUM 1587 and GUM 1588. UV/Vis spectra are shown for orsellinic acid (1), mitorubrinol acetate (2), rubiginosin A (3), an unknown rubiginosin A – like derivative (URg) and rubiginosin – like derivatives (UC 2 and UC 3). ESI mass spectra are shown for compounds URg and 2.
HPLC profiling of extracts from single and dual culture experiments (Figs 10, 11)
Figure 10.
HPLC-UV profiles at 210 nm derived from barley-malt agar (A–C, E) and stromal (E) extracts and compound standard (F). UV/Vis spectra are shown for identified compounds in mono- and dual culture (C) experiments of STMA 18166 (Hym. fraxineus, A) and DSM 107933 (H. texense, B; UC 2, 4 – unknown compounds); stromal metabolites (4 – mitorubrinol; URg – unknown rubiginosin A derivative; 3 – rubiginosin A; 2 – mitorubrinol acetate; 7 – mitorubrin; UC2 – Unknown compound 2 of GLM-F116101 (H. texense, D), and ... ESI mass spectra of 8 in positive and negative modes... of 8 8 (rickiol A, F) identified in the mono culture extract of MUCL 54624 (H. rubiginosum, E).
Figure 11.
HPLC-UV chromatograms at 210 nm from mono cultural barley-malt agar extracts of MUCL 47152 (H. rubiginosum), STMA 18166 (Hym. fraxineus), STMA 13090 (H. fuscum) and one dual culture experiment thereof. UV/Vis spectra are shown for phomopsidin (5), 10-hydroxyphomopsidin (6), orthosporin (9), daldinone B (10), 1,8-dimethoxynaphthalene (11), daldinin F (12), 5–methylmellein (13), viridiol (14) and an unidentifiable compound (UC 6) after comparison of data with internal databases. The UV signal of UC 6 was enhanced in the dual culture extract.
In total, 32 different Hypoxylon strains were screened for production of phomopsidin (5, Kobayashi et al. 2003) and 10-hydroxyphomopsidin (6, Halecker et al. 2020). Due to the availability of well-studied strains of H. rubiginosum, H. perforatum and H. petriniae in public culture collections, a pre-screening was conducted to confirm production of 5 and 6 (with 13, 7 and 4 strains each, respectively (cf. Table 3, Fig. 11H). Out of these 24 strains, 16 emerged as producers of compound 5 and partially 6 (12 strains). Compound 6 was not detected in the absence of 5. Out of those, two strains of H. rubiginosum (MUCL 47152 and MUCL 47970), one representative of H. perforatum (MUCL 47187) and one culture of H. petriniae (MUCL 53756) were selected for further testing against Hym. fraxineus. The results are illustrated, based on four examples in Fig. 7, showing the dual cultures after 1–4 weeks of incubation. The chemical structures are shown in Fig. 8 and selected chromatographic data are depicted in Figs 10, 11.
Table 3.
Identified secondary metabolites in axenic cultures on barley-malt medium of the surveyed strains. Strains in bold have been used concurrently against STMA 18166 (Hymenoscyphus fraxineus) in an antagonism assay. Identified compounds: 5: phomopsidin; 6: 10-hydroxyphomopsidin; 8: rickiol A; 9: orthosporin 10: daldinone B; 11: 1,8-dimethoxynaphtahlene; 13: 5-methyl-mellein. Identified stromal azaphilone groups detected in culture: MI = Mitorubrin type; NA = Naphthalene type; DA =Daldinin type. For chemical structures, see Fig. 8.
| Organism | Strain | Culture metabolites | Stromal metabolites | ||||
|---|---|---|---|---|---|---|---|
| 5 | 6 | Others | MI | NA | DA | ||
| Hypoxylon guilanense | MUCL 57726 | – | – | – | – | – | – |
| Hypoxylon aff. rubiginosum | MUCL 57724 | + | + | – | + | – | – |
| Hypoxylon rubiginosum | MUCL 57727 | + | – | – | – | – | – |
| Hypoxylon aff. rubiginosum | MUCL 57725 | + | + | – | – | – | – |
| Hypoxylon perforatum | MUCL 57728 | – | – | 10 | – | – | – |
| Hypoxylon perforatum | CBS 119011 | – | – | 10 | – | – | – |
| Hypoxylon perforatum | MUCL 47187 | + | + | – | – | – | – |
| Hypoxylon perforatum | MUCL 54798 | – | – | 10 | – | – | – |
| Hypoxylon perforatum | STMA 13041 | + | + | – | – | – | – |
| Hypoxylon perforatum | STMA 14051 | – | – | 10 | – | – | – |
| Hypoxylon perforatum | CBS 140779 | – | – | 10 | – | – | – |
| Hypoxylon petriniae | MUCL 53756 | + | + | – | – | – | – |
| Hypoxylon petriniae | STMA 12020 | – | – | – | – | – | – |
| Hypoxylon petriniae | STMA 13303 | – | – | – | – | – | – |
| Hypoxylon petriniae | STMA 13313 | – | – | 10 | – | – | – |
| Hypoxylon rubiginosum | MUCL 2354 | – | – | – | – | – | – |
| Hypoxylon rubiginosum | MUCL 47152 | + | + | 9, 10 | – | + | – |
| Hypoxylon rubiginosum | MUCL 47970 | + | + | 9, 10 | – | + | – |
| Hypoxylon rubiginosum | MUCL 47150 | + | – | – | + | – | – |
| Hypoxylon rubiginosum | MUCL 52672 | + | + | – | + | – | – |
| Hypoxylon rubiginosum | MUCL 54624 | – | – | 8 | – | – | – |
| Hypoxylon rubiginosum | MUCL 2709 | – | – | – | – | – | – |
| Hypoxylon rubiginosum | MUCL 34183 | + | + | 13 | – | – | – |
| Hypoxylon rubiginosum | MUCL 47147 | + | – | – | + | – | – |
| Hypoxylon rubiginosum | STMA 04040 | + | + | – | + | – | – |
| Hypoxylon rubiginosum | STMA 07027 | + | + | – | – | – | – |
| Hypoxylon rubiginosum | STMA 13346 | + | + | – | – | – | – |
| Hypoxylon rubiginosum | STMA 17058 | + | + | – | – | – | – |
| Hypoxylon cercidicola | MUCL 54180 | + | – | 13 | – | – | – |
| Hypoxylon fuscum | STMA 13090 | – | – | 11, 13 | – | + | + |
| Hypoxylon texense | DSM 107933 | – | – | – | + | – | – |
| Hypoxylon crocopeplum | CBS 119004 | – | – | – | + | – | – |
| Hypoxylon carneum | MUCL 54177 | – | – | 10 | – | – | – |
Figure 7.
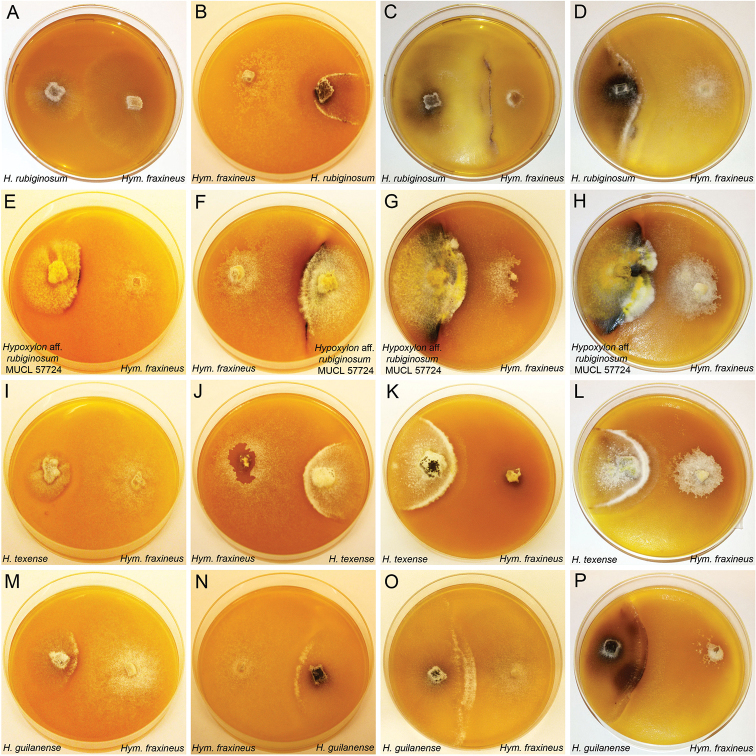
Illustration of antagonist test by dual culture technique of Hypoxlon spp. and Hymenoscyphus fraxineus on barley-malt agar in 9-cm diam. plates A dual culture of H. rubiginosum (MUCL 47152) against Hym. fraxineus (STMA 18166) after 1 wk of incubation B dual culture of H. rubiginosum (MUCL 47152) against Hym. fraxineus (STMA 18166) after 2 wk of incubation C dual culture of H. rubiginosum (MUCL 47152) against Hym. fraxineus (STMA 18166) after 3 wk of incubation D dual culture of H. rubiginosum (MUCL 47152) against Hym. fraxineus (STMA 18166) after 4 wk of incubation E–H (Hypoxylon aff. rubiginosumMUCL 57724) against Hym. fraxineus after 1, 2, 3, 4 wk I–LH. texense (DSM 107933) against Hym. fraxineus after 1, 2, 3, 4 wk M–PH. guilanense (MUCL 57726) against Hym. fraxineus after 1, 2, 3, 4 wk.
Figure 8.

Chemical structures of discussed secondary metabolites. Orsellinic acid (1); mitorubrinol acetate (2); rubiginosin A (3); mitorubrinol (4); phomopsidin (5); 10-hydroxyphomopsidin (6); mitorubrin (7); rickiol A (8); orthosporin (9); daldinone B (10); 1,8-dimethoxynaphthalene (11); daldinin F (12); 5-methyl mellein (13); viridiol (14).
Strikingly, during evaluation and comparison of the HPLC UV/Vis chromatograms with our internal database, the mitorubrin derivatives 2, 4 and 7 were identified by direct comparison of chromatograms derived from extracts of stromata and cultures of the ex-type strain and the holotype of H. texense (Sir et al. 2019; Figs 7 I–L, 11B, D). Beneath the aforementioned UC 2, another yet undescribed compound was revealed (UC 4). The main metabolite of the mono cultural extract of MUCL 54624 was identified by comparison of UV/Vis and MS data as rickiol A (8; Fig. 11E–F), previously described from H. rickii (Surup et al. 2018b). Orthosporin (9; Quang et al. 2002), daldinone B (10; Stadler et al. 2008) was identified by comparison with an internal database in several strains of H. rubiginosum, H. perforatum and H. petriniae (cf. Tables 3, 4, Fig. 11H). The mono cultural extract of H. fuscum (STMA 13090) revealed 1,8 dimethoxynaphthalene (11; Chang et al. 2014) and another unidentified peak (UC 6, Fig. 11J, K) with an identical UV/Vis spectrum as 11, as well as traces of Daldinin F (12; Quang et al. 2004) and 5- methylmellein (13; Stadler et al. 2005b) as the main product. Interestingly, the UV signal of UC 7 was visibly enhanced in the chromatogram derived from the dual culture extract. The phytotoxic compound viridiol (14; Figs 10A, 11I–K) was found in both mono and dual culture extracts of Hym. fraxineus (Andersson et al. 2010; Halecker et al. 2020).
Table 4.
Identified secondary metabolites in dual culture (barley-malt medium with Hymenoscyphus fraxineus) of the surveyed strains listed in Table 3. Identified compounds: 5: phomopsidin; 6: 10-hydroxyphomopsidin; 8: rickiol A; 9: orthosporin; 10: daldinone B; 11: 1,8-dimethoxynaphtahlene; 13: 5-methyl-mellein. Identified stromal azaphilone groups detected in culture: MI = Mitorubrin type; NA = Naphthalene type; DA = Daldinin type. For chemical structures, see Fig. 8.
| Organism | Strain | Culture metabolites | Stromal metabolites | |||
|---|---|---|---|---|---|---|
| 5 | 6 | Others | MI | NA | ||
| Hypoxylon cercidicola | MUCL 54180 | + | – | 13 | – | – |
| Hypoxylon fuscum | STMA 13090 | – | – | 11, 13 | – | + |
| Hypoxylon texense | DSM 107933 | – | – | – | + | – |
| Hypoxylon crocopeplum | CBS 119004 | – | – | – | + | – |
| Hypoxylon perforatum | MUCL 47187 | + | – | – | + | – |
| Hypoxylon petriniae | MUCL 53756 | + | – | – | – | – |
| Hypoxylon aff. rubiginosum | MUCL 57724 | + | + | – | + | – |
| Hypoxylon rubiginosum | MUCL 47152 | + | – | 9, 10 | – | + |
| Hypoxylon rubiginosum | MUCL 47970 | + | – | 9, 10 | – | + |
| Hypoxylon guilanense | MUCL 57726 | – | – | – | – | – |
| Hypoxylon carneum | MUCL 54177 | – | – | 10 | – | – |
Discussion
The present study dealt with the identification of Hypoxylon species from Northern Iran based on morphological, chemotaxonomic and phylogenetic data, focusing on the H. rubiginosum complex. The specimens encountered appeared morphologically and chemotaxonomically related to H. rubiginosumsensu stricto, as revealed from their morphology and secondary metabolite profiles. While the majority of specimens were assigned to typical H. rubiginosum, we have encountered a new taxon that significantly deviates from the complex in both stromatal and ascospore morphology and appears most closely related to a species that was so far only reported from the southern USA (Sir et al. 2019). Furthermore, we found two specimens that slightly differed in one or two characters from typical H. rubiginosum and also showed deviating positions in the phylogenetic trees, but are so far only known from single collections. Attempts should be made to encounter additional specimens of these fungi, which may eventually lead to their recognition as new species. The recent study on intragenomic polymorphisms in Hypoxylaceae has suggested that molecular data alone may be misleading in this family and new taxa should be based on multiple records sharing the same genotypic and phenotypic features (Stadler et al. 2020). Hsieh et al. (2005) have already established that protein-coding genes provide a better resolution in the Hypoxylaceae than ITS and finally even omitted this locus from the phylogeny and rather decided to focus on tub2 and alpha-actin sequences. Kuhnert et al. (2014) also found tub2 to be more suitable than ITS in their phylogeny, based on material from the Caribbean.
Our phylogenetic analyses confirmed previous results (Wendt et al. 2018; Lambert et al. 2019; Sir et al. 2019), suggesting that the genus Hypoxylon appears paraphyletic in Hypoxylaceae, with a relatively small clade comprising the type species H. fragiforme as “core group” to which members of the Hypoxylon rubiginosum complex form a sister clade. The genus will eventually need to be further subdivided, but molecular data for the majority of known species remain incomplete and such a task should only commence as the phylogenetic data matrix has increased. Our study further contributed to this monumental task by adding some data on representatives from the Middle East, a geographic area that has certainly not been as well explored as Western Europe and other parts of the world.
A main objective of this work was to assess the antagonistic potential of the newly isolated cultures and some strains of related species against an important pathogen, following the recent discovery that an endophytic isolate of H. rubiginosum from a resistant ash tree inhibited the growth of the alien pathogen, Hym. fraxineus (Halecker et al. 2020). Assessment of axenic cultures of the Hypoxylon species in a single medium (barley-malt) led to the detection of phomopsidin in one out of five strains of H. petriniae, two out of seven strains of H. perforatum and ten out of 13 strains of H. rubiginosum. The stromata of these three taxa have been frequently reported from Fraxinus and it is plausible that they all occur as endophytes in this host and only form the stromata on dead host tissues. On the other hand, phomopsidin was not detected in other related, but apparently rare species like H. texense, H. crocopeplum and H. carneum. Only the two latter species, however, were represented in our study by cultures that were isolated from stromata growing on Fraxinus wood. In addition, our results need to be further validated because we cannot exclude that some of the strains, which have been kept in culture collections for many years, may have degenerated. In any case, our results suggest that phomopsidin is not a specific marker for the species complex or for H. rubiginosumsensu stricto. As the compound is preferentially observed in dual cultures, its biosynthesis may be under control of epigenetic effectors. Therefore, in the future, it would be useful to evaluate a broader range of ascospore-derived cultures of Hypoxylon for their potential as biocontrol agents against the ash dieback pathogen and to define the genetic mechanisms encoding phomopsidin biosynthesis.
Last but not least, the current study also revealed some interesting aspects for potential follow-up projects. For instance, the examination of H. fuscum (a species that has never been isolated from Fraxinus, but is actually associated with Corylus and other Betulaceae) in the antagonism assay, revealed the production of several hitherto unknown compounds whose production was significantly enhanced in the presence of Hym. fraxineus. This observation suggests that it will be worthwhile to further study the secondary metabolism of Hypoxylon species in other scenarios using the dual culture approach. The first step would be to scale-up the production of the unknown molecules, isolating enough for structure elucidation and biological studies. This should not be expected to be a trivial task, but it appears doable using the methodology that is presently available.
The production of known and yet unidentified azaphilones (i.e. a compound class that is normally found in high concentrations in the stromata of various Hypoxylaceae, but was rarely observed in their mycelial cultures) in H. rubiginosum and allies, is another interesting observation relating to the differential expression of biosynthetic genes encoding secondary metabolites. It should be rewarding to evaluate the regulation mechanisms that lead to the production of the pigments, aided by genomic and transcriptomic studies.
Supplementary Material
Acknowledgements
This study was partially financed by the Ministry of Science, Research and Technology (MSRT) of Iran. Mohammad Javad wants to express his appreciation to all colleagues in the labs of Irmgard Krisai-Greilhuber (Vienna) and Marc Stadler (Braunschweig) for their support, especially Kathrin Wittstein and Silke Reinecke and to all colleagues in the University of Guilan. The authors want to express their gratitude to Manfred Rohde for recording the SEM pictures.
Citation
Pourmoghaddam MJ, Lambert C, Surup F, Khodaparast SA, Krisai-Greilhuber I, Voglmayr H, Stadler M (2020) Discovery of a new species of the Hypoxylon rubiginosum complex from Iran and antagonistic activities of Hypoxylon spp. against the Ash Dieback pathogen, Hymenoscyphus fraxineus, in dual culture. MycoKeys 66: 105–133. https://doi.org/10.3897/mycokeys.66.50946
Supplementary materials
Discovery of a new species of the Hypoxylon rubiginosum complex from Iran and antagonistic activities of Hypoxylon species towards the Ash Dieback pathogen, Hymenoscyphus fraxineus, in dual culture
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Mohammad Javad Pourmoghaddam, Christopher Lambert, Frank Surup, Seyed Akbar Khodaparast, Irmgard Krisai-Greilhuber, Hermann Voglmayr, Marc Stadler
Data type
Multimedia.
References
- Andersson PF, Johansson SBK, Stenlid J, Broberg A. (2010) Isolation, identification and necrotic activity of viridiol from Chalara fraxinea, the fungus responsible for dieback of ash. Forest Pathology 40: 43–46. 10.1111/j.1439-0329.2009.00605.x [DOI] [Google Scholar]
- Bills GF, Gonzalez-Menendez V, Platas G, Fournier J, Persoh D, Stadler M. (2012) Hypoxylon pulicicidum sp. nov. (Ascomycota, Xylariales), a pantropical insecticide-producing endophyte. PLoS ONE 7: e46687. 10.1371/journal.pone.0046687 [DOI] [PMC free article] [PubMed]
- Bitzer J, Læssøe T, Fournier J, Kummer V, Decock C, Tichy HV, Piepenbring M, Peršoh D, Stadler M. (2008) Affinities of Phylacia and the daldinoid Xylariaceae, inferred from chemotypes of cultures and ribosomal DNA sequences. Mycological Research 112: 251–270. 10.1016/j.mycres.2007.07.004 [DOI] [PubMed] [Google Scholar]
- Chang CW, Chang HS, Cheng MJ, Liu TW, Hsieh SY, Yuan GF, Chen IS. (2014) Inhibitory effects of constituents of an endophytic fungus Hypoxylon investiens on nitric oxide and interleukin-6 production in RAW264.7 macrophages. Chemistry & Biodiversity 11(6): 949–961. 10.1002/cbdv.201300364 [DOI] [PubMed] [Google Scholar]
- Chepkirui C, Stadler M. (2017) The genus Diaporthe: a rich source of diverse and bioactive metabolites. Mycological Progress 16: 477–494. 10.1007/s11557-017-1288-y [DOI] [Google Scholar]
- Daranagama DA, Camporesi E, Tian Q, Liu X, Chamyuang S, Stadler M, Hyde KD. (2015) Anthostomella is polyphyletic comprising several genera in Xylariaceae. Fungal Diversity 73: 203–238. 10.1007/s13225-015-0329-6 [DOI] [Google Scholar]
- Daranagama DA, Hyde KD, Sir EB, Thambugala KM, Tian Q, Samarakoon MC, McKenzie EHC, Jayasiri SC, Tibpromma S, Bhat JD, Liu XZ, Stadler M. (2018) Towards a natural classification and backbone tree for Graphostromataceae, Hypoxylaceae, Lopadostomataceae and Xylariaceae. Fungal Diversity 88: 1–165. 10.1007/s13225-017-0388-y [DOI] [Google Scholar]
- Fournier J, Köpcke B, Stadler M. (2010) New species of Hypoxylon from western Europe and Ethiopia. Mycotaxon 113: 209–235. 10.5248/113.209 [DOI] [Google Scholar]
- Fournier J, Flessa F, Peršoh D, Stadler M. (2011) Three new Xylaria species from southwestern Europe. Mycological Progress 10: 33–52. 10.5248/113.209 [DOI] [Google Scholar]
- Halecker S, Surup F, Kuhnert E, Mohr KI, Brock NL, Dickschat JS, Junker C, Schulz B, Stadler M. (2014) Hymenosetin, a 3-decalynoyl tetramic acid antibiotic from cultures of the ash dieback pathogen, Hymenoscyphus pseudoalbidus. Phytochemistry 100: 86–91. 10.1016/j.phytochem.2014.01.018 [DOI] [PubMed] [Google Scholar]
- Halecker S, Surup F, Solheim H, Stadler M. (2018) Albiducins A and B, salicylaldehyde antibiotics from the ash tree-associated saprotrophic fungus Hymenoscyphus albidus. Journal of Antibiotics (Tokyo) 71: 339–341. 10.1038/ja.2017.66 [DOI] [PubMed] [Google Scholar]
- Halecker S, Wennrich JP, Rodrigo S, Andrée N, Rabsch L, Baschien C, Steinert M, Stadler M, Surup F, Schulz B. (2020) Fungal endophytes for biocontrol of ash dieback: The antagonistic potential of Hypoxylon rubiginosum Fungal Ecology 45 (in press). 10.1016/j.funeco.2020.100918 [DOI]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Helaly SE, Thongbai B, Stadler M. (2018) Diversity of biologically active secondary metabolites from endophytic and saprotrophic fungi of the ascomycete order Xylariales. Natural Products Report 35: 992–1014. 10.1039/C8NP00010G [DOI] [PubMed] [Google Scholar]
- Hsieh HM, Ju YM, Rogers JD. (2005) Molecular phylogeny of Hypoxylon and closely related genera. Mycologia 97: 844–865. 10.1080/15572536.2006.11832776 [DOI] [PubMed] [Google Scholar]
- Ju YM, Rogers JD. (1996) A revision of the genus Hypoxylon. Mycologia Memoir n° 20. APS Press, St. Paul, 365 pp. [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics, 20(4): 1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Meguro S, Yoshimoto T, Namikoshi M. (2003) Absolute structure, biosynthesis, and anti-microtubule activity of phomopsidin, isolated from a marine- derived fungus Phomopsis sp. Tetrahedron 59: 455–459. 10.1016/S0040-4020(02)01566-1 [DOI] [Google Scholar]
- Koukol O, Kelnarová I, Černý K. (2015) Recent observations of sooty bark disease of sycamore maple in Prague (Czech Republic) and the phylogenetic placement of Cryptostroma corticale. Forest Pathology 45: 21–27. 10.1111/efp.12129 [DOI] [Google Scholar]
- Kuhnert E, Fournier J, Peršoh D, Luangsa-ard JJD, Stadler M. (2014) New Hypoxylon species from Martinique and new evidence on the molecular phylogeny of Hypoxylon based on ITS rDNA and β-tubulin data. Fungal Diversity 64: 181–203. 10.1007/s13225-013-0264-3 [DOI] [Google Scholar]
- Kuhnert E, Sir EB, Lambert C, Hyde KD, Hladki AI, Romero AI, Rohde M, Stadler M. (2017) Phylogenetic and chemotaxonomic resolution of the genus Annulohypoxylon (Xylariaceae) including four new species. Fungal Diversity 85: 1–43. 10.1007/s13225-016-0377-6 [DOI] [Google Scholar]
- Lambert C, Wendt L, Hladki AI, Romero AI, Stadler M, Sir EB. (2019) Hypomontagnella (Hypoxylaceae): a new genus segregated from Hypoxylon by a polyphasic taxonomic approach. Mycological Progress 18: 187–201. 10.1007/s11557-018-1452-z [DOI] [Google Scholar]
- Miller JH. (1961) A Monograph of the World Species of Hypoxylon A Monograph of the World Species of Hypoxylon. University of Georgia Press, Athens.
- Pažoutová S, Follert S, Bitzer J, Keck M, Surup F, Šrůtka P, Holuša J, Stadler M. (2013) A new endophytic insect-associated Daldinia species, recognised from a comparison of secondary metabolite profiles and molecular phylogeny. Fungal Diversity 60: 107–123. 10.1007/s13225-013-0238-5 [DOI] [Google Scholar]
- Petrini LE, Petrini O. (1985) Xylariaceous fungi as endophytes. Sydowia 38: 216–234. https://www.zobodat.at/pdf/Sydowia_38_0216-0234.pdf [Google Scholar]
- Quang DN, Hashimoto T, Tanaka M, Baumgartner M, Stadler M, Asakawa Y. (2002) Chemical constituents of the Ascomycete Daldinia concentrica. Journal of Natural Products, 65: 1869–1874. 10.1021/np020301h [DOI] [PubMed] [Google Scholar]
- Quang DN, Hashimoto T, Tanaka M, Stadler M, Asakawa Y. (2004) Cyclic azaphilones daldinins E and F from the ascomycete fungus Hypoxylon fuscum (Xylariaceae). Phytochemistry, 65(4): 469–473. 10.1016/j.phytochem.2003.09.022 [DOI] [PubMed] [Google Scholar]
- Rayner RW. (1970) A Mycological Colour Chart. Commonwealth Mycological Institute, Kew and British Mycological Society.
- Silvestro D, Michalak I. (2012) raxmlGUI: a graphical front-end for RAxML. Organisms Diversity & Evolution 12: 335–337. 10.1007/s13127-011-0056-0 [DOI] [Google Scholar]
- Sir EB, Kuhnert E, Lambert C, Hladki AI, Romero AI, Stadler M. (2016) New species and reports of Hypoxylon from Argentina recognized by a polyphasic approach. Mycological Progress 15: 1–42. 10.1007/s11557-016-1182-z [DOI] [Google Scholar]
- Sir EB, Becker K, Lambert C, Bills FG, Kuhnert E. (2019) Observations on Texas hypoxylons, including two new Hypoxylon species and widespread environmental isolates of the H. croceum complex identified by a polyphasic approach. Mycologia 111: 832–856. 10.1080/00275514.2019.1637705 [DOI] [PubMed] [Google Scholar]
- Stadler M, Wollweber H, Mühlbauer A, Asakawa Y, Hashimoto T, Rogers JD, Ju YM, Wetzstein HG, Tichy HV. (2001) Molecular chemotaxonomy of Daldinia and other Xylariaceae. Mycological Research 105: 1191–1205. 10.1016/S0953-7562(08)61990-5 [DOI] [Google Scholar]
- Stadler M, Wollweber H, Fournier J. (2004) A host-specific species of Hypoxylon from France, and notes on the chemotaxonomy of the “Hypoxylon rubiginosum complex”. Mycotaxon 90: 187–211. [Google Scholar]
- Stadler M, Hellwig V. (2005a) Chemotaxonomy of the Xylariaceae and remarkable bioactive compounds from Xylariales and their associated asexual stages. Recent Research Developments in Phytochemistry 9: 41–93. [Google Scholar]
- Stadler M, Laessoe T, Vasilyeva L. (2005b) The genus Pyrenomyxa and its affinities to other cleistocarpous Hypoxyloideae as inferred from morphological and chemical traits. Mycologia 97: 1129–1139. 10.1080/15572536.2006.11832761 [DOI] [PubMed] [Google Scholar]
- Stadler M, Fournier J, Granmo A, Beltrán-Tejera E. (2008) The “red Hypoxylons” of the temperate and subtropical Northern Hemisphere. North American Fungi 3: 1–73. 10.2509/naf2008.003.0075 [DOI] [Google Scholar]
- Stadler M, Fournier J, Læssøe T, Chlebicki A, Lechat C, Flessa F, Rambold G, Peršoh D. (2010) Chemotaxonomic and phylogenetic studies of Thamnomyces (Xylariaceae). Mycoscience 51: 189–207. 10.1007/S10267-009-0028-9 [DOI] [Google Scholar]
- Stadler M, Kuhnert E, Peršoh D, Fournier J. (2013) The Xylariaceae as model example for a unified nomenclature following the “one fungus-one name” (1F1N) concept. Mycology 4: 5–21. 10.3114/sim0016 [DOI] [Google Scholar]
- Stadler M, Læssøe T, Fournier J, Decock C, Schmieschek B, Tichy HV, Peršoh D. (2014) A polyphasic taxonomy of Daldinia (Xylariaceae). Studies in Mycology 77: 1–143. 10.3114/sim0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler M, Lambert C, Wibberg D, Kalinowski J, Cox RJ, Kolarik M, Kuhnert E. (2020) Intragenomic polymorphisms in the ITS region of high quality genomes of the Hypoxylaceae (Xylariales, Ascomycota). Mycological Progress, 19: 235–245. 10.1007/s11557-019-01552-9 [DOI] [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Steglich W, Klaar M, Furtner W. (1974) Mitorubrin derivatives from Hypoxylon fragiforme. Phytochemistry 13: 2874–2875. 10.1016/0031-9422(74)80262-1 [DOI] [Google Scholar]
- Surup F, Halecker S, Nimtz M, Rodrigo S, Schulz B, Steinert M, Stadler M. (2018a) Hyfraxins A and B, cytotoxic ergostane-type steroid and lanostane triterpenoid glycosides from the invasive ash dieback ascomycete Hymenoscyphus fraxineus. Steroids 135: 92–97. 10.1016/j.steroids.2018.03.007 [DOI] [PubMed] [Google Scholar]
- Surup F, Kuhnert E, Böhm A, Pendzialek T, Solga D, Wiebach V, Engler H, Berkessel A, Stadler M, Kalesse M. (2018b) The Rickiols: 20-, 22-, and 24-membered macrolides from the ascomycete Hypoxylon rickii. Chemistry-an European Journal 24: 2200–2213. 10.1002/chem.201704928 [DOI] [PubMed] [Google Scholar]
- Swofford DL. (2002) PAUP* 4.0b10: phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland.
- Triebel D, Peršoh D, Wollweber H, Stadler M. (2005) Phylogenetic relationships among Daldinia, Entonaema and Hypoxylon as inferred from ITS nrDNA sequences. Nova Hedwigia 80: 25–43. 10.1127/0029-5035/2005/0080-0025 [DOI] [Google Scholar]
- U’Ren JM, Miadlikowska J, Zimmerman NB, Lutzoni F, Stajich JE, Arnold AE. (2016) Contributions of North American endophytes to the phylogeny, ecology, and taxonomy of Xylariaceae (Sordariomycetes, Ascomycota). Molecular Phylogenetics and Evolution 98: 210–232. 10.1016/j.ympev.2016.02.010 [DOI] [PubMed] [Google Scholar]
- Wendt L, Sir EB, Kuhnert E, Heitkämper S, Lambert C, Hladki AI, Romero AI, Luangsa-ard JJ, Srikitikulchai P, Peršoh D, Stadler M. (2018) Resurrection and emendation of the Hypoxylaceae, recognized from a multigene phylogeny of the Xylariales. Mycological Progress 17: 115–154. 10.1007/s11557-017-1311-3 [DOI] [Google Scholar]
- Zhang N, Castlebury LA, Miller AN, Huhndorf SM, Schoch CL, Seifert KA, Rossman AY, Rogers JD, Kohlmeyer J, Volkmann-Kohlmeyer B, Sung GH. (2006) An overview of the systematics of the Sordariomycetes based on a four-gene phylogeny. Mycologia 98: 1076–1087. 10.1080/15572536.2006.11832635 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Discovery of a new species of the Hypoxylon rubiginosum complex from Iran and antagonistic activities of Hypoxylon species towards the Ash Dieback pathogen, Hymenoscyphus fraxineus, in dual culture
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Mohammad Javad Pourmoghaddam, Christopher Lambert, Frank Surup, Seyed Akbar Khodaparast, Irmgard Krisai-Greilhuber, Hermann Voglmayr, Marc Stadler
Data type
Multimedia.