Abstract
The mangrove Kandelia obovata (Rhizophoraceae) is an important coastal shelterbelt and landscape tree distributed in tropical and subtropical areas across East Asia and Southeast Asia. Herein, a chromosome-level reference genome of K. obovata based on PacBio, Illumina, and Hi-C data is reported. The high-quality assembled genome size is 177.99 Mb, with a contig N50 value of 5.74 Mb. A large number of contracted gene families and a small number of expanded gene families, as well as a small number of repeated sequences, may account for the small K. obovata genome. We found that K. obovata experienced two whole-genome polyploidization events: one whole-genome duplication shared with other Rhizophoreae and one shared with most eudicots (γ event). We confidently annotated 19,138 protein-coding genes in K. obovata and identified the MADS-box gene class and the RPW8 gene class, which might be related to flowering and resistance to powdery mildew in K. obovata and Rhizophora apiculata, respectively. The reference K. obovata genome described here will be very useful for further molecular elucidation of various traits, the breeding of this coastal shelterbelt species, and evolutionary studies with related taxa.
Subject terms: Genome, Evolution
Introduction
Mangrove forests are coastal ecosystems with unique biodiversity that provides many ecosystem services and functions1. Mangrove loss will increase the threat of coastal hazards (i.e., erosion, storm surges, and tsunamis) to human safety and shoreline development2. Specifically, this will reduce coastal water quality and biodiversity and threaten adjacent coastal habitats, thereby weakening the main resources on which the human community relies, including a large number of products and services provided by mangroves3,4. Therefore, detailed studies and analyses of the genome and evolution of mangroves are urgently required, especially in the context of frequent human disturbance and inevitable sea-level rise.
The mangrove species Kandelia obovata belongs to Rhizophoraceae, which is called “Qiuqie” in Chinese, with the Latin name of K. candel in “Flora Reipublicae Popularis Sinicae”5. Later, in 2008, its Latin name was changed to K. obovata in the “Flora of China”6. K. obovata is a woody plant predominantly found in tropical and subtropical tidal salt wetlands distributed from East Asia to Southeast Asia7. K. obovata adapts to transitional ecosystems where the land and ocean meet by overcoming periodic and aperiodic tidal effects, which induce high salinity, severe erosion, and anaerobic conditions8. K. obovata plays a crucial role in protecting biodiversity and combating erosion9,10. Specifically, the mangrove K. obovata can protect the embankment, accelerate the natural deposition of the beach, filter organic matter and pollutants from inland areas, and provide an ideal habitat for the marine flora and fauna11. At the same time, due to its beautiful shape, unique floral pattern and fragrance, K. obovata is an excellent coastal wetland landscape plant and horticultural ornamental plant (Fig. 1).
Fig. 1. Morphological features of the flower and fruit of K. obovata.
a K. obovata trees in a coastal wetland. b Flowers. c Young fruits. d Cone-like fruits
Here, the genome of the mangrove K. obovata was sequenced using PacBio sequencing as well as the Illumina next-generation sequencing platform. These data can help clarify the history of mangrove colonization and mangrove adaptation mechanisms in intertidal zones. Furthermore, this study will provide a basis for the conservation of mangrove diversity and in-depth development of genetic resources for mangroves, as well as the development and utilization of coastal horticultural plants.
Results and discussion
Genome sequence and assembly
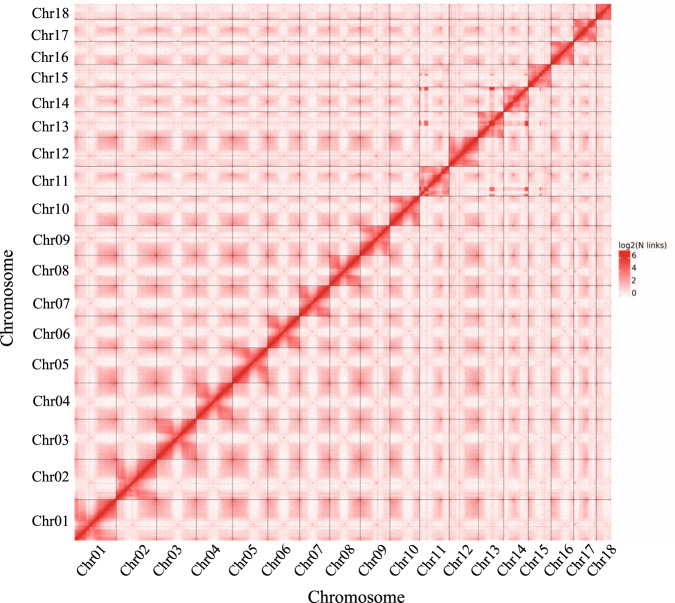
K. obovata contains 36 chromosomes (2n = 2x = 36)6. To assess genome size, survey sequencing was performed, and 65.27 Gb of clean data was obtained (Supplementary Table 1). The survey analysis indicated that the K. obovata genome size is 211.86 Mb and has a low level of heterozygosity of approximately 0.38% (Supplementary Fig. 1). The assembled genome is 178.44 Mb in size, with a scaffold N50 value of 279.55 kb obtained by using Illumina sequencing (Table 1). To improve K. obovata assembly quality, we conducted Pacific Biosciences RSII sequencing and obtained 25 Gb of single-molecule real-time long reads (average read length of 11.9 kb; Supplementary Fig. 2, Supplementary Table 1). The final assembled genome is 177.99 Mb in size, with a contig N50 value of 5.74 Mb (Table 1). The quality of the assembly was evaluated using Benchmarking Universal Single-Copy Orthologs (BUSCO)12. The results showed that the gene set completeness of the assembled genome is 97.3%, indicating that the K. obovata genome assembly is very complete and of high quality (Table 1). Finally, high-throughput/resolution chromosome conformation capture (Hi-C) technology was adopted to assess the chromosome-level diploid genome. The results showed that the lengths of the chromosomes ranged from 5.03 to 13.8 Mb (Supplementary Table 2), with a total length of 178.01 Mb and a scaffold N50 of 10.03 Mb (Fig. 2, Table 1).
Table 1.
The statistical results of Hi-C assembly
| Assembly | Size (bp) |
|---|---|
| Illumina sequencing assembly | |
| Scaffold N50 | 279,548 |
| Scaffold N90 | 28,239 |
| Longest Scaffold | 1,696,757 |
| Total Scaffold length | 178,438,058 |
| PacBio sequencing assembly | |
| Contig N50 | 5,743,053 |
| Contig N90 | 2,939,642 |
| Longest Contig | 13,452,090 |
| Total Contig length | 177,986,124 |
| BUSCO | 97.3% |
| Hi-C assembly | |
| Scaffold N50 | 10,026,007 |
| Scaffold N90 | 7,500,541 |
| Longest Contig | 13,797,742 |
| Total Contig length | 178,014,124 |
Fig. 2.
Intensity signal heat map of the Hi-C chromosome
Gene prediction and annotation
We confidently annotated 19,138 protein-coding genes in K. obovata (Supplementary Fig. 3, Supplementary Table 3), of which 19,136 (99.17%) were supported by de novo prediction, transcriptome data, and homolog prediction (Supplementary Table 4). The genome of Rhizophora apiculata, also belonging to Rhizophoreae, has 26,640 protein-coding genes, which is 7502 more than observed in K. obovata13. The BUSCO12 assessment indicated that the completeness of the gene set of the annotated genome was 90% for K. obovata (Supplementary Table 5). In addition, 105 microRNAs, 307 transfer RNAs, 167 ribosomal RNAs, and 199 small nuclear RNAs were identified in the K. obovata genome (Supplementary Table 6).
Using homology-based and de novo approaches to identify transposable elements (TEs), we estimated that 24.07% of the K. obovata genome consists of repetitive sequences (Supplementary Figs. 4 and 5 and Supplementary Tables 7 and 8) and 29% of the R. apiculata genome consists of repetitive sequences13. Compared with those of closely related nonmangrove plant genomes, the repetitive portions of the R. apiculata genome, comprising predominantly TE families, are significantly reduced, and the decrease in TE number largely resulted in a general decrease in genome size among true mangroves13. The small repetitive sequences may be one reason for the small genome of K. obovata. In addition, 18,266 genes were functionally annotated, among which 11,124 and 14,401 were annotated to Gene Ontology terms and Kyoto Encyclopedia of Genes and Genomes terms, respectively, and 12,491 genes were functionally annotated in all five databases (Supplementary Fig. 6, Supplementary Table 9).
Evolution of gene families
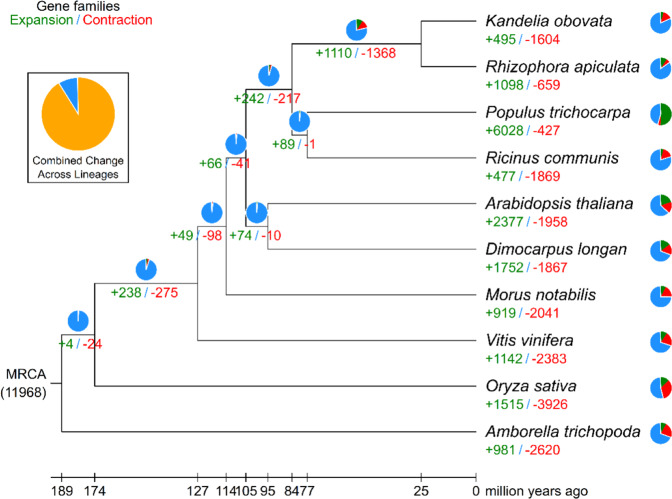
We constructed a phylogenetic tree and estimated the divergence times of K. obovata and nine other plant species based on genes extracted from a total of 1095 single-copy families (Supplementary Figs. 7 and 8, Supplementary Table 10). As expected, K. obovata was sister to R. apiculata (Supplementary Fig. 9). The estimated Rhizophoreae divergence time was 83.15 Mya, and the divergence time between K. obovata and R. apiculata was 24.63 Mya (Supplementary Fig. 9). Next, using CAFÉ 3 (ref. 14), we found that 1110 gene families were expanded in the lineage leading to the Rhizophoreae, whereas 1368 families were contracted (Fig. 3). Four hundred and ninety-five gene families were expanded in K. obovata, compared with the 1098 in R. apiculata (Fig. 3). At the same time, 1604 gene families were contracted in K. obovata, compared with the 659 in R. apiculata. K. obovata has more contracted gene families than R. apiculata and fewer expanded gene families than R. apiculata, which may be the reason that the genome of K. obovata is smaller than that of R. apiculata. For the expanded gene families, we conducted GO enrichment analysis and found enrichment for the GO terms “structural constituent of cytoskeleton” and “structural constituent of ribosome” (Supplementary Table 11). For the contracted gene families, enrichment was detected for the GO terms “protein kinase activity”, “terpene synthase activity”, “oxidoreductase activity”, “nutrient reservoir activity”, “defense response”, and “sulfotransferase activity” (Supplementary Table 12). Gene families with K. obovata-specific expansion and contraction might relate to adaptation to K. obovata-specific coastal niches. Further research is required to validate the function of these genes.
Fig. 3. The expansion and contraction of gene families.
The green number indicates the number of expanded gene families, and the red number indicates the number of contracted gene families. The blue color in the circle shows the gene families whose copy numbers are constant, while the orange color represents the proportion of 11,968 gene families in the most recent common ancestor that have expanded or contracted during late differentiation
Synteny analysis and an ancient polyploidization event
Whole-genome polyploidization events are a feature of many taxa and an efficient mechanisms of genome expansion15. To detect the occurrence of polyploidization events in Rhizophoreae, we used the default parameters of JCVI v0.9.14 (ref. 16) to analyze the protein sequences of K. obovata, R. apiculata, and Vitis vinifera and obtained the gene pairs in the collinear regions. The results showed that there were 11,010 collinear gene pairs between K. obovata and R. apiculata, 10,893 collinear gene pairs between K. obovata and V. vinifera, 3,840 collinear gene pairs within K. obovata and 4,646 collinear gene pairs within R. apiculata (Supplementary Table 13).
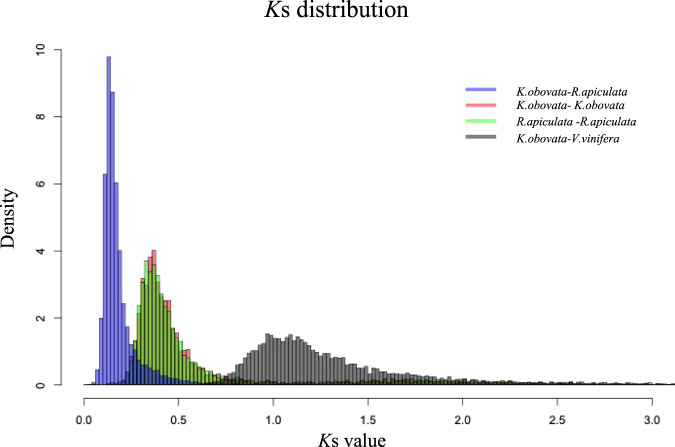
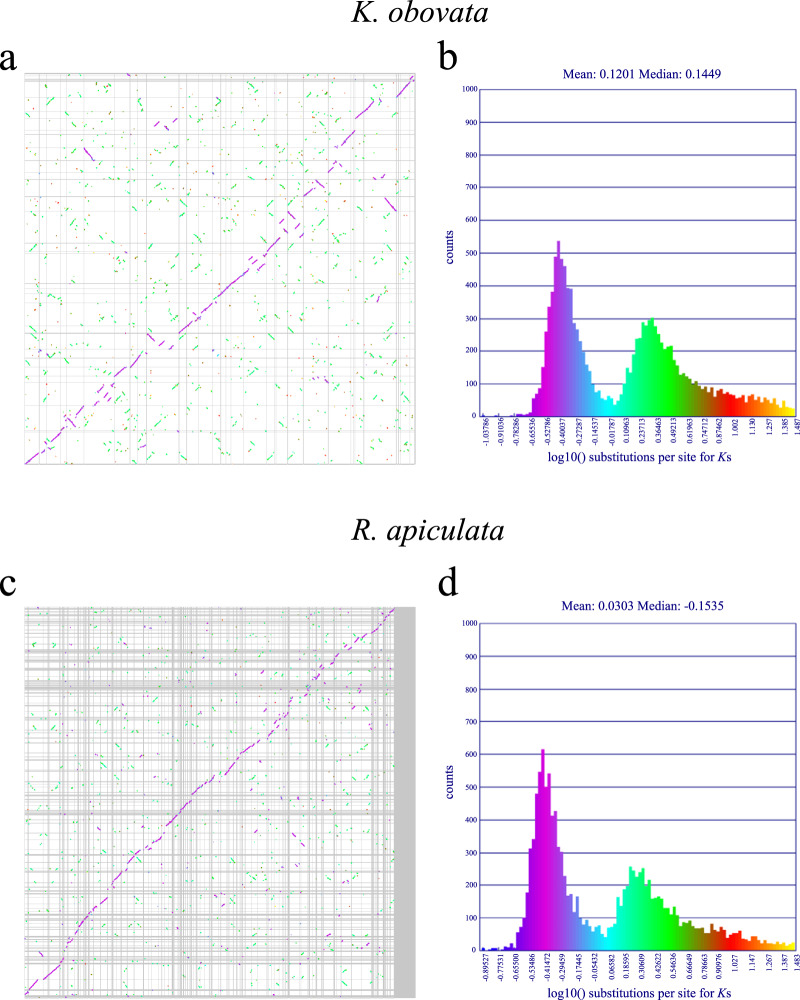
We estimated the distributions of synonymous substitutions per synonymous site (Ks) values to more precisely infer the timing of polyploidization events in the K. obovata genome. The distributions of Ks for paralogous K. obovata genes showed two peaks, one at Ks = 0.38 and the other at Ks = 1.5–1.9 (Fig. 4, Supplementary Fig. 10a). The Ks distribution of R. apiculata also had two peaks, one at Ks = 0.32 and the other at Ks = 1.5–1.9 (Fig. 4, Supplementary Fig. 10b). The results suggested that K. obovata and R. apiculata experienced two polyploidization events. To confirm these two polyploidization events, we further analyzed the Ks distribution of K. obovata and R. apiculata and that of K. obovata and V. vinifera. We observed that the Ks distribution of K. obovata and R. apiculata had one peak, at Ks = 0.1–0.16, which was smaller than the first peak in the Ks distributions within K. obovata (Ks = 0.38) and R. apiculata (Ks = 0.32) (Fig. 4). The first peak in the K. obovata Ks distribution (Ks = 0.38) indicates that K. obovata shares a whole-genome duplication (WGD) event with other Rhizophoreae. In addition, we found that the Ks distribution of K. obovata and V. vinifera had one peak, at Ks = 0.9–1.4, which was also smaller than the second peak in the Ks distributions within K. obovata (Ks = 1.5–1.9) and R. apiculata (Ks = 1.5–1.9) (Fig. 4). The second peak in the K. obovata Ks distribution (Ks = 1.5–1.9) indicates that the common ancestor of K. obovata and V. vinifera experienced an ancient polyploidization event. This event was shared by most eudicots, called the γ event, which is an ancient whole-genome triplication event17. Finally, we provide direct evidence of gene collinearity, as shown in Fig. 5; the purple peak corresponds to the first peak of the K. obovata Ks distribution (Ks = 0.38) and R. apiculata Ks distribution (Ks = 0.32) (Fig. 5b, d), and the green peak corresponds to the second peak of the K. obovata Ks distribution (Ks = 1.5–1.9) and R. apiculata Ks distribution (Ks = 1.5–1.9) (Fig. 5a, c). The purple collinear region is an extra copy of the genomes of K. obovata and R. apiculata, and the green collinear region is also an extra copy of the genes in the genomes of K. obovata and R. apiculata (Fig. 5). These copies correspond to two polyploidization events of K. obovata and R. apiculata. Therefore, our study verified that K. obovata experienced two polyploidization events: one WGD event shared with Rhizophoreae and one shared with most eudicots (γ event).
Fig. 4. Ks distributions between K. obovata and R. apiculata and K. obovata and V. vinifera and within K. obovata and R. apiculata.
Peaks of intraspecies Ks distributions indicate ancient whole-genome polyploidization events, and peaks of interspecies Ks distributions indicate speciation events
Fig. 5. Collinear point diagram and Ks values corresponding to the collinear blocks.
a The collinear point diagram of K. obovata. b Distribution of log10 (Ks) values of the collinear blocks in K. obovata. c The collinear point diagram of R. apiculata. d Distribution of log10 (Ks) values of the collinear blocks in R. apiculata. The ordinate of b, d is the number of gene pairs corresponding to the Ks value, and the abscissa is the log10 (Ks) value
MADS-box gene family analysis
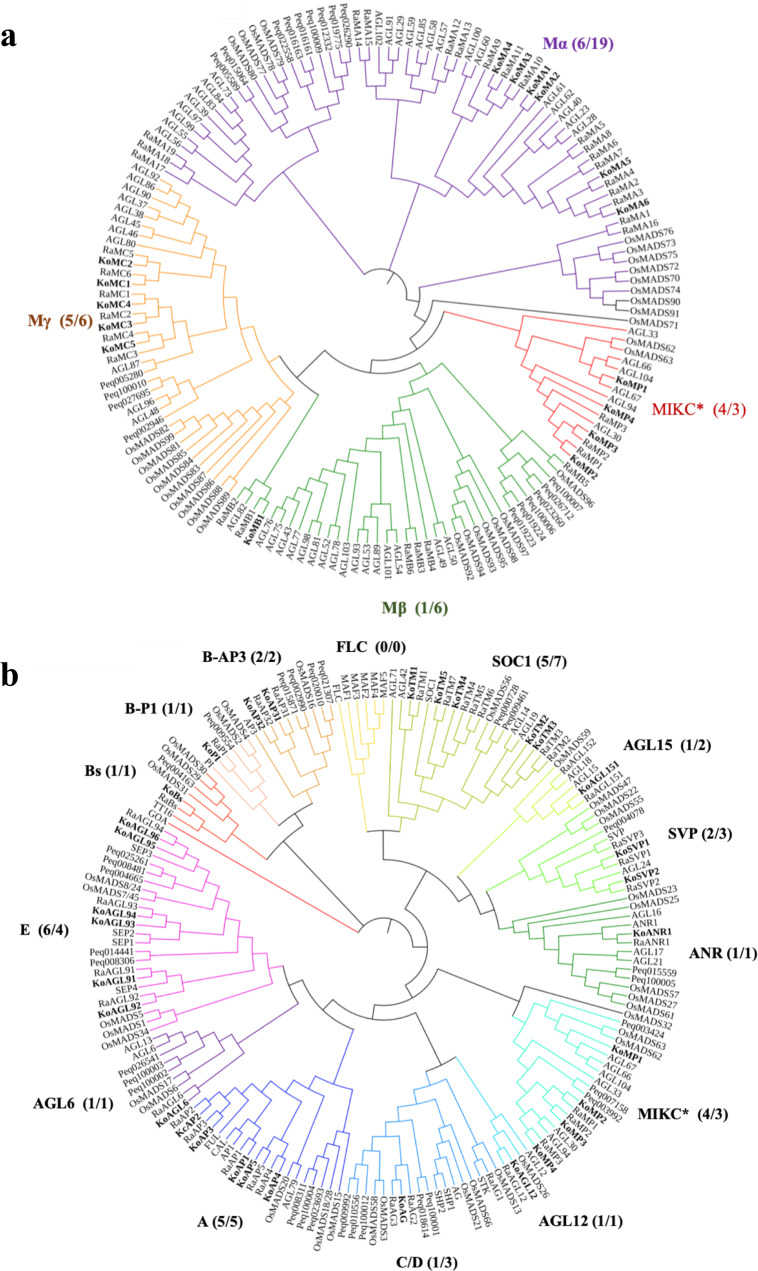
MADS-box genes play a key role in many important processes during plant development, especially during flower development18. We evaluated the MADS-box genes in K. obovata and R. apiculata. The K. obovata and R. apiculata genomes encode 43 and 65 MADS-box genes, respectively. There are 12 type I and 31 type II MADS-box genes in the K. obovata genome and 31 type I and 34 type II genes in the R. apiculata genome (Table 2, Supplementary Table 14). Interactions among type I MADS-box genes promote the initiation of endosperm development19. The type I genes of R. apiculata were approximately three times more numerous than those of K. obovata (Fig. 6a, Table 2). In addition, only 1 pseudogene type I genes were found in the K. obovata genome (Supplementary Table 14), suggesting that the type I MADS-box genes of K. obovata experienced a lower gain rate and higher loss rate than type II MADS-box genes.
Table 2.
MADS-box genes in Arabidopsis thaliana, Oryza sativa, Phalaenopsis equestris, K. obovata, and R. apiculata
| Category | A. thalianaa | O. sativab | P. equestrisc | K. obovata | R. apiculatad |
|---|---|---|---|---|---|
| Type II (total) | 45 | 44 | 29 | 31 | 34 |
| MIKCc | 39 | 39 | 28 | 27 | 31 |
| MIKC* | 6 | 5 | 1 | 4 | 3 |
| Type I (total) | 61 | 31 | 22 | 12 | 31 |
| Mα | 25 | 12 | 10 | 6 | 19 |
| Mβ | 20 | 9 | 0 | 1 | 6 |
| Mγ | 16 | 10 | 12 | 5 | 6 |
| Total | 106 | 75 | 51 | 43 | 65 |
aThe whole-genome sequence of A. thaliana was extracted from the NCBI database, BioProject: PRJNA477266 (ref. 14)
bThe whole-genome sequence of O. sativa was extracted from rice.plantbiology.msu.edu/
cThe whole-genome sequence of P. equestris was extracted from the NCBI database, BioProject: PRJNA192198 (ref. 15)
dThe whole-genome sequence of R. apiculata was extracted from http://evolution.sysu.edu.cn/Sequences.html
Fig. 6. Phylogenetic analysis of MADS-box genes from A. thaliana, O. sativa, P. equestris, K. obovata, and R. apiculata.
a Phylogenetic tree of type I MADS-box genes. b Phylogenetic tree of type II MADS-box genes. The number on the left in parentheses represents the homologous MADS genes of K. obovata, and the number on the right represents the homologous MADS genes of R. apiculata. The bolded gene ID numbers beginning with “Ko” represent the gene IDs of K. obovata; those beginning with “Ra” represent the gene IDs of R. apiculata
Type II MADS-box genes include two types: MIKCC and MIKC*20. MIKC*-type gene regulation has a major impact on pollen gene expression21,22. Plant MIKCC-type genes are the most widely studied MADS-box genes because they are essential for plant growth and development23,24. The K. obovata genome has four MIKC*-type genes and 27 MIKCC-type genes, while the R. apiculata genome has three MIKC*-type genes and 31 MIKCC-type genes (Fig. 6b, Table 2). Fewer C/D-class and AGL6 genes were found in K. obovata and R. apiculata than in rice, whereas more B-AP3-class and E-class genes were found in K. obovata than in rice (Fig. 6b). A-class, B-class, C/D-class, and E-class gene clades are well known for their roles in the specification of floral organ identity25, notably, the ABCDE flowering model26–28. K. obovata and R. apiculata have the same number of A-class and B-class genes (five members). K. obovata (six members) has more E-class genes than R. apiculata (four members), and R. apiculata (one member) has fewer C-class genes than K. obovata (three members) (Fig. 6b). The AGL12 gene is involved in root cell differentiation29, and the ANR1 gene is involved in the regulation of lateral root development30. Furthermore, the loss of the AGL12 gene may result in the loss of the ability to develop true roots for terrestrial growth29. K. obovata and R. apiculata each contain one AGL12-clade gene and one ANR1-clade gene (Fig. 6b), which may be because mangrove roots have adapted to environments at the interface of land and sea. SOC1, SVP, FLC, and AGL15 regulate flowering time31–34. SOC1 integrates multiple flowering signals related to photoperiod, temperature, hormones, and age34. Notably, we found that SOC1-like genes were expanded in both K. obovata (five members of SOC1) and R. apiculata (seven members of SOC1) (Fig. 6b). Sequence variation among these SOC1-like genes could be associated with the functional diversification of the SOC1 clade in K. obovata and R. apiculata.
Disease resistance-related genes
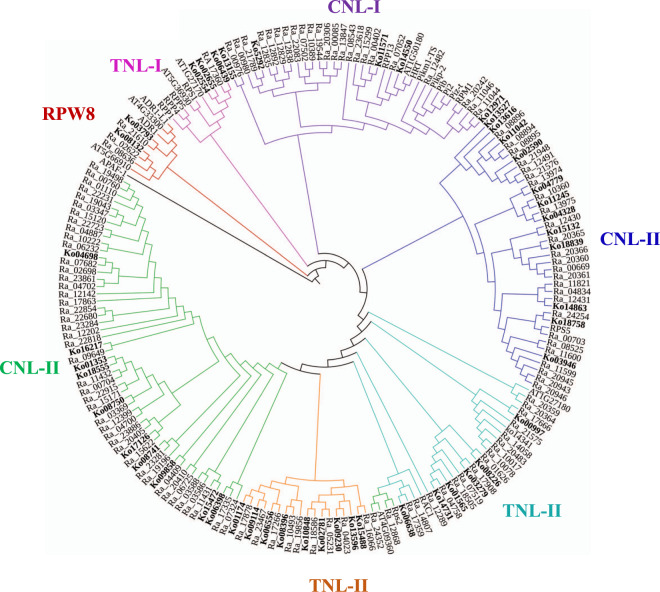
Plant resistance genes (R genes) exist in large families and usually contain a nucleotide-binding site (NBS) domain and a leucine-rich repeat (LRR) domain, denoted NLR35. According to the presence or absence of different domains in the N-terminal region, resistance genes encoding NBS domains can be divided into the TNL (TIR-NBS-LRR), CNL (CC-NBS-LRR), and RNL (RPW8-NBS-LRR) groups36. A total of 165 and 292 nucleotide-binding site (NBS)-containing R genes were identified in K. obovata and R. apiculata, respectively; this might be because the distribution of R. apiculata is wider than that of K. obovata (Fig. 7, Supplementary Table 15).
Fig. 7. Phylogenetic reconstruction of the NLR proteins in K. obovata and R. apiculata.
The NBS domain of human apoptotic protease-activating factor-1 (APAF-1) is located at the root of the tree. The bolded gene ID numbers beginning with “Ko” represent the gene IDs of K. obovata; those beginning with “Ra” represent the gene IDs of R. apiculata
We selected NLR candidate genes from K. obovata and R. apiculata with complete domains to construct a phylogenetic tree. The results showed that these candidate genes were divided into the TNL, RNL, and CNL families (Fig. 7). RPW8 is a family of genes with highly specifically expressed characteristics, including resistance to powdery mildew37. The phylogenetic tree showed that RPW8 genes were significantly separated from all other CNL genes (Fig. 7). The RPW8 clade contained two K. obovata and three R. apiculata genes and clustered with two ADR1 genes from Arabidopsis, indicating that RPW8 genes might be associated with resistance to powdery mildew (Fig. 7).
Conclusion
Although K. obovata is well known as a coastal shelterbelt and landscape tree in tropical and subtropical areas, research on this species has been hampered by a lack of genetic data. We obtained a chromosome-level reference genome of K. obovata, assembled a 177.99 Mb genome, and annotated 19,136 protein-coding genes. A large number of contracted gene families and a small number of expanded gene families, as well as a small number of repeated sequences, resulted in a smaller genome in K. obovata than in R. apiculata. Ks analysis revealed that K. obovata experienced two polyploidization events, namely, the recent WGD shared with other Rhizophoreae and the ancient polyploidization event shared with most eudicots (γ event). The Rhizophoreae divergence time was 83.15 Mya, and the divergence time between K. obovata and R. apiculata was 24.63 Mya. We identified MADS-box and RPW8 genes in K. obovata, which might be related to flowering and resistance to powdery mildew, respectively. The genomic sequence analysis of the mangrove K. obovata helped reveal its mechanisms of adaptation to the intertidal zone; this knowledge is critical for understanding its genetic evolution and reproduction.
Materials and methods
DNA preparation and sequencing
Fresh K. obovata tissues were collected from the Quanzhou Estuary Wetland Provincial Nature Reserve, Fujian Province, China. Genomic DNA was isolated from the fresh leaves of K. obovata for de novo sequencing and assembly. Paired-end libraries (500 bp) were constructed according to the Illumina protocol. Genome size and heterozygosity were measured using KmerFreq and GCE based on a 17-K-mer distribution. In addition, a 20 kb insert library was constructed according to the PacBio RSII protocol and subsequently sequenced on the PacBio platform (Supplementary Table 1). The transcriptomes of different tissues of K. obovata were sequenced on the Illumina platform.
Genome assembly
De novo assembly of the PacBio reads was performed. FALCON (https://github.com/PacificBiosciences/FALCON)38 was used to correct errors in the original data. Then, SMARTdenovo v1.0 was used to assemble the corrected data39, and Arrow software (https://github.com/PacificBiosciences/GenomicConsensus) was used to polish the assembly results. To further eliminate Indel and SNP errors in the assembly sequence, we compared the second-generation small-fragment data to the assembly results and corrected the assembly results again with Pilon v1.22 (ref. 40). To confirm the quality of the genome assembly, we performed a BUSCO v3 (ref. 12) (http://busco.ezlab.org/) assessment using single-copy orthologous genes.
Hi-C library construction and assembly of the chromosome
Fresh leaves of K. obovata were used to construct a Hi-C sequencing library, which was sequenced on the NovaSeq platform. SOAPnuke v1.5.3 (ref. 41) was used to filter the original data (filtration parameter: filter -n 0.01 -l 20 -q 0.4 -d -M 3 -A 0.3 -Q 2 -i -G --seqType 1) to obtain clean reads. Then, the clean data were compared with the genome using Juicer software42. The results were filtered, and misaligned reads were removed. The genome sequence was preliminarily clustered, sequenced, and directed using 3D-DNA43. Juicer-box42 was again used to adjust, reset, and cluster the genome sequence. Finally, we evaluated genome integrity using BUSCO v3 software12.
Identification of repetitive sequences
TEs contribute to genome dynamism in terms of both size and structure through insertions and eventual loss44. Tandem Repeats Finder (http://tandem.bu.edu/trf/trf.html, v4.07) was used to predict tandem repeats across the genome45. TEs were first identified using RepeatMasker v3.3.0 (http://www.repeatmasker.org) and RepeatProteinMask based on Repbase v21.12 (http://www.girinst.org/repbase)46. Then, two de novo prediction software programs, RepeatModeler (http://www.repeatmasker.org/RepeatModeler/)47 and LTR_FINDER v1.06 (http://tlife.fudan.edu.cn/ltr_finder/)48, were used to identify TEs in the genomes. Finally, repeat sequences with identities ≥50% were grouped into the same classes.
Gene prediction and annotation
Homology-based, de novo, and transcriptome-based predictions were integrated to predict high-quality protein-coding genes. For homology-based prediction, homologous proteins from five available whole-genome sequences, namely, those of Arabidopsis thaliana, Linum usitatissimum, Populus trichocarpa, Ricinus communis, and Salix purpurea, were aligned to the K. obovata genome sequence using Exonerate v2.0 (https://www.ebi.ac.uk/Tools/psa/genewise/)49. Gene structures were generated using GeneWise v2.4.1 (ref. 50). Three ab initio prediction software programs, namely, Augustus v3.0.2 (http://bioinf.uni-greifswald.de/augustus/)51, Fgenesh (https://omictools.com/fgenesh-tool)52, and GlimmerHMM53, were employed for de novo gene prediction. Then, the homology-based and ab initio gene structures were merged into a nonredundant gene model using Maker v2.31.8 (ref. 54). TopHat v2.0.11 was used to map RNA-seq reads to the assembly55, and Cufflinks v2.2.1 (ref. 56) was applied to combine the mapping results for transcript structural predictions.
The protein sequences of the consensus gene set were aligned to seven protein databases, including GO (The Gene Ontology Consortium)57, KEGG (http://www.genome.jp/kegg/)58, InterPro (https://www.ebi.ac.uk/interpro/)59, Swiss-Prot (http://www.uniprot.org)60, and TrEMBL (http://www.uniprot.org/)60, for predicted gene annotation. The rRNAs were identified by aligning the rRNA template sequences from the Rfam61 database against the genome using the BLASTN algorithm with an E-value cutoff of 1E–5. The tRNAs were predicted using tRNAscan-SE v1.3.1 (http://lowelab.ucsc.edu/tRNAscan-SE/)62, and other ncRNAs were predicted by Infernal software (http://infernal.janelia.org/) against the Rfam database.
Phylogenetic analysis
Genes from whole-genome sequences of ten species (K. obovata, Amborella trichopoda, Arabidopsis thaliana, Dimocarpus longan, Morus notabilis, Populus trichocarpa, Rhizophora apiculata, Ricinus communis, Vitis vinifera, and Oryza sativa) were used for gene-family clustering analysis. OrthoMCL v2.0.9 (ref. 63) was used to identify orthologous groups among the ten species. Pairwise similarities between all protein sequences were calculated using BLASTP with an E-value cutoff of 1E–5. To obtain reliable single-copy orthologous groups, we filtered out single-copy orthologous groups containing proteins of length <200 bp. MUSCLE v3.8.31 (ref. 64) was used to perform multisequence alignment of the protein sequences of the filtered single-copy orthologous group, and nucleotide alignment results were obtained by the corresponding relationship between protein sequences and nucleotide sequences. Finally, the nucleotide sequences of the single-copy orthologous group were connected to form a supergene, and then the data set was employed to construct a phylogenetic tree by using the GTR + gamma model in MrBayes65.
Estimation of divergence time
The Markov chain Monte Carlo algorithm for Bayesian estimation was employed to infer the divergence time of each tree node using the MCMCTree module of PAML v4.7 (ref. 66). The nucleic acid replacement model used was the GTR model, and the molecular clock model used was the independent rate model. The MCMC process included 100,000 burn-in iterations and 1,000,000 sampling iterations (with a sample taken every 100 iterations). To obtain a more stable result, the same parameter was executed twice. Calibration times were obtained from TimeTree (http://www.timetree.org).
Gene family expansion and contraction
We measured the expansion and contraction of orthologous gene families using CAFÉ 3 (https://github.com/hahnlab/CAFE)14. Based on maximum likelihood modeling of gene gain and loss, we analyzed gene families for signs of expansion or contraction using genomic data from the ten species.
Collinearity analysis
Within collinear segments, genes are conserved in function and sequence and remain highly conserved during the evolution of species. We used the default parameters of JCVI v0.9.14 (https://pypi.org/project/jcvi/)11 to analyze the protein sequences of K. obovata, R. apiculata, and V. vinifera and obtained the gene pairs in collinear regions. Then, we used COGE (https://genomevolution.org/coge/) for online analysis, examined the relationship between Ks peaks and collinear regions, and verified the WGD event experienced by the common ancestor of K. obovata and R. apiculata.
Whole-genome duplication
We used Ks distribution analysis to infer WGD events of K. obovata and R. apiculata. Diamond v0.9.24 (ref. 67) was used to conduct self-alignment of the protein sequences of the two species and then extract the mutual optimal alignment in the alignment results. Finally, Codeml in the PAML package was used to calculate the Ks values39,68.
MADS-box analysis
The hidden Markov model (HMM) profile of the MADS-box gene family (PF00319) was obtained from Pfam (http://pfam.xfam.org). MADS-box gene family proteins were separately searched with HMMER 3.1 (with the default parameters)69. InterProScan v 5.19 (ref. 70) was used to identify MADS-box gene family candidates in the genomes of K. obovata and R. apiculata. The genomic data of R. apiculata were downloaded from http://evolution.sysu.edu.cn/Sequences.html. MADS-box gene candidates were further confirmed with the 60 amino acid domains available from SMART71 and online BLAST analysis (https://www.ncbi.nlm.nih.gov). Specifically, the protein sequence set for the MADS-box gene candidates was subjected to BLAST analysis against the assembled transcriptomes of the roots, stems, leaves, flowers, and fruits of K. obovata with the TBLASTN program. A phylogenetic tree was then constructed using MEGA5 (ref. 72) with the default parameters.
Disease resistance genes
Predicted proteins from the K. obovata and R. apiculate genomes were scanned using HMMER v3.1 (E-value cut-off of 1 × 10−5)69 using the HMM corresponding to the Pfam NLR protein family (NB-ARC: PF00931; TIR: PF01582; RPW8: PF05659; LRR: PF00560, PF07723, PF07725 and PF12799). To remove false-positive NB-ARC domain hits, InterProScan v5.19 was used to check the protein domains of the extracted sequences70. The NBS domains of the genes confirmed by both HMMER and InterProScan were extracted according to InterProScan annotation and aligned using MAFFT v7.310 (ref. 63); the alignment was then input into FastTree73 with the JTT model and visualized using EvolView74.
Supplementary information
Chromosome-scale assembly of the Kandelia obovata genome
Acknowledgements
This research was jointly funded by the National Science Foundation of China (41801062), the Special Project for the Cultivation of Major Achievements in the Peak Discipline of Forestry Science (118/71201800709), the Special Subsidy for Leading Talents of Scientific and Technological Innovation in Fujian Province (118/KRC16006A), and the Fujian Forestry Science and Technology Research Project (Min[2019]6). The authors would like to thank Prof. Wenqing Wang (College of the Environment and Ecology, Xiamen University) and Dr. Jiafang Huang (School of Geographical Sciences, Fujian Normal University) for kindly providing the picture of K. obovata.
Data availability
Genome sequences have been submitted to the National Genomics Data Center (NGDC). PacBio whole-genome sequencing data and Illumina data have been deposited in BioProject/GSA (https://bigd.big.ac.cn/gsa.)75 under accession codes PRJCA002330/CRA002395 and the whole-genome assembly and annotation data have been deposited in BioProject/GWH (https://bigd.big.ac.cn/gwh)76 under accession codes PRJCA002330/GWHACBH00000000.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
These authors contributed equally: Min-Jie Hu, Wei-Hong Sun
Contributor Information
Shuang-Quan Zou, Email: zou@fafu.edu.cn.
Zhong-Jian Liu, Email: zjliu@fafu.edu.cn.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41438-020-0300-x).
References
- 1.Kauffman JB, et al. Shrimp ponds lead to massive loss of soil carbon and greenhouse gas emissions in northeastern Brazilian mangroves. Ecol. Evol. 2018;8:5530–5540. doi: 10.1002/ece3.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilman EL, Ellison JC, Duke NC, Field CD. Threats to mangroves from climate change and adaptation options: a review. Aquat. Bot. 2008;89:237–250. doi: 10.1016/j.aquabot.2007.12.009. [DOI] [Google Scholar]
- 3.Nagelkerken I, et al. The habitat function of mangroves for terrestrial and marine fauna: a review. Aquat. Bot. 2008;89:155–185. doi: 10.1016/j.aquabot.2007.12.007. [DOI] [Google Scholar]
- 4.Walters BB, et al. Ethnobiology, socio-economics and management of mangrove forests: a review. Aquat. Bot. 2008;89:220–236. doi: 10.1016/j.aquabot.2008.02.009. [DOI] [Google Scholar]
- 5.Wight et al. in Flora Reipublicae Popularis Sinicae (ed Delectis florae Reipublicae Popularis Sinicae agenda academiae sinicae) Vol. 52, 133–135 (Sciences Press, Beijing, 1983).
- 6.Qin, H. N. & David, E. B. in Flora of China (eds Wu, Z. Y., Peter, R.H. & Hong, D.) Vol. 13, 295–299 (Sciences Press, Beijing, 2009).
- 7.Sheue CR, Liu HY, Yong JWH. Kandelia obovata (Rhizophoraceae), a new mangrove species from Asia. Taxon. 2003;52:287–294. doi: 10.2307/3647398. [DOI] [Google Scholar]
- 8.Giri C, et al. Status and distribution of mangrove forests of the world using earth observation satellite data. Glob. Ecol. Biogeogr. 2011;20:154–159. doi: 10.1111/j.1466-8238.2010.00584.x. [DOI] [Google Scholar]
- 9.Wardiatno Y, Mardiansyah, Prartono T, Tsuchiya M. Possible food sources of macrozoobenthos in the manko mangrove ecosystem, Okinawa (Japan): a stable isotope analysis approach. Trop. Life Sci. Res. 2015;26:53–65. [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Q, et al. Characteristics and distribution of microplastics in the coastal mangrove sediments of China. Sci. Total Environ. 2019;31:134807. doi: 10.1016/j.scitotenv.2019.134807. [DOI] [PubMed] [Google Scholar]
- 11.Rogers A, Mumby PJ. Mangroves reduce the vulnerability of coral reef fisheries to habitat degradation. PLoS Biol. 2019;17:e3000510. doi: 10.1371/journal.pbio.3000510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simao FA, et al. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 13.Xu SH, et al. The origin, diversification and adaptation of a major mangrove clade (Rhizophoreae) revealed by whole-genome sequencing. Natl. Sci. Rev. 2017;4:721–734. doi: 10.1093/nsr/nwx065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han MV, Thomas GWC, Lugo-Martinez J, Hahn MW. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol. Biol. Evol. 2013;30:1987–1997. doi: 10.1093/molbev/mst100. [DOI] [PubMed] [Google Scholar]
- 15.McGrath, C. L. & Lynch, M. Evolutionary significance of whole-genome duplication. in Poly-ploidy and Genome Evolution (eds Soltis, P. S. & D. E., Soltis) 1–20 (Springer Berlin Heidelberg, Berlin, Heidelberg, 2012).
- 16.Tang, H. B. et al. JCVI v0.9.14, https://pypi.org/project/jcvi/ (2014).
- 17.Wu SD, Han BC, Jiao YN. Genetic contribution of Paleopolyploidy to adaptive evolution in angiosperms. Mol. Plant. 2019;13:59–71. doi: 10.1016/j.molp.2019.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, et al. Genome-wide identification, characterization of the MADS-box gene family in Chinese jujube and their involvement in flower development. Sci. Rep. 2017;7:1025. doi: 10.1038/s41598-017-01159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masiero S, Colombo L, Grini PE, Schnittger A, Kater MM. The emerging importance of type I MADS box transcription factors for plant reproduction. Plant Cell. 2011;23:865–872. doi: 10.1105/tpc.110.081737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henschel K, et al. Two ancient classes of MIKC-type MADS-box genes are present in the moss physcomitrella patens. Mol. Biol. Evol. 2002;19:801–804. doi: 10.1093/oxfordjournals.molbev.a004137. [DOI] [PubMed] [Google Scholar]
- 21.Adamczyk BJ, Fernandez DE. MIKC* MADS domain heterodimers are required for pollen maturation and tube growth in Arabidopsis. Plant Physiol. 2009;149:1713–1723. doi: 10.1104/pp.109.135806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, et al. Functional conservation of MIKC*-Type MADS box genes in Arabidopsis and rice pollen maturation. Plant Cell. 2013;25:1288–1303. doi: 10.1105/tpc.113.110049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theissen G, Melzer R. Molecular mechanisms underlying origin and diversification of the angiosperm flower. Ann. Bot. 2007;100:603–609. doi: 10.1093/aob/mcm143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li C, et al. Genome-wide characterization of the MADS-box gene family in radish (Rahpanus sativus L.) and assessment of its roles in flowering and floral organogenesis. Front. Plant Sci. 2016;7:1390. doi: 10.3389/fpls.2016.01390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheng XG, et al. Genome wide analysis of MADS-box gene family in Brassica oleracea reveals conservation and variation in flower development. BMC Plant Biol. 2019;19:106. doi: 10.1186/s12870-019-1717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coen ES, Meyerowita EM. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- 27.Zahn LM, Feng B, Ma H. Beyond the ABC-model: regulation of floral homeotic genes. Adv. Bot. Res. 2006;44:163–207. doi: 10.1016/S0065-2296(06)44004-0. [DOI] [Google Scholar]
- 28.Silva CS, et al. Evolution of the plant reproduction master regulators LFY and the MADS transcription factors: the role of protein structure in the evolutionary development of the flower. Front. Plant Sci. 2015;6:1193. doi: 10.3389/fpls.2015.01193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ibarra-Laclette E, et al. Architecture and evolution of a minute plant genome. Nature. 2013;498:94–98. doi: 10.1038/nature12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, Forde BG. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science. 1998;279:407–409. doi: 10.1126/science.279.5349.407. [DOI] [PubMed] [Google Scholar]
- 31.Searle I, et al. The transition factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006;20:898–912. doi: 10.1101/gad.373506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reeves PA, et al. Evolution conservation of the FLOWERING LOCUS C mediated vernalization response: evidence from the sugar beet (Bsta vulgaris) Genetics. 2007;176:295–307. doi: 10.1534/genetics.106.069336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee JH, et al. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 2007;21:397–402. doi: 10.1101/gad.1518407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J, Lee I. Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 2010;61:2247–2254. doi: 10.1093/jxb/erq098. [DOI] [PubMed] [Google Scholar]
- 35.Lozano R, Hamblin MT, Prochnik S, Jannink JL. Identification and distribution of the NBS-LRR gene family in the Cassava genome. BMC Genomics. 2015;16:360. doi: 10.1186/s12864-015-1554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiang LX, et al. Genome-wide comparative analysis of NBS-encoding genes in four Gossypium species. BMC Genomics. 2017;18:292. doi: 10.1186/s12864-017-3682-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao S, et al. The atypical resistance gene, RPW8, recruits components of basal defence for powdery mildew resistance in Arabidopsis. Plant J. 2005;42:95–110. doi: 10.1111/j.1365-313X.2005.02356.x. [DOI] [PubMed] [Google Scholar]
- 38.Chin CS, et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods. 2016;13:1050–1054. doi: 10.1038/nmeth.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blanc G, Wolfe KH. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004;16:1667–1678. doi: 10.1105/tpc.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walker BJ, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y, et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience. 2017;7:120. doi: 10.1093/gigascience/gix120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Durand NC, et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Sys. 2016;3:95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dudchenko O, et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017;356:92–95. doi: 10.1126/science.aal3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hawkins JS, Proulx SR, Rapp RA, Wendel JF. Rapid DNA loss as a counterbalance to genome expansion through retrotransposon proliferation in plants. Proc. Natl Acad. Sci. USA. 2009;106:17811–17816. doi: 10.1073/pnas.0904339106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benson G. Tandem Repeats Finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 47.Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21:351–358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- 48.Zhao X, Hao W. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:W265–W268. doi: 10.1093/nar/gkm456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Slater GSC, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stanke M, Schoffmann O, Morgenstern B, Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics. 2006;7:62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Majoros WH, Pertea M, Salzberg SL. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20:2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- 54.Holt C, Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 2011;12:491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq. experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ashburner M, et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ogata H, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones P, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boeckmann B, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Griffiths-Jones S, et al. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fischer, S. et al. in Current Protocols in Bioinformatics (eds Andreas, D. et al.) Vol. 6, Ch. 6 (Zhang, 2011).
- 64.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 66.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 67.Benjamin B, Chao C, Daniel HH. Fast and sensitive protein alignment using diamond. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 68.Wang K, et al. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 2012;44:1098–1103. doi: 10.1038/ng.2371. [DOI] [PubMed] [Google Scholar]
- 69.Eddy SR. Accelerated profile HMM searches. PLoS Comput. Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Finn RD, et al. InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res. 2017;45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43:D257–D260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Price MN, Dehal PS, Arkin AP. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;10:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.He Z, et al. Evolviewv2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016;44:W236–W241. doi: 10.1093/nar/gkw370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Y, et al. GSA: Genome Sequence Archive. Genomics Proteomics Bioinformatics. 2017;15:14–18. doi: 10.1016/j.gpb.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Z, et al. Database resources of the BIG Data Center in 2019. Nucleic Acids Res. 2019;47:D8–D14. doi: 10.1093/nar/gky993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Chromosome-scale assembly of the Kandelia obovata genome
Data Availability Statement
Genome sequences have been submitted to the National Genomics Data Center (NGDC). PacBio whole-genome sequencing data and Illumina data have been deposited in BioProject/GSA (https://bigd.big.ac.cn/gsa.)75 under accession codes PRJCA002330/CRA002395 and the whole-genome assembly and annotation data have been deposited in BioProject/GWH (https://bigd.big.ac.cn/gwh)76 under accession codes PRJCA002330/GWHACBH00000000.