Abstract
The human lifespan is a heritable trait, which is intricately linked to the development of disorders. Here, we show that genetic associations for the parental lifespan are enriched in open chromatin of blood cells. By using blood expression quantitative trait loci (eQTL) derived from 31,684 samples, we identified for the lifespan 125 cis- and 559 trans-regulated expressed genes (eGenes) enriched in adaptive and innate responses. Analysis of blood single-cell expression data showed that eGenes were enriched in dendritic cells (DCs) and the modelling of cell ligand-receptor interactions predicted crosstalk between DCs and a cluster of monocytes with a signature of cytotoxicity. In two-sample Mendelian randomization (MR), we identified 16 blood cis-eGenes causally associated with the lifespan. In MR, the majority of cis-eGene-disorder association pairs had concordant effects with the lifespan. The present work underlined that the lifespan is linked with the immune response and identifies eGenes associated with the lifespan and disorders.
Subject terms: Data mining, Quantitative trait
Arnaud Chignon et al. present an integrated analysis of single-cell expression data from blood and genome-wide studies to identify expressed genes (eGenes) associated with human lifespan. The 125 eGenes identified are enriched for cardiometabolic traits and disorders and 16 are causally related to lifespan, as shown by Mendelian Randomization analysis.
Introduction
Duration of life or lifespan is a complex trait, which is determined by environmental and genetic factors. The heritability of the human lifespan is estimated at ~15–30%1. A recent meta-analysis of genome-wide association data, including more than a million parental lifespans, has identified 12 loci associated with the lifespan2. Genome-wide association studies (GWAS) in different complex traits have highlighted that gene variants are enriched in noncoding regions with cis-regulatory activity3. These genomic regions are enriched in expression quantitative trait loci (eQTL)4,5. We hypothesized that a better knowledge of the functional consequences of regulatory variants on gene expression might also provide significant insights into mechanisms of human aging. In addition, using eQTLs to identify genetically regulated expressed genes (eGenes) may give us the opportunity to determine whether the variants are associated with the lifespan or directly causal by using Mendelian randomization (MR) techniques. The main assumption of MR is that the variables that are measured, referred to as instrumental variables, only affect the outcome through the exposure and without confounders. In other words, if the genetic variants only affect the lifespan through their role in modifying gene expression, then we can assess their likelihood of playing a causal role on this outcome. This strategy of considering independent gene variants in an allelic series as instrumental variables is a powerful tool for causal inference. However, there are some challenges. If a genetic variant is associated with the outcome through an alternative mechanism (often referred to as horizontal pleiotropy), it may lead to an inflation of type I error. However, different statistical approaches have been developed to assess the robustness of the association discovered by MR and mitigate false positives. The Cochran’s Q test for heterogeneity and the intercept test in Egger regression are routinely used to detect associations where the instrumental variables provide estimates that may violate the main assumption6. Also, the identification of outliers with the MR-PRESSO package is another tool to identify instrumental variables that may associate with the outcome through an alternative exposure and to provide corrected estimates7. By combining these approaches, we can perform a robust estimate of causation.
The human lifespan is intricately intertwined with the development of diseases. As such, the trajectory of aging is variable throughout a person’s lifespan and may be altered by the different risk factors and disorders. We hypothesized that some genetic variants may exhibit antagonist pleiotropy. That is, some variants may provide a survival advantage or reproductive success earlier in life, but predispose to disease later in life. Rather than focusing on the individual alleles, we were curious whether similar genetic pathways were involved in chronic disease and lifespan potential. Gene expression is controlled in tissue-specific dynamic networks, with some genes coordinating the activity of functional modules. By using networks and causal inference, we sought to examine if genetically determined gene expression identified for the lifespan was also involved with the risk of several chronic human diseases. Thus, by using a multi-pronged approach, the objectives of this work on the lifespan were to: assess the tissue enrichment of genetic association data, discover cis- and trans-regulated eGenes and assess causal associations in MR, identify loci under positive selection and showing antagonist pleiotropy, evaluate the cellular enrichment of eGenes and infer cell communication pathways by using single-cell gene expression data, and use network as a tool to document pathways and assess the link between lifespan-associated eGenes and disorders. In this work, we leveraged 27 GWAS in order to map eGenes associated with the parental lifespan and risk factors/disorders. We found that genetic regions potentially influencing the lifespan were enriched in open chromatin in blood cells, which regulate eGenes involved in the control of immunity. Analysis of blood single-cell expression data showed that eGenes were enriched in dendritic cells (DCs) and the modelling of cell ligand–receptor interactions predicted crosstalk between eGenes expressed in DCs and a cluster of monocytes with a signature of cytotoxicity. Three loci under positive selection had antagonist pleiotropy with the lifespan. In MR, we identified blood eGenes associated with the lifespan and a long-livedness. Lifespan-associated eGenes were linked to different disorders and causal inference showed that a majority of relationships were concordant (e.g., eGenes negatively associated with the lifespan were positively associated with the risk of disorders).
Results
Mapping and annotation of lifespan genetic association data
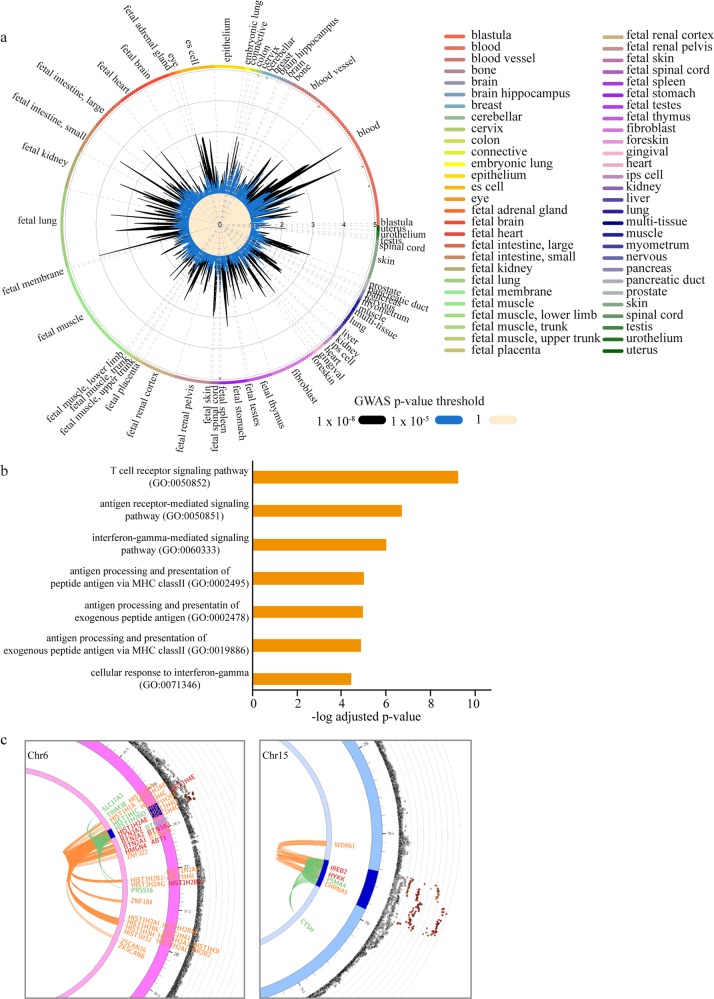
Supplementary Fig. 1 presents an overview schematic of the analysis pipeline. The parental lifespan GWAS is enriched in noncoding intergenic and intronic genomic regions (Supplementary Fig. 2). We were interested in understanding what tissues might be enriched in genetic association data for the parental lifespan. We implemented GARFIELD8, which uses summary statistics data to generate linkage disequilibrium (LD)-corrected annotations based on data from the Roadmap Epigenomics9 and the Encyclopedia of DNA Elements (ENCODE) projects10. Implementation of GARFIELD on summary statistics of genetic association data totaling 1,012,240 parental lifespans2 identified significant enrichments in CD19+ primary cells (marker of B cells) (P = 3.41 × 10−7) and GM12892 (a B cell line) (P = 1.59 × 10−5) (Fig. 1a and Supplementary Data 1). Considering the strong enrichment of lifespan genetic association data in immune cells, we leveraged summary statistics of blood cis-eQTL data derived from 31,684 samples to map genetically cis-regulated eGenes11 associated with the parental lifespan by using the Functional Mapping and Annotation of GWAS (FUMA) tool. In lifespan loci, individual significant single-nucleotide polymorphisms (SNPs) (PGWAS < 5 × 10−8, r2 < 0.6) and SNPs in LD (see Methods) were mapped to blood cis-eQTLs. SNP–gene pairs were filtered for multiple testing correction at false discovery rate (FDR) (PFDR < 0.05). In total, 4042 SNP–gene pairs (PFDR < 0.05) tagging 125 blood cis-regulated eGenes were mapped to lifespan genetic association data (Supplementary Data 2). By using EnrichR, we found that blood eGenes were enriched in gene ontology (GO) for T cell receptor (TCR)- (P = 5.65 × 10−10), antigen receptor- (P = 1.98 × 10−7), and interferon γ-mediated signaling pathways (P = 9.94 × 10−7) (Fig. 1b and Supplementary Data 3). Among the blood eGenes, 18 were listed in the database of Online Gene Essentiality12 (fold enrichment = 1.94, P = 0.009, hypergeometric test) (Supplementary Data 4). In the Open Targets database13, 13 drugs/antibodies targeting 8 eGenes (Supplementary Data 5) are/were evaluated in different phases of development (phases I–IV), whereas 19 eGenes are predicted to be tractable for the development of small molecules (Supplementary Data 6). As cis-regulation involves chromatin interactions, we also used FUMA to analyze chromatin contact between lifespan loci and genes by using chromatin conformation capture (Hi-C) data in GM12878 (B cell line). Lifespan loci were mapped to genes within a window region (250 and 50 bp upstream and downstream of the transcription start site (TSS), respectively). There were 56 individual significant SNPs located in 46 genomic regions involved in 205 intra-chromosomal loopings with distant regions. Among the distant regions interacting with the individual significant SNPs, 124 genes were mapped (Supplementary Data 7) and were enriched in GO for nucleosome (P = 5.44 × 10−10) and chromosome (P = 6.80 × 10−10) assemblies (Supplementary Data 8). Overall, 36 genes were mapped by both blood eQTLs and chromatin interactions (Supplementary Data 9). Figure 1c shows zoomed-in circos plots of genetic association data along with chromatin interactions in the extended histone locus in chromosome 6 and the 15q25.1 locus.
Fig. 1. Mapping and annotation of parental lifespan GWAS.
a Lifespan gene variant enrichment showed by radial lines (numbers correspond to fold enrichment value) at two GWAS P value thresholds (P < 1 × 10−5 in blue and P < 1 × 10−8 in black) in different tissues and cell types. Significant enrichments are represented by dots in the outer circle. b Gene ontology for 125 blood cis-regulated eGenes mapped from lifespan; graph showing the seven GO terms with the lowest P values. c Zoomed-in circos plots of genetic association data and chromatin interactions in the extended histone locus in chromosome 6 and in the 15q25.1 locus. Names of the genes are in orange for genes mapped only by chromatin interactions, in green for genes mapped only by eQTL, and in red for genes mapped by both chromatin interactions and eQTLs.
Network and regulons associated with cis-regulated eGenes
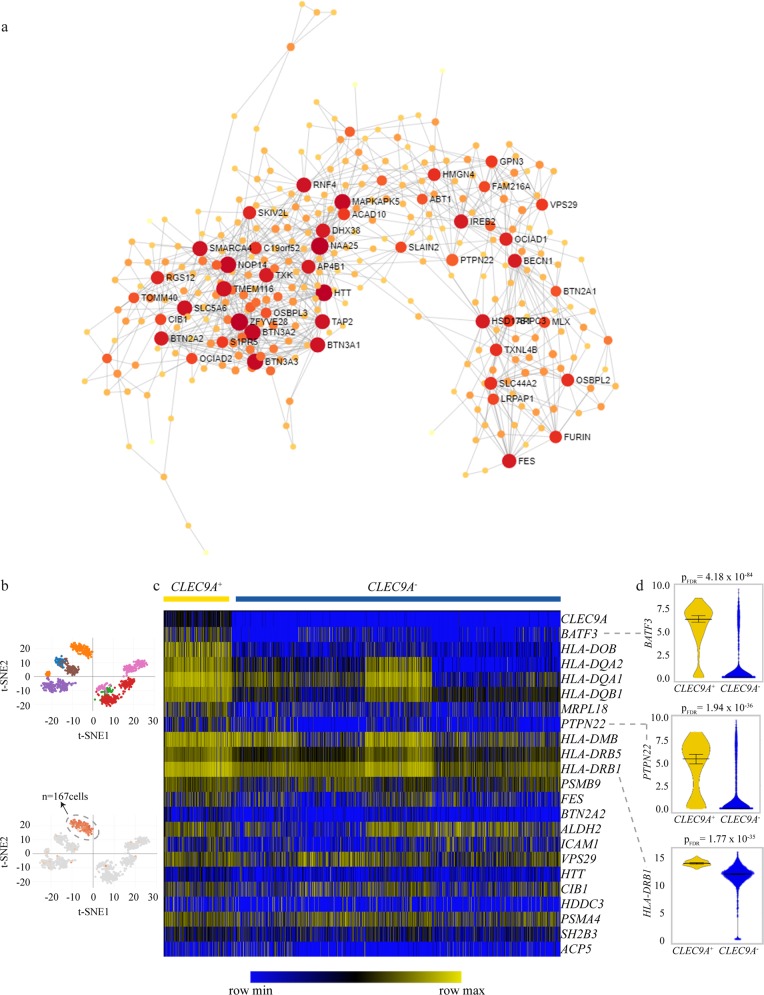
To identify cis-regulated eGenes with central functions, we used a network approach by assessing blood-specific co-expression data14. We extracted eGene co-expression networks in whole blood from the database of tissue and cancer specific biological networks. This resulted in a network with 4453 nodes and 7321 edges. For visualization purpose, Fig. 2a shows a reduced network topology of key central nodes and interactions. In this network, several eGenes had elevated central betweenness (Supplementary Data 10), which is a measure of nodes with the shortest path acting as bottlenecks between gene modules. The co-expression network in the blood was analyzed for pathway enrichment by using Reactome15. The network was enriched in gene expression (P = 8.08 × 10−23), immune system (P = 1.47 × 10−10), and RNA polymerase II transcription (P = 1.55 × 10−10) (Supplementary Data 11). Transcription factors (TFs) and their target genes (regulons) are involved in the control of cell fate16. We investigated the regulons of blood eGenes associated with the lifespan by using ChIP-X Enrichment Analysis 3 (ChEA317), which provides TF enrichment for genes by using an expanded list of different sources such as TF–gene co-expression, TF–gene co-occurence, and chromatin immunoprecipitation-sequencing (ChIP-seq) data. TFs were ranked according to their associations with cis-regulated eGenes. Most highly ranked TFs were: SP140, interferon regulatory factor (IRF) family and BATF3, an AP-1 member involved in DC differentiation18 (Supplementary Table 12). A list of blood eGene-derived regulons for SP140, IRF5, IRF8, and BATF3 is provided in Supplementary Data 13. We next wanted to identify immune cells that expressed BATF3 and if they were enriched with lifespan eGenes. Analysis of single-cell RNA-sequencing data of monocytes and DCs (GEO accession number GSE94820) showed that a cluster expressing CLEC9A, a marker for a subset of conventional DC (cDC1), was enriched in differentially eGenes including BATF3, as well as 21 cis-eGenes (fold enrichment = 2.8, P = 8.74 × 10−5, hypergeometric test) (Fig. 2b–d and Supplementary Data 14). Consistently, pathway analysis with Reactome showed that these eGenes were enriched for major histocompatibility complex class II antigen presentation (P = 6.55 × 10−9) (Supplementary Data 15). These data suggest that lifespan eGene-associated regulon may be involved in DC function, including the sensing of dead cell antigens and may thus bridge the innate with the adaptive immune responses.
Fig. 2. Blood eGenes network and cell enrichment.
a Reduced network topology of key central nodes and interactions. b–d Analysis of single-cell RNA-sequencing data of monocytes and dendritic cells (GEO accession number GSE94820). b t-SNE graphs showing a cell cluster expressing CLEC9A. c Heatmap of gene enrichment in CLEC9A− and CLEC9A+ cell clusters. d Violin plots showing data distribution (vertical bars illustrate median and 95% CI) and comparing expression in CLEC9A− and CLEC9A+ cell clusters.
MR for the lifespan
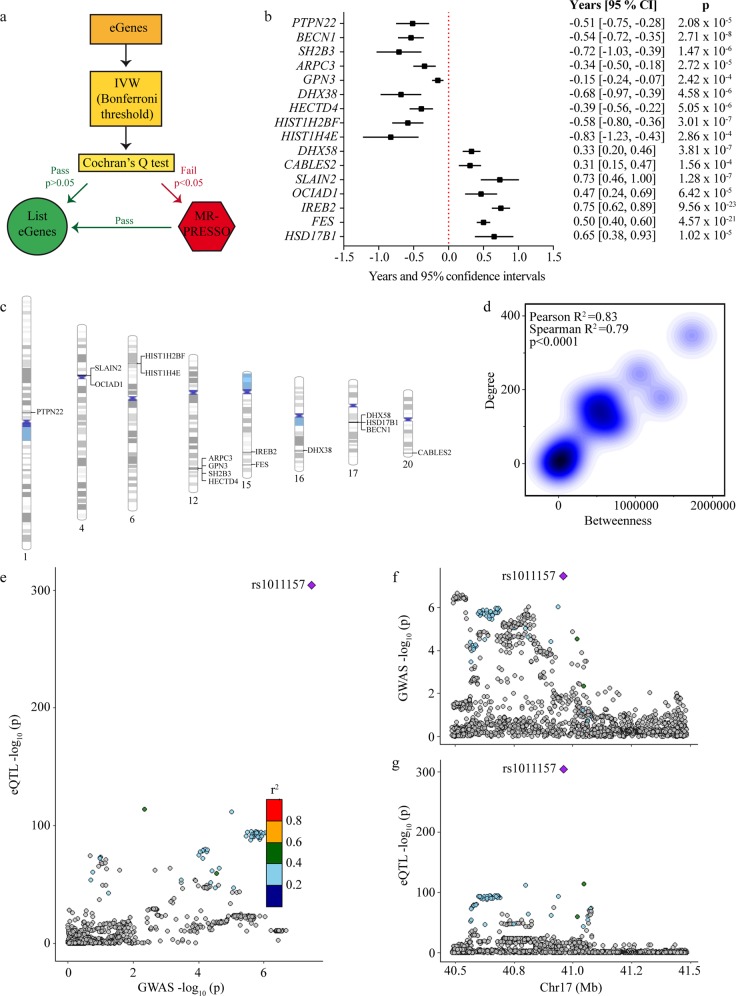
The lifespan GWAS combined fathers and mothers into one parental survival. Effect sizes were reported as the −loge(Cox hazard ratio), from which years gained or lost could be estimated (see Methods). Causal inference in MR was performed for the blood cis-regulated eGenes on the parental lifespan. Independent gene variants (r2 < 0.1) located within ±500 kb from the TSS and associated with the blood expression (P < 0.001 corresponds to ~F statistics > 10) were selected as instrumental variables19 (Supplementary Data 16). The strategy to perform MR analyses is illustrated in Fig. 3a. Enough instrumental variables (minimum 3) to perform MR were available for 116 genes (mean instrumental variables per gene 26) (Supplementary Data 16). After a correction for multiple testing (Bonferroni correction, Pcausal < 4.3 × 10−4, 0.05/116), 23 eGenes were significantly associated in inverse variance-weighted (IVW) MR with the lifespan (Supplementary Data 17). Among these eGenes, ten did not show heterogeneity on the Cochran’s Q test and were considered as lifespan causally associated eGenes (Supplementary Data 17). For the eGenes significant in IVW but with significant heterogeneity, we used the MR-PRESSO package, which tests for the presence of outliers and provides corrected analyses (see Methods). Using this strategy, MR-PRESSO identified six eGenes, for which outliers were detected and provided estimates without distortion after the removal of these outliers (Supplementary Data 18). Hence, by using IVW MR and MR-PRESSO we identified 16 blood eGenes (PTPN22, ARPC3, GPN3, HECTD4, DHX58, BECN1, CABLES2, SLAIN2, OCIAD1, HIST1H2BF, HIST1H4E, SH2B3, IREB2, FES, DHX38, HSD17B1), which were considered causally associated with the lifespan (Fig. 3b). Lifespan causally associated eGenes were located on chromosomes 1, 4, 6, 12, 15, 16, 17, and 20 (Fig. 3c). Among these eGenes, only FES and SH2B3 were previously mapped to the lifespan2. In model organisms, the deletions of BECN1 and OCIAD1 have been previously associated with increased and decreased lifespans, respectively20,21. These data including the directional effects are concordant with the present findings in human as the blood expression of BECN1 and OCIAD1 were negatively and positively associated with the lifespan, respectively. For IREB2 (Pcausal = 9.56 × 10−23), which was also mapped by chromatin interaction (Fig. 1c), an increase of 1 SD in the expression was associated with a gain of 0.75 year (9 months) across the lifespan (see Methods) (Fig. 3b). In the blood co-expression network, a kernel density function analysis showed that lifespan causally associated cis-eGenes were distributed along a positive gradient for the degree (hubness) and central betweenness (bottleneck), two metrics indicating prominence in network22 (Fig. 3d). Of the 16 causally associated eGenes, 10 genes including IREB2, OCIAD1, BECN1, and PTPN22 were among the top percentile (>99 percentile) nodes with highest betweenness centrality (fold enrichment = 61.7, P = 1.43 × 10−13, hypergeometric test) (Supplementary Data 10). In sensitivity analyses, we tested the 16 blood eGenes in Egger MR, which provides the intercept test as a mean to evaluate horizontal pleiotropy. In Egger MR for the lifespan, we found that PTPN22, DHX58, CABLES2, SLAIN2, OCIAD1, IREB2, FES, and BECN1 remained significant and without horizontal pleiotropy on the intercept test (Supplementary Data 19). The direction of the effect was consistent in IVW and Egger regressions. As an additional measure of sensitivity, we performed Bayesian colocalization analyses between blood cis-eQTLs and lifespan genetic association data. This analysis showed that BECN1 had a strong posterior probability (PP = 0.98) of shared genetic signal between gene expression and the lifespan. For BECN1, the colocalization signal is illustrated in Fig. 3e–g by using LocusCompare23 and shows that rs1011157 is the gene variant with the lowest P values for both cis-eQTL and lifespan genetic association data.
Fig. 3. Causal inference on lifespan cis-eQTL genes.
a Scheme of Mendelian randomization analysis strategy. b Forest plots indicating the effects (in years) on lifespan of the 16 blood eGenes considered to be causal. c Chromosomal ideograms for the genomic locations of the lifespan causally associated eGenes. d Kernel density plots between the degree and the betweenness of the 16 causally associated eGenes, Pearson’s and Spearman’s correlations. e–g Bayesian colocalization analysis between blood cis-eQTLs and lifespan genetic association data. e LocusCompare showing colocalization signal for BECN1, with rs1011157 having the lowest P value for both cis-eQTL and lifespan genetic association. f Genetic association signal for the GWAS (parental lifespan). g Genetic association signal for the blood cis-eQTL.
Identification of trans-eQTL genes
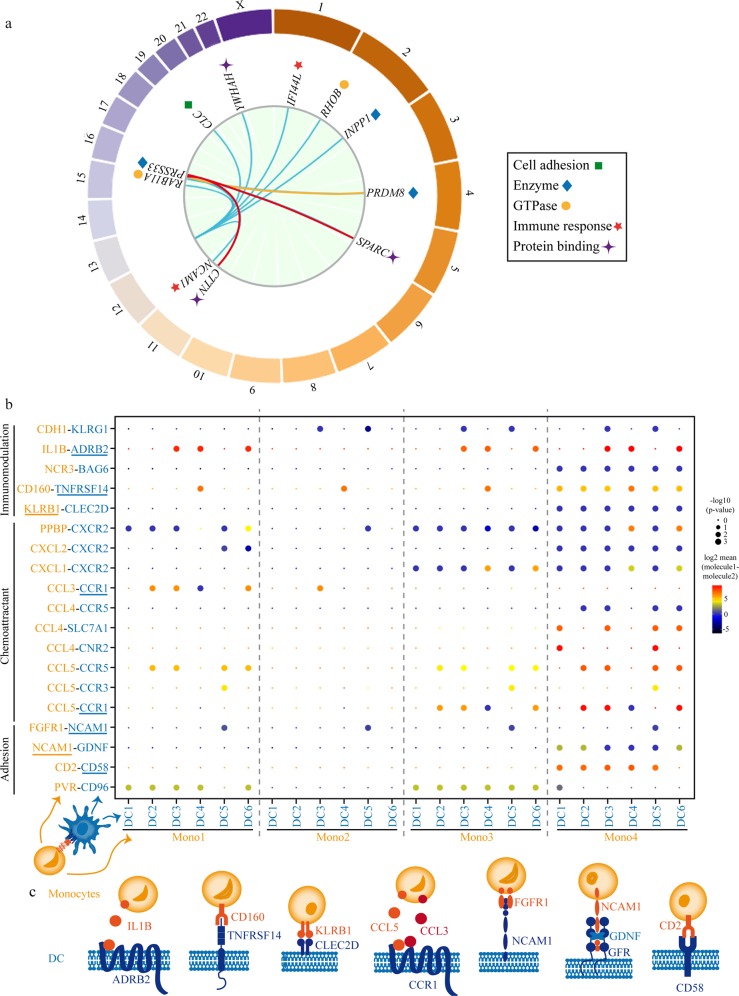
We next sought to identify trans-regulated genes11 (>5 Mb from risk loci and/or on a different chromosome) at lifespan causally associated cis-regulated blood eGenes loci. The identification of trans-regulated genes may provide insights about downstream pathways regulated by cis-eGenes. From the individual significant SNPs and variant in LD associated with the lifespan, 4641 SNP–gene pairs (PFDR < 0.05) tagging 567 trans-regulated blood genes were identified. In total, 559 trans-regulated genes were mapped to five lifespan loci, in which causally associated cis-regulated eGenes were identified (Supplementary Data 20). Lifespan sentinel variant rs597808 (PGWAS = 7.32 × 10−13), which is a cis-eQTL for SH2B3 (Pcis-eQTL = 7.48 × 10−68), was associated with 462 trans-eQTL genes. Overall, at SH2B3 locus, 548 trans-eQTL genes were mapped and enriched in GO for cytokine-mediated signaling (P = 2.66 × 10−9), which is consistent with the function of the cis-regulated eGene SH2B3, an adapter molecule for immune signaling24 (Supplementary Data 21). The trans-eQTL genes mapped by SNPs associated with the cis-expression of SH2B3 included 41 TFs curated in TFCheckpoint data25 for the human (fold enrichment = 1.42, P = 0.02, hypergeometric test) and included several regulators of the immune response, such as members of AP-1, STAT, and IRF signaling pathways (Supplementary Data 22). Also, trans-eQTL genes associated with the lifespan SNPs at SH2B3 were enriched in ligand–receptor encoding genes26 (fold enrichment = 2.27, P = 1.43 × 10−7, hypergeometric test) (Supplementary Data 23). In total, 57 ligand–receptor encoding genes were trans-eQTLs associated with the SH2B3 locus and were enriched in Reactome pathway for cytokine signaling in the immune system (P = 8.18 × 10−9) (Supplementary Data 24). Gene variant rs1230666 (lifespan PGWAS = 6.44 × 10−9) is a cis-eQTL for the lifespan causally associated eGene PTPN22 (Pcis-eQTL = 1.06 × 10−41) and also a trans-eQTL for seven genes (Supplementary Data 20). Among these trans-eQTL genes, CD6 (Ptrans-eQTL = 5.17 × 10−7), CTLA4 (Ptrans-eQTL = 5.33 × 10−7) and IL2RA (Ptrans-eQTL = 2.55 × 10−8) are involved in T cell activation, whereas ARID5B (Ptrans-eQTL = 3.21 × 0−6) is a transcriptional cofactor involved in B cell differentiation27. The trans-eQTL genes mapped by the lifespan variant rs1230666 were enriched in ARCHS4 for T lymphocyte (P = 0.001) (Supplementary Data 25), which is consistent with the high level and function of the cis-regulated eGene PTPN22 in these cells28–30. Next, we evaluated if the genetic signal between cis- and trans-eQTLs was shared by using colocalization analyses. We found strong evidence of shared genetic signal (PP > 0.8) between the region of cis-regulated eGene SH2B3 and trans-regulated genes YWHAH, RAB11A, PRSS33, CLC, NCAM1, IFI44L, RHOB, and INPP1 (Fig. 4a and Supplementary Data 26). PRSS33 is a serine protease with an immune function in eosinophils31, whereas IFI44L is involved in interferon type I response32. NCAM1 (also known as CD56) is aberrantly expressed in different malignancies and is a marker of natural killer (NK) cells33. Recently, NCAM1 was found to be expressed by a novel cluster of monocytes (Mono4) with a gene signature of cytotoxicity34. Figure 4b shows predicted ligand–receptor interactions derived from single-cell gene expression of peripheral blood mononuclear cells (GEO accession number GSE94820) where monocytes are predicted to interact with DCs through NCAM1 and different chemokines and immunomodulatory signals for cytotoxicity. Among the ligand–receptor interactions illustrated in Fig. 4b, there are five other lifespan trans-eQTL genes associated with the SH2B3 locus, which are expressed by DCs (ADRB2, TNFRSF14, CCR1, CD58) and Mono4 (KLRB1) (Supplementary Data 20). Also, we identified a colocalization signal between the cis-regulation of IREB2 and the trans-regulation of PRDM8, which encodes for a histone methyltransferase (Fig. 4a and Supplementary Data 26). Finally, the genetic signal for the cis-regulation of FES was shared with trans-regulated genes SPARC and CTTN (Fig. 4a and Supplementary Data 26). SPARC encodes for a cysteine-rich matrix protein involved the control of cell growth35–37, whereas CTTN is involved in the organization of actin38,39.
Fig. 4. Trans-eQTLs genes.
a Circos illustrating shared genetic signal between cis- and trans-regulated genes; trans-regulated genes showing shared genetic signal with cis-regulated SH2B3 (cyan), IREB2 (yellow), and FES (red). b Single-cell gene expression (GEO accession number GSE94820) analysis showing CellPhoneDB predicted ligand–receptor interactions between monocytes and dendritic cells (DCs); on the y-axis, molecules identified as lifespan trans-QTLs are underlined; molecules expressed by monocytes and DCs are represented in yellow and blue, respectively. c Graphic representation of predicted molecular interactions between monocytes and DCs and including lifespan trans-eQTLs.
Positive selection at cis-regulated eGene loci
Among the different risk loci associated with the cis-regulated eGenes, three overlapped with genomic regions under positive selection in a genome-wide scan (PopHumanScan40). Among these loci, two are well-documented regions under selective sweep: the human leukocyte antigen locus in chromosome 6, which has many blood cis-eQTLs, is under a balanced selection41, whereas the SH2B3 locus has been previously highlighted to be under positive selection42. The derived allele for the lifespan sentinel variant rs597808, which is a blood cis-eQTL for SH2B3 (Pcis-eQTL = 7.48 × 10−68), has an elevated prevalence in population with a European ancestry (frequency in CEU = 0.45) and is associated with a decreased lifespan (−0.28 year per allele, PGWAS = 7.32 × 10−13). Gene variant rs597808 is in strong LD with rs3184504 (r2 = 0.98), a coding variant previously associated at genome-wide level with cardiometabolic traits/disorders43,44 and autoimmune diseases45,46. The other region under positive selection in PopHumanScan is located at 17q21.31 where the lifespan index gene variant rs1011157 (PGWAS = 3.58 × 10−8) (Fig. 5a) is a strong cis-eQTL in the blood (Pcis-eQTL = 5.76 × 10−305) for the expression of BECN1, a gene involved in autophagy47. At this locus, the derived allele T is positively associated with the expression of BECN1 in the blood and negatively with the lifespan (−0.33 year per allele, PGWAS = 3.58 × 10−8). In the blood, an increase of 1 SD in genetically determined expression of BECN1 was associated with a reduction of 0.5 year (6 months) (Pcausal = 2.71 × 10−8) across the lifespan. The derived allele at rs1011157 is absent in African populations, whereas it is present in other populations with frequencies varying from 14% in CEU to 35% in Japanese in Tokyo, Japan.
Fig. 5. Positive selection at cis-regulated eGene loci.
Locus zoom of the region surrounding rs1011157, indicated to be under positive selection in PopHumanScan.
Lifespan cis-regulated eGenes and disorders
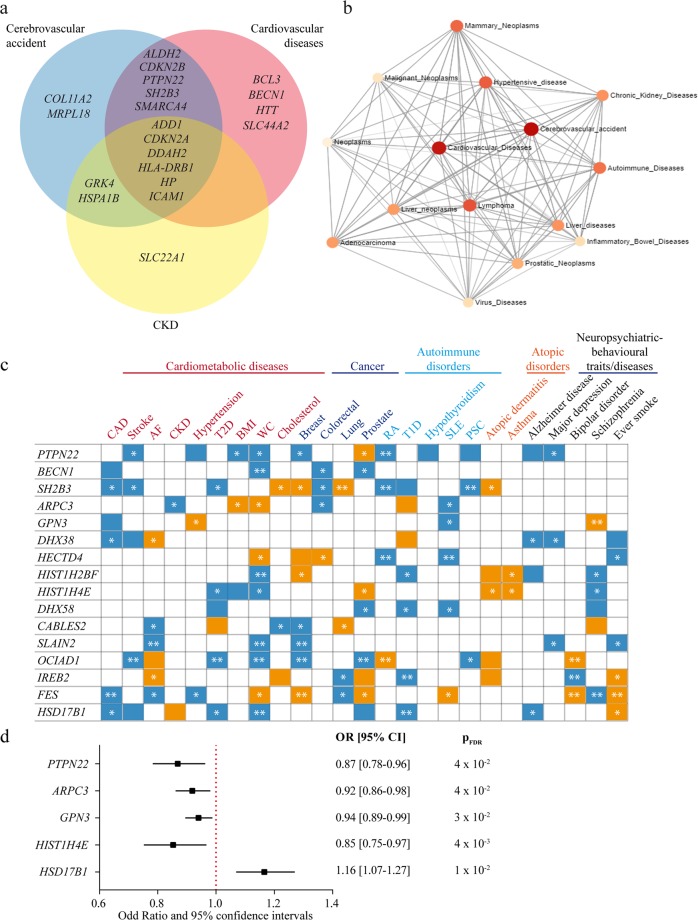
To assess how eGenes related to the lifespan are potentially connected to different disorders, we interrogated DisGenet48, which provides an expanded and curated database of gene–disease associations. Lifespan blood cis-eGenes were significantly enriched in cerebrovascular accident (fold enrichment = 2.16, P = 0.006, hypergeometric test), cardiovascular diseases (fold enrichment = 1.91, P = 0.01, hypergeometric test) and chronic kidney disease (CKD) (fold enrichment = 2.49, P = 0.01, hypergeometric test) (Fig. 6a). We generated a disease network using the DisGenet data, which showed that cerebrovascular accident and cardiovascular diseases were linked to different group disorders, such as lymphoma, hypertensive disease, CKD, autoimmune diseases, and neoplasms (Fig. 6b). We next examined whether blood cis-eGenes related to the lifespan were enriched in primary immunodeficiency disorders and cancer. Among the listed primary immunodeficiency disorder genes, we identified five blood cis-eGenes (fold enrichment = 3.01, P = 0.02, hypergeometric test) (TAP2, CIB1, ACP5, DCLRE1B, and SKIV2L) that were associated with the lifespan. In PhenoScanner49, TAP2, DCLRE1B, and SKIV2L have been previously mapped in GWAS to autoimmune disorders such as rheumatoid arthritis (RA) (Supplementary Data 27). Among the genes listed in the COSMIC50 database for cancer, seven blood lifespan cis-eGenes (ALDH2, BCL3, CDKN2A, FES, HIST1H3B, SH2B3, and SMARCA4) were identified (fold enrichment = 1.4, P = 0.22, hypergeometric test). Thus, these findings suggest that lifespan cis-regulated eGenes may be involved in different disease-related outcomes. To assess the role of lifespan causally associated cis-eGenes on different risk factors and diseases, we performed MR analyses. In total, causal inference using MR was performed by using summary statistics from 25 GWAS for risk factors/diseases englobing cardiometabolic diseases/traits, autoimmune disorders, atopic disorders, cancer, and neuropsychiatric-behavioral traits/diseases (Supplementary Data 28). Figure 6c illustrates the risk factors/diseases related with the lifespan causally associated cis-eGenes. In MR, 117 risk factors/disorders were associated with lifespan causally associated cis-eGenes (P < 0.05), whereas 28 associations remained significant after a Bonferroni correction (P < 1.27 × 10−4, 0.05/392 traits). Figure 6c provides a color chart for the consistency in the directional effect of the cis-regulation on risk factors/disorders and the lifespan. When considering associations remaining significant after the Bonferroni correction, 75% of eGene–disorder association pairs were concordant with the lifespan (e.g., the cis-regulated eGene that decreased the risk of disorder/risk factor increased the lifespan). Some cis-eGenes, such as SH2B3 and FES, showed both concordant and non-concordant associations between the lifespan and disorders/risk factors. On the other hand, eGenes such as BECN1, DHX58, SLAIN2, and PTPN22 were largely concordant in the direction for different risk factor/disease associations with the lifespan. A gain-of-function variant that change an amino acid (R620W) in PTPN22 (allele T at rs2476601) has been previously associated with several autoimmune disorders51,52. With regard to the lifespan, gain-of-function variant rs2476601 in PTPN22 was associated with a reduced lifespan (−0.27 year per allele, PGWAS = 1.63 × 10−5). For the expression, the data are consistent with these observations as MR-based causal inference showed that the expression of PTPN22 in the blood was negatively associated with the lifespan (−0.51 year per 1 SD, Pcausal = 2.08 × 10−5) and positively associated with the risk of RA (per 1 SD odds ratio (OR): 2.62, 95% CI: 2.25–3.06, Pcausal = 3.37 × 10−9) (Fig. 6c). Other causal associations with the cis-expression are in line with previous mapping in GWAS. For instance, genetically determined expression of FES was strongly associated with coronary artery disease (per 1 SD OR: 0.90, 95% CI: 0.88–0.92, Pcausal = 1.09 × 10−15), which is consistent with the genetic signal in GWAS by the tag SNP rs17514846 (PGWAS = 9.85 × 10−27) at the FURIN/FES locus. However, other strong relationships in MR, such as the associations between the cis-regulation of OCIAD1 with stroke (per 1 SD OR: 0.87, 95% CI: 0.82–0.91, Pcausal = 1.54 × 10−7) and breast cancer (per 1 SD OR: 0.78, 95% CI: 0.71–0.86, Pcausal = 1.25 × 10−6), were not mapped by previous GWAS. These data are in line with a recent study highlighting that a significant proportion of causal associations between cis-expression and traits/disorders have no genome-wide significant SNPs in GWAS53.
Fig. 6. Lifespan cis-regulated eGenes and disorders.
a Venn diagram of blood cis-eGenes associated with cerebrovascular accident, cardiovascular diseases, and chronic kidney diseases (CKDs). b Blood cis-eGenes disease network showing centrality of cerebrovascular accident and cardiovascular diseases. c Chart indicating the consistency in the directional effect of the cis-regulation on risk factors/disorders and the lifespan derived from Mendelian randomization analyses; blue: concordant (e.g. the cis-regulated eGene that decreased the risk of disorder/risk factor increased the lifespan), orange: non-concordant (e.g. the cis-regulated eGene that decreased the risk of disorder/risk factor decreased the lifespan). Blue and orange square P < 0.05, *P < 0.05 (FDR), **P < 1.27 × 10−4 (Bonferroni). AF: atrial fibrillation, CKD: chronic kidney disease, T2D: type 2 diabetes, BMI: body mass index, WC: waist circumference, RA: rheumatoid arthritis, T1D: type 1 diabetes, SLE: systemic lupus erythematosus, PSC: primary sclerosing cholangitis. d Forest plot of genes found to be causally associated with long-livedness (≥90 years).
Causal inference for long-livedness
Longevity as assessed by the parental lifespan does not address whether gene variants and cis-regulated eGenes are associated with long-livedness. We thus leveraged and imputed GWAS summary statistics totaling 9793 individuals and examining associations with a long-livedness54 (≥90 years) (methods) in order to perform MR analyses. Among the 16 blood cis-eGenes causally associated with the lifespan, we could perform MR analyses for long-livedness for 15 genes. In this small series, there were no eGenes significant at a Bonferroni threshold level. However, this analysis showed at an FDR significance level (PFDR < 0.05) that genetically determined expressions of PTPN22 (per 1 SD OR: 0.87, 95% CI: 0.78–0.96, Pcausal = 0.04), ARPC3 (per 1 SD OR: 0.92, 95% CI: 0.86–0.98, Pcausal = 0.04), GPN3 (per 1 SD OR: 0.94, 95% CI: 0.89–0.99, Pcausal = 0.03), HIST1H4E (per 1 SD OR: 0.85, 95% CI: 0.75–0.97, Pcausal = 0.04), and HSD17B1 (per 1 SD OR: 1.16, 95% CI: 1.07–1.27, Pcausal = 0.01) were causally associated with a long-livedness (≥90 years) (Fig. 6d). These association data for long-livedness were consistent with the directional effects of these eGenes on the lifespan.
Discussion
By using a multi-level approach, which integrated mapping of GWAS, eQTLs, pathway and cell enrichment, MR, and network analyses (Supplementary Fig. 1), we provide evidence that cis- and trans-regulated blood eGenes are linking the human lifespan with the immune response. Functional annotations of parental lifespan GWAS were enriched in the blood. In total, 16 blood eGenes were causally associated with the lifespan. In a co-expression network, causally associated cis-regulated blood eGenes with the lifespan were enriched in nodes with elevated central betweenness, which are shortest path nodes referred to as bottlenecks that link different expressed gene modules. Lifespan-associated eGenes were enriched in cardiovascular disorders, which were linked in a network to CKD, autoimmune disorders, and cancer. In MR, several lifespan-associated eGenes were also associated with the risk of disorders.
Analysis of genetic association data for the lifespan showed a strong enrichment of gene variants in the blood. We found a strong enrichment for TCR- and interferon γ-mediated signaling pathways for the blood eGenes mapped to the lifespan GWAS. These data are consistent with the hypothesis that control of inflammation is linked to the aging process55. However, the present data suggest that antigen presentation as well as the orchestration of the immune response by T cells may be involved in the human lifespan. We highlighted that lifespan-associated cis-eGenes were enriched in a subset of cDC1 expressing CLEC9A. Studies have underscored that CLEC9A is involved in the recognition and cross-presentation of dead cell antigens56. In addition, single-cell expression data revealed that six lifespan trans-eQTL genes (NCAM1, ADRB2, TNFRSF14, CCR1, CD58, and KLRB1) were expressed by DCs and/or a cluster of cytotoxic monocytes, which were predicted to interact. Monocytes/macrophages with a cytotoxic profile have been previously described in tumors57. Hence, follow-up studies could provide further mechanistic insights by examining the crosstalk between DCs and monocytes in the context of aging. Autophagy plays a significant role in the function of immune cells, including DCs58. We found a strong signal in both colocalization and MR analyses for the expression of BECN1, a key regulator of autophagy, on the lifespan. Recent data in Caenorhabditis elegans (C. elegans) showed that the inhibition of bec-1 (Beclin homolog) in adulthood extended the lifespan20. Conversely, in C. elegans the inhibition of bec-1 at the developmental stage decreased the lifespan. Present findings in human showed that lifelong genetically determined BECN1 level in the blood was causally and negatively associated with the lifespan. The expression of IREB2 and DHX58 in blood cells was positively associated with the lifespan. The control of iron metabolism by IREB2 could play a role in host defense against pathogens. DHX58 is a RIG-I-like receptor involved in the interferon γ pathway and host defense against viral infection59. Also, some eGenes causally associated with the lifespan, such as FES, SH2B3, and PTPN22, are well known for their role in signaling pathways in immune cells60–62. In blood cells, we identified that cis-regulation of SH2B3 was associated with 548 trans-eQTL genes, which were enriched in TFs as well as with ligands–receptors. These data are consistent with the role of the encoded protein as an adapter molecule in the immune signaling cascade. PTPN22 is a protein tyrosine phosphatase highly expressed in lymphocytes and it decreases the signaling mediated by the TCR63. Gain-of-function variant in PTPN22 (frequency in CEU = 0.12) was previously identified as being associated to different autoimmune disorders, such as RA, Hashimoto’s thyroiditis, Graves’ disease, type 1 diabetes (T1D), systemic sclerosis, and systemic lupus erythematosus64. Consistently, we underlined that genetically determined expression of PTPN22 was positively associated in MR with the development of RA, thyroid disorder, and primary sclerosing cholangitis. PTPN22 negatively regulates signaling by the TCR and experiments suggest that it may promote the selection of autoreactive clones and affect the function of regulatory T cells65.
We found that a subset of causally associated eGenes were also associated in MR with long-livedness (≥90 years). The expression of PTPN22 and ARPC3 in blood cells was negatively associated with long-livedness. ARPC3 encodes for a protein that regulates the polymerization of actin and in T cells it participates to the recycling of the TCR66. On the other hand, HSD17B1, which encodes for a β-hydroxysteroid dehydrogenase that regulates androgen and estrogen levels67, was positively associated with an extended lifespan. HSD17B1 is expressed by monocytes and the metabolism of sex steroid hormones could participate to immune senescence68.
Previous work underlined that genetic association data for cardiovascular diseases showed frequent antagonist pleiotropy with reproductive traits69. As such, selective pressure for the fitness operates on traits that occur during the first decades of life. Therefore, it is unlikely that traits and disorders that occur late during a lifetime are under selective pressure. To this effect, in the present study we found that loci identified in a scan for positive selection showed antagonist pleiotropy with the lifespan. At the SH2B3 locus, the derived allele at rs597808 increases the expression of SH2B3 and decreases the lifespan. One study reported that the SH2B3 rs3184504-derived allele, which is in high LD with rs597808 (r2 = 0.98), is associated with an increased production of IL1B through a NOD2 (nucleotide-binding oligomerization domain-containing protein 2) recognition pathway and may have been selected to protect against bacterial infection42. Also, the derived allele at rs1011157, which is in a region identified in a genome-wide scan for positive selection, is a strong cis-eQTL for the expression of BECN1 and is negatively associated with the human lifespan. Experimental evidence suggests that BECN1 may participate in host defense against viral infections70.
The lifespan is a complex trait characterized by a landscape of trajectories, which vary during a lifetime according to complex interactions between the genotype and environmental factors. We found that lifespan-associated blood eGenes were enriched in cardiovascular diseases. In a disease network, we observed that cardiometabolic traits and diseases were linked to autoimmune disorders and cancer. Together, these findings provide support to different observational data. For instance, RA, an autoimmune disorder, is associated with an elevated cardiovascular risk71. Hypertension, a cardiovascular risk factor, has been associated with the risk of breast cancer72. Lifespan-associated gene OCIAD1 is dysregulated in different cancers73–75 and recent findings suggest that it controls embryonic stem cell differentiation76. In mice, the deletion of OCIAD1 promoted the degradation of p53 along with the development of a myeloproliferative disorder and a reduced lifespan21. Consistently, we found that genetically predicted higher expression of OCIAD1 in the blood was associated with an increased lifespan and a reduced risk of breast and prostate cancers. Also, blood cis-regulated eGenes associated with the lifespan were enriched for genes involved in rare primary immunodeficiency disorders. Taken together, these findings highlight that cis-regulation of genes with important functions in immunity and cell fate determination are at the interface of a multimorbid space, which is shared with the lifespan. We found that the majority of the eGene–disorder association pairs significant in MR were concordant for their effects on the lifespan. For instance, positive associations for the expression of eGenes with coronary artery disease, stroke, and dementia were negatively associated with the lifespan. These chronic disorders are well known for their negative associations with the vital prognosis77,78.
The present work has some limitations. MR is a powerful inference tool; however, only randomized clinical trials can provide a confirmation of causality. The MR analysis performed for long-livedness had limited power. Hence, the associations with long-livedness should be considered exploratory at this stage and could be used as a resource to generate hypotheses for further investigations.
This work provides evidence that genetically determined expression of genes in blood cells is associated with the lifespan. The immune system likely plays a significant role in the trajectory of the human lifespan. Causally associated cis-regulated blood eGenes established connections between a landscape of morbid states and trajectories with the human lifespan and long-livedness. The identification of genetically regulated pathways involved in the lifespan and long-livedness may help develop strategies to provide a healthy aging.
Methods
Lifespan genetic associations
Full summary statistics of genome-wide association data for the lifespan were obtained from 1,012,240 parental lifespans2, including 691,621 parental lifespans from UK Biobank (excluding SNPs with MAF < 0.005) and 320,619 parental lifespans from LifeGen consortium79 (excluding SNPs with MAF < 0.01). Lifespan was derived from parental survival (age and alive/dead status) and offspring genotype by using an association test2. Parents who died below the age of 40 years were excluded. The association test was conducted under the Cox’s proportional hazards model as described in refs. 79,80. The Cox’s model used a hazard ratio, and to be consistent with the lifespan (which implies a positive effect size for a longer life), the effect sizes were expressed as −loge(Cox’s hazard ratio) corresponding to a loge(protection ratio). Years of life were estimated as loge(protection ratio) × 10 according to the average effect across cohorts of the hazard ratio on the lifespan79. GWAS summary statistics for the lifespan were publicly available2.
Annotation of lifespan genetic associations
GARFIELD8 was used to characterize functional, cellular and regulatory contribution of genetic variations for the lifespan. It provides enrichment of genome-wide summary association statistic, which is corrected for the LD structure, in tissue-specific functional elements. GARFIELD uses annotations from ENCODE10 and Roadmap9 epigenomics data (1005 features, including genomic annotations, chromatin states, histone modifications, DNaseI hypersensitive sites, and TF binding sites, in a number of cell lines and tissues). The software includes a C++ code for data pre-processing and a R code for fold enrichment, significant testing, and visualization. LD data are included as well as annotation data. P values for each SNP were extracted from the lifespan genome-wide association data summary statistics and default settings were used.
Mapping of lifespan genetic associations
GWAS for the lifespan was mapped to blood cis- and trans-regulated genes by using FUMA81. With the FUMA SNP2GENE function, blood cis- and trans-eQTLs data from 31,684 blood samples11 (data from https://www.eqtlgen.org/index.html) were used to map genetic associations for the lifespan. Genomic loci associated with the lifespan were defined using a pre-calculated LD structure of the 1000G EUR reference population. SNPs in genomic loci with LD r2 < 0.6, P value < 5 × 10−8 and MAF ≥ 0.01 were identified as independent significant SNPs (IndSigSNPs). SNPs that have LD r2 ≥ 0.6 and MAF ≥ 0.01 with one of the IndSigSNPs were included as candidate SNPs. These SNPs might not be available in the GWAS dataset, but are available in the 1000G EUR reference population. IndSigSNPs independent from each other (LD r2 < 0.1) were identified as lead SNPs. Genomic loci closely located (<250 kb based on the most right and left SNPs of each locus) were merged into one genomic locus. Gene annotation was based on Ensembl (build 85) and entrez ID yielding identification of 19,436 protein coding genes. Blood cis-eQTL mapping mapped IndSigSNPs to genes up to 1 Mb apart (called cis-regulated blood eGenes), and blood trans-eQTL mapping mapped IndSigSNPs to genes >5 Mb from genomic loci and/or on a different chromosome. Only significant SNP–gene pairs (PFDR < 0.05) were kept.
Chromatin interaction mapping for the lifespan was performed with the FUMA SNP2GENE function using Hi-C data in GM12878 (GEO accession number GSE8711282). IndSigSNPs located into significantly interacting regions were kept and then mapped to genes whose promoter regions (250 bp upstream and 500 bp downstream of the TSS) were located within other significantly interaction regions. Those SNP–gene loops formed frequently interacting regions has described previously82. Only significant SNP–gene contacts were kept (PFDR < 1 × 10−6). Circos zoom of genetic associations (eQTL and chromatin interaction) were generated with the integrated tool in FUMA.
Gene essentiality
Gene essentiality was defined by using the Online GEne Essentiality database12 (OGEE), which provides a list of human essential genes based on experimental data. The Homo sapiens dataset of essential genes was downloaded to identify essential eGenes. A hypergeometric test was performed to test overrepresentation of essentiality for the eGenes.
Drug-target identification
The Open Targets database13 provides drug-target identification and prioritization based on human genetic data and clinical information. The Open Targets database was used to identify small molecules and/or antibodies targeting the eGenes, their clinical trial status, and tractability for the development of small molecules.
Network analyses
A co-expression network analysis was performed to identify eGenes with central functions. NetworkAnalyst83 was used to generate a tissue-specific co-expression network of eGenes based on whole-blood expression data from the TCSBN database14. Pathway enrichment for the network was evaluated by using the Reactome database15. Metrics for centrality (degree and betweenness) were obtained from NetworkAnalyst. High betweenness centrality enrichment for the causal eGenes was evaluated by testing these genes against the top percentile (>99 percentile) nodes with the highest betweenness centrality with a hypergeometric test. The Minimum Network tool and the Graphopt layout were used to generate the visual representation of the network. The Origin software was used to make the kernel density plot.
DisGenet48 data were downloaded to assess the association of eGenes with disorders by using BeFree gene–disease associations, which provides a curated list from text mining in MEDLINE. eGenes associated with diseases available in the BeFree database were integrated in a gene–disease matrix. One gene connected with two diseases formed a disease–disease pair used to generate a edge list. Disease–disease pairs were thus obtained for each eGene and used to perform a disease network for the lifespan using NetworkAnalyst as a visualization tool83.
TFs and regulons analysis
ChEA317 was used to identify TFs associated with the expression of eGenes. ChEA3 provides TFs enrichment for genes based on ChIP-seq experiments (from ENCODE10, ReMap84, and data publicly available), co-expression data between TFs and genes (based on processed RNA-seq from GTEx85 and ARCHS486), and TF-gen87.
Single-cell analyses
Single-cell RNA-sequencing data of monocytes and DCs (GEO accession number GSE94820) were analyzed by using Automated Single-cell Analysis Pipeline (ASAP88). Data were filtered for the variance (threshold 50%), log-transformed, clustered by K-means, and reduction of dimension with t-SNE at perplexity 30. Differential expression was performed by using Limma89 at PFDR < 0.05. For heatmap representation, log-transformed data were used for Z-score distribution and visualized by using Morpheus.
To infer ligand–receptor interactions between monocytes and DCs, the publicly available CellPhoneDB v2.0 package90 was used. Single-cell RNA-sequencing data of monocytes and DCs (GEO accession number GSE94820) was log2 transformed and used as an input in CellPhoneDB using default parameters. Significant predicted ligand–receptor interactions were represented by using CellPhoneDB.
Mendelian randomization
Causal inference for eGenes on the lifespan was evaluated with two-sample MR by selecting independent SNPs (instrumental variables) associated with the expression of these eGenes. SNPs were analyzed within a window of 500 kb around the TSS of each eGene, then only SNPs strongly associated with the eGene expression (P < 0.001 corresponds to ~F statistics > 10) and independent (r2 < 0.1 based on the 1000G EUR reference panel) were selected as instrumental variables. For eQTLGen data, β (effect size) and SE were estimated for each instrumental variable from their Z-score (Z), allele frequency (p), and sample size (n) (data available from www.eqtlgen.org/cis-eqtls.html) using the following equation91:
where .
MR was performed by using the MR package92. Horizontal pleiotropy was estimated by using the heterogeneity test evaluated with the Cochran’s Q test and was considered significant when Pheterogeneity < 0.05. The MR-PRESSO package7 was performed for eGenes significant in IVW, but with significant heterogeneity on the Cochran’s Q test (Pheterogeneity < 0.05). MR-PRESSO performs an IVW MR and includes three components: detection of heterogeneity with the MR-PRESSO Global test, correction for heterogeneity via outlier removal with the MR-PRESSO Outlier test, and test of significant difference in the causal estimate before and after removal of outliers with the MR-PRESSO Distortion test. MR-PRESSO was performed if PGlobal test < 0.05 and Poutliers test < 0.05; the test was considered significant if PIVW-corrected < 0.05 and PDistortion test > 0.05. Egger regression and the intercept test to evaluate horizontal pleiotropy of instrumental variables were performed as sensitivity analyses. An absence of horizontal pleiotropy was considered if PIntercept > 0.05.
Genetic colocalization analysis
Shared genetic signals between the expression of cis-regulated eGenes (cis-eQTLs) and lifespan genetic associations or the expression of blood trans-eQTL genes were evaluated using the HyPrColoc package93. HyPrColoc provides a Bayesian colocalization analysis across traits in a genomic region in order to test for shared genetic signal. Genomic regions were defined as 500 kb downstream and 500 kb upstream of the TSS of each eGene. As described previously, β an SE for each SNPs associated with the expression of eGenes were estimated from Z-score, allele frequency, and sample size. A shared genetic signal between the expression of a cis-eGene and the lifespan or the expression of a blood trans-eQTL gene was considered if the PP was >0.8. LocusCompare23 was used to visualize the shared genetic signal between a cis-eGene and lifespan genetic associations at a locus. Biocircos94 was used to present colocalization signals between cis-eQTL and trans-eQTL genes.
GO, pathway, and enrichment analyses
GO and pathway enrichment were performed by using EnrichR87 and data were reported by using adjusted P values. The TFCheckpoint25 database, which provides experimental evidence of human TFs on gene transcription regulation, was downloaded. The primary immunodeficiency deficiency and ligand–receptor gene lists were downloaded from https://esid.org/Working-Parties/Registry-Working-Party/ESID-Registry/List-of-diseases-and-genes and https://www.nature.com/articles/ncomms8866, respectively26. The enrichment for eGenes was performed by using hypergeometric test.
Positive selection at cis-regulated eGene loci
The PopHumanScan40 catalog, which regroups regions of the human genome showing strong evidences of positive selection along the human lineage was downloaded. Lifespan loci genomic coordinates were intersected with the positive selection scan from PopHumanScan. Ancestral and derived alleles were identified from the Ensembl 92 VCF file (ftp://ftp.ensembl.org/pub/release-92/variation/vcf/homo_sapiens/homo_sapiens.vcf.gz). Variant allele frequencies were visualized by using The Geography of Genetic Variants Browser95 (GGV) and data from the 1000 Genomes.
Gene–phenotype associations
The PhenoScanner49 catalog containing human genotype–phenotype associations was used to identify immune disorders associated with eGenes enriched in primary immunodeficiency disorders.
Imputation from summary statistics
The RAISS96 package was used to impute summary statistics. It relies on a Gaussian imputation of summary-level data and empirical evidence indicates no increase in false-positive rate compared to imputation on individual data. First, LD-correlation matrix from the 1000G EUR reference population was generated with plink, and Z-scores were calculated from long-livedness GWAS summary statistics54. RAISS was then run with default settings. and were estimated from Z-scores as described above to perform MR.
Statistics
Hypergeometric tests were performed by using the hypergea R package. Spearman’s and Pearson’s correlations were performed with GraphPad Prism 5.0 (GraphPad Software, Inc., San Diego, CA).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work was supported by the Canadian Institutes of Health Research grants to P.M. (FRN148778, FRN159697) and the Quebec Heart and Lung Institute Fund. Y.B. holds a Canada Research Chair in Genomics of Heart and Lung Diseases. S.T. and B.J.A. hold a junior scholarship from Fonds de Recherche du Québec-Santé (FRQS). P.M. holds a Fonds de Recherche du Québec-Santé (FRQS) Research Chair.
Author contributions
A.C. and P.M. designed the study. A.C., V.B.-B., Z.L. conducted analyses of mapping, MR, and colocalization. Z.L. and P.M. conducted analyses for positive selection. Network analyses were performed by P.M., Z.L., and A.C. M.-C.B. performed graphs and integrative figure. Y.B., S.T., D.A., and B.J.A. provided important intellectual inputs. P.M. and A.C. drafted the manuscript. All the authors critically reviewed the manuscript and provided scientific inputs.
Data availability
We performed analyses based on publicly available GWAS summary statistics (Supplementary Information). As all analyses were based on publicly available GWAS summary statistics, no ethical approval was required. GWAS summary statistics can be found at the links below for the indicated phenotypes:
Lifespan: https://datashare.is.ed.ac.uk/handle/10283/3209; CAD: https://data.mendeley.com/datasets/gbbsrpx6bs/1; stroke: http://www.megastroke.org/download.html; AF: http://csg.sph.umich.edu/willer/public/afib2018/; CKD: https://ckdgen.imbi.uni-freiburg.de/; T2D: http://diagram-consortium.org/downloads.html; BMI: https://portals.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files; cholesterol: https://www.understandingsociety.ac.uk/; breast cancer: http://bcac.ccge.medschl.cam.ac.uk/bcacdata/icogs-complete-summary-results/; colorectal cancer: https://grasp.nhlbi.nih.gov/FullResults.aspx; prostate cancer: http://practical.icr.ac.uk/blog/; RA: http://plaza.umin.ac.jp/~yokada/datasource/software.htm; PSC: https://www.ipscsg.org/; atopic dermatitis: ftp://ftp.ebi.ac.uk/pub/databases/gwas/summary_statistics/PaternosterL_26482879_GCST003184; asthma: https://genepi.qimr.edu.au/staff/manuelf/gwas_results/main.html; Alzheimer: https://ctg.cncr.nl/software/summary_statistics; major depresssion: https://www.med.unc.edu/pgc/shared-methods/data-access-portal/; bipolar disorder: https://www.med.unc.edu/pgc/data-index/; schizophrenia: https://www.med.unc.edu/pgc/data-index/; ever smoke: https://www.thessgac.org/data; hypertension, waist circumference, lung cancer, T1D, hypothyroidism and SLE: http://www.nealelab.is/uk-biobank; long-livedness: https://grasp.nhlbi.nih.gov/FullResults.aspx.
Code availability
All software used in this analysis is publicly available at the URLs below:
GARFIELD package: https://www.ebi.ac.uk/birney-srv/GARFIELD/; FUMA: https://fuma.ctglab.nl/; eQTLGen: https://www.eqtlgen.org/index.html; Hypergea package: https://cran.r-project.org/web/packages/hypergea/index.html; Enrichr: https://amp.pharm.mssm.edu/Enrichr/; OGEE: http://ogee.medgenius.info/browse/; Open Targets: https://www.opentargets.org/; NetworkAnalyst: https://www.networkanalyst.ca/; Reactome: https://reactome.org/; ChEA3: https://amp.pharm.mssm.edu/chea3/; ASAP: https://asap.epfl.ch/; Limma: http://bioinf.wehi.edu.au/limma/; Morpheus: https://software.broadinstitute.org/morpheus/; Mendelian randomization package: https://cran.r-project.org/web/packages/MendelianRandomization/index.html; MR-PRESSO package: https://github.com/rondolab/MR-PRESSO; HyPrColoc: https://github.com/jrs95/hyprcoloc; TFcheckpoint: http://www.tfcheckpoint.org/; PopHumanScan: https://pophumanscan.uab.cat/; DisGeNET: https://www.disgenet.org/home/; COSMIC: https://cancer.sanger.ac.uk/cosmic; CellPhoneDB: https://github.com/Teichlab/cellphonedb; PhenoScanner: http://www.phenoscanner.medschl.cam.ac.uk/; RAISS package: https://gitlab.pasteur.fr/statistical-genetics/raiss; UK Biobank: http://www.nealelab.is/uk-biobank.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information is available for this paper at 10.1038/s42003-020-0937-x.
References
- 1.Herskind AM, et al. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900. Hum. Genet. 1996;97:319–323. doi: 10.1007/BF02185763. [DOI] [PubMed] [Google Scholar]
- 2.Timmers PR, et al. Genomics of 1 million parent lifespans implicates novel pathways and common diseases and distinguishes survival chances. eLife. 2019;8:e39856. doi: 10.7554/eLife.39856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morley M, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 6.Burgess S, Bowden J, Fall T, Ingelsson E, Thompson SG. Sensitivity analyses for robust causal inference from mendelian randomization analyses with multiple genetic variants. Epidemiology. 2017;28:30–42. doi: 10.1097/EDE.0000000000000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verbanck M, Chen C-Y, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018;50:693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iotchkova V, et al. GARFIELD classifies disease-relevant genomic features through integration of functional annotations with association signals. Nat. Genet. 2019;51:343–353. doi: 10.1038/s41588-018-0322-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roadmap Epigenomics Consortium, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Võsa, U. et al. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. bioRxiv10.1101/447367 (2018).
- 12.Chen W-H, Minguez P, Lercher MJ, Bork P. OGEE: an online gene essentiality database. Nucleic Acids Res. 2012;40:D901–D906. doi: 10.1093/nar/gkr986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carvalho-Silva D, et al. Open Targets Platform: new developments and updates two years on. Nucleic Acids Res. 2019;47:D1056–D1065. doi: 10.1093/nar/gky1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S, et al. TCSBN: a database of tissue and cancer specific biological networks. Nucleic Acids Res. 2018;46:D595–D600. doi: 10.1093/nar/gkx994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fabregat A, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46:D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stadhouders R, Filion GJ, Graf T. Transcription factors and 3D genome conformation in cell-fate decisions. Nature. 2019;569:345–354. doi: 10.1038/s41586-019-1182-7. [DOI] [PubMed] [Google Scholar]
- 17.Keenan AB, et al. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 2019;47:W212–W224. doi: 10.1093/nar/gkz446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glasmacher E, et al. A genomic regulatory element that directs assembly and function of immune-specific AP-1–IRF complexes. Science. 2012;338:975–980. doi: 10.1126/science.1228309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burgess S, Thompson SG. Avoiding bias from weak instruments in Mendelian randomization studies. Int. J. Epidemiol. 2011;40:755–764. doi: 10.1093/ije/dyr036. [DOI] [PubMed] [Google Scholar]
- 20.Wilhelm T, et al. Neuronal inhibition of the autophagy nucleation complex extends life span in post-reproductive C. elegans. Genes Dev. 2017;31:1561–1572. doi: 10.1101/gad.301648.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sinha S, et al. Asrij/OCIAD1 suppresses CSN5-mediated p53 degradation and maintains mouse hematopoietic stem cell quiescence. Blood. 2019;133:2385–2400. doi: 10.1182/blood.2019000530. [DOI] [PubMed] [Google Scholar]
- 22.Barabási A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu B, Gloudemans MJ, Rao AS, Ingelsson E, Montgomery SB. Abundant associations with gene expression complicate GWAS follow-up. Nat. Genet. 2019;51:768–769. doi: 10.1038/s41588-019-0404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takaki S, et al. Characterization of Lnk. An adaptor protein expressed in lymphocytes. J. Biol. Chem. 1997;272:14562–14570. doi: 10.1074/jbc.272.23.14562. [DOI] [PubMed] [Google Scholar]
- 25.Chawla K, Tripathi S, Thommesen L, Lægreid A, Kuiper M. TFcheckpoint: a curated compendium of specific DNA-binding RNA polymerase II transcription factors. Bioinformatics. 2013;29:2519–2520. doi: 10.1093/bioinformatics/btt432. [DOI] [PubMed] [Google Scholar]
- 26.Ramilowski JA, et al. A draft network of ligand–receptor-mediated multicellular signalling in human. Nat. Commun. 2015;6:7866. doi: 10.1038/ncomms8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Healy J, Richer C, Bourgey M, Kritikou EA, Sinnett D. Replication analysis confirms the association of ARID5B with childhood B-cell acute lymphoblastic leukemia. Haematologica. 2010;95:1608–1611. doi: 10.3324/haematol.2010.022459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bottini N, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat. Genet. 2004;36:337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 29.Begovich AB, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am. J. Hum. Genet. 2004;75:330–337. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orozco G, et al. Association of a functional single-nucleotide polymorphism of PTPN22, encoding lymphoid protein phosphatase, with rheumatoid arthritis and systemic lupus erythematosus. Arthritis Rheum. 2005;52:219–224. doi: 10.1002/art.20771. [DOI] [PubMed] [Google Scholar]
- 31.Toyama S, et al. A novel protease, PRSS33 (serine protease 33), is specifically and constitutively expressed in eosinophils. J. Allergy Clin. Immunol. 2017;139:AB163. doi: 10.1016/j.jaci.2016.12.535. [DOI] [Google Scholar]
- 32.Smeekens SP, et al. Functional genomics identifies type I interferon pathway as central for host defense against Candida albicans. Nat. Commun. 2013;4:1–10. doi: 10.1038/ncomms2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Acker, H. H., Capsomidis, A., Smits, E. L. & Van Tendeloo, V. F. CD56 in the immune system: more than a marker for cytotoxicity? Front. Immunol. 8, 10.3389/fimmu.2017.00892 (2017). [DOI] [PMC free article] [PubMed]
- 34.Villani, A.-C. et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science356, 10.1126/science.aah4573 (2017). [DOI] [PMC free article] [PubMed]
- 35.Rempel SA, Golembieski WA, Fisher JL, Maile M, Nakeff A. SPARC modulates cell growth, attachment and migration of U87 glioma cells on brain extracellular matrix proteins. J. Neurooncol. 2001;53:149–160. doi: 10.1023/A:1012201300188. [DOI] [PubMed] [Google Scholar]
- 36.Chen J, et al. SPARC is a key regulator of proliferation, apoptosis and invasion in human ovarian cancer. PLoS ONE. 2012;7:e42413. doi: 10.1371/journal.pone.0042413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alachkar H, et al. SPARC promotes leukemic cell growth and predicts acute myeloid leukemia outcome. J. Clin. Invest. 2014;124:1512–1524. doi: 10.1172/JCI70921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsubara T, et al. Critical role of cortactin in actin ring formation and osteoclastic bone resorption. J. Bone Miner. Metab. 2006;24:368–372. doi: 10.1007/s00774-006-0701-4. [DOI] [PubMed] [Google Scholar]
- 39.Ammer AG, Weed SA. Cortactin branches out: roles in regulating protrusive actin dynamics. Cell Motil. Cytoskeleton. 2008;65:687–707. doi: 10.1002/cm.20296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murga-Moreno J, Coronado-Zamora M, Bodelón A, Barbadilla A, Casillas S. PopHumanScan: the online catalog of human genome adaptation. Nucleic Acids Res. 2019;47:D1080–D1089. doi: 10.1093/nar/gky959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solberg OD, et al. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Hum. Immunol. 2008;69:443–464. doi: 10.1016/j.humimm.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhernakova A, et al. Evolutionary and functional analysis of celiac risk loci reveals SH2B3 as a protective factor against bacterial infection. Am. J. Hum. Genet. 2010;86:970–977. doi: 10.1016/j.ajhg.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schunkert H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dichgans M, et al. Shared genetic susceptibility to ischemic stroke and coronary artery disease: a genome-wide analysis of common variants. Stroke. 2014;45:24–36. doi: 10.1161/STROKEAHA.113.002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barrett JC, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stahl EA, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Menon, M. B. & Dhamija, S. Beclin 1 phosphorylation—at the center of autophagy regulation. Front. Cell Dev. Biol. 6, 10.3389/fcell.2018.00137 (2018). [DOI] [PMC free article] [PubMed]
- 48.Piñero J, et al. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database J. Biol. Databases Curation. 2015;2015:bav028. doi: 10.1093/database/bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kamat, M. A. et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics (Oxf., Engl.) 10.1093/bioinformatics/btz469 (2019). [DOI] [PMC free article] [PubMed]
- 50.Tate JG, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steck AK, et al. rs2476601 T allele (R620W) defines high-risk PTPN22 type I diabetes-associated haplotypes with preliminary evidence for an additional protective haplotype. Genes Immun. 2009;10:S21–S26. doi: 10.1038/gene.2009.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elshazli R, Settin A. Association of PTPN22 rs2476601 and STAT4 rs7574865 polymorphisms with rheumatoid arthritis: a meta-analysis update. Immunobiology. 2015;220:1012–1024. doi: 10.1016/j.imbio.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 53.Porcu E, et al. Mendelian randomization integrating GWAS and eQTL data reveals genetic determinants of complex and clinical traits. Nat. Commun. 2019;10:1–12. doi: 10.1038/s41467-019-10936-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Broer L, et al. GWAS of longevity in CHARGE consortium confirms APOE and FOXO3 candidacy. J. Gerontol. A. 2015;70:110–118. doi: 10.1093/gerona/glu166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosa M, et al. A Mendelian randomization study of IL6 signaling in cardiovascular diseases, immune-related disorders and longevity. Npj Genom. Med. 2019;4:23. doi: 10.1038/s41525-019-0097-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sancho D, et al. Identification of a dendritic cell receptor that couples sensing of necrosis to immunity. Nature. 2009;458:899–903. doi: 10.1038/nature07750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsung K, Dolan JP, Tsung YL, Norton JA. Macrophages as effector cells in interleukin 12-induced T cell-dependent tumor rejection. Cancer Res. 2002;62:5069–5075. [PubMed] [Google Scholar]
- 58.Ghislat G, Lawrence T. Autophagy in dendritic cells. Cell. Mol. Immunol. 2018;15:944–952. doi: 10.1038/cmi.2018.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reikine, S., Nguyen, J. B. & Modis, Y. Pattern recognition and signaling mechanisms of RIG-I and MDA5. Front. Immunol. 5, 10.3389/fimmu.2014.00342 (2014). [DOI] [PMC free article] [PubMed]
- 60.Greer P. Closing in on the biological functions of fps/fes and fer. Nat. Rev. Mol. Cell. Biol. 2002;3:278–289. doi: 10.1038/nrm783. [DOI] [PubMed] [Google Scholar]
- 61.Mori, T. et al. Lnk/Sh2b3 controls the production and function of dendritic cells and regulates the induction of IFN-γ-producing T cells. J. Immunol. (Baltim. Md 1950)193, 1728–1736 (2014). [DOI] [PubMed]
- 62.Clarke F, et al. The protein tyrosine phosphatase PTPN22 negatively regulates presentation of immune complex derived antigens. Sci. Rep. 2018;8:1–11. doi: 10.1038/s41598-017-17765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brownlie RJ, Zamoyska R, Salmond RJ. Regulation of autoimmune and anti-tumour T-cell responses by PTPN22. Immunology. 2018;154:377–382. doi: 10.1111/imm.12919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gregersen PK. Gaining insight into PTPN22 and autoimmunity. Nat. Genet. 2005;37:1300–1302. doi: 10.1038/ng1205-1300. [DOI] [PubMed] [Google Scholar]
- 65.Maine CJ, et al. PTPN22 alters the development of T regulatory cells in the thymus. J. Immunol. (Baltim, Md 1950) 2012;188:5267–5275. doi: 10.4049/jimmunol.1200150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Galgano D, et al. The T cell IFT20 interactome reveals new players in immune synapse assembly. J. Cell Sci. 2017;130:1110–1121. doi: 10.1242/jcs.200006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hilborn E, Stål O, Jansson A. Estrogen and androgen-converting enzymes 17β-hydroxysteroid dehydrogenase and their involvement in cancer: with a special focus on 17β-hydroxysteroid dehydrogenase type 1, 2, and breast cancer. Oncotarget. 2017;8:30552–30562. doi: 10.18632/oncotarget.15547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hammer F, et al. Sex steroid metabolism in human peripheral blood mononuclear cells changes with aging. J. Clin. Endocrinol. Metab. 2005;90:6283–6289. doi: 10.1210/jc.2005-0915. [DOI] [PubMed] [Google Scholar]
- 69.Byars SG, et al. Genetic loci associated with coronary artery disease harbor evidence of selection and antagonistic pleiotropy. PLOS Genet. 2017;13:e1006328. doi: 10.1371/journal.pgen.1006328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Münz C. Beclin-1 targeting for viral immune escape. Viruses. 2011;3:1166–1178. doi: 10.3390/v3071166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.England BR, Thiele GM, Anderson DR, Mikuls TR. Increased cardiovascular risk in rheumatoid arthritis: mechanisms and implications. BMJ. 2018;361:k1036. doi: 10.1136/bmj.k1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Han, H. et al. Hypertension and breast cancer risk: a systematic review and meta-analysis. Sci. Rep. 7, 10.1038/srep44877 (2017). [DOI] [PMC free article] [PubMed]
- 73.Yang A-H, Chen J-Y, Lee C-H, Chen J-Y. Expression of NCAM and OCIAD1 in well-differentiated thyroid carcinoma: correlation with the risk of distant metastasis. J. Clin. Pathol. 2012;65:206–212. doi: 10.1136/jclinpath-2011-200416. [DOI] [PubMed] [Google Scholar]
- 74.De Marchi T, et al. Targeted MS assay predicting tamoxifen resistance in estrogen-receptor-positive breast cancer tissues and sera. J. Proteome Res. 2016;15:1230–1242. doi: 10.1021/acs.jproteome.5b01119. [DOI] [PubMed] [Google Scholar]
- 75.Ji W, et al. OCIAD1 promoted pancreatic ductal adenocarcinoma migration by regulating ATM. Pancreatol. J. Int. Assoc. Pancreatol. IAP Al. 2019;19:751–759. doi: 10.1016/j.pan.2019.01.009. [DOI] [PubMed] [Google Scholar]
- 76.Shetty DK, Kalamkar KP, Inamdar MS. OCIAD1 controls electron transport chain complex I activity to regulate energy metabolism in human pluripotent stem cells. Stem Cell Rep. 2018;11:128–141. doi: 10.1016/j.stemcr.2018.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Benjamin Emelia J, et al. Heart disease and stroke statistics—2019 update: a report from the American Heart Association. Circulation. 2019;139:e56–e528. doi: 10.1161/CIR.0000000000000659. [DOI] [PubMed] [Google Scholar]
- 78.Kramarow EA, Tejada-Vera B. Dementia mortality in the United States, 2000–2017. Natl Vital. Stat. Rep. 2019;68:1–29. [PubMed] [Google Scholar]
- 79.Joshi PK, et al. Genome-wide meta-analysis associates HLA-DQA1/DRB1 and LPA and lifestyle factors with human longevity. Nat. Commun. 2017;8:910. doi: 10.1038/s41467-017-00934-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cox DR. Regression models and life-tables. J. R. Stat. Soc. Ser. B. 1972;34:187–220. [Google Scholar]
- 81.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schmitt AD, et al. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep. 2016;17:2042–2059. doi: 10.1016/j.celrep.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou G, et al. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47:W234–W241. doi: 10.1093/nar/gkz240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chèneby J, Gheorghe M, Artufel M, Mathelier A, Ballester B. ReMap 2018: an updated atlas of regulatory regions from an integrative analysis of DNA-binding ChIP-seq experiments. Nucleic Acids Res. 2018;46:D267–D275. doi: 10.1093/nar/gkx1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carithers LJ, et al. A novel approach to high-quality postmortem tissue procurement: the GTEx project. Biopreserv. Biobank. 2015;13:311–319. doi: 10.1089/bio.2015.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lachmann A, et al. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 2018;9:1–10. doi: 10.1038/s41467-018-03751-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gardeux V, David FPA, Shajkofci A, Schwalie PC, Deplancke B. ASAP: a web-based platform for the analysis and interactive visualization of single-cell RNA-seq data. Bioinformatics (Oxf., Engl.) 2017;33:3123–3125. doi: 10.1093/bioinformatics/btx337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47–e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Efremova, M., Vento-Tormo, M., Teichmann, S. A. & Vento-Tormo, R. CellPhoneDB v2.0: Inferring cell–cell communication from combined expression of multi-subunit receptor-ligand complexes. bioRxiv10.1101/680926 (2019). [DOI] [PubMed]
- 91.Zhu Z, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 92.Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int. J. Epidemiol. 2017;46:1734–1739. doi: 10.1093/ije/dyx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Foley, C. N. et al. A fast and efficient colocalization algorithm for identifying shared genetic risk factors across multiple traits. bioRxiv10.1101/592238 (2019). [DOI] [PMC free article] [PubMed]
- 94.Cui Y, et al. BioCircos.js: an interactive Circos JavaScript library for biological data visualization on web applications. Bioinformatics. 2016;32:1740–1742. doi: 10.1093/bioinformatics/btw041. [DOI] [PubMed] [Google Scholar]
- 95.Marcus JH, Novembre J. Visualizing the geography of genetic variants. Bioinformatics (Oxf., Engl.) 2017;33:594–595. doi: 10.1093/bioinformatics/btw643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Julienne H, Shi H, Pasaniuc B, Aschard H. RAISS: robust and accurate imputation from summary statistics. Bioinformatics (Oxf. Engl.) 2019;35:4837–4839. doi: 10.1093/bioinformatics/btz466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
We performed analyses based on publicly available GWAS summary statistics (Supplementary Information). As all analyses were based on publicly available GWAS summary statistics, no ethical approval was required. GWAS summary statistics can be found at the links below for the indicated phenotypes:
Lifespan: https://datashare.is.ed.ac.uk/handle/10283/3209; CAD: https://data.mendeley.com/datasets/gbbsrpx6bs/1; stroke: http://www.megastroke.org/download.html; AF: http://csg.sph.umich.edu/willer/public/afib2018/; CKD: https://ckdgen.imbi.uni-freiburg.de/; T2D: http://diagram-consortium.org/downloads.html; BMI: https://portals.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files; cholesterol: https://www.understandingsociety.ac.uk/; breast cancer: http://bcac.ccge.medschl.cam.ac.uk/bcacdata/icogs-complete-summary-results/; colorectal cancer: https://grasp.nhlbi.nih.gov/FullResults.aspx; prostate cancer: http://practical.icr.ac.uk/blog/; RA: http://plaza.umin.ac.jp/~yokada/datasource/software.htm; PSC: https://www.ipscsg.org/; atopic dermatitis: ftp://ftp.ebi.ac.uk/pub/databases/gwas/summary_statistics/PaternosterL_26482879_GCST003184; asthma: https://genepi.qimr.edu.au/staff/manuelf/gwas_results/main.html; Alzheimer: https://ctg.cncr.nl/software/summary_statistics; major depresssion: https://www.med.unc.edu/pgc/shared-methods/data-access-portal/; bipolar disorder: https://www.med.unc.edu/pgc/data-index/; schizophrenia: https://www.med.unc.edu/pgc/data-index/; ever smoke: https://www.thessgac.org/data; hypertension, waist circumference, lung cancer, T1D, hypothyroidism and SLE: http://www.nealelab.is/uk-biobank; long-livedness: https://grasp.nhlbi.nih.gov/FullResults.aspx.
All software used in this analysis is publicly available at the URLs below:
GARFIELD package: https://www.ebi.ac.uk/birney-srv/GARFIELD/; FUMA: https://fuma.ctglab.nl/; eQTLGen: https://www.eqtlgen.org/index.html; Hypergea package: https://cran.r-project.org/web/packages/hypergea/index.html; Enrichr: https://amp.pharm.mssm.edu/Enrichr/; OGEE: http://ogee.medgenius.info/browse/; Open Targets: https://www.opentargets.org/; NetworkAnalyst: https://www.networkanalyst.ca/; Reactome: https://reactome.org/; ChEA3: https://amp.pharm.mssm.edu/chea3/; ASAP: https://asap.epfl.ch/; Limma: http://bioinf.wehi.edu.au/limma/; Morpheus: https://software.broadinstitute.org/morpheus/; Mendelian randomization package: https://cran.r-project.org/web/packages/MendelianRandomization/index.html; MR-PRESSO package: https://github.com/rondolab/MR-PRESSO; HyPrColoc: https://github.com/jrs95/hyprcoloc; TFcheckpoint: http://www.tfcheckpoint.org/; PopHumanScan: https://pophumanscan.uab.cat/; DisGeNET: https://www.disgenet.org/home/; COSMIC: https://cancer.sanger.ac.uk/cosmic; CellPhoneDB: https://github.com/Teichlab/cellphonedb; PhenoScanner: http://www.phenoscanner.medschl.cam.ac.uk/; RAISS package: https://gitlab.pasteur.fr/statistical-genetics/raiss; UK Biobank: http://www.nealelab.is/uk-biobank.