Abstract
Acquired resistance to chemotherapy is an important clinical problem and can also occur without detectable cytogenetic aberrations or gene mutations. Chronic lymphocytic leukemia (CLL) is molecularly well characterized and has been elemental for establishing central paradigms in oncology. This prompted us to check whether specific epigenetic changes at the level of DNA methylation might underlie development of treatment resistance. We used Illumina Infinium HumanMethylation450 BeadChips to obtain DNA methylation profiles of 71 CLL patients with differential responses. Thirty-six patients were categorized as relapsed/refractory after treatment with fludarabine or bendamustine and 21 of them had genetic aberrations of TP53. The other 35 patients were untreated at the time of sampling and 15 of them had genetic aberration of TP53. Although we could not correlate chemoresistance with epigenetic changes, the patients were comprehensively characterized regarding relevant prognostic and molecular markers (e.g. IGHV mutation status, chromosome aberrations, TP53 mutation status, clinical parameters), which makes our dataset a unique and valuable resource that can be used by researchers to test alternative hypotheses.
Subject terms: Chronic lymphocytic leukaemia, DNA methylation
| Measurement(s) | DNA methylation |
| Technology Type(s) | methylation profiling by array |
| Factor Type(s) | TP53 mutation status • response to fludarabine or bendamustine • chromosomal aberration • IGHV mutation status |
| Sample Characteristic - Organism | Homo sapiens |
Machine-accessible metadata file describing the reported data: 10.6084/m9.figshare.12006624
Background & Summary
Chronic lymphocytic leukemia (CLL) is the most common leukemia in the Western world and mainly affects elderly patients1. Its incidence rate was 8.3 cases per 100 000 men and 5.8 cases per 100 000 women in Germany in 20142. CLL is characterized by accumulation of small B lymphocytes with a mature appearance in blood, bone marrow, lymph nodes and other lymphoid tissues3. The clinical course of CLL differs depending on the biological characteristics of the disease (hypermutation status of the immunoglobulin heavy-chain genes (IGHV), presence of specific genomic aberrations and/or recurrent mutations in oncogenes and tumor suppressor genes)4–6. Some of these genetic features are associated with distinct epigenetic profiles, e.g. CLL tumours with high level of IGHV somatic hypermutation (M-CLL) have distinct DNA methylation patterns compared to CLL tumours with a low or absent IGHV mutational load (U-CLL)7.
Chemoimmunotherapeutic regimens like fludarabine, cyclophosphamide and rituximab (FCR) or bendamustine and rituximab (BR) achieve durable remissions in the majority of treatment-naïve CLL patients8–11. Although novel targeted and effective treatments for CLL were introduced in the past five years, FCR is not inferior to them as first-line therapy in the subgroup of young and fit patients with M-CLL without 17p deletion and/or TP53 mutation (del(17p)/TP53mut)12,13. Additionally, the high cost of novel targeted drugs limits their use in developing countries where conventional cytotoxic chemotherapy is still a viable option14,15. Thus, drugs like fludarabine and bendamustine will continue to be used in the future for treatment of CLL and development of resistance to these classical chemotherapeutics remains an important problem to study.
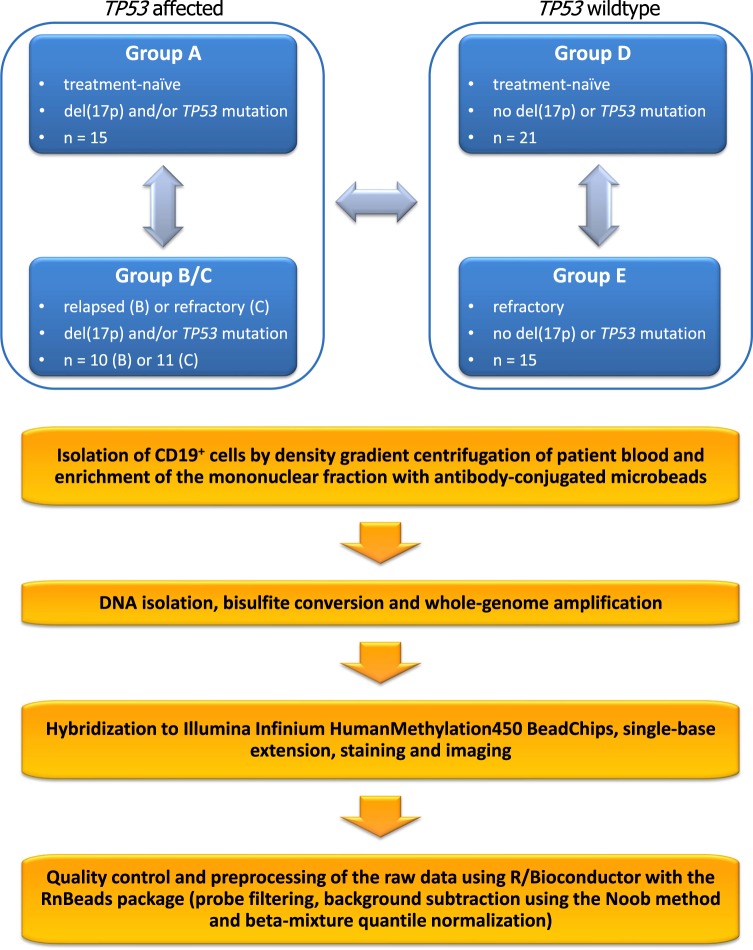
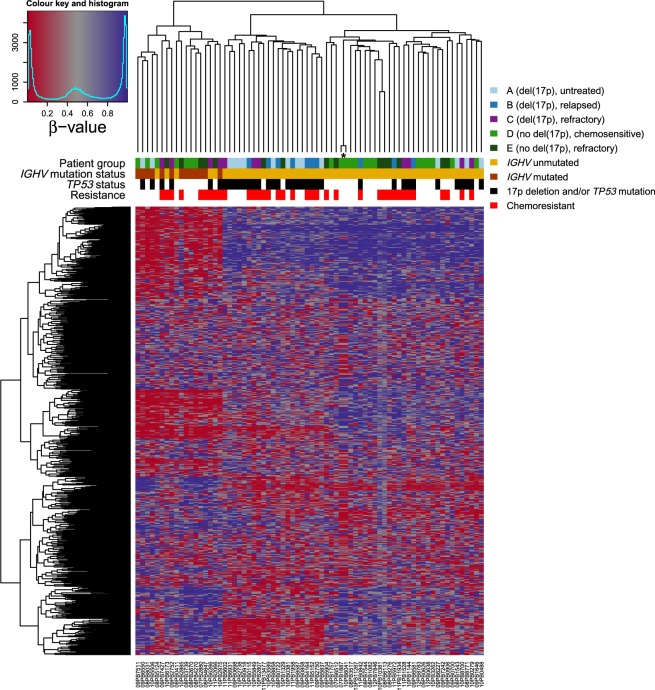
Chemorefractoriness of CLL is most often caused by functional impairment of the ATM-p53 DNA damage response pathway, mostly as a result of cytogenetic aberrations or mutations16,17. Del(17p) is found in 5% to 10% of patients at diagnosis but in up to 40% of patients relapsing after fludarabine-based treatment regimens18. Del(17p) causes loss of one allele of the tumour suppressor TP53 but in about 80% of the cases the other allele is also inactivated by somatic mutation6,18. Nevertheless, even monoallelic aberrations of TP53 confer poor prognosis. Interestingly, some cases of chemorefractory CLL show dysfunction of the ATM-p53 pathway without respective genetic lesions16,17. Additional genes and pathways have been implicated in development of resistance to fludarabine, although also in these cases mutations are not always detectable17,19,20. These observations leave the possibility that chemoresistance in CLL can also be driven by epigenetic mechanisms. In order to find epigenetic changes associated with chemoresistance, we selected samples from patients that were relapsed/refractory after treatment with fludarabine or bendamustine and/or had del(17p)/TP53mut, as well as samples from CLL patients without del(17p)/TP53mut who had treatment-naïve disease or who achieved prolonged remission after treatment with fludarabine- or bendamustine-based regimens. The grouping of the samples is shown in Fig. 1. This selection of samples allows comparing relapsed/refractory patients to untreated patients after stratification for the presence or absence of aberrations affecting the TP53 locus. In our opinion, this stratification is important because presence of TP53 aberrations could obscure the effect of epimutations, as TP53 aberrations themselves are a strong determinant of chemoresistance8,21,22. On the other hand, the chosen design of the study could allow to detect epimutations that additionally occur in the subgroup of TP53-disrupted CLL tumours to further reduce their sensitivity to chemotherapy. Genome-wide DNA methylation in all selected samples (N = 72) was quantified using Illumina Infinium HumanMethylation450 BeadChips. The resulting raw signal data and a normalized data matrix are provided here as a resource for studying relationships between epigenetics and chemoresistance in CLL. Basic clustering and principal component analyses did not intuitively show grouping of samples according to chemoresistance status. However, we cannot exclude that more sophisticated analyses will be able to extract relevant differences and correlations. Notably, the dataset is unique with the high proportion of patients with del17p and/or mutated TP53. This dataset thus allows comparison of epigenetic profiles of CLL patients with negative prognostic markers to profiles of patients with chemosensitive CLL and CLL not harbouring TP53 defects.
Fig. 1.
Schematic overview of the study design and experimental procedure.
Methods
Patient sample selection and molecular characterization
The biological and molecular characteristics of the 71 CLL patients included in the study are listed in Table 1 and Online-only Table 1. Fifty-one of the patients were subjects of the multi-centre CLL2O clinical trial (clinicaltrials.gov: NCT01392079) and were subdivided here into 4 subgroups depending on their del(17p)/TP53mut and treatment/response statuses as follows: groups A (N = 15), B (N = 10) and C (N = 11) consisted of patients with del(17p) and/or TP53 mutation and group E (N = 15) consisted of patients without del(17p) or TP53 mutation. Patients in group A were not treated previously but required treatment, patients in group B had relapsed after treatment with fludarabine- or bendamustine-containing regimens and patients in groups C and E were refractory to fludarabine or bendamustine. Additional 20 cases (group D) were patients whose tumours did not harbour del(17p) or TP53 mutation and who were not previously treated but some of whom required treatment and responded to subsequent therapy with fludarabine- or bendamustine-containing regimens (N = 6, Online-only Table 1). All patients had a confirmed diagnosis of CLL by flow cytometry; their IGHV mutational status and cytogenetics were also determined during the diagnostic workup. Unmutated IGHV gene (≥98% homology to germline) was detected by sequencing in 58 patients (81.7%). Fluorescent in-situ hybridization (FISH) analysis revealed the presence of del(17p) in 35 of a total 36 patients in groups A, B and C, as well as the absence of such an aberration in all patients from groups D and E. Mutated TP53 was detected in 30 of total 36 patients in groups A, B and C, and in none of the patients in groups D and E. One of the patients in group D had two consecutive samples taken with a time difference of 40 months (Online-only Table 1). All patients provided informed consent to subsequent analysis and research in accordance with the Declaration of Helsinki and under a protocol approved by the ethical committee of the University of Ulm.
Table 1.
Biological and molecular features of CLL patients included in the study.
| Characteristic | All patients (N = 71) | A (del(17p), untreated, N = 15) | B (del(17p), relapsed, N = 10) | C (del(17p), refractory, N = 11) | D (no del(17p), chemosensitive, N = 20) | E (no del(17p), refractory, N = 15) | P Value* |
|---|---|---|---|---|---|---|---|
| Age at sampling, years | 0.27 | ||||||
| Median | 64 | 66 | 62 | 67 | 62 | 63 | |
| Range | 38–84 | 49–77 | 53–68 | 54–76 | 38–84 | 38–71 | |
| IGHV mutational status, number (%) | 0.45 | ||||||
| Mutated (<98% homology) | 13 (18.3) | 2 (13.3) | 0 (0) | 3 (27.3) | 5 (25) | 3 (20) | |
| Unmutated (≥98% homology) | 58 (81.7) | 13 (86.7) | 10 (100) | 8 (72.7) | 15 (75) | 12 (80) | |
| FISH analysis, number (%) | P < 0.0001 | ||||||
| Normal karyotype | 12 (16.9) | 0 (0) | 0 (0) | 0 (0) | 10 (50) | 2 (13.3) | |
| Del(13q14) | 46 (64.8) | 9 (60) | 6 (60) | 8 (72.7) | 10 (50) | 13 (86.7) | |
| Trisomy 12 | 7 (9.9) | 3 (20) | 1 (10) | 3 (27.3) | 0 (0) | 0 (0) | |
| Del(11q22) | 13 (18.3) | 3 (20) | 3 (33.3) | 3 (27.3) | 0 (0) | 4 (26.7) | |
| Del(17p13) | 35 (49.3) | 15 (100) | 10 (100) | 10 (90.9) | 0 (0) | 0 (0) | |
| TP53 mutational status, number (%) | P < 0.0001 | ||||||
| Mutated | 30 (42.3%) | 10 (66.7) | 9 (90) | 11 (100) | 0 (0) | 0 (0) | |
| Unmutated | 41 (57.7%) | 5 (33.3) | 1 (10) | 0 (0) | 20 (100) | 15 (100) |
*All P values are for comparisons across all five groups and are two-sided. P values for numerical variables were calculated with the use of the Kruskal–Wallis test, and P values for categorical variables were calculated with the use of the chi-square test or Fisher’s exact test. FISH profiles were summarized according to the hierarchical risk model4 before performing the test.
Online-only Table 1.
Clinical and molecular characteristics of CLL patients included in the study.
| Experimental group | Sample ID | Patient sex^ | Predicted sex^ | Patient age, years | Treated | Response to therapy | IGHV mutation status (% homology) | del13q14 | Nuclei with del13q14 (%) | +12p | Nuclei with trisomy 12 (%) | del14q32 | Nuclei with del14q32 (%) | del11q22 | Nuclei with del11q22 (%) | del17p13 | Nuclei with del17p13 (%) | TP53 mutation | TP53 mutation type | TP53 variant allele frequency (%) | Protocol 1 | Protocol 2 | Protocol 3 | Protocol 4 | Protocol 5 | Output files | Sample name in GEO | Alternative sample title in GEO | Protocol 6 | Final processed output |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A (del(17p), untreated) | 09PB8587 | NK | Female | 68 | No | NA | 100 | No | 4.5 | No | 0.0 | No | 1.5 | No | 1.50 | Yes | 18.00 | c.830G>T | missense | 5 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491005_R01C01_Grn.idat & 9422491005_R01C01_Red.idat | GSM4056697 | CLL_A_del17p_untreated_1 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| A (del(17p), untreated) | 10PB5506 | NK | Female | 71 | No | NA | 82.62 | Yes | 86.50 | No | 4.5 | No | 2.0 | No | 4.50 | Yes | 56.50 | c.527G>C; 528C>T | missense | 50 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491007_R05C02_Grn.idat & 9422491007_R05C02_Red.idat | GSM4056718 | CLL_A_del17p_untreated_6 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| A (del(17p), untreated) | 10PB8858 | NK | Female | 77 | No | NA | 100 | Yes | 87.00 | No | 1.5 | No | 2.0 | No | 2.50 | Yes | 89.00 | c.731G>T | missense | 90 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491034_R05C02_Grn.idat & 9422491034_R05C02_Red.idat | GSM4056742 | CLL_A_del17p_untreated_10 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| A (del(17p), untreated) | 09PB6227 | NK | Male | 61 | No | NA | 100 | No | 1.5 | No | 2.00 | No | 1.50 | Yes | 14.50 | Yes | 19.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491005_R06C01_Grn.idat & 9422491005_R06C01_Red.idat | GSM4056707 | CLL_A_del17p_untreated_3 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| A (del(17p), untreated) | 09PB0858 | NK | Female | 66 | No | NA | 100 | Yes | 24.50 | No | 2.0 | No | 5.5 | No | 1.00 | Yes | 94.50 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491020_R01C01_Grn.idat & 9422491020_R01C01_Red.idat | GSM4056721 | CLL_A_del17p_untreated_7 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| A (del(17p), untreated) | 08PB1543 | Female | Female | 49 | No | NA | 99.5 | No | 5.5 | No | 0.5 | No | 1.5 | No | 4.00 | Yes | 85.50 | c.734G>A | missense | 90 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491041_R03C01_Grn.idat & 9422491041_R03C01_Red.idat | GSM4056749 | CLL_A_del17p_untreated_13 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| A (del(17p), untreated) | 10PB0381 | NK | Female | 61 | No | NA | 100 | No | 2.0 | No | 2.50 | No | 2.50 | Yes | 75.00 | Yes | 84.50 | c.722C>T | missense | 50 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491005_R05C02_Grn.idat & 9422491005_R05C02_Red.idat | GSM4056706 | CLL_A_del17p_untreated_2 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| A (del(17p), untreated) | 10PB0918 | NK | Male | 51 | No | NA | 99.53 | Yes | 94.00 | No | 5.0 | No | 3.0 | No | 1.50 | Yes | 98.00 | c.742C>T | missense | 100 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491020_R06C01_Grn.idat & 9422491020_R06C01_Red.idat | GSM4056731 | CLL_A_del17p_untreated_8 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| A (del(17p), untreated) | 10PB0488 | NK | Male | 51 | No | NA | 99.52 | Yes | 84.00 | No | 0.5 | No | 2.0 | No | 3.00 | Yes | 92.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491041_R02C02_Grn.idat & 9422491041_R02C02_Red.idat | GSM4056748 | CLL_A_del17p_untreated_12 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| A (del(17p), untreated) | 09PB6550 | Female | Female | 51 | No | NA | 92.64 | Yes | 89.50 | No | 1.0 | No | 3.0 | No | 2.00 | Yes | 23.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491007_R03C01_Grn.idat & 9422491007_R03C01_Red.idat | GSM4056713 | CLL_A_del17p_untreated_5 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| A (del(17p), untreated) | 08PB8771 | Male | Male | 69 | No | NA | 100 | Yes | 86.0 | No | 1.0 | No | 1.0 | Yes | 90.50 | Yes | 92.50 | c.329G>T | missense | 80 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491034_R01C01_Grn.idat & 9422491034_R01C01_Red.idat | GSM4056733 | CLL_A_del17p_untreated_9 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| A (del(17p), untreated) | 09PB5837 | NK | Male | 65 | No | NA | 99.5 | No | 3.0 | Yes | 53.5 | No | 2.5 | No | 0.50 | Yes | 59.50 | c.413C>T | missense | 45 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491055_R01C01_Grn.idat & 9422491055_R01C01_Red.idat | GSM4056757 | CLL_A_del17p_untreated_14 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| A (del(17p), untreated) | 10PB2738 | Male | Male | 68 | No | NA | 99.52 | Yes | 30.0 | Yes | 58.0 | No | 1.0 | No | 3.50 | Yes | 92.50 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491007_R02C02_Grn.idat & 9422491007_R02C02_Red.idat | GSM4056712 | CLL_A_del17p_untreated_4 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| A (del(17p), untreated) | 11PB6931 | NK | Female | 67 | No | NA | 99.05 | No | 3.5 | No | 0.00 | Yes | 86.0 | No | 0.50 | Yes | 64.00 | c.743G>A | missense | 40 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491034_R06C01_Grn.idat & 9422491034_R06C01_Red.idat | GSM4056743 | CLL_A_del17p_untreated_11 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| A (del(17p), untreated) | 09PB0698 | Female | Female | 72 | No | NA | 100 | Yes | 53.0 | Yes | 16.5 | No | 5.0 | No | 3.50 | Yes | 73.50 | c.809T>G | missense | 80 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491055_R06C01_Grn.idat & 9422491055_R06C01_Red.idat | GSM4056767 | CLL_A_del17p_untreated_15 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| B (del(17p), relapsed) | 11PB5152 | NK | Male | 68 | Yes | Relapsed | 100 | Yes | 8.5 | No | 2.0 | No | 0.0 | No | 1.50 | Yes | 24.00 | c.536A>G; c.733G>T | missense; missense | 50; 10 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491005_R02C01_Grn.idat & 9422491005_R02C01_Red.idat | GSM4056699 | CLL_B_del17p_relapsed_2 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| B (del(17p), relapsed) | 09PB6581 | NK | Male | 59 | Yes | Relapsed | 100 | Yes | 30.5 | No | 1.5 | No | 1.0 | No | 3.50 | Yes | 87.00 | c.794T>C | missense | 60 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491020_R01C02_Grn.idat & 9422491020_R01C02_Red.idat | GSM4056722 | CLL_B_del17p_relapsed_4 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| B (del(17p), relapsed) | 09PB2750 | NK | Male | 54 | Yes | Relapsed | 99.65 | No | 2.0 | Yes | 74.0 | No | 6.0 | No | 3.00 | Yes | 15.50 | c.535C>T | missense | 60 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491041_R04C01_Grn.idat & 9422491041_R04C01_Red.idat | GSM4056751 | CLL_B_del17p_relapsed_8 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| B (del(17p), relapsed) | 10PB0115 | NK | Female | 60 | Yes | Relapsed | 99.1 | No | 5.0 | No | 0.5 | Yes | 61.0 | Yes | 91.00 | Yes | 64.50 | c.632C>T; c.675–683del9bp | missense; in frame deletion | 55; 5 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491005_R01C02_Grn.idat & 9422491005_R01C02_Red.idat | GSM4056698 | CLL_B_del17p_relapsed_1 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| B (del(17p), relapsed) | 10PB3589 | NK | Male | 64 | Yes | Relapsed | 100 | Yes | 53.5 | No | 3.5 | Yes | 87.0 | No | 4.00 | Yes | 91.50 | c.652–654del3bp | in frame deletion | 15 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491034_R02C01_Grn.idat & 9422491034_R02C01_Red.idat | GSM4056735 | CLL_B_del17p_relapsed_7 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| B (del(17p), relapsed) | 11PB1958 | NK | Male | 64 | Yes | Relapsed | 100 | Yes | 42.5 | No | 1.0 | No | 2.0 | No | 3.00 | Yes | 67.00 | c.584T>C; c.673-2A>C; c.842A>T | missence; splice site; missense | 30; 20; 20 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491055_R02C01_Grn.idat & 9422491055_R02C01_Red.idat | GSM4056759 | CLL_B_del17p_relapsed_9 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| B (del(17p), relapsed) | 11PB5972 | NK | Male | 67 | Yes | Relapsed | 100 | Yes | 88.5 | No | 1.5 | No | 1.5 | Yes | 14.50 | Yes | 20.50 | c.742C>T | missense | 30 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491007_R04C01_Grn.idat & 9422491007_R04C01_Red.idat | GSM4056715 | CLL_B_del17p_relapsed_3 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| B (del(17p), relapsed) | 08PB0722 | Male | Male | 58 | Yes | Relapsed | 100 | No | 4.0 | No | 0.0 | No | 2.0 | Yes | 89.50 | Yes | 88.50 | c.452C>A | missense | 90 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491034_R01C02_Grn.idat & 9422491034_R01C02_Red.idat | GSM4056734 | CLL_B_del17p_relapsed_6 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| B (del(17p), relapsed) | 11PB0842 | NK | Female | 67 | Yes | Relapsed | 100 | Yes | 88.0 | No | 0.5 | No | 4.5 | No | 2.50 | Yes | 13.50 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491055_R02C02_Grn.idat & 9422491055_R02C02_Red.idat | GSM4056760 | CLL_B_del17p_relapsed_10 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| B (del(17p), relapsed) | 09PB4164 | Male | Male | 53 | Yes | Relapsed | 100 | No | 3.5 | No | 1.00 | No | 2.50 | No | 4.50 | Yes | 83.00 | c.557-559+5del8bp | in frame deletion | 20 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491020_R02C01_Grn.idat & 9422491020_R02C01_Red.idat | GSM4056723 | CLL_B_del17p_relapsed_5 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| C (del(17p), refractory) | 09PB7427 | NK | Male | 73 | Yes | Refractory | 94.6 | Yes | 74.5 | Yes | 65.5 | No | 3.5 | No | 0.50 | Yes | 86.50 | c.394A>G | missense | 60 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491005_R03C01_Grn.idat & 9422491005_R03C01_Red.idat | GSM4056701 | CLL_C_del17p_refractory_2 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| C (del(17p), refractory) | 11PB1528 | NK | Male | 76 | Yes | Refractory | 100 | No | 8.5 | No | 4.50 | No | 1.50 | No | 2.50 | Yes | 52.00 | c.785G>T | missense | 50 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491020_R02C02_Grn.idat & 9422491020_R02C02_Red.idat | GSM4056724 | CLL_C_del17p_refractory_4 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| C (del(17p), refractory) | 10PB8700 | NK | Male | 64 | Yes | Refractory | 100 | Yes | 95.5 | No | 1.5 | No | 4.0 | No | 6.50 | Yes | 98.50 | c.429_430ins3bp | in frame insertion | 100 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491041_R05C02_Grn.idat & 9422491041_R05C02_Red.idat | GSM4056754 | CLL_C_del17p_refractory_9 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| C (del(17p), refractory) | 10PB2807 | Female | Female | 75 | Yes | Refractory | 100 | Yes | 25.5 | No | 1.0 | No | 0.5 | No | 2.00 | Yes | 94.00 | c.733G>T | missense | 90 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491005_R02C02_Grn.idat & 9422491005_R02C02_Red.idat | GSM4056700 | CLL_C_del17p_refractory_1 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| C (del(17p), refractory) | 10PB1144 | NK | Male | 76 | Yes | Refractory | 100 | Yes | 88.5 | No | 1.5 | No | 1.0 | No | 3.50 | Yes | 30.50 | c.734G>T | missense | 20 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491034_R03C01_Grn.idat & 9422491034_R03C01_Red.idat | GSM4056737 | CLL_C_del17p_refractory_7 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| C (del(17p), refractory) | 10PB5849 | NK | Female | 64 | Yes | Refractory | 99.03 | No | 5.0 | Yes | 60.0 | No | 1.5 | No | 2.00 | Yes | 90.00 | c.672+1G>T | splice site | 85 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491055_R03C01_Grn.idat & 9422491055_R03C01_Red.idat | GSM4056761 | CLL_C_del17p_refractory_10 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| C (del(17p), refractory) | 09PB2752 | NK | Male | 54 | Yes | Refractory | 97.65 | Yes | 88.0 | No | 1.5 | Yes | 63.5 | No | 5.00 | Yes | 43.50 | c.375+1G>T; c.523C>G | splice site; missense | 45; 50 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491007_R05C01_Grn.idat & 9422491007_R05C01_Red.idat | GSM4056717 | CLL_C_del17p_refractory_3 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| C (del(17p), refractory) | 11PB5602 | NK | Female | 67 | Yes | Refractory | 100 | No | 1.5 | Yes | 69.5 | No | 2.5 | No | 3.00 | Yes | 77.50 | c.176_188del13bp; c.669_672+10del14bp | frameshift; frameshift | 5; 5 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491034_R02C02_Grn.idat & 9422491034_R02C02_Red.idat | GSM4056736 | CLL_C_del17p_refractory_6 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| C (del(17p), refractory) | 10PB6266 | Male | Male | 60 | Yes | Refractory | 98.3 | Yes | 92.0 | No | 3.0 | No | 2.5 | Yes | 91.50 | No | 3.00 | c.386ins1bp | frameshift | 90 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491055_R04C02_Grn.idat & 9422491055_R04C02_Red.idat | GSM4056764 | CLL_C_del17p_refractory_11 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| C (del(17p), refractory) | 10PB0279 | NK | Male | 72 | Yes | Refractory | 100 | Yes | 73.0 | No | 1.0 | No | 3.5 | Yes | 95.50 | Yes | 94.50 | c.277–277delC; c.594–599del6bp | frameshift; in frame deletion | 90; 10 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491020_R03C01_Grn.idat & 9422491020_R03C01_Red.idat | GSM4056725 | CLL_C_del17p_refractory_5 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| C (del(17p), refractory) | 10PB2975 | NK | Male | 58 | Yes | Refractory | 95.71 | Yes | 10.0 | No | 0.5 | No | 2.5 | Yes | 53.50 | Yes | 39.00 | c.159_185del27bp; c.833C>G | in frame deletion; missense | 2; 45 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491041_R05C01_Grn.idat & 9422491041_R05C01_Red.idat | GSM4056753 | CLL_C_del17p_refractory_8 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| D (no del(17p), chemosensitive) | 08PB0411 | Female | Female | 62 | No | NA | 98.2 | No | 4.0 | No | 0.0 | No | 12.0 | No | 2.00 | No | 4.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491005_R05C01_Grn.idat & 9422491005_R05C01_Red.idat | GSM4056705 | CLL_D_nodel17p_chemosensitive_2 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| D (no del(17p), chemosensitive) | 10PB3262 | Male | Male | 56 | No | NA | 100 | Yes | 26.0 | No | 1.0 | No | 1.0 | No | 1.00 | No | 5.50 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491020_R05C01_Grn.idat & 9422491020_R05C01_Red.idat | GSM4056729 | CLL_D_nodel17p_chemosensitive_9 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| D (no del(17p), chemosensitive) | 08PB1082 | Male | Male | 66 | No | NA | 99.53 | Yes | 87.5 | No | 1.0 | No | 2.5 | No | 11.00 | No | 4.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491041_R01C02_Grn.idat & 9422491041_R01C02_Red.idat | GSM4056746 | CLL_D_nodel17p_chemosensitive_14 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| D (no del(17p), chemosensitive) | 09PB0626 | Male | Male | 59 | Yes | Responded | 100 | No | 6.5 | No | 1.5 | No | 2.5 | No | 0.50 | No | 1.50 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491005_R04C02_Grn.idat & 9422491005_R04C02_Red.idat | GSM4056704 | CLL_D_nodel17p_chemosensitive_1 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| D (no del(17p), chemosensitive) | 09PB0700 | Male | Male | 65 | No | NA | 100 | Yes | 79.0 | No | 1.5 | No | 7.0 | No | 1.50 | No | 2.50 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491020_R04C02_Grn.idat & 9422491020_R04C02_Red.idat | GSM4056728 | CLL_D_nodel17p_chemosensitive_8 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| D (no del(17p), chemosensitive) | 08PB2670 | Female | Female | 73 | No | NA | 94.06 | Yes | 56.0 | No | 1.0 | No | 1.0 | No | 0.50 | No | 4.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491041_R04C02_Grn.idat & 9422491041_R04C02_Red.idat | GSM4056752 | CLL_D_nodel17p_chemosensitive_16 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| D (no del(17p), chemosensitive) | 09PB1646 | Male | Male | 52 | No | NA | 100 | No | 1.5 | No | 1.0 | No | 5.0 | No | 2.00 | No | 4.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491005_R06C02_Grn.idat & 9422491005_R06C02_Red.idat | GSM4056708 | CLL_D_nodel17p_chemosensitive_3 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| D (no del(17p), chemosensitive) | 08PB3739 | Male | Male | 38 | No | NA | 93.81 | No | 5.5 | No | 4.0 | No | 4.0 | No | 4.00 | No | 4.50 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491020_R06C02_Grn.idat & 9422491020_R06C02_Red.idat | GSM4056732 | CLL_D_nodel17p_chemosensitive_10 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| D (no del(17p), chemosensitive) | 09PB1757 | Male | Male | 72 | Yes | Responded | 100 | No | 4.0 | No | 2.5 | No | 2.5 | No | 1.50 | No | 2.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491041_R06C02_Grn.idat & 9422491041_R06C02_Red.idat | GSM4056756 | CLL_D_nodel17p_chemosensitive_17 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| D (no del(17p), chemosensitive) | 08PB4270 | Female | Female | 43 | No | NA | 94.59 | Yes | 20.0 | No | 2.5 | No | 4.5 | No | 1.50 | No | 5.50 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491007_R02C01_Grn.idat & 9422491007_R02C01_Red.idat | GSM4056711 | CLL_D_nodel17p_chemosensitive_5 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| D (no del(17p), chemosensitive) | 08PB5124 | Female | Male | 84 | No | NA | 100 | No | 3.0 | No | 3.5 | No | 4.5 | No | 3.50 | No | 3.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491034_R05C01_Grn.idat & 9422491034_R05C01_Red.idat | GSM4056741 | CLL_D_nodel17p_chemosensitive_12 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| D (no del(17p), chemosensitive) | 08PB7544 | Male | Male | 41 | No | NA | 100 | Yes | 19.0 | No | 0.0 | No | 3.0 | No | 4.50 | No | 5.50 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491055_R05C01_Grn.idat & 9422491055_R05C01_Red.idat | GSM4056765 | CLL_D_nodel17p_chemosensitive_19 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| D (no del(17p), chemosensitive) | 09PB7511 | Male | Male | 71 | No | NA | 87.39 | Yes | 83.0 | No | 0.0 | No | 0.0 | No | 2.00 | No | 3.50 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491007_R01C02_Grn.idat & 9422491007_R01C02_Red.idat | GSM4056710 | CLL_D_nodel17p_chemosensitive_4 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| D (no del(17p), chemosensitive) | 08PB8031 | Female | Female | 63 | No | NA | 81.48 | No | 5.0 | No | 0.0 | No | 2.5 | No | 3.00 | No | 3.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491034_R04C02_Grn.idat & 9422491034_R04C02_Red.idat | GSM4056740 | CLL_D_nodel17p_chemosensitive_11 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| D (no del(17p), chemosensitive) | 08PB3517 | Male | Male | 68 | Yes | Responded | 99.55 | Yes | 82.0 | No | 4.0 | No | 0.0 | No | 5.00 | No | 3.50 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491055_R01C02_Grn.idat & 9422491055_R01C02_Red.idat | GSM4056758 | CLL_D_nodel17p_chemosensitive_18 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| D (no del(17p), chemosensitive) | 12PB0638 | Male | Male | 51 | Yes | Responded | 100 | Yes | 9.5 | No | 0.5 | No | 1.5 | No | 2.00 | No | 3.50 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491007_R04C02_Grn.idat & 9422491007_R04C02_Red.idat | GSM4056716 | CLL_D_nodel17p_chemosensitive_6 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| D (no del(17p), chemosensitive) | 07PB1887* | Male | Male | 58 | Yes | Responded | 100 | No | 6.0 | No | 1.0 | No | 3.0 | No | 0.50 | No | 4.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491034_R06C02_Grn.idat & 9422491034_R06C02_Red.idat | GSM4056744 | CLL_D_nodel17p_chemosensitive_13_timepoint1 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| D (no del(17p), chemosensitive) | 10PB6041* | Male | Male | 61 | Yes | Responded | 100 | Yes | 16.0 | No | 2.5 | No | 1.5 | No | 5.50 | No | 5.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491055_R03C02_Grn.idat & 9422491055_R03C02_Red.idat | GSM4056762 | CLL_D_nodel17p_chemosensitive_13_timepoint2 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| D (no del(17p), chemosensitive) | 12PB8783 | Female | Female | 62 | No | NA | 100 | No | 4.5 | No | 2.0 | No | 0.0 | No | 4.00 | No | 7.50 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491007_R06C02_Grn.idat & 9422491007_R06C02_Red.idat | GSM4056720 | CLL_D_nodel17p_chemosensitive_7 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| D (no del(17p), chemosensitive) | 13PB11267 | Male | Male | 64 | Yes | Responded | 100 | No | 4.5 | No | 0.0 | No | 8.0 | No | 5.00 | No | 4.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491041_R02C01_Grn.idat & 9422491041_R02C01_Red.idat | GSM4056747 | CLL_D_nodel17p_chemosensitive_15 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| D (no del(17p), chemosensitive) | 12PB7846 | Male | Male | 45 | No | NA | 100 | Yes | 67.0 | No | 1.0 | No | 0.5 | No | 0.00 | No | 3.50 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491055_R06C02_Grn.idat & 9422491055_R06C02_Red.idat | GSM4056768 | CLL_D_nodel17p_chemosensitive_20 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| E (no del(17p), refractory) | 08PB4647 | Male | Male | 60 | Yes | Refractory | 97.7 | Yes | 91.0 | No | 2.5 | No | 1.0 | No | 0.50 | No | 2.50 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491005_R04C01_Grn.idat & 9422491005_R04C01_Red.idat | GSM4056703 | CLL_E_nodel17p_refractory_2 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| E (no del(17p), refractory) | 08PB8276 | Male | Male | 66 | Yes | Refractory | 99.57 | Yes | 96.0 | No | 1.0 | No | 1.5 | No | 2.50 | No | 2.50 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491020_R04C01_Grn.idat & 9422491020_R04C01_Red.idat | GSM4056727 | CLL_E_nodel17p_refractory_7 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| E (no del(17p), refractory) | 09PB7242 | NK | Male | 46 | Yes | Refractory | 99.53 | No | 4.0 | No | 2.5 | No | 6.5 | No | 1.50 | No | 5.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491041_R01C01_Grn.idat & 9422491041_R01C01_Red.idat | GSM4056745 | CLL_E_nodel17p_refractory_11 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| E (no del(17p), refractory) | 09PB0906 | Male | Male | 68 | Yes | Refractory | 98.98 | Yes | 38.0 | No | 5.0 | No | 2.5 | Yes | 94.00 | No | 4.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491005_R03C02_Grn.idat & 9422491005_R03C02_Red.idat | GSM4056702 | CLL_E_nodel17p_refractory_1 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| E (no del(17p), refractory) | 11PB2066 | NK | Male | 71 | Yes | Refractory | 98.3 | Yes | 80.5 | No | 1.5 | No | 0.5 | No | 2.00 | No | 4.50 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491020_R03C02_Grn.idat & 9422491020_R03C02_Red.idat | GSM4056726 | CLL_E_nodel17p_refractory_6 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| E (no del(17p), refractory) | 11PB11934 | NK | Female | 70 | Yes | Refractory | 100 | Yes | 31.5 | No | 1.0 | No | 0.0 | Yes | 96.50 | No | 4.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491041_R06C01_Grn.idat & 9422491041_R06C01_Red.idat | GSM4056755 | CLL_E_nodel17p_refractory_13 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| E (no del(17p), refractory) | 10PB6173 | NK | Female | 57 | Yes | Refractory | 99.54 | Yes | 14.0 | No | 2.5 | No | 2.5 | No | 1.00 | No | 4.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491007_R01C01_Grn.idat & 9422491007_R01C01_Red.idat | GSM4056709 | CLL_E_nodel17p_refractory_3 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| E (no del(17p), refractory) | 10PB10381 | Male | Male | 46 | Yes | Refractory | 100 | Yes | 76.5 | No | 1.0 | No | 3.0 | Yes | 60.00 | No | 3.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491020_R05C02_Grn.idat & 9422491020_R05C02_Red.idat | GSM4056730 | CLL_E_nodel17p_refractory_8 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values | |
| E (no del(17p), refractory) | 10PB1329 | NK | Male | 52 | Yes | Refractory | 100 | Yes | 13.5 | No | 2.5 | Yes | 55.5 | No | 7.50 | No | 5.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491041_R03C02_Grn.idat & 9422491041_R03C02_Red.idat | GSM4056750 | CLL_E_nodel17p_refractory_12 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values | |
| E (no del(17p), refractory) | 10PB2885 | NK | Female | 65 | Yes | Refractory | 96.13 | Yes | 84.5 | No | 4.5 | No | 2.0 | No | 4.00 | No | 6.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491007_R06C01_Grn.idat & 9422491007_R06C01_Red.idat | GSM4056719 | CLL_E_nodel17p_refractory_5 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| E (no del(17p), refractory) | 08PB6113 | Female | Female | 38 | Yes | Refractory | 100 | No | 3.5 | No | 1.0 | No | 0.0 | No | 4.50 | No | 7.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491034_R04C01_Grn.idat & 9422491034_R04C01_Red.idat | GSM4056739 | CLL_E_nodel17p_refractory_10 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values | |
| E (no del(17p), refractory) | 08PB5904 | Male | Male | 69 | Yes | Refractory | 99.06 | Yes | 87.0 | No | 1.5 | No | 0.5 | Yes | 89.50 | No | 4.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491055_R04C01_Grn.idat & 9422491055_R04C01_Red.idat | GSM4056763 | CLL_E_nodel17p_refractory_14 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| E (no del(17p), refractory) | 11PB13477 | Male | Male | 66 | Yes | Refractory | 100 | Yes | 91.5 | No | 0.0 | No | 1.5 | No | 3.00 | No | 3.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491007_R03C02_Grn.idat & 9422491007_R03C02_Red.idat | GSM4056714 | CLL_E_nodel17p_refractory_4 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| E (no del(17p), refractory) | 07PB2880 | Male | Male | 63 | Yes | Refractory | 95.95 | Yes | 97.0 | No | 0.0 | No | 0.5 | No | 3.00 | No | 2.50 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491034_R03C02_Grn.idat & 9422491034_R03C02_Red.idat | GSM4056738 | CLL_E_nodel17p_refractory_9 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
| E (no del(17p), refractory) | 07PB6512 | Male | Male | 59 | Yes | Refractory | 100 | Yes | 44.0 | No | 5.0 | No | 7.0 | No | 7.50 | No | 5.00 | none detected | NA | 0 | Isolation of peripheral blood mononuclear cells (PBMCs) from blood samples via density gradient centrifugation (Pancoll human) | Purification of CD19-positive cell fraction using CD19 MicroBeads | Genomic DNA extraction (Qiagen AllPrep kit) | Bisulfite DNA conversion, whole-genome amplification, fragmentation and hybridization to Illumina Infinium HumanMethylation450 BeadChips (standard Illumina protocols) | Single-base extension, staining and scanning (standard Illumina protocols) | 9422491055_R05C02_Grn.idat & 9422491055_R05C02_Red.idat | GSM4056766 | CLL_E_nodel17p_refractory_15 | Raw data filtering, background subtraction (Noob) and normalization (BMIQ) | Data matrix of beta-values |
NA = not applicable, NK = Not known.
*Samples 07PB1887 and 10PB6041 were taken from the same patient with a time difference of 40 months. There was cytogenetic progression from normal karyotype to del13q14. Only the first sample (07PB1887) is included in the summary characteristics provided in Table 1.
^Gender was not available for a part of the anonymized patients' samples.
We used the gender prediction algorithm in RnBeads to predict patients' sex.
Sample preparation
Blood samples from CLL patients were subjected to density gradient centrifugation (Pancoll human, #P04-60500, PAN-Biotech, Germany) to isolate peripheral blood mononuclear cells (PBMCs), which were then enriched for CD19+ B cells using CD19 MicroBeads (#130-050-301, Miltenyi Biotec, Germany) and LS columns (#130-042-401, Miltenyi Biotec). The purity of the enriched cell fractions was confirmed using a FACSCalibur flow cytometer (Becton Dickinson & Co.) and a monoclonal mouse anti-human CD19 antibody (clone HD37, DakoCytomation, Denmark). Purified cell samples were flash frozen and stored as dry cell pellets at −80 °C for further analysis.
DNA extraction, bisulfite conversion and methylation level quantification
The 72 frozen cell pellets were processed in 6 batches, taking care that samples from each of the 5 subgroups (A-E) were approximately equally divided among the 6 batches to mitigate possible batch effects. DNA was extracted from the cell pellets by the Qiagen AllPrep kit (#80204) and quantification and quality control were performed using a NanoDrop ND-1000 UV-Vis Spectrophotometer (Thermo Scientific, USA). One and a half micrograms of DNA from each sample were sent to the Genomics and Proteomics Core Facility of the German Cancer Research Center (DKFZ) for bisulfite conversion and hybridization to Illumina Infinium HumanMethylation450 BeadChips, according to the manufacturer’s instructions. The bisulfite conversion was performed using the EZ DNA Methylation Kit (Zymo Research) and then the converted DNA was whole-genome amplified and fragmented. The processed samples were distributed randomly among 6 Illumina Infinium HumanMethylation450 BeadChips. The core facility was blinded regarding the identity of the samples and the experimental groups to which they belonged. After hybridization, single-base extension and staining, BeadChips were scanned using an Illumina iScan reader, and the fluorescence intensity raw data for each sample was recorded as two IDAT files, one for the green (Cy3) and one for the red (Cy5) channel23. Quality control of the whole procedure was performed using the Methylation Module of Illumina’s GenomeStudio software.
Data processing and statistics
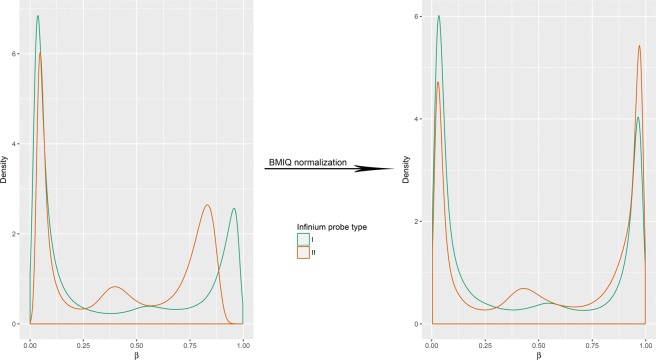
After acquiring the raw data, we performed quality control, preprocessing and basic analysis using R/Bioconductor with the RnBeads package24. Illumina probes known to be cross-reactive or overlapping known SNPs25 were excluded from analysis. This was also done for probes giving unreliable measurements as determined by the Greedycut algorithm implemented in RnBeads. The data from the remaining probes were subjected to background subtraction using the Noob method26 and beta-mixture quantile normalization (BMIQ)27. In a subsequent step, probes of non-CpG context, probes binding to sequences on sex chromosomes and probes with low standard deviation were filtered out. CpG sites on the sex chromosomes were excluded to avoid gender-specific methylation bias, as groups within our study did not contain equal numbers of males and females. CpG sites with low standard deviation are generally not informative and removing them from the analysis is a common approach to increase power for detection of differentially methylated CpGs and to improve sensitivity of clustering28,29. The data obtained by the remaining probes23 were used in downstream analyses. Methylation levels of CpG sites were calculated as β-values (β = intensity of the methylated allele (M)/[intensity of the unmethylated allele (U) + intensity of the methylated allele (M) + 100].
Both multidimensional scaling (MDS) and principal-component analysis (PCA) were used as dimension reduction techniques. Hierarchical clustering was carried out using the Manhattan distance metric and complete linkage criteria.
Data records
The complete DNA methylation microarray dataset has been deposited in the NCBI Gene Expression Omnibus (GEO) database and consists of the raw data in the form of 72 pairs (red/green fluorescence) of raw Intensity Data files (.idat), the processed data matrix and a metadata table describing the samples and their groups23. For convenience, Online-only Table 1 lists all patients and samples with their characteristics, as well as experimental and analytical procedures and output data file names.
Technical Validation
Quality control of genomic DNA
Genomic DNA 260 nm/280 nm absorbance ratios were determined using a NanoDrop ND-1000 UV-Vis Spectrophotometer (Thermo Scientific, USA). All samples had ratios in the range 1.8–2.0, as expected for DNA of high purity (Online-only Table 2).
Online-only Table 2.
DNA concentrations and quality control parameters measured using a NanoDrop ND-1000 UV-Vis Spectrophotometer. Each sample was measured in triplicates.
| Sample ID | ng/µl | A260 | A280 | 260/280 | 260/230 | Constant | Cursor Pos. | Cursor abs. | 340 raw |
|---|---|---|---|---|---|---|---|---|---|
| 09PB8587 | 237.8 | 4.756 | 2.472 | 1.92 | 2.04 | 50 | 230 | 2.328 | −0.006 |
| 09PB8587 | 242.24 | 4.845 | 2.531 | 1.91 | 2.01 | 50 | 230 | 2.414 | 0.02 |
| 09PB8587 | 243.92 | 4.878 | 2.53 | 1.93 | 2.05 | 50 | 230 | 2.383 | 0.006 |
| 11PB5152 | 94.52 | 1.89 | 0.997 | 1.9 | 1.98 | 50 | 230 | 0.957 | −0.01 |
| 11PB5152 | 93.13 | 1.863 | 1.015 | 1.84 | 2.05 | 50 | 230 | 0.908 | 0.001 |
| 11PB5152 | 87.66 | 1.753 | 0.942 | 1.86 | 1.99 | 50 | 230 | 0.88 | −0.022 |
| 09PB7427 | 167.37 | 3.347 | 1.74 | 1.92 | 2.12 | 50 | 230 | 1.578 | 0.022 |
| 09PB7427 | 164.15 | 3.283 | 1.706 | 1.92 | 2.17 | 50 | 230 | 1.511 | 0.03 |
| 09PB7427 | 162.06 | 3.241 | 1.654 | 1.96 | 2.16 | 50 | 230 | 1.501 | 0.043 |
| 08PB4647 | 141.64 | 2.833 | 1.473 | 1.92 | 2 | 50 | 230 | 1.415 | 0.044 |
| 08PB4647 | 137.1 | 2.742 | 1.432 | 1.91 | 2 | 50 | 230 | 1.368 | 0.02 |
| 08PB4647 | 137.33 | 2.747 | 1.418 | 1.94 | 2.03 | 50 | 230 | 1.356 | 0.018 |
| 10PB5506 | 169.28 | 3.386 | 1.781 | 1.9 | 2.12 | 50 | 230 | 1.598 | 0.015 |
| 10PB5506 | 158.03 | 3.161 | 1.632 | 1.94 | 2.17 | 50 | 230 | 1.455 | 0.007 |
| 10PB5506 | 154.57 | 3.091 | 1.615 | 1.91 | 2.09 | 50 | 230 | 1.48 | 0.012 |
| 09PB6581 | 170.2 | 3.404 | 1.776 | 1.92 | 2.07 | 50 | 230 | 1.644 | 0.012 |
| 09PB6581 | 177.76 | 3.555 | 1.866 | 1.91 | 2.11 | 50 | 230 | 1.683 | −0.001 |
| 09PB6581 | 168.37 | 3.367 | 1.763 | 1.91 | 2.1 | 50 | 230 | 1.606 | 0.007 |
| 11PB1528 | 297.15 | 5.943 | 3.105 | 1.91 | 2.06 | 50 | 230 | 2.887 | −0.054 |
| 11PB1528 | 296.77 | 5.935 | 3.088 | 1.92 | 2.07 | 50 | 230 | 2.871 | −0.052 |
| 11PB1528 | 266.53 | 5.331 | 2.795 | 1.91 | 2.03 | 50 | 230 | 2.625 | −0.057 |
| 08PB8276 | 663.84 | 13.277 | 7.061 | 1.88 | 2.23 | 50 | 230 | 5.941 | 0.206 |
| 08PB8276 | 652.39 | 13.048 | 6.983 | 1.87 | 2.21 | 50 | 230 | 5.906 | 0.171 |
| 08PB8276 | 675.04 | 13.501 | 7.219 | 1.87 | 2.21 | 50 | 230 | 6.108 | 0.151 |
| 10PB8858 | 169.94 | 3.399 | 1.786 | 1.9 | 2.01 | 50 | 230 | 1.687 | −0.033 |
| 10PB8858 | 166.87 | 3.337 | 1.746 | 1.91 | 2.08 | 50 | 230 | 1.606 | −0.029 |
| 10PB8858 | 165.86 | 3.317 | 1.712 | 1.94 | 2.1 | 50 | 230 | 1.576 | −0.022 |
| 09PB2750 | 59.26 | 1.185 | 0.635 | 1.87 | 0.68 | 50 | 230 | 1.736 | −0.037 |
| 09PB2750 | 58.97 | 1.179 | 0.636 | 1.85 | 0.7 | 50 | 230 | 1.68 | −0.032 |
| 09PB2750 | 60.67 | 1.213 | 0.613 | 1.98 | 0.72 | 50 | 230 | 1.693 | −0.034 |
| 08PB0411 | 129.81 | 2.596 | 1.35 | 1.92 | 2.04 | 50 | 230 | 1.272 | 0.003 |
| 08PB0411 | 124.6 | 2.492 | 1.298 | 1.92 | 1.97 | 50 | 230 | 1.267 | 0.09 |
| 08PB0411 | 127.42 | 2.548 | 1.337 | 1.91 | 1.99 | 50 | 230 | 1.281 | 0.01 |
| 09PB3261 | 225.27 | 4.505 | 2.341 | 1.92 | 1.69 | 50 | 230 | 2.661 | −0.03 |
| 09PB3261 | 225.73 | 4.515 | 2.331 | 1.94 | 1.7 | 50 | 230 | 2.655 | −0.038 |
| 09PB3261 | 228.23 | 4.565 | 2.374 | 1.92 | 1.68 | 50 | 230 | 2.722 | −0.029 |
| 09PB7242 | 130.02 | 2.6 | 1.374 | 1.89 | 2.15 | 50 | 230 | 1.208 | 0.023 |
| 09PB7242 | 124.29 | 2.486 | 1.305 | 1.9 | 2.16 | 50 | 230 | 1.15 | −0.001 |
| 09PB7242 | 145.46 | 2.909 | 1.536 | 1.89 | 2.21 | 50 | 230 | 1.316 | −0.007 |
| 08PB1082 | 124.97 | 2.499 | 1.33 | 1.88 | 2.03 | 50 | 230 | 1.233 | 0 |
| 08PB1082 | 123.44 | 2.469 | 1.32 | 1.87 | 2.01 | 50 | 230 | 1.229 | 0.026 |
| 08PB1082 | 125.76 | 2.515 | 1.336 | 1.88 | 2.06 | 50 | 230 | 1.219 | −0.009 |
| 09PB6227 | 130.08 | 2.602 | 1.366 | 1.9 | 2.07 | 50 | 230 | 1.254 | 0 |
| 09PB6227 | 131.68 | 2.634 | 1.386 | 1.9 | 2.08 | 50 | 230 | 1.267 | −0.008 |
| 09PB6227 | 135.02 | 2.7 | 1.442 | 1.87 | 2.01 | 50 | 230 | 1.344 | −0.02 |
| 10PB0115 | 160.62 | 3.212 | 1.713 | 1.88 | 2.14 | 50 | 230 | 1.504 | −0.018 |
| 10PB0115 | 154.96 | 3.099 | 1.663 | 1.86 | 2.11 | 50 | 230 | 1.468 | −0.009 |
| 10PB0115 | 138.74 | 2.775 | 1.452 | 1.91 | 2.14 | 50 | 230 | 1.298 | −0.018 |
| 10PB8700 | 184.12 | 3.682 | 1.915 | 1.92 | 2.11 | 50 | 230 | 1.749 | −0.007 |
| 10PB8700 | 185.67 | 3.713 | 1.932 | 1.92 | 2.16 | 50 | 230 | 1.723 | −0.013 |
| 10PB8700 | 189.23 | 3.785 | 1.976 | 1.91 | 2.18 | 50 | 230 | 1.739 | −0.022 |
| 09PB0906 | 103.3 | 2.066 | 1.101 | 1.88 | 1.76 | 50 | 230 | 1.175 | −0.022 |
| 09PB0906 | 100.68 | 2.014 | 1.069 | 1.88 | 1.73 | 50 | 230 | 1.161 | −0.029 |
| 09PB0906 | 103.97 | 2.079 | 1.114 | 1.87 | 1.75 | 50 | 230 | 1.187 | −0.014 |
| 09PB0626 | 178.39 | 3.568 | 1.88 | 1.9 | 2.12 | 50 | 230 | 1.68 | −0.006 |
| 09PB0626 | 174.59 | 3.492 | 1.806 | 1.93 | 2.06 | 50 | 230 | 1.691 | −0.016 |
| 09PB0626 | 197.27 | 3.945 | 2.076 | 1.9 | 2.19 | 50 | 230 | 1.804 | −0.008 |
| 09PB0858 | 314.84 | 6.297 | 3.299 | 1.91 | 2.19 | 50 | 230 | 2.875 | −0.014 |
| 09PB0858 | 294.16 | 5.883 | 3.107 | 1.89 | 2.12 | 50 | 230 | 2.776 | −0.012 |
| 09PB0858 | 274.05 | 5.481 | 2.909 | 1.88 | 2.13 | 50 | 230 | 2.577 | −0.011 |
| 10PB2807 | 138.46 | 2.769 | 1.437 | 1.93 | 2.13 | 50 | 230 | 1.297 | −0.002 |
| 10PB2807 | 136.71 | 2.734 | 1.427 | 1.92 | 2.07 | 50 | 230 | 1.322 | 0.005 |
| 10PB2807 | 136.26 | 2.725 | 1.45 | 1.88 | 2.06 | 50 | 230 | 1.324 | −0.019 |
| 11PB2066 | 185.75 | 3.715 | 1.944 | 1.91 | 2.22 | 50 | 230 | 1.675 | 0.001 |
| 11PB2066 | 151.89 | 3.038 | 1.582 | 1.92 | 2.13 | 50 | 230 | 1.424 | −0.018 |
| 11PB2066 | 152.21 | 3.044 | 1.621 | 1.88 | 2.08 | 50 | 230 | 1.466 | −0.001 |
| 09PB0700 | 171.36 | 3.427 | 1.815 | 1.89 | 1.91 | 50 | 230 | 1.797 | 0.015 |
| 09PB0700 | 167.51 | 3.35 | 1.755 | 1.91 | 2.08 | 50 | 230 | 1.607 | −0.002 |
| 09PB0700 | 189.73 | 3.795 | 1.991 | 1.91 | 2.08 | 50 | 230 | 1.825 | −0.015 |
| 10PB3589 | 131.42 | 2.628 | 1.371 | 1.92 | 1.92 | 50 | 230 | 1.369 | 0.006 |
| 10PB3589 | 123.4 | 2.468 | 1.295 | 1.91 | 1.91 | 50 | 230 | 1.292 | 0.006 |
| 10PB3589 | 122.48 | 2.45 | 1.289 | 1.9 | 1.88 | 50 | 230 | 1.306 | 0.003 |
| 10PB1144 | 269.53 | 5.391 | 2.876 | 1.87 | 2.16 | 50 | 230 | 2.498 | 0.032 |
| 10PB1144 | 268.84 | 5.377 | 2.829 | 1.9 | 2.26 | 50 | 230 | 2.379 | 0.036 |
| 10PB1144 | 267.25 | 5.345 | 2.851 | 1.87 | 2.2 | 50 | 230 | 2.429 | 0.028 |
| 11PB11934 | 147.75 | 2.955 | 1.543 | 1.92 | 1.79 | 50 | 230 | 1.649 | 0.028 |
| 11PB11934 | 179 | 3.58 | 1.89 | 1.89 | 1.79 | 50 | 230 | 1.998 | 0.041 |
| 11PB11934 | 177.37 | 3.547 | 1.884 | 1.88 | 1.88 | 50 | 230 | 1.886 | 0.021 |
| 09PB1646 | 150.98 | 3.02 | 1.576 | 1.92 | 2.05 | 50 | 230 | 1.473 | 0.025 |
| 09PB1646 | 146.39 | 2.928 | 1.538 | 1.9 | 2.08 | 50 | 230 | 1.409 | 0.022 |
| 09PB1646 | 144.76 | 2.895 | 1.521 | 1.9 | 2.03 | 50 | 230 | 1.429 | 0.019 |
| 10PB0381 | 136.57 | 2.731 | 1.448 | 1.89 | 2.08 | 50 | 230 | 1.314 | 0.024 |
| 10PB0381 | 129.38 | 2.588 | 1.36 | 1.9 | 1.96 | 50 | 230 | 1.321 | 0.097 |
| 10PB0381 | 135.79 | 2.716 | 1.462 | 1.86 | 2.02 | 50 | 230 | 1.347 | 0.032 |
| 10PB5849 | 123.27 | 2.465 | 1.271 | 1.94 | 1.88 | 50 | 230 | 1.312 | −0.003 |
| 10PB5849 | 117.2 | 2.344 | 1.241 | 1.89 | 1.88 | 50 | 230 | 1.244 | 0.026 |
| 10PB5849 | 122.95 | 2.459 | 1.304 | 1.89 | 1.92 | 50 | 230 | 1.283 | 0.025 |
| 10PB6173 | 144.45 | 2.889 | 1.527 | 1.89 | 2.12 | 50 | 230 | 1.362 | 0.031 |
| 10PB6173 | 134.68 | 2.694 | 1.397 | 1.93 | 2.05 | 50 | 230 | 1.315 | 0.036 |
| 10PB6173 | 136.93 | 2.739 | 1.438 | 1.91 | 2.1 | 50 | 230 | 1.301 | −0.004 |
| 08PB3739 | 140.6 | 2.812 | 1.455 | 1.93 | 1.96 | 50 | 230 | 1.433 | 0.006 |
| 08PB3739 | 186.3 | 3.726 | 1.97 | 1.89 | 2.09 | 50 | 230 | 1.784 | 0.02 |
| 08PB3739 | 170.65 | 3.413 | 1.779 | 1.92 | 2.03 | 50 | 230 | 1.684 | 0.024 |
| 10PB0918 | 106.88 | 2.138 | 1.129 | 1.89 | 1.87 | 50 | 230 | 1.14 | 0.007 |
| 10PB0918 | 105.58 | 2.112 | 1.105 | 1.91 | 1.86 | 50 | 230 | 1.133 | 0.017 |
| 10PB0918 | 108.59 | 2.172 | 1.152 | 1.89 | 1.87 | 50 | 230 | 1.164 | 0.014 |
| 10PB10381 | 90.36 | 1.807 | 0.945 | 1.91 | 1.9 | 50 | 230 | 0.95 | 0.008 |
| 10PB10381 | 91.38 | 1.828 | 0.98 | 1.86 | 1.89 | 50 | 230 | 0.968 | 0.018 |
| 10PB10381 | 93.91 | 1.878 | 0.958 | 1.96 | 1.95 | 50 | 230 | 0.966 | 0.963 |
| 09PB1757 | 107.61 | 2.152 | 1.135 | 1.9 | 1.74 | 50 | 230 | 1.238 | 0.009 |
| 09PB1757 | 122.51 | 2.45 | 1.279 | 1.92 | 1.82 | 50 | 230 | 1.35 | 0.006 |
| 09PB1757 | 118.49 | 2.37 | 1.248 | 1.9 | 1.83 | 50 | 230 | 1.292 | 0.026 |
| 10PB0488 | 117.75 | 2.355 | 1.213 | 1.94 | 2.08 | 50 | 230 | 1.13 | 0.01 |
| 10PB0488 | 116.92 | 2.338 | 1.196 | 1.96 | 2.12 | 50 | 230 | 1.101 | 0.043 |
| 10PB0488 | 117.08 | 2.342 | 1.207 | 1.94 | 2.09 | 50 | 230 | 1.118 | 0.005 |
| 11PB1958 | 128.52 | 2.57 | 1.317 | 1.95 | 2.06 | 50 | 230 | 1.248 | −0.184 |
| 11PB1958 | 131.26 | 2.625 | 1.362 | 1.93 | 2.1 | 50 | 230 | 1.252 | 0.03 |
| 11PB1958 | 132.73 | 2.655 | 1.381 | 1.92 | 2.04 | 50 | 230 | 1.304 | 0.021 |
| 09PB2752 | 120.89 | 2.418 | 1.251 | 1.93 | 1.94 | 50 | 230 | 1.244 | 0.238 |
| 09PB2752 | 112.77 | 2.255 | 1.177 | 1.92 | 1.93 | 50 | 230 | 1.17 | 0.019 |
| 09PB2752 | 119.5 | 2.39 | 1.24 | 1.93 | 1.79 | 50 | 230 | 1.334 | 0.026 |
| 10PB1329 | 135.09 | 2.702 | 1.416 | 1.91 | 2 | 50 | 230 | 1.348 | 0.021 |
| 10PB1329 | 133.02 | 2.66 | 1.372 | 1.94 | 2.08 | 50 | 230 | 1.281 | 0.017 |
| 10PB1329 | 133.4 | 2.668 | 1.378 | 1.94 | 2.11 | 50 | 230 | 1.263 | 0.02 |
| 08PB4270 | 136.03 | 2.721 | 1.423 | 1.91 | 2.03 | 50 | 230 | 1.34 | 0.02 |
| 08PB4270 | 133.77 | 2.675 | 1.374 | 1.95 | 2.09 | 50 | 230 | 1.28 | 0.03 |
| 08PB4270 | 122.93 | 2.459 | 1.269 | 1.94 | 1.98 | 50 | 230 | 1.242 | 0.176 |
| 09PB6550 | 164.06 | 3.281 | 1.706 | 1.92 | 2.07 | 50 | 230 | 1.582 | 0.029 |
| 09PB6550 | 162.45 | 3.249 | 1.681 | 1.93 | 2.11 | 50 | 230 | 1.54 | 0.034 |
| 09PB6550 | 159.09 | 3.182 | 1.647 | 1.93 | 2.11 | 50 | 230 | 1.507 | 0.048 |
| 11PB5972 | 201.88 | 4.038 | 2.093 | 1.93 | 1.72 | 50 | 230 | 2.349 | 0.037 |
| 11PB5972 | 204.9 | 4.098 | 2.161 | 1.9 | 1.72 | 50 | 230 | 2.388 | 0.041 |
| 11PB5972 | 207.18 | 4.144 | 2.139 | 1.94 | 1.71 | 50 | 230 | 2.42 | 0.042 |
| 11PB5602 | 255.45 | 5.109 | 2.659 | 1.92 | 2.25 | 50 | 230 | 2.269 | 0.041 |
| 11PB5602 | 250.73 | 5.015 | 2.634 | 1.9 | 2.22 | 50 | 230 | 2.261 | −0.056 |
| 11PB5602 | 244.12 | 4.882 | 2.569 | 1.9 | 2.11 | 50 | 230 | 2.315 | 0.053 |
| 10PB2885 | 107.15 | 2.143 | 1.142 | 1.88 | 1.99 | 50 | 230 | 1.079 | 0.044 |
| 10PB2885 | 103.55 | 2.071 | 1.066 | 1.94 | 1.96 | 50 | 230 | 1.058 | 0.057 |
| 10PB2885 | 104.68 | 2.094 | 1.076 | 1.94 | 1.96 | 50 | 230 | 1.066 | 0.051 |
| 08PB5124 | 125.87 | 2.517 | 1.295 | 1.94 | 1.79 | 50 | 230 | 1.404 | 0.057 |
| 08PB5124 | 120.39 | 2.408 | 1.26 | 1.91 | 1.76 | 50 | 230 | 1.364 | 0.054 |
| 08PB5124 | 121.32 | 2.426 | 1.272 | 1.91 | 1.68 | 50 | 230 | 1.448 | 0.051 |
| 08PB7544 | 220.26 | 4.405 | 2.313 | 1.9 | 1.94 | 50 | 230 | 2.269 | 0.055 |
| 08PB7544 | 221.37 | 4.427 | 2.326 | 1.9 | 2.07 | 50 | 230 | 2.137 | 0.061 |
| 08PB7544 | 201.62 | 4.032 | 2.132 | 1.89 | 1.97 | 50 | 230 | 2.042 | 0.061 |
| 09PB7511 | 145.22 | 2.904 | 1.494 | 1.94 | 2.08 | 50 | 230 | 1.398 | 0.017 |
| 09PB7511 | 146.83 | 2.937 | 1.534 | 1.91 | 2.14 | 50 | 230 | 1.373 | 0.018 |
| 09PB7511 | 149.23 | 2.985 | 1.549 | 1.93 | 2.09 | 50 | 230 | 1.425 | 0.041 |
| 08PB8771 | 129.23 | 2.585 | 1.316 | 1.96 | 2.16 | 50 | 230 | 1.198 | 0.021 |
| 08PB8771 | 128.1 | 2.562 | 1.325 | 1.93 | 2.15 | 50 | 230 | 1.193 | 0.021 |
| 08PB8771 | 146.59 | 2.932 | 1.55 | 1.89 | 2.04 | 50 | 230 | 1.439 | 0.085 |
| 08PB0722 | 129.23 | 2.585 | 1.353 | 1.91 | 2.07 | 50 | 230 | 1.247 | 0.011 |
| 08PB0722 | 126.97 | 2.539 | 1.323 | 1.92 | 2.06 | 50 | 230 | 1.232 | 0.009 |
| 08PB0722 | 121.42 | 2.428 | 1.244 | 1.95 | 1.99 | 50 | 230 | 1.22 | 0.016 |
| 10PB6266 | 108.39 | 2.168 | 1.112 | 1.95 | 1.54 | 50 | 230 | 1.403 | 0.004 |
| 10PB6266 | 106.26 | 2.125 | 1.088 | 1.95 | 1.5 | 50 | 230 | 1.417 | 0.007 |
| 10PB6266 | 112.6 | 2.252 | 1.145 | 1.97 | 1.6 | 50 | 230 | 1.405 | 0.019 |
| 08PB6113 | 166.43 | 3.329 | 1.737 | 1.92 | 2.09 | 50 | 230 | 1.594 | −0.026 |
| 08PB6113 | 149.03 | 2.981 | 1.554 | 1.92 | 1.99 | 50 | 230 | 1.497 | 0.011 |
| 08PB6113 | 189.34 | 3.787 | 1.963 | 1.93 | 2.15 | 50 | 230 | 1.762 | 0.003 |
| 08PB8031 | 137.03 | 2.741 | 1.4 | 1.96 | 1.99 | 50 | 230 | 1.379 | 0.002 |
| 08PB8031 | 141.31 | 2.826 | 1.453 | 1.95 | 1.98 | 50 | 230 | 1.429 | 0.008 |
| 08PB8031 | 137.14 | 2.743 | 1.418 | 1.93 | 2.02 | 50 | 230 | 1.358 | 0.216 |
| 09PB5837 | 515.13 | 10.303 | 5.412 | 1.9 | 2.22 | 50 | 230 | 4.64 | −0.007 |
| 09PB5837 | 562.42 | 11.248 | 5.943 | 1.89 | 2.22 | 50 | 230 | 5.074 | 0.004 |
| 09PB5837 | 560.8 | 11.216 | 5.987 | 1.87 | 2.18 | 50 | 230 | 5.139 | −0.004 |
| 09PB4164 | 109.06 | 2.181 | 1.147 | 1.9 | 2.14 | 50 | 230 | 1.021 | −0.007 |
| 09PB4164 | 112.32 | 2.246 | 1.154 | 1.95 | 2.16 | 50 | 230 | 1.04 | −0.002 |
| 09PB4164 | 117.47 | 2.349 | 1.216 | 1.93 | 2.11 | 50 | 230 | 1.115 | −0.002 |
| 10PB0279 | 152.93 | 3.059 | 1.604 | 1.91 | 2.1 | 50 | 230 | 1.456 | 0.004 |
| 10PB0279 | 163.34 | 3.267 | 1.69 | 1.93 | 2.13 | 50 | 230 | 1.532 | 0.004 |
| 10PB0279 | 144.31 | 2.886 | 1.487 | 1.94 | 2.12 | 50 | 230 | 1.365 | −0.009 |
| 08PB5904 | 171.4 | 3.428 | 1.795 | 1.91 | 2.09 | 50 | 230 | 1.643 | −0.03 |
| 08PB5904 | 181.86 | 3.637 | 1.921 | 1.89 | 2.09 | 50 | 230 | 1.739 | 0.004 |
| 08PB5904 | 180.91 | 3.618 | 1.875 | 1.93 | 2.11 | 50 | 230 | 1.718 | −0.002 |
| 08PB3517 | 181.36 | 3.627 | 1.89 | 1.92 | 2.18 | 50 | 230 | 1.661 | −0.002 |
| 08PB3517 | 163.03 | 3.261 | 1.682 | 1.94 | 2.16 | 50 | 230 | 1.506 | 0.001 |
| 08PB3517 | 171.92 | 3.438 | 1.775 | 1.94 | 2.18 | 50 | 230 | 1.579 | −0.003 |
| 12PB0638 | 182 | 3.64 | 1.892 | 1.92 | 2.02 | 50 | 230 | 1.804 | −0.012 |
| 12PB0638 | 139.74 | 2.795 | 1.445 | 1.93 | 1.94 | 50 | 230 | 1.439 | −0.004 |
| 12PB0638 | 150.93 | 3.019 | 1.571 | 1.92 | 1.96 | 50 | 230 | 1.54 | 0.008 |
| 10PB2738 | 252.44 | 5.049 | 2.66 | 1.9 | 2.14 | 50 | 230 | 2.356 | 0.079 |
| 10PB2738 | 243.53 | 4.871 | 2.584 | 1.88 | 2.05 | 50 | 230 | 2.375 | 0.021 |
| 10PB2738 | 251.59 | 5.032 | 2.645 | 1.9 | 2.21 | 50 | 230 | 2.28 | 0.011 |
| 10PB2975 | 105.21 | 2.104 | 1.106 | 1.9 | 1.29 | 50 | 230 | 1.637 | 0.011 |
| 10PB2975 | 91.55 | 1.831 | 0.951 | 1.93 | 1.9 | 50 | 230 | 0.962 | −0.018 |
| 10PB2975 | 97.22 | 1.944 | 1.009 | 1.93 | 1.98 | 50 | 230 | 0.984 | 0.035 |
| 11PB13477 | 231.6 | 4.632 | 2.419 | 1.92 | 2.21 | 50 | 230 | 2.096 | 0.018 |
| 11PB13477 | 239.65 | 4.793 | 2.506 | 1.91 | 2.15 | 50 | 230 | 2.234 | 0.011 |
| 11PB13477 | 226.53 | 4.531 | 2.348 | 1.93 | 2.22 | 50 | 230 | 2.044 | 0.064 |
| 07PB1887 | 198.82 | 3.976 | 2.109 | 1.89 | 2.12 | 50 | 230 | 1.88 | 0.013 |
| 07PB1887 | 187.69 | 3.754 | 1.959 | 1.92 | 2.16 | 50 | 230 | 1.739 | −0.004 |
| 07PB1887 | 171.17 | 3.423 | 1.788 | 1.92 | 2.17 | 50 | 230 | 1.576 | −0.003 |
| 11PB6931 | 157.53 | 3.151 | 1.625 | 1.94 | 2.28 | 50 | 230 | 1.384 | 0 |
| 11PB6931 | 156.69 | 3.134 | 1.632 | 1.92 | 2.2 | 50 | 230 | 1.425 | 0.019 |
| 11PB6931 | 154.11 | 3.082 | 1.641 | 1.88 | 2.25 | 50 | 230 | 1.368 | 0.016 |
| 07PB2880 | 200.73 | 4.015 | 2.11 | 1.9 | 2.14 | 50 | 230 | 1.877 | 0.007 |
| 07PB2880 | 198.35 | 3.967 | 2.036 | 1.95 | 2.19 | 50 | 230 | 1.811 | 0.004 |
| 07PB2880 | 202.41 | 4.048 | 2.119 | 1.91 | 2.18 | 50 | 230 | 1.858 | 0.013 |
| 10PB6041 | 171.75 | 3.435 | 1.808 | 1.9 | 2.11 | 50 | 230 | 1.629 | 0.009 |
| 10PB6041 | 190.53 | 3.811 | 2.015 | 1.89 | 2.15 | 50 | 230 | 1.774 | 0.004 |
| 10PB6041 | 174.07 | 3.481 | 1.826 | 1.91 | 2.1 | 50 | 230 | 1.661 | −0.008 |
| 09PB0698 | 174.57 | 3.491 | 1.825 | 1.91 | 2.21 | 50 | 230 | 1.582 | −0.006 |
| 09PB0698 | 139.6 | 2.792 | 1.451 | 1.92 | 2.23 | 50 | 230 | 1.252 | −0.018 |
| 09PB0698 | 156.91 | 3.138 | 1.623 | 1.93 | 2.24 | 50 | 230 | 1.4 | 0.025 |
| 07PB6512 | 175.31 | 3.506 | 1.818 | 1.93 | 2.11 | 50 | 230 | 1.661 | 0.001 |
| 07PB6512 | 192.66 | 3.853 | 2.02 | 1.91 | 2.13 | 50 | 230 | 1.812 | −0.015 |
| 07PB6512 | 174.94 | 3.499 | 1.837 | 1.9 | 2.12 | 50 | 230 | 1.654 | −0.008 |
| 12PB8783 | 156.29 | 3.126 | 1.611 | 1.94 | 1.68 | 50 | 230 | 1.856 | −0.013 |
| 12PB8783 | 153.44 | 3.069 | 1.593 | 1.93 | 1.69 | 50 | 230 | 1.82 | 0.004 |
| 12PB8783 | 153.83 | 3.077 | 1.588 | 1.94 | 1.67 | 50 | 230 | 1.844 | 0.002 |
| 13PB11267 | 241.17 | 4.823 | 2.508 | 1.92 | 2.21 | 50 | 230 | 2.18 | −0.014 |
| 13PB11267 | 247.17 | 4.943 | 2.593 | 1.91 | 2.21 | 50 | 230 | 2.242 | −0.006 |
| 13PB11267 | 240.73 | 4.815 | 2.502 | 1.92 | 2.2 | 50 | 230 | 2.186 | −0.01 |
| 11PB0842 | 188.39 | 3.768 | 1.949 | 1.93 | 2.18 | 50 | 230 | 1.729 | 0.011 |
| 11PB0842 | 179.38 | 3.588 | 1.864 | 1.92 | 2.13 | 50 | 230 | 1.686 | 0.006 |
| 11PB0842 | 192 | 3.84 | 1.999 | 1.92 | 2.16 | 50 | 230 | 1.775 | 0.006 |
| 12PB7846 | 157.68 | 3.154 | 1.636 | 1.93 | 2.06 | 50 | 230 | 1.532 | −0.038 |
| 12PB7846 | 168.2 | 3.364 | 1.765 | 1.91 | 2.09 | 50 | 230 | 1.613 | −0.03 |
| 12PB7846 | 154.06 | 3.081 | 1.593 | 1.93 | 2.07 | 50 | 230 | 1.488 | −0.032 |
| 08PB1543 | 84.01 | 1.68 | 0.863 | 1.95 | 2 | 50 | 230 | 0.839 | −0.001 |
| 08PB1543 | 84.08 | 1.682 | 0.903 | 1.86 | 1.93 | 50 | 230 | 0.873 | 0.006 |
| 08PB1543 | 83.55 | 1.671 | 0.859 | 1.94 | 1.97 | 50 | 230 | 0.849 | 0.022 |
| 08PB2670 | 104.94 | 2.099 | 1.093 | 1.92 | 1.72 | 50 | 230 | 1.219 | −0.027 |
| 08PB2670 | 101.87 | 2.037 | 1.047 | 1.95 | 1.79 | 50 | 230 | 1.14 | −0.015 |
| 08PB2670 | 108.54 | 2.171 | 1.146 | 1.89 | 1.74 | 50 | 230 | 1.245 | 0.361 |
Quality control of bisulfite conversion and Infinium 450k data
Quality control of bisulfite conversion and of data obtained by the Illumina Infinium HumanMethylation450 BeadChips was performed independently by the team of the core facility using the Methylation Module of Illumina’s GenomeStudio software (Supplementary File 1 and Online-only Table 3) and by us using the rnb.run.qc command of the RnBeads package (Fig. 2). Both analyses ascertained the correct execution of the separate steps of the whole experimental procedure: bisulfite conversion, hybridization, single-base extension and stripping. Figure 2a,b demonstrate bisulfite conversion efficiency as reported by control probes of Infinium I or II design, respectively. Overall hybridization performance was assessed using synthetic reference targets that are present in the hybridization buffer at three concentrations (low, medium and high) and that resulted in signals with well separable intensity intervals, as expected (Fig. 2c). The extension controls showed high efficiency of extension with any of the 4 nucleotides (Fig. 2d) and the staining controls demonstrated high efficiency and sensitivity of the staining step (Fig. 2e). The overall performance of the assay from amplification to detection is summarized by the signal from probes that query non-polymorphic bases in the genome – one probe for each nucleotide (Fig. 2f).
Online-only Table 3.
Quality control of signals from the Illumina Infinium HumanMethylation450 BeadChips.
| Index | Sample ID | Sentrix Barcode | Sample Section | Detected CpG (0.01) | DetectRate (0.01) | Loci NOT detected (0.01) | Detected CpG (0.05) | Signal Average GRN | Signal Average RED | Signal P05 GRN | Signal P05 RED | Signal P25 GRN | Signal P25 RED | Signal P50 GRN | Signal P50 RED | Signal P75 GRN | Signal P75 RED | Signal P95 GRN | Signal P95 RED | DNA concentration (ng/µl) | DNA volume (µl) | DNA quantity loaded (ng) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 09PB8587 | 9422491005 | R01C01 | 485085 | 99.90% | 492 | 485165 | 3651 | 8821 | 211 | 411 | 468 | 1308 | 1234 | 4781 | 6569 | 15293 | 11915 | 26452 | 29.1 | 34.3 | 1000 |
| 2 | 10PB0115 | 9422491005 | R01C02 | 485131 | 99.91% | 446 | 485203 | 3729 | 8353 | 239 | 476 | 504 | 1355 | 1371 | 4890 | 6632 | 14167 | 12040 | 24642 | 35.1 | 28.5 | 1000 |
| 3 | 11PB5152 | 9422491005 | R02C01 | 485445 | 99.97% | 132 | 485494 | 3957 | 8787 | 248 | 487 | 544 | 1407 | 1495 | 4956 | 6994 | 14999 | 12769 | 26193 | 31.7 | 31.6 | 1000 |
| 4 | 10PB2807 | 9422491005 | R02C02 | 485141 | 99.91% | 436 | 485213 | 4030 | 8682 | 265 | 519 | 551 | 1414 | 1418 | 4976 | 7249 | 14806 | 13005 | 25570 | 39.3 | 25.4 | 1000 |
| 5 | 09PB7427 | 9422491005 | R03C01 | 485435 | 99.97% | 142 | 485480 | 4209 | 9271 | 265 | 524 | 578 | 1462 | 1549 | 5041 | 7582 | 15980 | 13405 | 27712 | 37.6 | 26.6 | 1000 |
| 6 | 09PB0906 | 9422491005 | R03C02 | 485418 | 99.97% | 159 | 485480 | 4418 | 8669 | 263 | 504 | 567 | 1331 | 1551 | 4307 | 8127 | 15071 | 13857 | 26491 | 38 | 26.3 | 1000 |
| 7 | 08PB4647 | 9422491005 | R04C01 | 485267 | 99.94% | 310 | 485347 | 4502 | 9217 | 285 | 579 | 610 | 1433 | 1608 | 4310 | 8235 | 16153 | 14142 | 28523 | 41.7 | 24 | 1000 |
| 8 | 09PB0626 | 9422491005 | R04C02 | 485414 | 99.97% | 163 | 485473 | 4655 | 9046 | 294 | 604 | 626 | 1461 | 1820 | 4346 | 8487 | 15754 | 14258 | 27851 | 32.2 | 31 | 1000 |
| 9 | 08PB0411 | 9422491005 | R05C01 | 485084 | 99.90% | 493 | 485181 | 4437 | 9338 | 275 | 549 | 605 | 1459 | 1661 | 4635 | 8053 | 16226 | 13947 | 28595 | 39.6 | 25.3 | 1000 |
| 10 | 10PB0381 | 9422491005 | R05C02 | 485102 | 99.90% | 475 | 485175 | 4362 | 9486 | 288 | 569 | 610 | 1597 | 1620 | 5574 | 7800 | 16175 | 13948 | 27647 | 46.4 | 21.6 | 1000 |
| 11 | 09PB6227 | 9422491005 | R06C01 | 485382 | 99.96% | 195 | 485425 | 4410 | 9549 | 281 | 553 | 589 | 1482 | 1604 | 4927 | 7906 | 16364 | 14151 | 29340 | 42 | 23.8 | 1000 |
| 12 | 09PB1646 | 9422491005 | R06C02 | 485366 | 99.96% | 211 | 485428 | 4407 | 9354 | 289 | 562 | 594 | 1427 | 1541 | 4707 | 7874 | 16145 | 14491 | 28790 | 53.1 | 18.8 | 1000 |
| 13 | 10PB6173 | 9422491007 | R01C01 | 485081 | 99.90% | 496 | 485198 | 4169 | 9048 | 276 | 577 | 564 | 1525 | 1434 | 5155 | 7476 | 15427 | 13526 | 26653 | 37.2 | 26.9 | 1000 |
| 14 | 09PB7511 | 9422491007 | R01C02 | 485401 | 99.96% | 176 | 485457 | 4103 | 8717 | 270 | 600 | 549 | 1400 | 1516 | 4274 | 7427 | 15087 | 12850 | 26602 | 38.4 | 26 | 1000 |
| 15 | 08PB4270 | 9422491007 | R02C01 | 485133 | 99.91% | 444 | 485222 | 4416 | 8742 | 254 | 523 | 553 | 1277 | 1545 | 3909 | 8126 | 15388 | 13836 | 27270 | 35.8 | 27.9 | 1000 |
| 16 | 10PB2738 | 9422491007 | R02C02 | 485447 | 99.97% | 130 | 485484 | 4170 | 8703 | 244 | 500 | 524 | 1274 | 1418 | 4146 | 7634 | 15196 | 13255 | 26902 | 39.8 | 25.1 | 1000 |
| 17 | 09PB6550 | 9422491007 | R03C01 | 485121 | 99.91% | 456 | 485190 | 4487 | 9178 | 252 | 491 | 567 | 1402 | 1645 | 4942 | 8185 | 15855 | 14144 | 27522 | 37.2 | 26.9 | 1000 |
| 18 | 11PB13477 | 9422491007 | R03C02 | 485445 | 99.97% | 132 | 485482 | 4493 | 9192 | 254 | 489 | 563 | 1372 | 1585 | 4874 | 8242 | 15934 | 14150 | 27585 | 40.4 | 24.8 | 1000 |
| 19 | 11PB5972 | 9422491007 | R04C01 | 485271 | 99.94% | 306 | 485370 | 4624 | 9269 | 245 | 460 | 571 | 1359 | 1611 | 4755 | 8513 | 16163 | 14666 | 28159 | 40 | 25 | 1000 |
| 20 | 12PB0638 | 9422491007 | R04C02 | 485429 | 99.97% | 148 | 485475 | 4686 | 9088 | 248 | 465 | 577 | 1313 | 1808 | 4322 | 8591 | 15933 | 14474 | 28215 | 36.4 | 27.4 | 1000 |
| 21 | 09PB2752 | 9422491007 | R05C01 | 485385 | 99.96% | 192 | 485447 | 4260 | 9676 | 245 | 452 | 545 | 1471 | 1365 | 5767 | 7630 | 16552 | 14303 | 28418 | 45.4 | 22 | 1000 |
| 22 | 10PB5506 | 9422491007 | R05C02 | 485065 | 99.89% | 512 | 485147 | 4459 | 9472 | 259 | 478 | 568 | 1441 | 1486 | 5264 | 8187 | 16322 | 14343 | 28140 | 40.9 | 24.4 | 1000 |
| 23 | 10PB2885 | 9422491007 | R06C01 | 485093 | 99.90% | 484 | 485164 | 4520 | 9316 | 254 | 498 | 560 | 1371 | 1642 | 4558 | 8242 | 16026 | 14329 | 29148 | 41.3 | 24.2 | 1000 |
| 24 | 12PB8783 | 9422491007 | R06C02 | 485129 | 99.91% | 448 | 485196 | 4712 | 9357 | 269 | 514 | 592 | 1406 | 1796 | 4578 | 8646 | 16181 | 14538 | 29097 | 41 | 24.4 | 1000 |
| 25 | 09PB0858 | 9422491020 | R01C01 | 485018 | 99.88% | 559 | 485139 | 4272 | 8893 | 253 | 492 | 569 | 1451 | 1563 | 4823 | 7669 | 15287 | 13647 | 26617 | 25.8 | 37 | 953 |
| 26 | 09PB6581 | 9422491020 | R01C02 | 485384 | 99.96% | 193 | 485475 | 4184 | 8923 | 286 | 608 | 591 | 1511 | 1627 | 4542 | 7545 | 15345 | 12978 | 27054 | 36.6 | 27.4 | 1000 |
| 27 | 09PB4164 | 9422491020 | R02C01 | 485391 | 99.96% | 186 | 485452 | 4513 | 9004 | 282 | 548 | 614 | 1455 | 1640 | 4721 | 8206 | 15598 | 14233 | 27006 | 38.5 | 26 | 1000 |
| 28 | 11PB1528 | 9422491020 | R02C02 | 485392 | 99.96% | 185 | 485441 | 4659 | 9155 | 325 | 664 | 679 | 1684 | 1939 | 5263 | 8352 | 15416 | 14308 | 26900 | 30.7 | 32.6 | 1000 |
| 29 | 10PB0279 | 9422491020 | R03C01 | 485433 | 99.97% | 144 | 485494 | 4537 | 9291 | 278 | 519 | 614 | 1470 | 1639 | 4958 | 8257 | 16096 | 14340 | 27738 | 35.1 | 28.5 | 1000 |
| 30 | 11PB2066 | 9422491020 | R03C02 | 485413 | 99.97% | 164 | 485448 | 4706 | 9292 | 309 | 616 | 647 | 1518 | 1716 | 4654 | 8620 | 16096 | 14600 | 28190 | 40.8 | 24.5 | 1000 |
| 31 | 08PB8276 | 9422491020 | R04C01 | 485409 | 99.97% | 168 | 485471 | 4648 | 9277 | 275 | 507 | 618 | 1445 | 1748 | 4706 | 8502 | 16177 | 14433 | 28127 | 34.6 | 28.9 | 1000 |
| 32 | 09PB0700 | 9422491020 | R04C02 | 485424 | 99.97% | 153 | 485490 | 4720 | 9423 | 297 | 580 | 644 | 1473 | 1719 | 4545 | 8669 | 16485 | 14694 | 28790 | 35.5 | 28.2 | 1000 |
| 33 | 10PB3262 | 9422491020 | R05C01 | 485444 | 99.97% | 133 | 485488 | 4615 | 9232 | 289 | 547 | 638 | 1492 | 1745 | 4801 | 8327 | 15984 | 14533 | 27880 | 31.2 | 32.1 | 1000 |
| 34 | 10PB10381 | 9422491020 | R05C02 | 485446 | 99.97% | 131 | 485488 | 5023 | 9383 | 293 | 585 | 684 | 1620 | 2617 | 4853 | 9064 | 15967 | 14648 | 28812 | 37.5 | 26.7 | 1000 |
| 35 | 10PB0918 | 9422491020 | R06C01 | 485257 | 99.93% | 320 | 485348 | 4475 | 10116 | 276 | 605 | 611 | 1577 | 1647 | 4880 | 8149 | 17638 | 14105 | 31126 | 41.2 | 24.3 | 1000 |
| 36 | 08PB3739 | 9422491020 | R06C02 | 485239 | 99.93% | 338 | 485319 | 4613 | 10049 | 301 | 647 | 627 | 1595 | 1617 | 4957 | 8447 | 17388 | 14584 | 30731 | 31.5 | 31.7 | 1000 |
| 37 | 08PB8771 | 9422491034 | R01C01 | 485386 | 99.96% | 191 | 485453 | 3758 | 7873 | 255 | 509 | 528 | 1333 | 1372 | 4074 | 6866 | 13575 | 11652 | 23793 | 36 | 27.8 | 1000 |
| 38 | 08PB0722 | 9422491034 | R01C02 | 485361 | 99.96% | 216 | 485437 | 3801 | 7933 | 269 | 543 | 541 | 1357 | 1417 | 4319 | 6913 | 13526 | 11730 | 23654 | 34.8 | 28.8 | 1000 |
| 39 | 10PB3589 | 9422491034 | R02C01 | 485209 | 99.92% | 368 | 485286 | 4115 | 8595 | 304 | 606 | 611 | 1487 | 1550 | 4665 | 7498 | 14742 | 12619 | 25440 | 39.8 | 25.1 | 1000 |
| 40 | 11PB5602 | 9422491034 | R02C02 | 484874 | 99.86% | 703 | 485029 | 4161 | 8577 | 316 | 643 | 638 | 1522 | 1758 | 4559 | 7531 | 14611 | 12501 | 25709 | 29.6 | 33.8 | 1000 |
| 41 | 10PB1144 | 9422491034 | R03C01 | 485424 | 99.97% | 153 | 485472 | 4185 | 8568 | 292 | 604 | 604 | 1493 | 1646 | 4622 | 7619 | 14666 | 12746 | 25441 | 40.2 | 24.9 | 1000 |
| 42 | 07PB2880 | 9422491034 | R03C02 | 485306 | 99.94% | 271 | 485380 | 4248 | 8646 | 298 | 609 | 612 | 1425 | 1585 | 4136 | 7802 | 15084 | 12997 | 26260 | 36 | 27.8 | 1000 |
| 43 | 08PB6113 | 9422491034 | R04C01 | 485165 | 99.92% | 412 | 485224 | 4396 | 8633 | 282 | 597 | 636 | 1532 | 2331 | 4566 | 7855 | 14713 | 12801 | 26047 | 41.2 | 24.3 | 1000 |
| 44 | 08PB8031 | 9422491034 | R04C02 | 485024 | 99.89% | 553 | 485111 | 4244 | 9164 | 304 | 628 | 623 | 1563 | 1559 | 4902 | 7748 | 15745 | 13186 | 27171 | 40 | 25 | 1000 |
| 45 | 08PB5124 | 9422491034 | R05C01 | 485023 | 99.89% | 554 | 485113 | 4302 | 9383 | 314 | 673 | 643 | 1682 | 1621 | 5324 | 7816 | 16015 | 13332 | 27359 | 38.3 | 26.1 | 1000 |
| 46 | 10PB8858 | 9422491034 | R05C02 | 484942 | 99.87% | 635 | 485047 | 4480 | 9434 | 312 | 641 | 651 | 1589 | 1715 | 4925 | 8146 | 16221 | 13748 | 28290 | 25.7 | 37 | 952 |
| 47 | 11PB6931 | 9422491034 | R06C01 | 485002 | 99.88% | 575 | 485084 | 4388 | 8936 | 295 | 611 | 625 | 1507 | 1749 | 4663 | 7931 | 15268 | 13493 | 27067 | 39.3 | 25.5 | 1000 |
| 48 | 07PB1887 | 9422491034 | R06C02 | 485257 | 99.93% | 320 | 485386 | 4430 | 8773 | 312 | 644 | 624 | 1457 | 1597 | 4110 | 8133 | 15201 | 13742 | 27127 | 28.6 | 34.9 | 1000 |
| 49 | 09PB7242 | 9422491041 | R01C01 | 485475 | 99.98% | 102 | 485515 | 4030 | 8303 | 253 | 505 | 541 | 1328 | 1472 | 4189 | 7230 | 14294 | 12917 | 25500 | 49.4 | 20.2 | 1000 |
| 50 | 08PB1082 | 9422491041 | R01C02 | 485440 | 99.97% | 137 | 485500 | 3896 | 8119 | 243 | 499 | 516 | 1203 | 1404 | 3523 | 6986 | 14179 | 12587 | 26004 | 65.5 | 15.3 | 1000 |
| 51 | 13PB11267 | 9422491041 | R02C01 | 485461 | 99.98% | 116 | 485508 | 4358 | 8579 | 286 | 591 | 601 | 1309 | 1674 | 3600 | 7836 | 14966 | 13797 | 27693 | 96.2 | 10.4 | 1000 |
| 52 | 10PB0488 | 9422491041 | R02C02 | 485393 | 99.96% | 184 | 485440 | 4063 | 8812 | 278 | 607 | 572 | 1390 | 1506 | 4066 | 7269 | 15170 | 12999 | 27791 | 79.5 | 12.6 | 1000 |
| 53 | 08PB1543 | 9422491041 | R03C01 | 485184 | 99.92% | 393 | 485277 | 4430 | 9034 | 289 | 568 | 606 | 1466 | 1603 | 4817 | 7994 | 15546 | 14115 | 27126 | 43.2 | 23.1 | 1000 |
| 54 | 10PB1329 | 9422491041 | R03C02 | 485435 | 99.97% | 142 | 485494 | 4350 | 9460 | 297 | 654 | 628 | 1635 | 1822 | 5190 | 7772 | 16154 | 13517 | 28100 | 42.5 | 23.5 | 1000 |
| 55 | 09PB2750 | 9422491041 | R04C01 | 485507 | 99.99% | 70 | 485529 | 4335 | 8977 | 265 | 503 | 579 | 1381 | 1569 | 4791 | 7802 | 15540 | 13885 | 27005 | 66.3 | 15.1 | 1000 |
| 56 | 08PB2670 | 9422491041 | R04C02 | 485179 | 99.92% | 398 | 485260 | 4422 | 8870 | 272 | 549 | 588 | 1326 | 1714 | 3944 | 8009 | 15522 | 13844 | 28010 | 71.5 | 14 | 1000 |
| 57 | 10PB2975 | 9422491041 | R05C01 | 485372 | 99.96% | 205 | 485425 | 4494 | 9474 | 307 | 631 | 625 | 1523 | 1591 | 4834 | 8146 | 16368 | 14238 | 28682 | 44.2 | 22.6 | 1000 |
| 58 | 10PB8700 | 9422491041 | R05C02 | 485307 | 99.94% | 270 | 485386 | 4312 | 9582 | 300 | 636 | 603 | 1563 | 1487 | 5059 | 7783 | 16472 | 13897 | 28798 | 43.1 | 23.2 | 1000 |
| 59 | 11PB11934 | 9422491041 | R06C01 | 485104 | 99.90% | 473 | 485200 | 4608 | 9536 | 299 | 610 | 642 | 1561 | 1874 | 4846 | 8241 | 16270 | 14388 | 29437 | 64.4 | 15.5 | 1000 |
| 60 | 09PB1757 | 9422491041 | R06C02 | 485400 | 99.96% | 177 | 485459 | 4431 | 9729 | 306 | 659 | 624 | 1553 | 1613 | 4741 | 8004 | 16875 | 14067 | 29859 | 53.4 | 18.7 | 1000 |
| 61 | 09PB5837 | 9422491055 | R01C01 | 485459 | 99.98% | 118 | 485502 | 3217 | 7693 | 204 | 424 | 445 | 1208 | 1194 | 3928 | 5629 | 13163 | 10600 | 23798 | 88.3 | 11.3 | 1000 |
| 62 | 08PB3517 | 9422491055 | R01C02 | 485439 | 99.97% | 138 | 485489 | 3929 | 8063 | 258 | 525 | 533 | 1302 | 1388 | 3914 | 7164 | 14029 | 12406 | 24669 | 46.1 | 21.7 | 1000 |
| 63 | 11PB1958 | 9422491055 | R02C01 | 485462 | 99.98% | 115 | 485507 | 4087 | 8327 | 270 | 518 | 571 | 1360 | 1529 | 4477 | 7245 | 14202 | 13178 | 25164 | 58.5 | 17.1 | 1000 |
| 64 | 11PB0842 | 9422491055 | R02C02 | 485106 | 99.90% | 471 | 485194 | 4338 | 8399 | 277 | 551 | 589 | 1367 | 1715 | 4122 | 7854 | 14462 | 13482 | 25922 | 56.5 | 17.7 | 1000 |
| 65 | 10PB5849 | 9422491055 | R03C01 | 485223 | 99.93% | 354 | 485289 | 4128 | 8836 | 260 | 475 | 558 | 1395 | 1404 | 5096 | 7368 | 15149 | 13647 | 25970 | 51.9 | 19.3 | 1000 |
| 66 | 10PB6041 | 9422491055 | R03C02 | 485437 | 99.97% | 140 | 485494 | 4513 | 8485 | 265 | 509 | 581 | 1273 | 1584 | 3899 | 8305 | 14945 | 14174 | 26289 | 45 | 22.2 | 1000 |
| 67 | 08PB5904 | 9422491055 | R04C01 | 485423 | 99.97% | 154 | 485482 | 4330 | 8915 | 275 | 544 | 594 | 1372 | 1587 | 4219 | 7778 | 15481 | 13952 | 27851 | 89.1 | 11.2 | 1000 |
| 68 | 10PB6266 | 9422491055 | R04C02 | 485433 | 99.97% | 144 | 485477 | 4590 | 8741 | 274 | 523 | 590 | 1309 | 1560 | 4033 | 8490 | 15369 | 14462 | 27039 | 45.3 | 22.1 | 1000 |
| 69 | 08PB7544 | 9422491055 | R05C01 | 485395 | 99.96% | 182 | 485454 | 4255 | 8991 | 278 | 580 | 602 | 1429 | 1638 | 4295 | 7594 | 15540 | 13576 | 28130 | 64.5 | 15.5 | 1000 |
| 70 | 07PB6512 | 9422491055 | R05C02 | 485423 | 99.97% | 154 | 485487 | 4583 | 9045 | 278 | 553 | 614 | 1441 | 1680 | 4550 | 8299 | 15701 | 14566 | 27590 | 77.3 | 12.9 | 1000 |
| 71 | 09PB0698 | 9422491055 | R06C01 | 485074 | 99.90% | 503 | 485153 | 4220 | 9005 | 276 | 579 | 580 | 1397 | 1635 | 4270 | 7567 | 15469 | 13418 | 28391 | 76 | 13.2 | 1000 |
| 72 | 12PB7846 | 9422491055 | R06C02 | 485416 | 99.97% | 161 | 485470 | 4699 | 8945 | 288 | 575 | 622 | 1402 | 1741 | 4189 | 8546 | 15426 | 14862 | 28138 | 23.3 | 37 | 863 |
Fig. 2.
Distribution (median and range) of signal intensity for quality control probes on Illumina Infinium HumanMethylation450 arrays across all samples and in each of the colour channels (green/red). (a,b) Bisulfite conversion efficiency as reported by control probes of Infinium design I (a) or II (b). (c) Hybridization performance using synthetic reference targets present in the hybridization buffer at three concentrations. (d) Efficiency of extension of A, T, C and G nucleotides from hairpin probes (sample-independent). Probe 1 is specific for A, probe 2 for T, probe 3 for C and probe 4 for G. (e) Efficiency and sensitivity of the staining step (independent of the hybridization and extension steps). (f) Overall efficiency of the procedure estimated by querying non-polymorphic bases in the genome – one probe for each nucleotide. In all plots, labels give the expected intensity level: high, medium (Med), low or background (Bgnd).
The Infinium 450k BeadChip contains 65 genotyping probes that are useful for identification of sample mix-ups. These probes produced highly similar signal patterns in two of our samples, which was expected as these two samples (07PB1887 and 10PB6041) came from the same patient (Fig. 3).
Fig. 3.
Heatmap of signal from the 65 probes on the methylation arrays that distinguish single nucleotide polymorphisms (SNPs). Two samples show highly similar patterns, as they originate from the same patient.
Quality control of normalization procedure
Preprocessing of the raw data was performed using R/Bioconductor with the RnBeads package24. Probes known to be cross-reactive (43 230) or overlapping known single-nucleotide polymorphisms (SNPs; 8 704)25, as well as probes giving unreliable measurements (884 as determined by the Greedycut algorithm) were excluded from analysis. The data from the remaining 432 759 probes were subjected to background subtraction using the Noob method26 and beta-mixture quantile normalization (BMIQ)27. This normalization strategy successfully mitigated the inherent bias in β-value distributions between the two different types of probes (Infinium I and II) that are present on the HumanMethylation450 BeadChip30, as shown in Fig. 4. In a subsequent step, probes of non-CpG context (1 251), probes binding to sequences on sex chromosomes (9 917) and probes with standard deviation <0.005 (69 867) were filtered out. Thus, the data obtained by the remaining 351 724 probes qualified for downstream analysis.
Fig. 4.
Density plots of the β-values distribution for the Infinium I and II probes before and after background subtraction and beta-mixture quantile normalization (BMIQ).
Check for batch effects
Dimension reduction techniques are a powerful way of visualizing associations between different variables and global trends in DNA methylation data24. Applying PCA on the 10000 most variable CpGs in our data did not result in visible grouping of samples according to the BeadChip that they were applied on (Fig. 5). The lack of batch effects was further verified by Kruskal-Wallis one-way analysis of variance taking into account the first 8 primary components (Table 2).
Fig. 5.
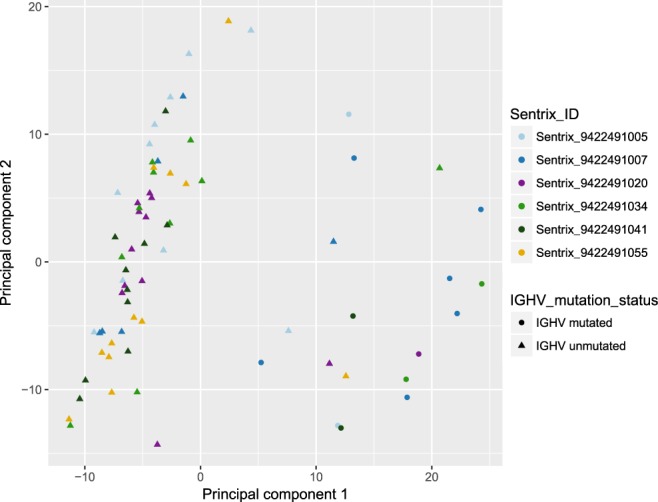
Principal component analysis (PCA) showing grouping of the 72 samples based on the 10000 most variable CpGs within the dataset. Samples are coloured according to the BeadChip (Sentrix_ID) that they were applied on. Samples of any given colour (batch) do not form separate clusters, whereas principal component 1 distinguishes M-CLL from U-CLL patients (P = 7.15 × 10−8, Wilcoxon rank sum test).
Table 2.
Table of p-values for associations of the first 8 principal components with IGHV mutation status as an intrinsic characteristic of the samples or with the BeadChip that the samples were applied on as an extrinsic variable (batch).
| Trait | Principal component | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| BeadChip number (Sentrix_ID)* | 0.15 | 0.26 | 0.21 | 0.22 | 0.24 | 0.68 | 0.65 | 0.27 |
| IGHV mutation status^ | 7.15 × 10–8 | 0.08 | 0.15 | 0.73 | 0.78 | 0.65 | 0.33 | 0.62 |
*P-values from a Kruskal-Wallis one-way analysis of variance.
^P-values from a two-sided Wilcoxon rank sum test.
Biological validation of the DNA methylation data
A technically sound dataset would allow confirmation of known facts. Using our dataset, we could replicate the finding that M-CLL and U-CLL are associated with distinct DNA methylation profiles7. In addition to the PCA in Fig. 5 and Table 2, we performed unsupervised hierarchical clustering using the 5000 most variable CpG sites. The resulting dendrograms and heatmap of β-values are presented in Fig. 6. In both analyses, samples were well separated according to IGHV mutation status, with the bigger cluster consisting only of U-CLL cases and the smaller cluster comprising all M-CLL cases plus five U-CLL cases, three of which had a considerable level of IGHV hypermutation (98.2–98.3% homology to germline; see also Online-only Table 1), thus possibly belonging to the intermediate CLL group as defined by Kulis et al.7.
Fig. 6.
Unsupervised hierarchical clustering of the 72 CLL samples based on the β-values of the 5000 most variable CpG sites. Clustering was based on Manhattan distance with complete linkage. Columns represent samples and rows CpGs. Two of the samples originate from the same patient (see the main text) and are marked by an asterisk. The high similarity of DNA methylation patterns between them affirms the reproducibility of the methodology.
The highly similar DNA methylation profiles of the two serial samples from one patient (Fig. 6) demonstrated the reproducibility and robustness of the whole analytical procedure (the two samples were hybridized to different BeadChips) and in addition supported previous observations that evolution of DNA methylation in CLL is limited to cases that acquire high-risk genetic alterations31.
Usage notes
In our exploratory analysis, we could not observe obvious clustering of samples based on patients’ sensitivity or resistance to chemotherapy (Figs. 5 and 6). Accordingly, a differential methylation test using the limma method with the threshold for difference of β-values set to 0.1 and the false discovery rate set to 0.05 could not find any differentially methylated CpGs between chemoresistant and untreated/chemosensitive patients, neither in the subgroup of patients with del(17p)/TP53mut (groups B and C vs. group A), nor in the subgroup without these aberrations (group E vs. group D). Nevertheless, as multiple testing correction inflates the type II error rate considerably, we cannot exclude that some of the tested CpGs have truly different methylation between the groups and a causative role in chemoresistance development. In this regard, our dataset can be a valuable resource for conducting hypothesis-driven research addressing questions of chemoresistance in CLL.
Additionally, thanks to the rich annotation of the samples, our dataset can be used to explore associations of DNA methylation with other markers of prognostic or predictive value in CLL, e.g. presence of specific chromosome aberrations (see Table 1 and Online-only Table 1). Such analyses should be performed with the necessary caution and subsequent validation of the findings in an independent clinical cohort32. Conversely, our dataset can serve as the validation dataset for findings originating from other clinical cohorts.
DNA methylation is highly informative when analyzed together with additional layers of the epigenomic regulatory landscape. That is why we recommend that any CpGs of interest found to be differentially methylated between groups be analyzed in the context of published whole-genome maps of histone modifications, chromatin accessibility and chromatin states of CLL and normal B cells33. Data from the ENCODE project can also be useful if parallels with a broader variety of cell types are sought34,35. Additional insights can be gained if tools like HOMER are used to identify transcription factor recognition motifs around CpGs of interest.
Supplementary information
Acknowledgements
This work was supported by a grant from the Virtual Helmholtz Institute “Resistance against Apoptosis and Therapy” (VH-VI-404). We are immensely thankful to Prof. Reiner Siebert (Ulm University, Germany) and Prof. Jose I. Martin-Subero (Institut d’Investigacions Biomèdiques August Pi i Sunyer, Barcelona, Spain) for scientific discussions, as well as to Drs. Yassen Assenov, Murat Iskar and Martin Sill at the German Cancer Research Center (DKFZ) for discussions and suggestions regarding data analysis. We are grateful to Elisa Woinikanis and Sabrina Schrell at the University Hospital in Ulm for the excellent technical assistance. We thank the microarray unit of the DKFZ Genomics and Proteomics Core Facility for providing the Illumina Human Methylation arrays and related services. We are especially thankful to the patients for trial participation and donation of samples.
Online-only Tables
Author contributions
Study design: D.M., J.B., S.S. and D.Y.Y. Laboratory work: D.Y.Y. Data processing and analysis: D.Y.Y., J.B. and D.M. Data interpretation: all authors. Manuscript drafting: D.Y.Y. and D.M. Review of manuscript content: all authors. Approval of the final version: all authors.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41597-020-0456-0.
References
- 1.Rai KR, Jain P. Chronic lymphocytic leukemia (CLL)-Then and now. Am J Hematol. 2016;91:330–340. doi: 10.1002/ajh.24282. [DOI] [PubMed] [Google Scholar]
- 2.German Population-based Cancer Registry. German Centre for Cancer Registry Data (ZfKD) at the Robert Koch Institute, https://www.krebsdaten.de/Krebs/EN/Database/databasequery_step1_node.html (2017).
- 3.Zenz T, Mertens D, Kuppers R, Dohner H, Stilgenbauer S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer. 2010;10:37–50. doi: 10.1038/nrc2764. [DOI] [PubMed] [Google Scholar]
- 4.Dohner H, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 5.Nadeu F, et al. Clinical impact of the subclonal architecture and mutational complexity in chronic lymphocytic leukemia. Leukemia. 2018;32:645–653. doi: 10.1038/leu.2017.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zenz T, et al. TP53 mutation and survival in chronic lymphocytic leukemia. J Clin Oncol. 2010;28:4473–4479. doi: 10.1200/JCO.2009.27.8762. [DOI] [PubMed] [Google Scholar]
- 7.Kulis M, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44:1236–1242. doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
- 8.Eichhorst B, et al. First-line chemoimmunotherapy with bendamustine and rituximab versus fludarabine, cyclophosphamide, and rituximab in patients with advanced chronic lymphocytic leukaemia (CLL10): an international, open-label, randomised, phase 3, non-inferiority trial. Lancet Oncol. 2016;17:928–942. doi: 10.1016/S1470-2045(16)30051-1. [DOI] [PubMed] [Google Scholar]
- 9.Fischer K, et al. Long-term remissions after FCR chemoimmunotherapy in previously untreated patients with CLL: updated results of the CLL8 trial. Blood. 2016;127:208–215. doi: 10.1182/blood-2015-06-651125. [DOI] [PubMed] [Google Scholar]
- 10.Fischer K, et al. Bendamustine in combination with rituximab for previously untreated patients with chronic lymphocytic leukemia: a multicenter phase II trial of the German Chronic Lymphocytic Leukemia Study Group. J Clin Oncol. 2012;30:3209–3216. doi: 10.1200/JCO.2011.39.2688. [DOI] [PubMed] [Google Scholar]
- 11.Keating MJ, et al. Early results of a chemoimmunotherapy regimen of fludarabine, cyclophosphamide, and rituximab as initial therapy for chronic lymphocytic leukemia. J Clin Oncol. 2005;23:4079–4088. doi: 10.1200/JCO.2005.12.051. [DOI] [PubMed] [Google Scholar]
- 12.Shanafelt TD, et al. Ibrutinib–Rituximab or Chemoimmunotherapy for Chronic Lymphocytic Leukemia. N Engl J Med. 2019;381:432–443. doi: 10.1056/NEJMoa1817073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yosifov DY, Wolf C, Stilgenbauer S, Mertens D. From Biology to Therapy: The CLL Success Story. HemaSphere. 2019;3:e175. doi: 10.1097/HS9.0000000000000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilal T, Betcher JA, Leis JF. Economic Impact of Oral Therapies for Chronic Lymphocytic Leukemia-the Burden of Novelty. Curr Hematol Malig Rep. 2018;13:237–243. doi: 10.1007/s11899-018-0461-y. [DOI] [PubMed] [Google Scholar]
- 15.Shanafelt TD, et al. Impact of ibrutinib and idelalisib on the pharmaceutical cost of treating chronic lymphocytic leukemia at the individual and societal levels. J Oncol Pract. 2015;11:252–258. doi: 10.1200/JOP.2014.002469. [DOI] [PubMed] [Google Scholar]
- 16.te Raa GD, et al. Assessment of p53 and ATM functionality in chronic lymphocytic leukemia by multiplex ligation-dependent probe amplification. Cell Death Dis. 2015;6:e1852. doi: 10.1038/cddis.2015.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zenz T, et al. Detailed analysis of p53 pathway defects in fludarabine-refractory chronic lymphocytic leukemia (CLL): dissecting the contribution of 17p deletion, TP53 mutation, p53-p21 dysfunction, and miR34a in a prospective clinical trial. Blood. 2009;114:2589–2597. doi: 10.1182/blood-2009-05-224071. [DOI] [PubMed] [Google Scholar]
- 18.Yu L, et al. Survival of Del17p CLL Depends on Genomic Complexity and Somatic Mutation. Clin Cancer Res. 2017;23:735–745. doi: 10.1158/1078-0432.CCR-16-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moussay E, et al. Determination of genes and microRNAs involved in the resistance to fludarabine in vivo in chronic lymphocytic leukemia. Mol Cancer. 2010;9:115. doi: 10.1186/1476-4598-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pandzic T, et al. Transposon Mutagenesis Reveals Fludarabine Resistance Mechanisms in Chronic Lymphocytic Leukemia. Clin Cancer Res. 2016;22:6217–6227. doi: 10.1158/1078-0432.CCR-15-2903. [DOI] [PubMed] [Google Scholar]
- 21.Hallek M, et al. Addition of rituximab to fludarabine and cyclophosphamide in patients with chronic lymphocytic leukaemia: a randomised, open-label, phase 3 trial. Lancet. 2010;376:1164–1174. doi: 10.1016/S0140-6736(10)61381-5. [DOI] [PubMed] [Google Scholar]
- 22.Malcikova J, et al. Monoallelic and biallelic inactivation of TP53 gene in chronic lymphocytic leukemia: selection, impact on survival, and response to DNA damage. Blood. 2009;114:5307–5314. doi: 10.1182/blood-2009-07-234708. [DOI] [PubMed] [Google Scholar]
- 23.Yosifov DY, 2020. DNA methylation of chronic lymphocytic leukemia with differential response to chemotherapy. Gene Expression Omnibus. GSE136724 [DOI] [PMC free article] [PubMed]
- 24.Assenov Y, et al. Comprehensive analysis of DNA methylation data with RnBeads. Nat Methods. 2014;11:1138–1140. doi: 10.1038/nmeth.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price ME, et al. Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics Chromatin. 2013;6:4. doi: 10.1186/1756-8935-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Triche TJ, Jr., Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res. 2013;41:e90. doi: 10.1093/nar/gkt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teschendorff AE, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sartor MA, et al. Genome-wide methylation and expression differences in HPV(+) and HPV(−) squamous cell carcinoma cell lines are consistent with divergent mechanisms of carcinogenesis. Epigenetics. 2011;6:777–787. doi: 10.4161/epi.6.6.16216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Laird PW, Hinoue T, Groshen S, Siegmund KD. Non-specific filtering of beta-distributed data. BMC Bioinformatics. 2014;15:199. doi: 10.1186/1471-2105-15-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dedeurwaerder S, et al. Evaluation of the Infinium Methylation 450K technology. Epigenomics. 2011;3:771–784. doi: 10.2217/epi.11.105. [DOI] [PubMed] [Google Scholar]
- 31.Oakes CC, et al. Evolution of DNA methylation is linked to genetic aberrations in chronic lymphocytic leukemia. Cancer Discov. 2014;4:348–361. doi: 10.1158/2159-8290.CD-13-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kraft P, Zeggini E, Ioannidis JP. Replication in genome-wide association studies. Stat Sci. 2009;24:561–573. doi: 10.1214/09-STS290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beekman R, et al. The reference epigenome and regulatory chromatin landscape of chronic lymphocytic leukemia. Nat Med. 2018;24:868–880. doi: 10.1038/s41591-018-0028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Yosifov DY, 2020. DNA methylation of chronic lymphocytic leukemia with differential response to chemotherapy. Gene Expression Omnibus. GSE136724 [DOI] [PMC free article] [PubMed]