Specifications table
| Subject | Aquatic Science |
| Type of data | Table Image |
| How data were acquired | Compositional analysis by microscopy of Lugol's iodine-preserved samples. ELISA-based toxin analysis using microcystins-ADDA polyclonal antibody – enhanced ELISA kit (Product No. 520,011, Abraxis, Warminster, PA, USA) and anatoxin-a ELISA Microtiter Plate Kit (Product No. 520,060, Abraxis) Toxin analysis by LC-MS (Xevo G2-XS QTof MS, Waters Corp., Milford, MA, USA) |
| Data source location | City/Town/Region: Ontario Country: Canada Name of Body of Water: Thames River Latitude and longitude (and GPS coordinates) for collected samples/data: Station MECP 26 (ECCC ON02GE0006 [T2]): 42.31714; −82.44847 Station MECP 32 (ECCC ON02GE1005 [JR]): 42.35453; −82.321 Station MECP 35 (ECCC ON02GE9007 [CA]): 42.39289; −82.21069 Station MECP 36 (LTVCA): 42.406603; −82.185703 Station MECP 39 (ECCC ON02GE1000 [TT]): 42.545; −81.968 Sampling conducted from bridges over the river at approximately mid-stream and with samples acquired from 0–1 m depth. Physico-chemical parameters measured using a model 600QS sonde (YSI Inc., Yellow Springs, OH, USA). Daily samples were collected using autosamplers (Teledyne ISCO, Lincoln, NE, USA) located at the Thames River mouth (ECCC ON02GE0006) and Thamesville, ON (ECCC ON02GE1000) with samples collected at 8 h intervals. |
| Data format | Raw Analyzed |
| Identification | Whole water samples preserved with Lugol's iodine were analyzed by counting phytoplankton in a measured aliquot using a modified inverted microscope for the Utermöhl method. |
| Strain Characteristics | N/A |
| Data accessibility | Repository name: Mendeley Data Direct URL to data: http://doi.org/10.17632/85gy8ct4yn.3 |
Compositional Profile of the Strain's Biomass
| Lipid Profile | Not Available |
| HNO Analysis (if available) | Not Available |
| Protein, Carbohydrate, Lipid, Ash Content (if available) | Not Available |
| Protein and Amino Acid Profile (if available) | Not Available |
| Carbohydrate Profile (if available) | Not Available |
| Toxin Concentrations (ug/L) (if available) | Total Microcystin Concentration Station MECP 32 (ECCC ON02GE1005 [JR]): < Method Detection Limit (MDL) 0.1 ug/L Station MECP 35 (ECCC ON02GE9007 [CA]): < MDL 0.1 ug/L Station MECP 36 (LTCVA): < MDL 0.1 ug/L Station MECP 39 (ECCC ON02GE1000 [TT]): ≤ 1.4 ug/L Station MECP 26 (ECCC ON02GE0006 [T2]): ≤ 0.52 ug/L ELISA MDL is 0.1 ug/L total microcystins and 0.2 ug/L anatoxin-a. All sites exhibited anatoxin-a levels < MDL 0.2 ug/L. Samples analysed using LC MS/MS method indicated cyanotoxin levels < MDL 0.05 ug/L. |
| References to Methods Used for Profiling | Not Available |
1. Introduction
The Thames River is a priority tributary of the Lake Erie watershed, as identified in Annex 4 of the Great Lakes Water Quality Agreement. The river flows into Lake St. Clair in southwestern Ontario with land use in the watershed dominated by row crop agriculture.
In September 2019, a cyanobacterial bloom was observed in the lower Thames River. First reports of the bloom were communicated on September 23 by the Lower Thames Valley Conservation Authority to the Ontario Ministry of the Environment, Conservation and Parks (MECP) with synoptic sampling by our collaborative team commencing immediately and continuing through October 15 along a 50 km stretch of the river extending from Thamesville, ON to Prairie Siding, ON (Fig. 1). Additional daily sampling was provided by autosamplers located at the river mouth and at Thamesville.
Fig. 1.
Samples from near surface of Thames River acquired on September 23, 2019. Survey extended from Thamesville, ON (far left; 64 km from river mouth) to Prairie Siding, ON (far right; 14 km from river mouth). Samples in middle with distinct green discolouration were acquired from a 9 km stretch of the river through Chatham, ON.
Strong green discolouration was evident over a 9 km stretch of river through Chatham, ON located 29 km from the river mouth (Fig. 2). Sampling in Chatham identified Aphanizomenon flos-aquae as the dominant cyanobacterium with peak cell abundance surpassing 3.2 × 109 cells/L [1]. Also abundant within the bloom was Planktothrix agardhii [1].
Fig. 2.
Photo of Thames River in Chatham, ON acquired on September 23, 2019 near the time of bloom onset. Distinct green discolouration of the river is evident.
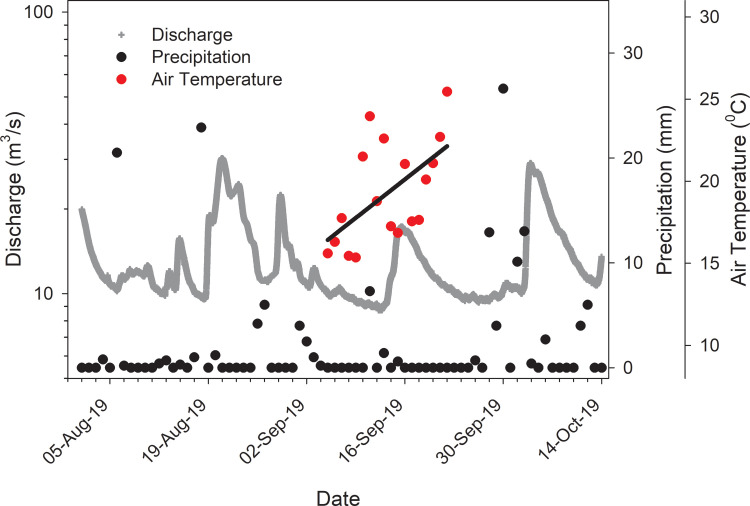
Low river discharge coincident with negligible rainfall and rapid warming were identified as factors responsible for a recent early spring bloom of P. agardhii in the Maumee River [2]. Similar conditions were recorded for the Thames River preceding the 2019 bloom (Fig. 3). By October 1, the bloom had been transported downriver [1] with remnants of the bloom persisting in the river through October 15. Whereas the Thames River represents a source of nutrients that promotes phytoplankton growth in Lake St. Clair, there is no evidence that the river directly seeds lake blooms. The September 2019 Thames River bloom and a previous documented bloom in the river [3] were dominated by filamentous cyanobacteria whereas colonial cyanobacteria such as Microcystis spp. typically bloom along the southern shore of Lake St. Clair [4].
Fig. 3.
Relationship between rainfall recorded at Chatham plotted with Thames River discharge measured 35 km upstream at Thamesville (ECCC hydrometric gaging station 02GE003). Median river discharge recorded for 3 weeks prior to the bloom was low (10.9 m3/s). Mean air temperature showed warming by 5 °C over this same period (r2 = 0.33).
In Chatham where the bloom was most visually intense, total microcystins and anatoxin-a were below ELISA detection limits. Low levels of total microcystins (< 1.5 ug/L) were detected by ELISA at the autosampler sites throughout the bloom period. Taxa present are known to produce other bioactive compounds and additional screening is recommended.
2. Environmental impact
Unknown at this time
3. Toxicity information
Analysis by ELISA did not detect total microcystins in the blooms samples from Chatham where the bloom appeared most intense, however low levels of total microcystins were detected in samples collected by autosampler at the Thames River mouth and at Thamesville. Analysis by LC-MS did not detect presence of microcystins (microcystin -LR, -dmLR, -RR, dmRR, -LA, -LF, -LY, -HilR, -LW, -YR, -HtyR, -WR), anatoxin-a or homo-anatoxin, however the taxa present are known to produce other bioactive compounds and additional screening is recommended.
4. Economic impact
We anticipate the economic impact of the bloom was negligible for the following reasons:
-
1.
No municipalities draw water from the mouth of the Thames River to Thamesville, the eastern extent of our survey, hence there were no costs associated with water treatment as a result of the bloom.
-
2.
Onset of the bloom was in early fall at a time when recreational use of the river had declined. Likewise, the early fall timing of the bloom did not coincide with major riverfront events in the city of Chatham which might have been negatively impacted by the bloom.
5. Experimental design, materials, and methods
Sampling of the Thames River was conducted from bridges over the river at approximately mid-stream and with samples acquired from 0–1 m depth. Four locations were sampled over four sampling dates between 18 August – 15 October 2019. Prior to collection of samples, physico-chemical parameters (pH, conductivity, temperature, dissolved oxygen) were measured using a 600QS sonde (YSI Inc., Yellow Springs, OH, USA). Samples were processed on site within 30 min of collection for determination of cyanotoxins. Sampling extended from morning (09:30 h) to late afternoon (17:00 h) with the majority of sampling conducted midday between 10:00–14:00 h. Whereas cyanobacteria are known to regulate gene expression under circadian influences [5], a recent transcriptomic analysis of diel metabolic functions of a cyanobacterial bloom in Lake Erie offered no evidence of altered expression of toxin genes between day or night [6]. Thus, time of sampling was not expected to influence our ability to measure toxins.
Sampling was augmented by automated samplers (Teledyne ISCO, Lincoln, NE, USA) maintained by Environment and Climate Change Canada (ECCC) which collected water at 8 h intervals from the Thames River mouth and at Thamesville, ON between 23 September – 15 October 2019. Daily integrated samples collected by autosampler were analyzed for cyanotoxins, turbidity, pH, and conductivity by the Ontario Ministry of Environment, Conservation and Parks (MECP; Toronto, ON, Canada).
For analysis of microcystins and anatoxin-a, 500 mL samples of whole water were collected and stored in amber glass bottles which were maintained chilled until analysis. Samples for analysis of anatoxin-a were additionally preserved with 600 mg sodium bisulfate and 50 mg of ascorbic acid. Cyanotoxins were anayzed by ELISA and select samples were additionally analysed by LC-MS. For ELISA, Abraxis Microcystins-ADDA ELISA and Anatoxin-a ELISA kits were used following manufacturer's instructions (Warminster, PA, USA). Twelve different microcystin variants and anatoxin-a were analyzed by LC-MS using on-line solid phase extraction (SPE). In brief, 5 ml aliquots of the homogenized samples were lysed by three freeze-thawing cycles. 15N-labelled microcystin-LR, -RR, -LA and -YR were added as internal standards. Syringe filtered samples (500 µl) were directly injected in the on-line SPE LC-QToF MS for instrumental analysis [7].
Whole water samples preserved with Lugol's iodine were analyzed by counting phytoplankton in a measured aliquot using a modified inverted microscope for the Utermöhl method plus a small magnification modification of the stratified counting technique of Munawar and Munawar [8]. A measured aliquot of mixed sample was placed into an inverted microscope counting chamber and allowed to settle a minimum of 4 h per centimeter of overlying water depth. Larger and recognizable rare cells were counted at 400 × along a minimum of one transect across the entire counting chamber. Smaller algae were counted at 1000 × along a measured transect until a minimum of 300 cells were enumerated. Phytoplankton were counted as individual cells. Cyanobacterial taxonomy reported here conforms to Anagnostidis and Komárek [9,10].
Acknowledgments
Acknowledgments
We thank Jessica Robson, Tina Mamone, Amanda Swiniarski and the MECP Great Lakes Unit for assistance in the field. Marianne Racine, Maria Dolores Molina, Carly Charon, Julianne Radford, L. Cynthia Watson, Vi Richardson and Leah Peacock provided assistance in the laboratory.
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Funding Information
This study was supported in part by Environment and Climate Change Canada ‘Great Lakes Renewal’ contract #3000692841 to RMM. Additional support was provided by the NIH (1P01ES028939-01) and NSF (OCE-1840715) to the Great Lakes Center for Fresh Waters and Human Health at Bowling Green State University.
Author Statement
R. Michael McKay: Project administration, Supervision, Investigation, Writing – Original draft, Funding acquisition Thijs Frenken: Investigation, Writing – Review and editing Ngan Diep: Conceptualization, Project administration, Writing – Review and editing, Data curation, Funding acquisition William R. Cody: Formal analysis Sophie Crevecoeur: Supervision, Investigation, Writing – Review and editing, Funding acquisition Alice Dove: Conceptualization, Project administration, Data curation, Funding acquisition Kenneth G. Drouillard: Supervision, Investigation Xavier Ortiz: Formal analysis Jason Wintermute: Investigation Arthur Zastepa: Conceptualization, Project administration, Supervision, Investigation, Writing – Review and editing, Funding acquisition.
References
- 1.McKay R.M., Frenken T., Diep N., Cody W., Crevecoeur S., Dove A., Drouillard K., Ortiz X., Wintermute J., Zastepa A. Thames river (Canada) cyanobacterial bloom. Mendeley Data. 2019 doi: 10.17632/85gy8ct4yn.3. v3, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKay R.M.L., Tuttle T., Reitz L.A., Bullerjahn G.S., Cody W.R., McDowell A.J., Davis T.W. Early onset of a microcystin-producing cyanobacterial bloom in an agriculturally-influenced Great Lakes tributary. J. Oceanol. Limnol. 2018;36:1112–1125. doi: 10.1007/s00343-018-7164-z. [DOI] [Google Scholar]
- 3.Lower Thames Conservation, “Blue-green algae in Thames River” thames river cyanobacteria bloom in chatham. https://www.lowerthames-conservation.on.ca/blue-green-algae-in-thames-river-thames-river-cyanobacteria-bloom-in-chatham/(accessed 26 March 2020).
- 4.Davis T.W., Watson S.B., Rozmarynowycz M.J., Ciborowski J.J.H., McKay R.M., Bullerjahn G.S. Phylogenies of microcystin-producing cyanobacteria in the lower Laurentian Great Lakes suggest extensive genetic connectivity. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0106093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Golden S.S., Ishiura M., Johnson C.H., Kondo T. Cyanobacterial circadian rhythms. Ann. Rev. Plant Physiol. Plant Mol. Biol. 1997;48:327–354. doi: 10.1146/annurev.arplant.48.1.327. [DOI] [PubMed] [Google Scholar]
- 6.Davenport E.J., Neudeck M.J., Matson P.G., Bullerjahn G.S., Davis T.W., Wilhelm S.W., Denny M.K., Krausfeldt L.E., Stough J.M.A., Meyer K.A., Dick G.J., Johengen T.H., Lindquist E., Tringe S.G., McKay R.M.L. Metatranscriptomic analyses of diel metabolic functions during a microcystis bloom in western Lake Erie (USA) Front. Microbiol. 2019;10:2081. doi: 10.3389/fmicb.2019.02081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ortiz X., Korenkova E., Jobst K., MacPherson K., Reiner E. A high throughput targeted and non-targeted method for the analysis of microcystins and anatoxin-A using on-line solid phase extraction coupled to liquid chromatography–quadrupole time-of-flight high resolution mass spectrometry. Anal. Bioanal. Chem. 2017;409:4959–4969. doi: 10.1007/s00216-017-0437-0. [DOI] [PubMed] [Google Scholar]
- 8.Munawar M., Munawar I.F. A lakewide study of phytoplankton biomass and its species composition in Lake Erie, April–December 1970. J. Fish. Res. Board Can. 1976;33:581–600. [Google Scholar]
- 9.Anagnostidis K., Komárek J. Modern approach to the classification system of cyanophytes. 1-Introduction. Arch. Hydrobiol. Suppl. 1985;71:291–302. [Google Scholar]
- 10.Anagnostidis K., Komárek J. Modern approach to the classification system of cyanophytes. 3- Oscillatoriales. Arch. Hydrobiol. Suppl. 1988;80:327–472. [Google Scholar]