Abstract
Background
Human tuberculosis (TB) is caused by a plethora of Mycobacterium tuberculosis complex (MTBC) strains belonging to seven phylogenetic branches. Lineages 2, 3 and 4 are considered “modern” branches of the MTBC responsible for the majority of worldwide TB. Since the current BCG vaccine confers variable protection against pulmonary TB, new candidates are investigated. MTBVAC is the unique live attenuated vaccine based on M. tuberculosis in human clinical trials.
Methods
MTBVAC was originally constructed by unmarked phoP and fadD26 deletions in a clinical isolate belonging to L4. Here we construct new vaccines based on isogenic gene deletions in clinical isolates of the L2 and L3 modern lineages. These three vaccine candidates were characterized at molecular level and also in animal experiments of protection and safety.
Findings
Safety studies in immunocompromised mice showed that MTBVAC-L2 was less attenuated than BCG Pasteur, while the original MTBVAC was found even more attenuated than BCG and MTBVAC-L3 showed an intermediate phenotype. The three MTBVAC candidates showed similar or superior protection compared to BCG in immunocompetent mice vaccinated with each MTBVAC candidate and challenged with three representative strains of the modern lineages.
Interpretation
MTBVAC vaccines, based on double phoP and fadD26 deletions, protect against TB independently of the phylogenetic linage used as template strain for their construction. Nevertheless, lineage L4 confers the best safety profile.
Funding
European Commission (TBVAC2020, H2020-PHC-643381), Spanish Ministry of Science (RTI2018-097625-B-I00), Instituto de Salud Carlos III (PI18/0336), Gobierno de Aragón/Fondo Social Europeo and the French National Research Council (ANR-10-LABX-62-IBEID, ANR-16-CE35-0009, ANR-16-CE15-0003).
Keywords: Tuberculosis, Virulence, Live vaccines, Epidemiology, Pathogen evolution, PDIM, ESAT-6, CFP-10
Graphical abstract
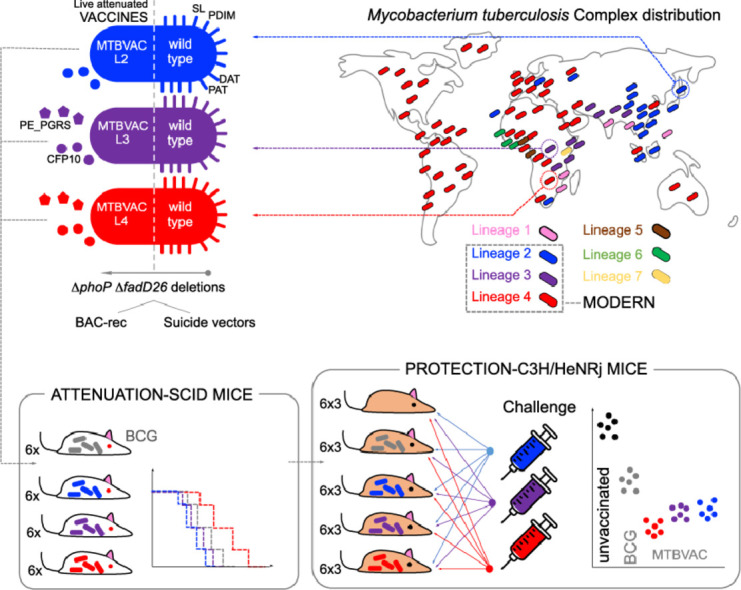
Research in context.
Evidence before this study
Current vaccines against pneumococci, meningococci, influenza or polio are designed taking into account the existing pathogen variability. However, to our knowledge, none of the existing TB vaccine candidates that aim to improve the efficacy of the current vaccine BCG, have addressed vaccine efficacy taking into consideration the evolutionary landscape of TB-causing bacteria. Indeed, TB vaccine candidates under clinical development have exclusively demonstrated protection against M. tuberculosis strains belonging to a single lineage, of the seven existing phylogenetic lineages, of this pathogen. Since 2012, a live vaccine based on attenuated M. tuberculosis (MTBVAC) is in Phase 1 and Phase 2 clinical trials in newborns and adults and in 2021 will be ready for efficacy studies.
Added value of this study
In this manuscript, we reappraise TB vaccine efficacy in the context of the evolutionary genomics of M. tuberculosis. Here, we construct and characterize live attenuated vaccines based on MTBVAC in the three modern lineages of M. tuberculosis, which represent the majority of circulating strains worldwide. This MTBVAC set is subsequently tested in mouse models of safety and protective efficacy against challenge with three representative strains from each modern lineage. Notably, this is the first time demonstrating that a live attenuated vaccine in clinical development is able to protect mice against pathogens from the most prevalent lineages, a result translationally relevant for the future efficacy trials with MTBVAC. Additionally, this is the first time to study how the genetic background impacts on safety and protective efficacy of live attenuated TB vaccines.
Implications of all the available evidence
With this study, we aim to anticipate future TB efficacy trials. This study provides proof-of-concept with MTBVAC and we aim to stablish recommendations for other vaccine candidates in the pipeline.
Alt-text: Unlabelled box
1. Introduction
Tuberculosis (TB) is the most devastating disease caused by a single infectious agent over the last 200 years [1]. In 2018, the WHO estimated 10 million new TB cases and more than 1.4 million deaths caused by TB [2].
TB is principally caused in humans by Mycobacterium tuberculosis and Mycobacterium africanum, classified within the M. tuberculosis complex (MTBC) as human-adapted mycobacteria [3], [4], [5]. Occasionally, zoonotic TB in humans caused by animal-adapted members of the MTBC is also observed [6]. Human-adapted mycobacteria can be classified in “ancestral“ or “modern” lineages based on the presence or absence of the M. tuberculosis specific deletion region TbD1 [7,8]. Lineages that harbor the TbD1 region include lineages 1 and 7 of M. tuberculosis, and lineages 5 and 6 of M. africanum. These are referred as ancient lineages and they are geographically restricted to specific areas, except for lineage 1, which shows an intermediate distribution, with main prevalence in South India and South East Asia [4,9]. By contrast, M. tuberculosis lineage 2, which are also known as representing Beijing strains, lineage 3 strains, known also as CAS/Dehli strains and lineage 4, corresponding to the Euro-American strain families are considered as modern lineages and they include worldwide distributed strains [9]. The widespread distribution of modern lineages probably reflects the adaptive evolution of the MTBC to transmit and cause disease in crowded, dense and urbanized populations, a hypothesis that was recently supported by data on increased resistance of modern lineages to oxidative stress and hypoxia relative to L1 strains [8]. As such, there is converging evidence from epidemiologic and experimental data, which suggests that strains from lineages 2, 3 and 4, have become more successful in terms of their geographical distribution, being responsible for a large proportion of the global TB burden [9].
The only vaccine licensed against TB is the Bacille Calmette and Guerin (BCG), which was obtained after in vitro passaging of Mycobacterium bovis -the causative agent of bovine TB- for 13 years at the beginning of the 20th century [10]. Loss of Region of Difference 1 (RD1) with respect to the MTBC members, substantially contributes to BCG attenuation [11,12]. This region codes for genes of the ESX-1 protein complex responsible for the co-secretion of ESAT-6 and CFP-10. Both proteins are major antigens of M. tuberculosis. Specifically, ESAT-6, secreted by ESX-1 type VII secretion system, has been extensively documented as a putative virulence and immunogenicity factor of MTBC pathogens. Among the various putative functions of ESAT-6, its implication in phagosomal rupture within infected phagocytes, is of particular interest, and enables ESX-1 proficient strains to get in contact with the host cytosol [13,14]. BCG was administered for the first time in 1921 and during the next decades BCG sub-strains emerged as a consequence of the parallel in vitro sub-cultivation of the original BCG in different laboratories [10,15]. BCG confers protection from the severe forms of the disease in children although protection against pulmonary TB in adolescents and adults is considerably variable [16]. Consequently, this main limitation of BCG to stop TB transmission imposes a priority in the research of new vaccines candidates. A rationale vaccine research requires to understand the MTBC virulence factors and immunogenicity conferred by mycobacterial components [17]. Today, a wide pipeline of vaccine candidates targeting different populations is currently in clinical trials including live attenuated vaccines, adjuvanted protein subunit vaccines, viral-vectored vaccines and whole cell inactivated vaccines [18].
MTBVAC is currently the unique live vaccine candidate in clinical development which is based on an attenuated strain of the human pathogen M. tuberculosis. MTBVAC consists on the unmarked deletion of phoP and fadD26 genes in the clinical M. tuberculosis isolate Mt103, representing a lineage 4 strain [19] .
The gene phoP encodes the transcription factor of the Two-Component System PhoPR, which controls approximately 2% of the genome content of M. tuberculosis [20], [21], [22], [23]. The PhoP regulon orchestrates diverse networks in M. tuberculosis, including the secretion of the ESAT-6 [24], the synthesis of sulfolipid and di- and polyacyltrehaloses [25], the expression of the non-coding RNA mcr7, impacting the activity of the Twin Arginine Translocation (Tat) secretion apparatus [23] and production of phosphatidylinositol mannosides [26]. The gene fadD26 is the first gene in an operon required for the synthesis of phthiocerol dimycocerosates (PDIM) [27], mycobacterial virulence lipids that are involved in phagosomal rupture in concert with ESAT-6 [28], [29], [30]. Since MTBVAC is derived from an M. tuberculosis clinical isolate, it conserves the whole T cell epitope repertoire described for MTBC pathogens including the major immunodominant antigens ESAT-6, CFP-10 and PPE68 absent in BCG as a consequence of the RD1 deletion. These three proteins, albeit small, are unusually immunogenic and contain 285 of the total 1.603 epitopes described in M. tuberculosis [31]. In line with this observation, our recent preclinical studies have demonstrated that immunity against ESAT-6 and CFP-10 conferred after MTBVAC vaccination remarkably correlate with improved vaccine efficacy relative to BCG Danish [32].
After an extensive preclinical testing in animal models [19,[33], [34], [35], [36]], MTBVAC was the first and unique live attenuated M. tuberculosis vaccine approved to enter into clinical trials in 2012 [37]. After successfully completed phases Ia (NCT02013245) [38] and Ib (NCT02729571) [39], currently, two phases IIa trials are ongoing in South Africa, one in newborns using MTBVAC as a prime vaccine (NCT03536117) and the other on in adults where MTBVAC is tested as a boost for BCG (NCT02933281).
In this study, we reappraise vaccine efficacy of newly constructed MTBVAC variants in the context of the global genetic diversity of the MTBC. The main aims of the study are linked to the question whether the original MTBVAC is able to protect against the three modern lineages of the MTBC or whether a vaccine candidate constructed in a specific lineage might lead to lineage-dependent improved protection.
2. Material and methods
2.1. Bacteria and culture conditions
M. tuberculosis strains used in this work are detailed in Table S1. Briefly, GC1237 [40], HMS13037 (a clinical isolate from our laboratory collection belonging to lineage 3 of M. tuberculosis), Mt103 [41], their respective phoP-fadD26 mutants, H37Rv [42], W4 (kindly provided by Gilla Kaplan) [43], HCU3524 (a clinical isolate from our laboratory collection belonging to lineage 3) and BCG Pasteur 1173P2, were grown in liquid media at 37 °C with Middlebrook 7H9 broth (Difco) supplemented with 10% (vol/vol) of ADC (0.5% bovine serum albumin, 0.2% dextrose, 0.085% NaCl and 0.0003% beef catalase) and 0.05% (vol/vol) Tween-80. For solid media, Middlebrook 7H10 (Difco) supplemented with ADC, except for the HMS13037 strain, supplemented with OADC (ADC with 0.005% (vol/vol) oleic acid) was used. When required, hygromycin (Hyg) 20 µg/ml or Kanamycin (Km) 20 µg/ml were added. Escherichia coli strains (MC1061, DH5α, DH10B, XL1-BLUE or BW25141) were grown in liquid media at 37 °C in Luria-Bertani (LB) broth. When required, ampicillin (Amp) 100 µg/ml, Km 20 µg/ml, Hyg, 50 µg/ml or chloramphenicol (Cm), 12.5 µg/ml were added. On solid media, LB-agar was used.
2.2. Construction of fadD26 and phoP mutants
Isogenic deletions in fadD26 and phoP genes present in the original MTBVAC vaccine [19] were obtained in GC1237 and HMS13037 strains using two different approaches: suicide plasmids and BAC-rec (see Table S2 for extended information about the vectors used). Suicide plasmid pAZ5 was used to obtain the fadD26 deletion in GC1237 and pAZ18 was used to construct the phoP deletion in GC1237 and HMS13037 as described in Arbués and colleagues [19]. pAZ5 and pAZ18 harbor fadD26 and phoP genes interrupted with the res-Hyg-res cassette respectively, Gentamicin resistance gene cassette, reporter gene xylE and the counter selectable marker sacB. Single recombinants were selected by plating transformants in Hyg-containing plates and testing positive XylE activity by addition of a catechol solution (0.55 g of catechol dissolved in 47.5 ml of water and 2.5 ml of PBS). Single recombinants were grown in liquid media and serial dilutions were plated onto 7H10 (ADC or OADC) containing 2% of sucrose (Suc) and Hyg to select double recombinants. XylE negative clones were confirmed by PCR using primers 5´upstream of 3´downstream of the region cloned in the plasmid, in res sites or in the gene deleted region. To remove the Hyg cassette, pAZ20 replicative plasmid harbouring γδ-resolvase, Km- and Gentamicin (Gm)-cassette resistance and the counter selectable marker sacB was used [19]. Unmarked deletion was confirmed by replica plating in Km, Hyg and w/o antibiotic plates and by PCR using the appropriate primers (Table S3). BAC-rec strategy was used to obtain fadD26 deletion in HMS13037. A M. tuberculosis H37Rv bacterial artificial chromosome (BAC) library obtained in the pBeloBAC11 vector contained in E. coli DH10B was used [44]. The clone DH10B carrying the Rv209 BAC containing the fadD26 (Rv2930) gene was selected and the thermosensitive plasmid pKD46 carrying red recombinase from lambda phage was co-transformed [45]. DH10 Rv209 pKD46 induced with arabinose 0.15% was transformed with a PCR product (using fadD26-P1-Fw and fadD26-P2 primers) containing the FRT-Km-FRT-cassette from pKD4 flanked with 40 bp of identity arms to target the fadD26 gene. Gene deletion in the BAC was confirmed by PCR amplification using specific primers. BAC Rv209-ΔfadD26::Km was used as template to obtain the allelic exchange substrate (AES) using KO-fadD26-Fw and KO-fadD26-Rv primers to transform in mycobacteria. AES consists on the FRT-Km-FRT cassette flanked with 700 pb of identity arms of the specific site of recombination and was transformed in HMS13037 strain harbouring pJV53H (induced with 0.2% of acetamide) [46]. Recombinants were plated onto Km-containing plates and were confirmed using appropriate primers. Loss of pJV53H plasmid was confirmed by replica plating onto Hyg and Km. To unmark the deletion, pRES-FLP-Mtb plasmid (HygR) harbouring the flp recombinase from Saccharomyces cerevisiae with adapted codons for M. tuberculosis [47]. Transformants were selected in Hyg-containing plates and the removal of the cassette was confirmed by replica plating and PCR amplification using fadD26F and fadD26R primers. Loss of pRES-FLP-Mtb plasmid was confirmed by replica plating.
2.3. Protein extraction and Western blotting
Cultures were grown in 7H9-0.05% Tween-80 supplemented with dextrose, NaCl and catalase to avoid albumin contamination in the secreted fraction. Bacteria were grown until an OD of 0.6–0.8 at 37 °C and pelleted. Supernatant fractions were filtered through a 0.22 µm-pore-size filter and incubated on ice for 2 h with 10% (vol/vol) of trichloroacetic acid. Sampled were centrifuged for 1 h at 4 °C and pellets were washed with acetone. Supernatants were discarded after centrifugation and pellets were air dried and dissolved in Tris 150 mM pH 8.8. For whole-cell protein extractions, bacteria were resuspended in PBS containing 1% Triton X-100 and transferred into tubes containing glass beads (MP Biomedicals). Suspensions were disrupted by Fast-Prep (6.5 m/s, 45 s) twice and samples were cooled on ice between the cycles. Supernatants containing soluble proteins were filtered through a 0.22 µm-pore-size filter were after centrifugation. Protein extractions were quantified using QuantiPro BCA assay (Sigma Aldrich). Samples were heated for 10 min at 100 °C after adding Laemmli buffer and samples were loaded in 12–17% polyacrylamide gels containing 0.1% SDS. Then, proteins were transferred to a PDFV membrane using a semi-dry electrophoretic transfer cell (Trans-Blot® Semi-Dry Transfer cell, Bio-Rad). Proteins were blocked with 5% (w/v) skimmed milk in TBS-T buffer (25 mM Tris pH = 7.5, 150 mM NaCl, 0.05% Tween 20) for 30 min and incubated overnight with the primary antibody. Membranes were then washed with TBS-T buffer before incubation with secondary antibodies for 1 h. Membranes were washed and signals were detected using chemiluminescent substrates (Western Bright™ Quantum, Advansta). Immunodetection was carried out using PhoP-antiserum (1:5000), antibodies anti-sigA and anti-CFP-10 (Thermo Scientific) (both at 1:5000) followed with the incubation with secondary antibody anti-rabbit IgG human serum adsorbed conjugate (1:5000) (KPL) or incubation with monoclonal antibody anti-ESAT-6 (1:2500) (abcam), anti-GroEL2 (Hsp65) (1:2500) (Invitrogen) or anti-PE_PGRS (1:2000) (described in [48,49]), followed by incubation with an anti-mouse IgG human serum adsorbed conjugate (1:5000) (KPL). To reprove blots, ReBlot Plus Strong Antibody Stripping Solution (Millipore) was used following the specification sheet.
2.4. RNA extraction and qRT-PCR
Bacteria were pelleted and resuspended in 250 µl in wash buffer (aqueous solution containing 0.137 M NaCl and 0.5% Tween 80) and 500 µl of RNA protect reagent (Quiagen) to avoid RNA degradation. Suspensions were centrifuged after 5-minute incubation at room temperature. Pellets were dissolved in lysis buffer (20 mM sodium acetate, 0.5% SDS, 0.1 mM EDTA) and 1 ml of phenol:chloroform (5:1), pH = 4.5. Bacterial suspensions were transferred into tubes containing glass beads (MP Biomedicals) and cells were lysed by Fast-Prep (2 cycles, 45 s at speed 6.5 m/s, samples were cooled on ice between cycles). Tubes were centrifuged and aqueous phases were transferred to a tube containing chloroform:isoamylic alcohol (24:1). After centrifugation upper phases were transferred to a tube containing isopropanol and 0.3 M sodium acetate (pH = 5.5) and tubes were incubated at −20 °C overnight. After centrifugation at 4 °C, precipitated nucleic acids were collected and pellets were washed with ethanol 70%. Samples were again centrifuged and pellets were dissolved in diethylpyrocarbonate (DEPC)-treated water (RNase free). DNA was removed by two consecutive incubations of 1 h at 37 °C with Turbo DNA-free (Ambion) by addition of 1 µl of DNase. Later, RNA was purified by adding phenol:chloroform (5:1) pH = 4.5, and previous steps were repeated to precipitate, collect, dry and dissolve the RNA in DEPC-treated water. RNA integrity was confirmed by agarose gel electrophoresis and absence of DNA was confirmed by absence of amplification products after PCR. Reverse transcription was performed using PrimeScript™ RT Reagent Kit as detailed in the product manual. qRT-PCR reaction was performed in the StepOne Plus Real Time PCR System (Applied Biosystems) using TB Green Premix Ex Taq™ (Tli Rnase H Plus) (Takara) kit. For the reaction, primers at a final concentration of 0.25 µM and cDNA diluted 1:10 were used. Normalization was calculated using the sigA housekeeping gene in each sample. Absence of unspecific PCR products were confirmed after examination of melting curves in each sample.
2.5. Neutral red staining
Bacteria grown on solid media were transferred to a tube containing 50% of methanol in water. Supernatants were discarded after 1 h of incubation. Pellets were resuspended in 750 µl of barbital buffer (1% sodium barbital in 5% NaCl, pH 9.8). After centrifugation, bacteria were resuspended in 4 ml of barbital buffer. 150 µl of a solution of 0.05% neutral red in barbital buffer were added and incubated at 37 °C for 1 h. After incubation, supernatants were discarded to evaluate the staining of the pelleted bacteria.
2.6. Mouse infection
All mice were observed and kept under controlled conditions. For protective efficacy experiments, eight-week old immunocompetent C3H/HeNRj female mice were vaccinated subcutaneously with 106 CFU in 100 µl of PBS. Mice were vaccinated with BCG Pasteur, MTBVAC, MTBVAC-L2::hyg or MTBVAC-L3::hyg, or unvaccinated as control group. Eight weeks later, mice were challenged by intranasal route with 200 CFU in 40 µl of PBS of each M. tuberculosis strain belonging to one of the modern lineages (W4, HCU3524 or H37Rv belonging to lineages 2, 3 and 4 respectively). Four weeks post-challenge, bacterial burden was evaluated in lungs and spleen. Serial dilutions were plated onto 7H10-ADC. For attenuation experiment, eight-week old immunocompromised SCID female mice were inoculated by intraperitoneal route with 106 bacteria in 100 µl PBS, equivalent to 2 times the dose of BCG recommended for humans. Mice were controlled and their weight was followed during the experiment. Experimental endpoint was defined when loss of weight was more than 20%.
2.7. Statistical analysis
For protection experiments, statistical analysis of CFUs in the organs was performed using One-way ANOVA, Bonferroni post-test. For safety experiments, the statistical analysis was calculated applying a Log-rank (Mantel-Cox) test.
2.8. Ethics statement
Experimental animal studies were performed in agreement with European and national directives for the protection of animal for experimental purposes. All procedures were carried out under Project Licenses PI50/14 (protective efficacy) and PI33/15 (attenuation) approved by the Ethic Committee for Animal Experiments from the University of Zaragoza.
2.9. Role of the funding source
The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication
3. Results
3.1. Construction of two new vaccine candidates based on isogenic phoP and fadD26 deletions present in MTBVAC
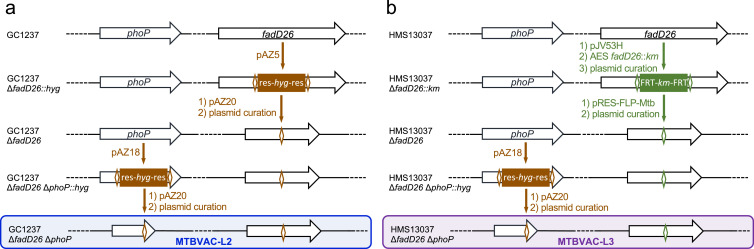
Based on previous knowledge acquired during the preclinical and clinical development of MTBVAC [37], [38], [39], a live attenuated M. tuberculosis strain belonging to lineage 4 [19], we have introduced simultaneous phoP and fadD26 deletions in two clinical isolates belonging to lineages 2 and 3 of M. tuberculosis. Our objective was to obtain a set of isogenic MTBVAC-like vaccine candidates constructed in the genetic backgrounds of the three modern lineages of M. tuberculosis. M. tuberculosis GC1237 [40], which is a Beijing strain classified to the Asian-Ancestral 3 sub-branch of lineage 2 strains [50] and the HMS13037 clinical isolate, a lineage 3 isolate showing a CAS-1 Delhi spoligotype, were selected as parental strains for the genetic constructions (Figure S1 and Table S1). To obtain double unmarked gene deletions, two alternative and complementary strategies based on suicide plasmids [19] or BAC-rec [32] were used. In the GC1237 strain, the fadD26 deletion was obtained using the suicide plasmid pAZ5 followed by the phoP deletion using pAZ18 (Fig. 1a). In the HMS13037 strain, BAC-rec was used to delete fadD26 and the suicide plasmid pAZ18 was used to construct the phoP deletion (Fig. 1b). The antibiotic markers were subsequently eliminated using γδ and FLP recombinases suitable for the suicide plasmids and BAC-rec strategies respectively (Fig. 1). Gene deletions were confirmed by PCR using two pairs of primers flanking the mutation (Figure S2, S3 and S4, Table S2), and the phoP deletion was additionally confirmed by Western-blot (Figure S4). Altogether, two new double unmarked phoP and fadD26 mutants were obtained and named MTBVAC-L2 and MTBVAC-L3, for lineage 2 and 3 respectively (Fig. 1).
Fig. 1.
Sequential genetic steps to construct unmarked ΔphoP ΔfadD26 mutants in M. tuberculosis strains from lineages 2 and 3. (a) Construction of MTBVAC-L2. (b) Construction of MTBVAC-L3. Those steps involving the use of suicide plasmids pAZ5 or pAZ18 are colored in brown and utilization of the BAC-rec strategy is indicated by green colors. The res and FRT sites used for resolution of the hygromycin and kanamycin cassettes are indicated by brown and green open diamonds respectively. Each genetic step was confirmed by PCR and/or Western-Blot (see Supplementary Material).
3.2. Molecular characterization of MTBVAC-L2 and MTBVAC-L3 demonstrate equivalent phenotypes described in MTBVAC
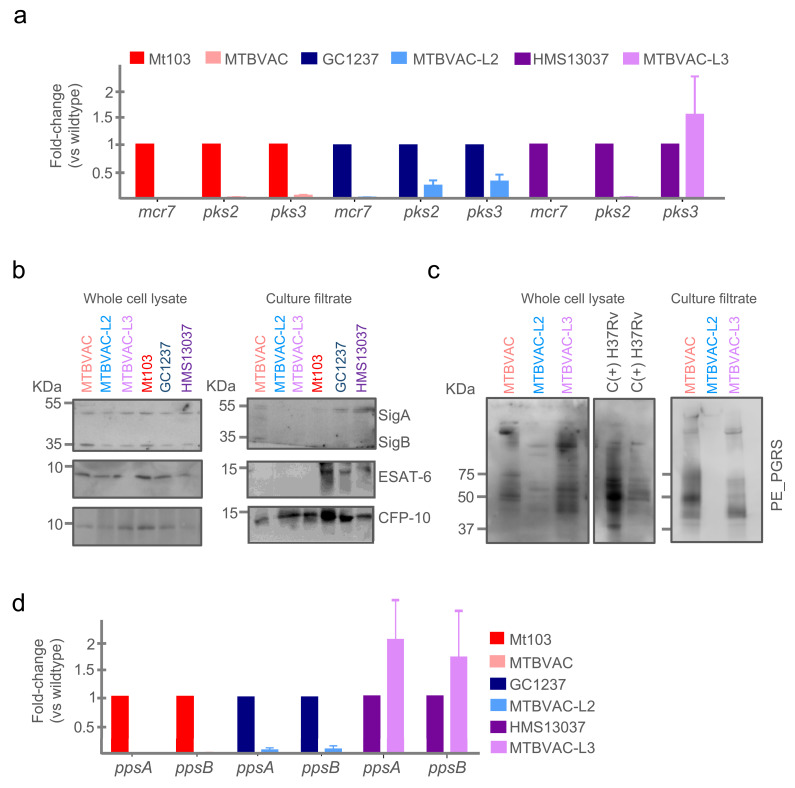
At first, PhoP-dependent phenotypes previously described for MTBVAC were evaluated in the new vaccine candidates. A transcriptomics approach was used to evaluate whether specific downregulation of representative transcripts of the PhoP-regulon (mcr7, pks2 and pks3) [20,23] existed in MTBVAC-L2, MTBVAC-L3 and MTBVAC, compared to their parental strains. For mcr7 and pks2 we observed the expected downregulation in the three MTBVAC variants. Exceptionally, expression of pks3 in MTBVAC-L3 was slightly increased compared to the parental HMS13037 strain (Fig. 2a), which might indicate that subtle inter-lineage differences could exist in the PhoP regulon. We also confirmed that MTBVAC, MTBVAC-L2 and MTBVAC-L3 are unable to fix neutral red, contrasting to their parental strains (Figure S5a). This phenotype is putatively related to the altered cell envelope absent in PDIM and acyltrehalose-derived lipids, whose synthesis involves fadD26 and phoP, respectively [19,25].
Fig. 2.
Molecular characterization of MTBVAC, MTBVAC-L2 and MTBVAC-L3. (a) Expression of representative genes from the PhoP-regulon (mcr7, pks2 and pks3) measured by qRT-PCR in MTBVAC, MTBVAC-L2 and MTBVAC-L3 compared to their respective parental strains. Each gene was normalized against sigA expression in each sample. Bars represent the mean and standard deviation from three independent experiments. (b) Western-blot of ESAT-6 and CFP-10 proteins in whole-cell lysates (left panel) and secreted fractions (right panel) in the MTBVAC vaccines and their wild type strains. Note the absence of ESAT-6 secretion as a consequence of the phoP mutation in all vaccine strains. Also note the presence of CFP-10 in the secreted fraction in all MTBVAC strains. Detection of SigA and SigB serves as loading control in the whole-cell lysate and also as cell integrity control in the secreted fraction. (c) Western-blot of PE_PGRS in total (left panels) and secreted fractions (right panel) from the MTBVAC vaccine set. Note the differential absence of PE_PGRS secretion in MTBVAC-L2. (d) Expression of the ppsAB genes belonging to the PDIM biosynthetic operon measured by qRT-PCR. Bars represent the mean and standard deviation from three independent experiments. Each sample was normalized relative to the endogenous control sigA.
One of the best characterized phenotypes linked to PhoP, is its involvement in regulation of the secretion of ESAT-6 [22,24,51]. Western-blot analyses showed that ESAT-6 was only secreted in the parental strains in contrast to MTBVAC, MTBVAC-L2 and MTBVAC-L3. Nevertheless, ESAT-6 was detected in the whole-cell lysate of both the parental strains and the fadD26- and phoP-deleted strains (Fig. 2b). Despite the inability of phoP-deletion mutants to secrete ESAT-6, it has been recently described that MTBVAC is able to secrete CFP-10 [32], although the mechanism of the process relative to the previously defined ESAT-6 and CFP-10 co-secretion [52], still needs to be defined. Here, we demonstrate that MTBVAC-L2 and MTBVAC-L3, similar to classical MTBVAC, does secrete CFP-10 into the supernatant (Fig. 2b).
RNA-seq of MTBVAC revealed that the operon formed by fadD26 and downstream genes (ppsA-D) is substantially less expressed compared to the Mt103 wild-type strain (Figure S6). We tried to confirm this result by qRT-PCR of ppsA and ppsB genes in MTBVAC-L2 and MTBVAC-L3 compared to their parental strains. The expected absence of the ppsAB transcripts was observed in MTBVAC and MTBVAC-L2; however, we observed similar levels of ppsAB in MTBVAC-L3 relative to the HMS13037 parental strain (Fig. 2d). This unexpected result might be related to the genetic scar left in the chromosome after resolution of the antibiotic resistance marker and will be discussed below.
3.3. MTBVAC-L2 produces but does not secrete PE_PGRS proteins and this phenotype is related to the lineage background used in genetic constructions
The ppe38 locus is polymorphic in the Beijing family resulting in deletion of the ppe38 gene at the branching point of the “modern” Beijing sub-strains [53]. Deletion of ppe38 completely blocks the ability to secrete PE_PGRS proteins (a subfamily of PE proteins). Interestingly, lack of secretion of these proteins has been associated with an increased virulence of M. tuberculosis strains in mouse models [53]. Moreover, BCG strains were also shown to lack PE_PGRS secretion due to the deletion of the RD5 region, although this absence had little impact on the protective efficacy of BCG strains in mice [49]. To further characterize the vaccine candidates MTBVAC, MTBVAC-L2 and MTBVAC-L3, the organization of the ppe38 locus was studied according to McEvoy and colleagues [54], indicating the putative absence of the ppe38 gene in GC1237 (Figure S5). Indeed, PE_PGRS proteins were detected by Western-blotting in the whole-cell fractions of the three strains (Fig. 2c) using a specific antibody, whereas PE_PGRS proteins were only detected in the supernatants of MTBVAC and MTBVAC-L3 strains, and not in MTBVAC-L2 supernatant fractions. Accordingly, here we confirm a differential phenotype between MTBVAC-L2 and MTBVAC/MTBVAC-L3 strains, which is related to the specific lineage used as background for vaccine construction.
3.4. MTBVAC-L2 and MTBVAC-L3 are attenuated in SCID mice although both showed less attenuation than MTBVAC
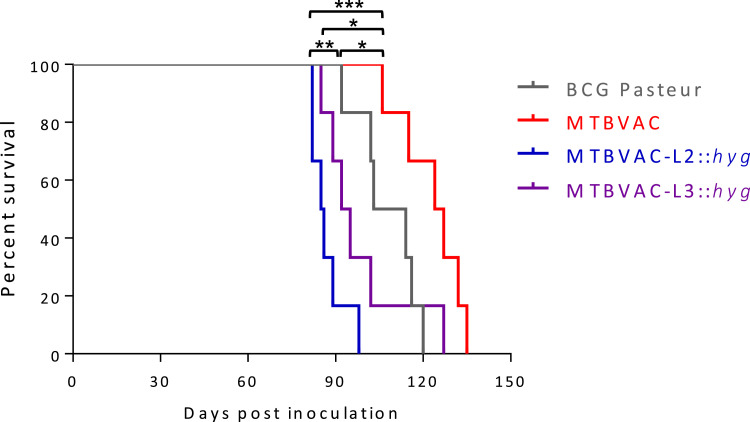
To evaluate the safety profile of the new MTBVAC candidates, a survival experiment was performed in immunocompromised SCID mice. To discard differences due to in vitro fitness of different vaccines, we first confirmed that MTBVAC-L2 and MTBVAC-L3 as well as their parental strains exhibited comparable in vitro growth curves (Figure S7).
Next, mice were inoculated with MTBVAC-L2 or MTBVAC-L3. BCG Pasteur and the original MTBVAC strains were used as comparators. MTBVAC-L2 and MTBVAC-L3 showed an attenuated profile in SCID mice (mean of survival of 85.5 days in the former and 93.5 days in the latter). Thus, MTBVAC-L3 exhibits comparable attenuation to BCG Pasteur but MTBVAC-L2 is less attenuated than the currently licensed BCG. Notably, of the three double phoP- and fadD26-deletion mutants tested, the original MTBVAC was the most attenuated and even safer than BCG Pasteur (Fig. 3).
Fig. 3.
Attenuation of MTBVAC, MTBVAC-L2 and MTBVAC-L3 in the SCID mouse model. Survival curves from groups of 6 SCID mice inoculated by intraperitoneal route with MTBVAC, MTBVAC-L2, MTBVAC-L3 and BCG Pasteur (as control) are shown. Asterisks indicate * p < 0.05, **p < 0.01 and *** p<0.001 [Log-rank (Mantel-Cox) test].
3.5. MTBVAC, MTBVAC-L2 and MTBVAC-L3 confer protection in mice against modern strains of M. tuberculosis and this phenotype is unrelated to the lineage used for vaccine construction
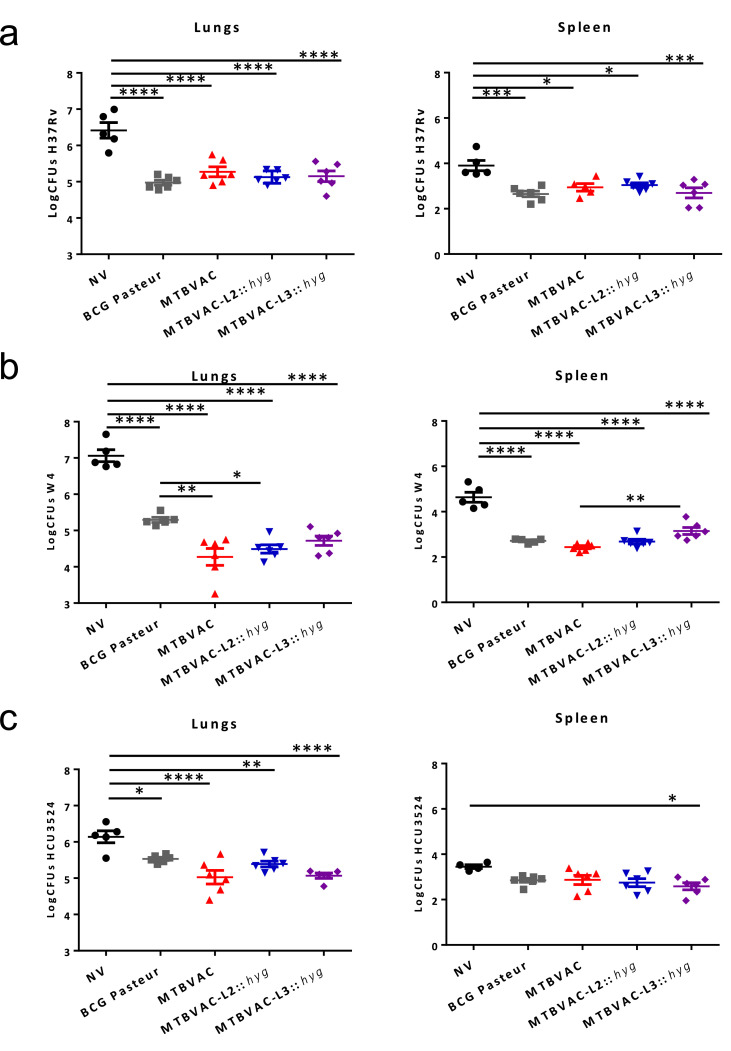
According to previous research, greater protection of MTBVAC relative to BCG Danish was observed in C3H/HeNRj mice with respect to other commonly used mouse strains such as C57BL/6 or BALB/c 32. This phenotype is likely attributable to the H-2k haplotype of the Major Histocompatibility Complex present in the C3H/HeNRj mouse strain, which is able to recognize both ESAT-6 and CFP-10 immunogens, in contrast to the H-2b haplotype in C57BL/6 mice that only recognizes ESAT-6, and/or the H-2d haplotype in BALB/c mice which does neither recognizes ESAT-6, nor CFP-10. Accordingly, the C3H/HeNRj model was considered as useful to discriminate the protective efficacy of MTBVAC compared to BCG [32]. In this study, we used the C3H/HeNRj mouse model to compare protective efficacy of MTBVAC, MTBVAC-L2 and MTBVAC-L3 against challenge with M. tuberculosis strains belonging to the modern lineages of the MTBC. For intranasal challenge, M. tuberculosis strains H37Rv (lineage 4), W4-Beijing (lineage 2) and HCU3524 (lineage 3) were used. The licensed vaccine BCG Pasteur 1173P2 was used as comparator, since this strain is widely used in animal experiments. Enumeration of bacterial burden in lungs and spleens showed that the three MTBVAC strains conferred similar protection against challenge with isolates belonging to modern lineages. After challenge with H37Rv, all vaccines tested conferred similar protection against lineage 4 (Fig. 4a). Challenge with a Beijing strain resulted in significant increased protection of animals previously vaccinated with MTBVAC and MTBVAC-L2 compared to BCG Pasteur (Fig. 4b). Variable protection was observed against challenge with lineage 3, the lowest bacterial burden in lungs was observed for vaccination with MTBVAC and MTBVAC-L3, followed by MTBVAC-L2 and BCG Pasteur (Fig. 4c). Noteworthy, the three MTBVAC vaccines exhibited comparable protection against each challenge strain, indicating that protective efficacy phenotypes are independent of the lineage used as MTBVAC background.
Fig. 4.
Protective efficacy of MTBVAC, MTBVAC-L2 and MTBVAC-L3 in C3H/HeNRj mice against challenge with strains from modern lineages of M. tuberculosis. The graphs represent bacterial burden in lungs (left panels) and spleen (right panels) from groups of 6 mice subcutaneously immunized with the corresponding vaccine and after a 4 weeks intranasal challenge against (a) H37Rv (lineage 4 strain), (b) W4-Bejiing (lineage 2 strain) and (c) HCU3524 (lineage 3 strain). All data are mean ± SEM. Asterisks indicate * p < 0.05, ** p < 0.01, *** p < 0.001 and **** p < 0.0001 [One-way ANOVA, Bonferroni post-test].
4. Discussion
To achieve the end of TB, new tools including new effective vaccines, better diagnostics and new drugs are needed [55]. Despite the diversity in vaccine strategies currently in clinical trials, preclinical research on new live vaccines continues in order to keep the vaccine pipeline filled. Recently, live vaccine candidates based on BCG [56,57] or attenuated M. tuberculosis [58] have been reported. Live attenuated M. tuberculosis vaccines are expected to confer an effective T-cell immune response since they maintain those antigens missing in RD regions from BCG [31]. MTBVAC is one of the two live attenuated vaccine candidates currently in clinical trials, representing the unique candidate based on genetic background of the human pathogen M. tuberculosis. MTBVAC confer higher and more durable T-cell responses than BCG in a vaccinated human cohort [39] and this finding might be related to the fact that MTBVAC contains approximately a 50% broader antigenic repertoire of T-cell epitopes compared to BCG [31]. However, the possible lineage-dependent protection of MTBVAC remains unknown and its protective efficacy in animal models has been exclusively evaluated against M. tuberculosis H37Rv or M. tuberculosis Erdman strains, both belonging to lineage 4 [19,36].
To evaluate lineage-dependent protection, we have constructed two new vaccine candidates based on isogenic deletions of MTBVAC in the lineages 2 and 3 of M. tuberculosis (MTBVAC-L2 and MTBVAC-L3). As a result, we have obtained the complete set of MTBVAC vaccines in the three modern lineages of M. tuberculosis. Molecular characterization of this vaccine set revealed that most PhoP- and FadD26-dependent phenotypes are maintained across lineages, although some differences were observed. The most prominent PhoP-regulated genes (mcr7, pks2 and pks3) are overall downregulated in the MTBVAC set. However, pks3 expression remained unchanged in MTBVAC-L3 compared to the parental strain. This unexpected finding opens new perspectives to decipher whether the PhoP-regulon might be lineage-dependent. Indeed, similar inter-strain differences have been recently observed in the PhoP regulon. The whiB6 gene, which belongs to the PhoP regulatory network, is positively regulated by PhoP in Mt103 and GC1237 strains while negatively regulated in H37Rv. This differential regulation is caused by a single nucleotide insertion exclusively present in H37Rv and results in lower ESAT-6 secretion in H37Rv relative to other M. tuberculosis strains [51].
Another marked differential phenotype was observed in the expression of the fadD26-ppsAD operon. The ppsAB genes are similarly expressed in MTBVAC-L3 relative to the HMS13037 parental strain in contrast to MTBVAC and MTBVAC-L2 when compared to their respective parental strains. A closer inspection of the genetic constructions suggests that the chromosomal “scar” after resolution of the antibiotic markers is different between MTBVAC-L3 (FRT site) and MTBVAC/MTBVAC-L2 (res site) (Fig. 1). Thus, the res site would cause a polar effect in expression of the 3´ fadD26 downstream region. By contrast, the FRT site contained in the fadD26 deleted gene in MTBVAC-L3 is designed to create nonpolar effects, as described by Datsenko and colleagues [45].
The two MTBVAC derivatives constructed here robustly reproduce the presence of CFP-10 and the absence of ESAT-6 in the secreted protein fraction, previously documented for MTBVAC [32]. It is important to mention that this phenotype could interfere with the diagnostic potential of current interferon gamma release assays (IGRA) -which contain ESAT-6 and CFP-10- as diagnostic antigens, as observed for MTBVAC-vaccinated newborns [39] and consequently, specific diagnostic tools able to differentiate between MTBVAC-immunized and M. tuberculosis-infected individuals are currently under study.
Preclinical safety experiments in immunodeficient mice revealed that albeit the three MTBVAC derivatives were attenuated, inter-lineage differences were found. Beijing-strain-derived MTBVAC-L2 was found to be the least attenuated candidate and only MTBVAC constructed from lineage 4 was more attenuated than BCG Pasteur. Some phenotypes specific to Beijing strains might have contributed to this finding. The presence of phenolic glycolipids in Beijing strains, which is different to L4 strains due to a frameshift mutation in the pks15/1 gene [59], are linked to inhibition of innate immune responses and higher virulence of the Beijing family [60]. Moreover, in this work, we explored another differential phenotype between the three vaccine candidates, linked to PE_PGRS secretion. Indeed, MTBVAC-L2 failed to secrete PE_PGRS proteins in contrast to MTBVAC and MTBVAC-L3. Since lack of secretion of these proteins has been associated with increased virulence in mice [53], this phenotype might also contribute to the lower attenuation of MTBVAC-L2 observed in SCID mice.
Zhang and colleagues recently demonstrated a marked heterogeneity in attenuation and protective efficacy among 13 different BCG sub-strains in mice [61]. Results from this study suggest a correlation between a higher protection conferred by those BCG strains which were less attenuated and vice versa [61]. Supporting differences in protective efficacy across BCG strains, we also observed that BCG Danish conferred less protection than MTBVAC in the C3H/HeNRj strain [32], albeit BCG Pasteur induces equivalent protection as MTBVAC vaccines in this model (Fig. 4a). However, our results obtained with the three MTBVAC derivatives, show that different levels of attenuation in SCID mice do not necessarily predict vaccine efficacy in immune-competent mice, as no clear association between attenuation and protection was found. It is important to mention nevertheless, that the dosage and inoculation routes differ between both studies, which might explain the differences in the time-to-humane endpoint durations after vaccination with BCG sub-strains or MTBVAC derivatives.
Our protective efficacy experiments against modern M. tuberculosis strains suggest no lineage-dependent protection in C3H/HeNRj mice, although additional protection experiments using different vaccination regimes or additional animal models might provide a more detailed insight on this topic. This similar protective efficacy might be related to the presence of the complete antigenic repertoire of M. tuberculosis across MTBVAC vaccines, which is thought to result in a wide and complete immune stimulation. Remarkably, higher protection was conferred by MTBVAC and MTBVAC-L2 compared to BCG Pasteur against challenge with W4-Beijing strain. Beijing strains have been associated to recent TB outbreaks, hyper-virulent phenotypes and drug resistance [62]. One hypothesis for this successfully emergence of L2 strains could be the ability of these strains to escape from BCG vaccination, although this remains elusive because of the controversial conclusions in different epidemiological studies [63]. It is noteworthy that MTBVAC vaccines conferred improved protection against a challenge with the W4-Beijing strain, which is an important finding, even in the light that the MTBVAC-L2 vaccine strain appeared slightly less attenuated than other vaccines in mouse experiments.
Our previous preclinical studies demonstrated that MTBVAC protected against challenge with H37Rv in mice [19] or with Erdman in non-human primates [36], both strains belonging to the lineage 4. Importantly, here we show for the first time that MTBVAC vaccine candidates are able to protect against different representatives of the modern lineages of M. tuberculosis in mice. This finding represents an important detail for the clinical evaluation of MTBVAC and putative new live attenuated vaccines based on phoP and fadD26 deletions.
Since the three MTBVAC vaccine candidates provide different relative levels of protection against challenge with modern M. tuberculosis strains, this suggests that the properties of the infecting strain or their distribution in different parts of the world may determine the efficacy of the vaccine used in humans. MTBVAC is today in Phase IIa in adults and newborns and the results presented here further support its progress into clinical efficacy trials. In such efficacy studies, we should be prepared for two possible scenarios. Either the original MTBVAC constructed in the lineage 4 background might confer equivalent protection independently of the vaccinated population; or alternatively, MTBVAC or the new MTBVAC-L2 and MTBVAC-L3 constructed in this work might confer efficacious protection in populations where strains from lineages 4, 2 or 3 are the most prevalent, respectively. In this latter scenario, the three MTBVAC vaccines could be combined into a polyvalent vaccine that affords a worldwide protection against TB. Altogether, our results not only expand the current knowledge of the MTBVAC vaccine but anticipate novel MTBVAC-based strategies for the future.
Author contribution
Conceptualization, J.G-A, C.M., and I. P; Methodology, J.G-A., C.M., R.B., N.A., W.F., F.S., S.U., and I.P.; Reagents, A.A., S.S.; Investigation, W.F., F.S., S. U. and I.P.; Writing – Original Draft, J.G-A, C.M. and I.P.; Writing–Review & Editing, J.G-A, C.M. and R.B.; Funding Acquisition, C.M. and R.B.; Supervision, J.G-A., C.M., R.B., N.A., W.F., and F.S. All authors reviewed the final report.
Funding sources
This work was supported by the European Commission Horizon 2020 (TBVAC2020, H2020-PHC-643381), the Spanish Ministry of Science (RTI2018-097625-B-I00), Instituto de Salud Carlos III (PI18/0336) and Gobierno de Aragón/Fondo Social Europeo, as well as by the French National Research Council ANR (ANR-10-LABX-62-IBEID, ANR-16-CE35-0009, ANR-16-CE15-0003). IP was recipient of a “DGA-Fondo Social Europeo” grant. IP was also recipient of a “Programa CAI-Ibercaja Estancias de Investigación” (CM3/18) and a “FEMS Research and Training grant” (FEMS-GO-2018-118) to perform an internship at Integrated Mycobacterial Pathogenomics Unit at the Institut Pasteur of Paris.
Declaration Competing of Interest
C.M., A.A. and J.G.-A. are co-inventors on the patent “Tuberculosis vaccine”. N.A., C.M. and J. G-A. are co-inventors on the patent “Compositions for use as a prophylactic agent to those at risk of infection of tuberculosis, or as secondary agents for treating infected tuberculosis patients”. Both patents were filled by the University of Zaragoza
Acknowledgments
We are grateful to Wilbert Bitter, Michael Brennan, and Ida Rosenkrands for antibodies.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102761.
Appendix. Supplementary materials
References
- 1.Scully T. Tuberculosis. Nature. 2013;502(7470):S1. doi: 10.1038/502S1a. [DOI] [PubMed] [Google Scholar]
- 2.WHO. Global Tuberculosis Report2019. 2019
- 3.Boritsch E.C., Brosch R. Evolution of mycobacterium tuberculosis: new insights into pathogenicity and drug resistance. Microbiol Spectr. 2016;4(5) doi: 10.1128/microbiolspec.TBTB2-0020-2016. [DOI] [PubMed] [Google Scholar]
- 4.Brites D., Gagneux S. The nature and evolution of genomic diversity in the mycobacterium tuberculosis complex. Adv Exp Med Biol. 2017;1019:1–26. doi: 10.1007/978-3-319-64371-7_1. [DOI] [PubMed] [Google Scholar]
- 5.Pai M., Behr M.A., Dowdy D. Tuberculosis. Nat Rev Dis Primers. 2016;2:16076. doi: 10.1038/nrdp.2016.76. [DOI] [PubMed] [Google Scholar]
- 6.Malone K.M., Gordon S.V. Mycobacterium tuberculosis complex members adapted to wild and domestic animals. Adv Exp Med Biol. 2017;1019:135–154. doi: 10.1007/978-3-319-64371-7_7. [DOI] [PubMed] [Google Scholar]
- 7.Brosch R., Gordon S.V., Marmiesse M. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci USA. 2002;99(6):3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bottai D., Frigui W., Sayes F. TbD1 deletion as a driver of the evolutionary success of modern epidemic Mycobacterium tuberculosis lineages. Nat Commun. 2020;11(1):684. doi: 10.1038/s41467-020-14508-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gagneux S., DeRiemer K., Van T. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103(8):2869–2873. doi: 10.1073/pnas.0511240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brosch R., Gordon S.V., Garnier T. Genome plasticity of BCG and impact on vaccine efficacy. Proc Natl Acad Sci USA. 2007;104(13):5596–5601. doi: 10.1073/pnas.0700869104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis K.N., Liao R., Guinn K.M. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette–Guerin attenuation. J Infect Dis. 2003;187(1):117–123. doi: 10.1086/345862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pym A.S., Brodin P., Brosch R., Huerre M., Cole S.T. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol Microbiol. 2002;46(3):709–717. doi: 10.1046/j.1365-2958.2002.03237.x. [DOI] [PubMed] [Google Scholar]
- 13.Simeone R., Bobard A., Lippmann J. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog. 2012;8(2) doi: 10.1371/journal.ppat.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Wel N., Hava D., Houben D. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell. 2007;129(7):1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- 15.Behr M.A., Small P.M. A historical and molecular phylogeny of BCG strains. Vaccine. 1999;17(7–8):915–922. doi: 10.1016/s0264-410x(98)00277-1. [DOI] [PubMed] [Google Scholar]
- 16.Ritz N., Hanekom W.A., Robins-Browne R., Britton W.J., Curtis N. Influence of BCG vaccine strain on the immune response and protection against tuberculosis. FEMS Microbiol Rev. 2008;32(5):821–841. doi: 10.1111/j.1574-6976.2008.00118.x. [DOI] [PubMed] [Google Scholar]
- 17.Kroesen V.M., Madacki J., Frigui W. Mycobacterial virulence: impact on immunogenicity and vaccine research [version 1; peer review: 4 approved] F1000Res. 2019;8(F1000 Faculty Rev) doi: 10.12688/f1000research.20572.1. 2025https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sable S.B., Posey J.E., Scriba T.J. Tuberculosis vaccine development: progress in clinical evaluation. Clin Microbiol Rev. 2019;33(1) doi: 10.1128/CMR.00100-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arbues A., Aguilo J.I., Gonzalo-Asensio J. Construction, characterization and preclinical evaluation of MTBVAC, the first live-attenuated M. tuberculosis-based vaccine to enter clinical trials. Vaccine. 2013;31(42):4867–4873. doi: 10.1016/j.vaccine.2013.07.051. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalo-Asensio J., Mostowy S., Harders-Westerveen J. PhoP: a missing piece in the intricate puzzle of Mycobacterium tuberculosis virulence. PLoS One. 2008;3(10):e3496. doi: 10.1371/journal.pone.0003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walters S.B., Dubnau E., Kolesnikova I., Laval F., Daffe M., Smith I. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol Microbiol. 2006;60(2):312–330. doi: 10.1111/j.1365-2958.2006.05102.x. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalo-Asensio J., Malaga W., Pawlik A. Evolutionary history of tuberculosis shaped by conserved mutations in the PhoPR virulence regulator. Proc Natl Acad Sci USA. 2014;111(31):11491–11496. doi: 10.1073/pnas.1406693111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Solans L., Gonzalo-Asensio J., Sala C. The PhoP-dependent ncRNA MCR7 modulates the TAT secretion system in Mycobacterium tuberculosis. PLoS Pathog. 2014;10(5) doi: 10.1371/journal.ppat.1004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frigui W., Bottai D., Majlessi L. Control of M. tuberculosis ESAT-6 secretion and specific t cell recognition by PhoP. PLoS Pathog. 2008;4(2):e33. doi: 10.1371/journal.ppat.0040033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalo Asensio J., Maia C., Ferrer N.L. The virulence-associated two-component PhoP-PhoR system controls the biosynthesis of polyketide-derived lipids in Mycobacterium tuberculosis. J Biol Chem. 2006;281(3):1313–1316. doi: 10.1074/jbc.C500388200. [DOI] [PubMed] [Google Scholar]
- 26.Diaz C., Perez Del Palacio J., Valero-Guillen P.L. Comparative metabolomics between Mycobacterium tuberculosis and the MTBVAC vaccine candidate. ACS Infect Dis. 2019;5(8):1317–1326. doi: 10.1021/acsinfecdis.9b00008. [DOI] [PubMed] [Google Scholar]
- 27.Camacho L.R., Constant P., Raynaud C. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J Biol Chem. 2001;276(23):19845–19854. doi: 10.1074/jbc.M100662200. [DOI] [PubMed] [Google Scholar]
- 28.Cox J.S., Chen B., McNeil M., Jacobs W.R., Jr. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 1999;402(6757):79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 29.Infante E., Aguilar L.D., Gicquel B., Pando R.H. Immunogenicity and protective efficacy of the Mycobacterium tuberculosis fadD26 mutant. Clin Exp Immunol. 2005;141(1):21–28. doi: 10.1111/j.1365-2249.2005.02832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Augenstreich J., Arbues A., Simeone R. ESX-1 and phthiocerol dimycocerosates of Mycobacterium tuberculosis act in concert to cause phagosomal rupture and host cell apoptosis. Cell Microbiol. 2017;19(7) doi: 10.1111/cmi.12726. [DOI] [PubMed] [Google Scholar]
- 31.Gonzalo-Asensio J., Marinova D., Martin C., Aguilo N. MTBVAC: attenuating the human pathogen of tuberculosis (TB) toward a promising vaccine against the TB epidemic. Front Immunol. 2017;8:1803. doi: 10.3389/fimmu.2017.01803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aguilo N., Gonzalo-Asensio J., Alvarez-Arguedas S. Reactogenicity to major tuberculosis antigens absent in BCG is linked to improved protection against Mycobacterium tuberculosis. Nat Commun. 2017;8:16085. doi: 10.1038/ncomms16085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aguilar D., Infante E., Martin C., Gormley E., Gicquel B., Hernandez Pando R. Immunological responses and protective immunity against tuberculosis conferred by vaccination of BALB/C mice with the attenuated Mycobacterium tuberculosis (phoP) SO2 strain. Clin Exp Immunol. 2007;147(2):330–338. doi: 10.1111/j.1365-2249.2006.03284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cardona P.J., Asensio J.G., Arbues A. Extended safety studies of the attenuated live tuberculosis vaccine SO2 based on phoP mutant. Vaccine. 2009;27(18):2499–2505. doi: 10.1016/j.vaccine.2009.02.060. [DOI] [PubMed] [Google Scholar]
- 35.Martin C., Williams A., Hernandez-Pando R. The live Mycobacterium tuberculosis phoP mutant strain is more attenuated than BCG and confers protective immunity against tuberculosis in mice and guinea pigs. Vaccine. 2006;24(17):3408–3419. doi: 10.1016/j.vaccine.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 36.Verreck F.A., Vervenne R.A., Kondova I. MVA.85A boosting of BCG and an attenuated, phoP deficient M. tuberculosis vaccine both show protective efficacy against tuberculosis in rhesus macaques. PLoS One. 2009;4(4):e5264. doi: 10.1371/journal.pone.0005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marinova D., Gonzalo-Asensio J., Aguilo N., Martin C. MTBVAC from discovery to clinical trials in tuberculosis-endemic countries. Expert Rev Vaccines. 2017;16(6):565–576. doi: 10.1080/14760584.2017.1324303. [DOI] [PubMed] [Google Scholar]
- 38.Spertini F., Audran R., Chakour R. Safety of human immunisation with a live-attenuated Mycobacterium tuberculosis vaccine: a randomised, double-blind, controlled phase I trial. Lancet Respiratory Med. 2015;3(12):953–962. doi: 10.1016/S2213-2600(15)00435-X. [DOI] [PubMed] [Google Scholar]
- 39.Tameris M., Mearns H., Penn-Nicholson A. Live-attenuated Mycobacterium tuberculosis vaccine MTBVAC versus BCG in adults and neonates: a randomised controlled, double-blind dose-escalation trial. Lancet Respiratory Med. 2019;7(9):757–770. doi: 10.1016/S2213-2600(19)30251-6. [DOI] [PubMed] [Google Scholar]
- 40.Alonso H., Aguilo J.I., Samper S. Deciphering the role of IS6110 in a highly transmissible Mycobacterium tuberculosis Beijing strain, GC1237. Tuberculosis. 2011;91(2):117–126. doi: 10.1016/j.tube.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 41.Jackson M., Raynaud C., Laneelle M.A. Inactivation of the antigen 85C gene profoundly affects the mycolate content and alters the permeability of the Mycobacterium tuberculosis cell envelope. Mol Microbiol. 1999;31(5):1573–1587. doi: 10.1046/j.1365-2958.1999.01310.x. [DOI] [PubMed] [Google Scholar]
- 42.Cole S.T., Brosch R., Parkhill J. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393(6685):537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 43.Bifani P.J., Mathema B., Liu Z. Identification of a W variant outbreak of Mycobacterium tuberculosis via population-based molecular epidemiology. JAMA. 1999;282(24):2321–2327. doi: 10.1001/jama.282.24.2321. [DOI] [PubMed] [Google Scholar]
- 44.Brosch R., Gordon S.V., Billault A. Use of a Mycobacterium tuberculosis H37Rv bacterial artificial chromosome library for genome mapping, sequencing, and comparative genomics. Infect Immun. 1998;66(5):2221–2229. doi: 10.1128/iai.66.5.2221-2229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Kessel J.C., Hatfull G.F. Recombineering in Mycobacterium tuberculosis. Nat Methods. 2007;4(2):147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 47.Song H., Niederweis M. Functional expression of the FLP recombinase in Mycobacterium bovis BCG. Gene. 2007;399(2):112–119. doi: 10.1016/j.gene.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abdallah A.M., Verboom T., Weerdenburg E.M. PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX-5. Mol Microbiol. 2009;73(3):329–340. doi: 10.1111/j.1365-2958.2009.06783.x. [DOI] [PubMed] [Google Scholar]
- 49.Ates L.S., Sayes F., Frigui W. RD5-mediated lack of PE_PGRS and PPE-MPTR export in BCG vaccine strains results in strong reduction of antigenic repertoire but little impact on protection. PLoS Pathog. 2018;14(6) doi: 10.1371/journal.ppat.1007139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merker M., Blin C., Mona S. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat Genet. 2015;47(3):242–249. doi: 10.1038/ng.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Solans L., Aguilo N., Samper S. A specific polymorphism in Mycobacterium tuberculosis H37Rv causes differential ESAT-6 expression and identifies WHIB6 as a novel ESX-1 component. Infect Immun. 2014;82(8):3446–3456. doi: 10.1128/IAI.01824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Renshaw P.S., Panagiotidou P., Whelan A. Conclusive evidence that the major T-cell antigens of the Mycobacterium tuberculosis complex ESAT-6 and CFP-10 form a tight, 1:1 complex and characterization of the structural properties of ESAT-6, CFP-10, and the ESAT-6*CFP-10 complex. Implications for pathogenesis and virulence. J Biol Chem. 2002;277(24):21598–21603. doi: 10.1074/jbc.M201625200. [DOI] [PubMed] [Google Scholar]
- 53.Ates L.S., Dippenaar A., Ummels R. Mutations in ppe38 block PE_PGRS secretion and increase virulence of Mycobacterium tuberculosis. Nat Microbiol. 2018;3(2):181–188. doi: 10.1038/s41564-017-0090-6. [DOI] [PubMed] [Google Scholar]
- 54.McEvoy C.R., van Helden P.D., Warren R.M., Gey van Pittius N.C. Evidence for a rapid rate of molecular evolution at the hypervariable and immunogenic Mycobacterium tuberculosis PPE38 gene region. BMC Evol Biol. 2009;9:237. doi: 10.1186/1471-2148-9-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.WHO . World Health Organization; 2015. WHO end TB strategy. [Google Scholar]
- 56.Groschel M.I., Sayes F., Shin S.J. Recombinant BCG expressing ESX-1 of mycobacterium marinum combines low virulence with cytosolic immune signaling and improved TB protection. Cell Rep. 2017;18(11):2752–2765. doi: 10.1016/j.celrep.2017.02.057. [DOI] [PubMed] [Google Scholar]
- 57.Moliva J.I., Hossfeld A.P., Sidiki S. Selective delipidation of Mycobacterium bovis BCG enables direct pulmonary vaccination and enhances protection against Mycobacterium tuberculosis. Mucosal Immunol. 2019;12(3):805–815. doi: 10.1038/s41385-019-0148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Levillain F., Kim H., Woong Kwon K. Preclinical assessment of a new live attenuated Mycobacterium tuberculosis Beijing-based vaccine for tuberculosis. Vaccine. 2019 doi: 10.1016/j.vaccine.2019.11.085. [DOI] [PubMed] [Google Scholar]
- 59.Constant P., Perez E., Malaga W. Role of the pks15/1 gene in the biosynthesis of phenolglycolipids in the Mycobacterium tuberculosis complex. evidence that all strains synthesize glycosylated p-hydroxybenzoic methyl esters and that strains devoid of phenolglycolipids harbor a frameshift mutation in the pks15/1 gene. J Biol Chem. 2002;277(41):38148–38158. doi: 10.1074/jbc.M206538200. [DOI] [PubMed] [Google Scholar]
- 60.Reed M.B., Domenech P., Manca C. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature. 2004;431(7004):84–87. doi: 10.1038/nature02837. [DOI] [PubMed] [Google Scholar]
- 61.Zhang L., Ru H.W., Chen F.Z. Variable virulence and efficacy of bcg vaccine strains in mice and correlation with genome polymorphisms. Mol Therapy. 2016;24(2):398–405. doi: 10.1038/mt.2015.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parwati I., van Crevel R., van Soolingen D. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infectious Dis. 2010;10(2):103–111. doi: 10.1016/S1473-3099(09)70330-5. [DOI] [PubMed] [Google Scholar]
- 63.Hanekom M., Gey van Pittius N.C., McEvoy C., Victor T.C., Van Helden P.D., Warren R.M. Mycobacterium tuberculosis Beijing genotype: a template for success. Tuberculosis. 2011;91(6):510–523. doi: 10.1016/j.tube.2011.07.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.