Abstract
Background
Interleukin (IL) 28B polymorphisms encoding pro-inflammatory and anti-inflammatory cytokines trigger diverse clinical outcome of hepatitis virus infection. However, there is controversy concerning the association of IL28B polymorphisms with the outcome of hepatitis B virus (HBV) infection, with several studies obtaining inconsistent results. We performed a meta-analysis to evaluate the role of 3 single nucleotide polymorphisms (SNPs) rs12979860, rs12980275 and rs8099917 in the progression of HBV infection, overall and by ethnicity.
Methods
Searched PubMed, Embase and Wiley Online Library electronic databases using ‘interleukin 28B’, ‘IL 28B’, ‘IL 28B polymorphism’, ‘hepatitis B virus’, ‘HBV’, and performed meta- analysis for rs12979860, rs12980275 and rs8099917 in Asian and Caucasian populations under the dominant recessive and allele model.
Results
Eighteen studies were found in total and used for this meta-analysis, including 5587 cases and 4295 controls. The IL28B polymorphism rs12979860 had no association with HBV persistence (CC vs CT + TT: OR = 0.86, 95% CI = 0.76–1.00; TT vs CT + CC: OR = 1.14, 95% CI = 0.76–1.70; T vs C: OR = 1.03, 95% CI = 0.94–1.13). Similarly, neither rs12980275 nor rs8099917 had associations with HBV persistence (rs12980275 in AA vs AG + AA: OR = 1.15, 95% CI = 0.96–1.38; rs8099917 in TT vs GT + GG: OR = 1.15, 95% CI = 0.96–1.39). There was also no significant association of IL28B polymorphisms with persistent HBV infection in Asians or Chinese. There was no evidence of an association of rs12979860 with the HBV-related hepatocellular carcinoma susceptibility (T vs C: OR = 1.53, 95% CI = 0.96–2.43).
Conclusion
IL28B polymorphisms had no association with the outcome of HBV infection overall, nor in the Asians and the Chinese. These 3 SNPs might not be relevant to the development of HBV infection.
Keywords: Interleukin 28B polymorphisms, Persistent HBV infection, Hepatitis B virus-related hepatocellular carcinoma, Meta-analysis
Background
Hepatitis B virus (HBV) is a serious public health issue which contributes to the global burden of disease. According to the World Health Organization, HBV claimed 887,000 lives in 2015 and resulted in 257 million chronic carriers in 2017 [1]. Moreover, approximately 2.7 million chronic carriers are also infected with human immunodeficiency virus (HIV) because HBV and HIV can both be transmitted by unprotected sex and injection drug use [2], living conditions that preclude access to running water [3], as well as from mother to child during pregnancy and birth [4]. Approximately 2 million disability-adjusted life-years (DALYs) are attributed to HBV infection [5]. However, the global burden of chronic HBV is unequally distributed by region. Incidence is high in Africa and Asia, and the endemic high prevalence of HBV and hepatitis C virus (HCV) infection is attributed to persistent infection [6]. HBV infection contributes a greater burden of disease in many developing countries. A safe and effective vaccine has been available since the 1980s, however, vaccination strategies differ from country to country due to cost [7]. It is still a great challenge for developing countries to prevent HBV infection.
HBV is a contagious liver disease that can be acute or chronic. Some people who get HBV recover completely from the acute illness, while others may develop a lifelong infection, which increases the risk of liver cirrhosis, liver failure and hepatitis hepatocellular carcinoma (HHC). HCC is the third leading cause of cancer death globally [8], and 78% of HCC is attributed to HBV or HCV [5]. The natural history of HBV infection varies from spontaneous recovery post-infection, to chronic asymptomatic carrier, to decompensated cirrhosis and liver cancer [9]. However, the underlying biological mechanisms that induce the different outcomes of HBV infection remain to be discovered, and hopefully exploited as a target of intervention or incorporated into a risk prediction tool.
Cytokines and regulatory molecules made a major contribution to the immune-pathogenesis of HBV infection. The cytokine Interferon-λ3 (IFN-λ3) coded by interleukin (IL) 28B polymorphisms has an anti-viral effect and could impede the HBV replication in hepatocyte cell lines. In recent years, several genome-wide association studies (GWAS) indicate the 3 single-nucleotide polymorphisms (SNPs) rs12979860 C/T, rs12980275 A/G and rs8099917 T/G, located on IL28B are associated with liver diseases [10, 11]. Furthermore, IL28B polymorphisms predict the serological response to Pegylated interferon-α (PEG-IFN-α) in terms of HBeAg sero-conversion that relate to HBV infection [12]. The association of genotypic variations in IL28B with HBV infection suggests a potential therapeutic target.
Currently, associations of IL28B with HBV infection are not completely consistent. For instance, the SNP rs12979860 was reported to be strongly related to HBV persistence under the allelic and dominant models [13]. Conversely, Song found there was no association of rs12979860 with the outcome of HBV infection [14]. In addition, several studies suggest a strong association of the IL28B gene with the HBV/HCV-induced HCC [15–17]. Nevertheless, few studies have specifically explored the relationship of the IL28B gene with HBV-related HCC.
Methods
Search strategy
We followed the PRISMA guidelines to perform this systematic review and meta-analysis. A systematic research of PubMed, Embase, Wiley Online Library databases was made with restriction to the English language from January 1, 2010 to June 1, 2018. The search terms included ‘interleukin 28B’, ‘IL 28B’, ‘IL 28B polymorphism’, and these terms in combination with ‘hepatitis B virus’ or ‘HBV’. Reference lists of the identified studies were also searched manually for additional eligible studies.
Selection criteria
The inclusion criteria were as follows: (i) studies of persistent HBV infection patients, i.e. chronic carriers with chronic hepatitis or liver cirrhosis or hepatocellular carcinoma as cases, and healthy participants without HBV infection or HBV recovered patients as controls; (ii) studies with precise IL28B genotypes in case and controls; (iii) studies providing odds ratios (OR) and 95% confidence intervals (CI) for the dominant model (CC vs CT + TT for rs12979860; AA vs AG + GG for rs12980275; TT vs GT + GG for rs8099917), recessive model (TT vs CT + CC for rs12979860; GG vs AG + AA for rs12980275; GG vs GT + TT for rs8099917), and allelic model (T vs C for rs12979860; G vs A for rs12980275; G vs T for rs8099917); (iv) case-control study design; (v) diagnosis of chronic HBV carriers based on seropositive results for hepatitis B surface antigen (HBsAg) for more than 6 months; diagnosis of HBV recovery based on seropositive results for hepatitis B core antibody (anti-HBc) and hepatitis B surface antibody (anti-HBs) without HBsAg for at least 6 months. The exclusion criteria were: (i) studies lacking healthy controls or HBV recovered controls; (ii) studies with inaccurate or insufficient information on IL28B genotypes and the genetic models of interest; (iii) studies not designed as a case-control study; (iv) studies including participants testing positive for antibodies against HCV and HIV.
Data extraction and quality assessment
We reviewed and identified all relevant publications according to the selection criteria, and extracted all available data for the eligible studies. The extracted information for each study included: name of the first author, year of publications, country of origin, ethnicity, SNP, number of genotyped cases and controls, and P-value derived from Hardy-Weinberg equilibrium (HWE) in controls.
The quality of each study was evaluated based on the Newcastle-Ottawa Scale (NOS), and 7–9 stars was perceived as high quality [18]. The appraisal items included factors, such as the comparability and representativeness of participants; ascertainment method of exposure; ascertainment method of cases and controls.
Statistical analysis
We assessed the association of IL28 polymorphisms rs12979860, rs12980275, rs8099917 with persistent HBV infection from which we reported ORs and 95% CIs from 3 genetic models. We also compared HCC patients as cases with healthy controls or recovery controls, used ORs and 95% CIs derived from the allelic model to examine the association of rs12979860 with the occurrence of HBV-related HCC. Stratified analysis was performed by ethnicity. Heterogeneity between studies was assessed by the chi-square-based Cochran’s Q-test and Higgins (I2) statistics, and the significance level was set at P = 0.1. Sequential omission of individual studies was conducted as a sensitivity analysis to test the heterogeneity. The impact of the 3 SNPs throughout meta-analysis was combined using a fixed-effects model (P ≥ 0.1 and I2 ≤ 0.25) and a random-effects model (P < 0.1 or I2 > 0.25). Visual inspection of asymmetry in the funnel plot was used to assess publication bias. Furthermore, Begg’s test and Egger’s test were also performed as a sensitivity analysis to examine publication bias. A P-value < 0.05 indicated publication bias was significant across studies. Trim and fill analysis was also conducted as a sensitivity analysis to diagnose publication bias in this meta-analysis. The statistical analysis was conducted by using R software, Version 3.3.3.
Results
Study characteristics
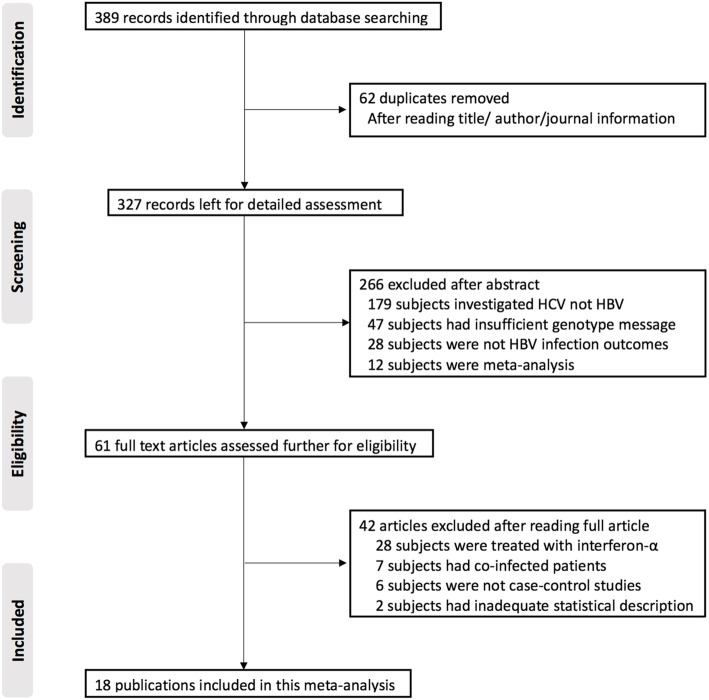
The study selection and inclusion process are illustrated in Fig. 1. A total of 389 publications were identified by the search, and 62 duplicates were removed based on title, author and journal information. This gave 327 papers for detailed assessment from which 266 publications were excluded. Among those excluded articles, 179 publications focused on HCV infection rather than HBV; 47 publications lacked precise information on genotype; 28 publications lacked relevant HBV infection outcome and 12 publications were meta-analysis. The remaining 61 publications were assessed based on the above selection criteria, and 42 were excluded. Among them, 28 publications concentrated on interferon-α treatment, 8 publications recruited co-infected participants, and 6 publications were cohort studies. Finally, 18 full published studies were included in this systematic review and meta-analysis after this screening.
Fig. 1.
Flow-chart of literature search. HBV: hepatitis B virus; IL28B: interleukin 28B
These 18 publications (Table 1) were published from 2010 to 2017, with the language restricted to English. There were 11 studies in Asia populations including Chinese Han, Korean and Thais, and the other 8 studies focused on other groups including American, Iranian, Italian, Spanish, Saudi Arabian and Turkish. All studies used persistent HBV infection patients as cases, and used healthy participants or patients recovered from prior HBV infection as controls. The 3 SNPs were genotyped using PCR, PCR-RELP or TaqMan assays. Minor allele frequencies of these 3 SNPs in Asians were significantly lower than those of the Caucasians (Additional file 1). All studies were hospital-based or population-based case-control studies. In total the 18 studies had 5587 cases and 4295 controls. There were 15 studies comprising 4913 cases and 3865 controls that considered rs12979860 C/T; 8 studies comprising 3765 cases and 2506 controls that considered rs12980275 A/G; and 11 studies comprising 3912 cases and 2900 controls that considered rs8099917 T/G. The distribution of genotypes in the controls of all studies was in agreement with HWE except for the study of Ren S [19]. The median NOS score of these studies was 7, and all studies had a high methodological quality according to NOS score. As shown in Table 1, 12 studies obtain 7 stars and 6 studies obtain 8 stars. Majority of the 8-star publications had well-matched cases and controls for gender, HBV copies and other relative factors. The common of most 7-star publications was that their data sources were mainly from the hospital, which might also lead to selection bias. In addition, some of them failed to fulfill the standard of comparability, i.e. study controls lack of description on matching for an additional factor.
Table 1.
Characteristics of included studies in the meta-analysis
| Author | Years/countries | Ethnicity | Sources | SNPs | Genotypes | Sample size | HWE in control | NOS score | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Healthy control | HBV recovery | HBV persistence | ||||||||
| Martin MP [20] | 2010/United States | Caucasian | Hospital-based | rs12979860 |
CC/CT/TT T vs C |
– |
157/175/52 279 vs 489 |
99/94/33 160 vs 292 |
0.77 | 7 |
| L.J.Peng [21] | 2011/China | Asian | Population-based | rs12979860 | CC/CT + TT | – | 206/22 | 574/77 | > 0.05 | 8 |
| Li WY [22] | 2011/China | Asian | Hospital-based | rs12979860 | CC/CT + TT | 179/24 | 180/23 | 178/25 | > 0.05 | 8 |
| rs12980275 | AA/AG + GG | 178 vs 25 | 181/22 | 179/24 | > 0.05 | |||||
| rs8099917 | TT/GT + GG | 182/21 | 182/21 | 180/23 | > 0.05 | |||||
| Fabris C [23] | 2011/Italy | Caucasian | Population-based | rs12979860 |
CC/CT/TT T vs C |
164/145/35 215 vs 473 |
– |
36/35/4 43 vs 107 |
0.386 | 8 |
| Chen J [24] | 2012/China | Asian | Population-based | rs12979860 |
CC/CT/TT T vs C |
213/29/2 33 vs 455 |
– |
1043/152/10 172 vs 2238 |
0.37 | 8 |
| rs12980275 |
AA/AG/GG G vs A |
210/33/1 35 vs 453 |
– |
1036/162/8 178 vs 2234 |
> 0.05 | |||||
| rs8099917 |
GG/GT/TT G vs T |
218/25/1 27 vs 461 |
– |
1075/126/4 130 vs 2276 |
> 0.05 | |||||
| Ren S [19] | 2012/China | Asian | Hospital-based | rs12979860 |
CC/CT/TT T vs C |
43/4/0 4 vs 90 |
33/7/3 13 vs 73 |
177/46/16 78 vs 400 |
0.01 | 8 |
| rs12980275 |
AA/AG/GG G vs A |
43/3/1 5 vs 89 |
37/5/1 7 vs 79 |
205/30/4 38 vs 440 |
0.01 | |||||
| rs8099917 |
GG/GT/TT G vs T |
44/2/1 4 vs 90 |
33/6/4 14 vs 72 |
197/31/11 53 vs 425 |
0.07 | |||||
| Luz MC [25] | 2012/Spain | Caucasian | Hospital-based | rs12979860 |
CC/CT/TT T vs C |
– |
22/21/6 33 vs 65 |
29/17/3 23 vs 75 |
0.78 | 7 |
| Lee DH [26] | 2013/South Korea | Asian | Hospital-based | rs12979860 | T vs C | – | 24 vs 380 | 62 vs 973 | > 0.05 | 7 |
| rs12980275 | G vs A | – | 27 vs 377 | 70 vs 965 | > 0.05 | |||||
| rs8099917 | G vs T | – | 22 vs 382 | 56 vs 979 | > 0.05 | |||||
| Al-Qahtani [27] | 2013/Saudi Arabia | Caucasian | Hospital-based | rs12979860 |
CC/CT/TT T vs C |
– |
174/91/20 131 vs 439 |
450/274/92 458 vs 1174 |
> 0.05 | 7 |
| rs12980275 |
AA/AG/GG G vs A |
– |
175/93/36 165 vs 443 |
548/229/44 317 vs 1325 |
> 0.05 | |||||
| rs8099917 |
GG/GT/TT G vs T |
– |
15/73/216 103 vs 505 |
31/187/606 249 vs 1399 |
> 0.05 | |||||
| Kim SU [13] | 2013/South Korea | Asian | Hospital-based | rs12979860 |
CC/CT/TT T vs C |
85/15/0 15 vs 185 |
189/31/0 31 vs 409 |
144/10/0 10 vs 298 |
> 0.05 | 7 |
| rs12980275 |
AA/AG/GG G vs A |
90/16/0 16 vs 196 |
208/34/1 36 vs 450 |
185/18/0 18 vs 388 |
> 0.05 | |||||
| rs8099917 |
GG/GT/TT G vs T |
90/16/0 16 vs 196 |
215/26/0 26 vs 456 |
192/12/0 12 vs 396 |
> 0.05 | |||||
| Seto WK [28] | 2013/China | Asian | Hospital-based | rs12979860 | T vs C | – | 11 vs 192 | 14 vs 189 | > 0.05 | 7 |
| rs8099917 | G vs T | – | 13 vs 190 | 15 vs 188 | > 0.05 | |||||
| Senem CK [29] | 2014/Turkey | Caucasian | Population-based | rs12979860 |
CC/CT/TT T vs C |
– |
46/65/6 77 vs 157 |
215/201/57 315 vs 631 |
> 0.05 | 7 |
| rs12980275 |
AA/AG/GG G vs A |
– |
36/67/15 97 vs 139 |
191/207/58 323 vs 589 |
> 0.05 | |||||
| Akkiz H [30] | 2014/Turkey | Caucasian | Hospital-based | rs12979860 |
CC/CT/TT T vs C |
92/91/25 141 vs 275 |
– |
43/50/17 84 vs 136 |
0.30 | 7 |
| Liao Y [31] | 2014/ China | Asian | Hospital-based | rs12979860 |
CC/CT + TT T vs C |
– |
420/66 66 vs 906 |
391/66 70 vs 844 |
> 0.05 | 8 |
| rs12980275 |
AA/AG + GG G vs A |
– |
418/68 68 vs 904 |
388/69 77 vs 842 |
> 0.05 | |||||
| rs8099917 |
GG/GT + TT G vs T |
– |
470/16 16 vs 956 |
433/24 24 vs 890 |
> 0.05 | |||||
| Shi XD [32] | 2015/China | Asian | Population-based | rs12979860 |
CC/CT/TT T vs C |
19/0/0 0 vs 38 |
– |
114/23/0 23 vs 251 |
> 0.05 | 7 |
| Kimkong [33] | 2015/Thailand | Asian | Hospital-based | rs12979860 |
CC/CT/TT T vs C |
– |
140/24/0 24 vs 304 |
116/12/0 12 vs 244 |
> 0.05 | 7 |
| rs8099917 |
GG/GT/TT G vs T |
– |
0/25/139 25 vs 303 |
0/8/120 8 vs 248 |
> 0.05 | |||||
| Song YZ [14] | 2017/China | Asian | Hospital-based | rs12979860 |
CC/CT/TT T vs C |
400/57/3 63 vs 857 |
– |
439/52/2 56 vs 930 |
0.79 | 7 |
| rs12980275 |
AA/AG/GG G vs A |
398/58/4 66 vs 854 |
– |
437/55/1 57 vs 929 |
0.50 | |||||
| rs8099917 |
GG/GT/TT G vs T |
2/52/406 56 vs 864 |
– |
1/49/443 51 vs 935 |
> 0.05 | |||||
| Baghbani JM [34] | 2017/Iran | Caucasian | Hospital-based | rs12979860 |
CC/CT/TT T vs C |
– |
28/49/17 83 vs 105 |
31/44/8 60 vs 106 |
> 0.05 | 7 |
| rs8099917 |
GG/GT/TT G vs T |
– |
9/32/53 50 vs 138 |
4/17/62 25 vs 141 |
> 0.05 | |||||
Association of IL28B polymorphisms with HBV persistence
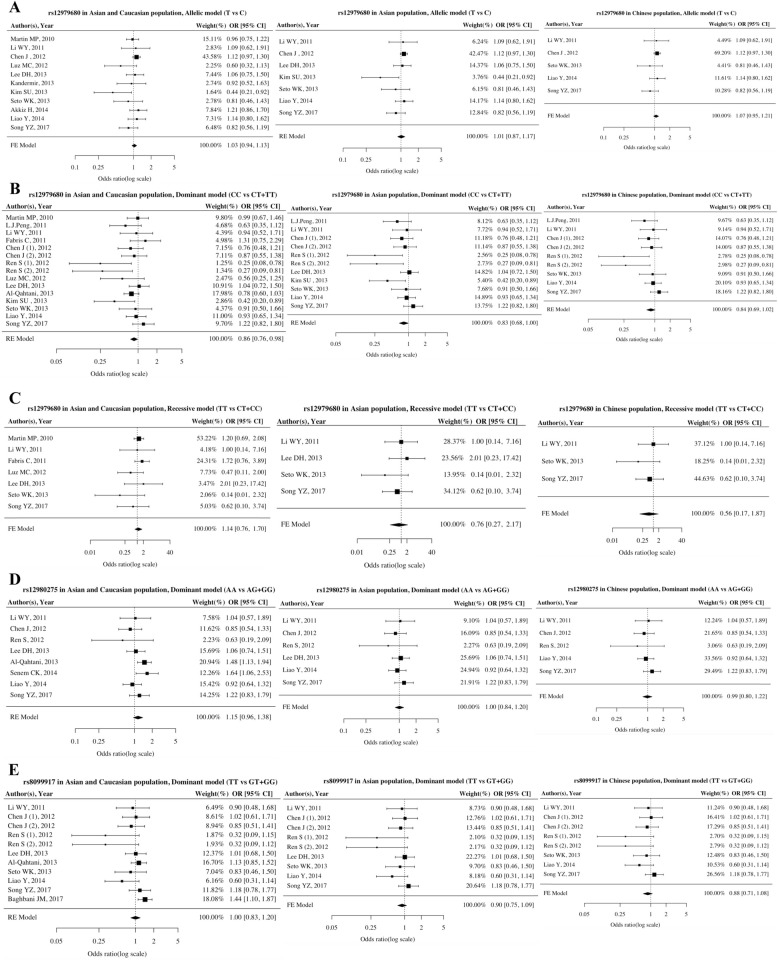
The association between IL28B polymorphisms and HBV infection are shown in Fig. 2. There was no association of the SNP rs12979860 with the risk of HBV persistence based on the allelic model (T vs C: OR = 1.03, 95% CI = 0.94–1.13, P = 0.53), the dominant model (CC vs CT + TT: OR = 0.86, 95% CI = 0.76–1.00, P = 0.05), or the recessive model (TT vs CT + CC: OR = 1.14, 95% CI = 0.76–1.70, P = 0.53). As few studies provided sufficient information on rs12980275 and rs8099917, these 2 SNPs were only considered in a dominant model. There was no association of these genotypes with HBV persistent infection (rs12980275 in AA vs AG + GG: OR = 1.15, 95% CI = 0.96–1.38, P = 0.17; rs8099917 in TT vs GT + GG: OR = 1.00, 95% CI = 0.83–1.20, P = 0.99).
Fig. 2.
Forest plots of association between IL28B polymorphisms and HBV persistence. a Overall analysis and subgroup analysis of rs12979860 under the allelic model. b Overall analysis and subgroup analysis of rs12979860 under the dominant model. c Overall analysis and subgroup analysis of rs12979860 under the recessive model. d Overall analysis and subgroup analysis of rs12980275 under the dominant model. e Overall analysis and subgroup analysis of rs8099917 under the dominant model
Subgroup analysis was performed for Asian and Chinese populations. There was no evidence that IL28B polymorphisms (rs12979860, rs12980275 and rs8099917) were associated with persistent HBV infection in Asian or Chinese populations. For Asians, rs12979860 had no association with HBV infection in the allelic, dominant or recessive model (T vs C: OR = 1.01, 95% CI = 0.87–1.17, P = 0.86; CC vs CT + TT: OR = 0.83, 95% CI = 0.68–1.00, P = 0.05; TT vs CT + CC: OR = 0.76, 95% CI = 0.27–2.17, P = 0.61). SNPs rs12980275 and rs8099917 also had no association with HBV infection in Asians using a fixed-effects model (rs12980275 in AA vs AG + GG: OR = 1.00, 95% CI = 0.84–1.20, P = 0.99; rs8099917 in TT vs GT + GG: OR = 0.90, 95% CI = 0.75–1.09, P = 0.28). In Chinese, none of these 3 SNPs were associated with HBV persistence (rs12979860 in T vs C: OR = 1.07, 95% CI = 0.95–1.21, P = 0.25; CC vs CT + TT: OR = 0.84, 95% CI = 0.69–1.02, P = 0.10; TT vs CT + CC: OR = 0.56, 95% CI = 0.17–1.87, P = 0.35; rs12980275 in AA vs AG + GG: OR = 0.98, 95% CI = 0.80–1.22, P = 0.89; rs8099917 in TT vs GT + GG: OR = 0.87, 95% CI = 0.71–1.08, P = 0.28) (Table 2).
Table 2.
Overall meta-analysis of the association of IL28B polymorphisms with persistent HBV infection
| SNP | Model | Ethnicity | Studies Number | OR | Heterogeneity | Publication bias | ORS | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | POR | I2 (%) | PH | M | PBegg | PEgger | |||||
| rs12979860 | Allelic | Total | 11 | 1.03 (0.94, 1.13) | 0.528 | 24.6 | 0.210 | F | 0.010 | 0.015 | 1.06 (0.97, 1.17) |
| Asian | 7 | 1.01 (0.87, 1.17) | 0.863 | 33.1 | 0.175 | R | 0.069 | 0.032 | 1.07 (0.96, 1.21) | ||
| Chinese | 5 | 1.07 (0.95,1.21) | 0.245 | 0.0 | 0.476 | F | 0.233 | 0.237 | Not necessary | ||
| Dominant | Total | 15 | 0.86 (0.76, 1.00) | 0.050 | 37.2 | 0.073 | R | 0.016 | 0.003 | 0.91 (0.78, 1.06) | |
| Asian | 11 | 0.83 (0.68, 1.00) | 0.051 | 44.7 | 0.053 | R | 0.006 | < 0.001 | 0.95 (0.73, 1.26) | ||
| Chinese | 9 | 0.84 (0.69, 1.02) | 0.081 | 40.8 | 0.095 | R | 0.045 | 0.001 | 0.98 (0.76, 1.26) | ||
| Recessive | Total | 7 | 1.14 (0.76, 1.70) | 0.526 | 0.0 | 0.504 | F | 0.562 | 0.200 | Not necessary | |
| Asian | 4 | 0.76 (0.27, 2.17) | 0.611 | 0.0 | 0.513 | F | 1.000 | 0.415 | Not necessary | ||
| Chinese | 3 | 0.56 (0.17, 1.87) | 0.350 | 0.0 | 0.526 | F | 1.000 | 0.327 | Not necessary | ||
| rs12980275 | Dominant | Total | 8 | 1.15 (0.96, 1.38) | 0.135 | 32.5 | 0.168 | R | 0.399 | 0.177 | Not necessary |
| Asian | 6 | 1.00 (0.84, 1.20) | 0.966 | 0.0 | 0.795 | F | 0.469 | 0.453 | Not necessary | ||
| Chinese | 5 | 0.98 (0.80, 1.22) | 0.890 | 0.0 | 0.689 | F | 0.483 | 0.495 | Not necessary | ||
| rs8099917 | Dominant | Total | 11 | 1.00 (0.83, 1.20) | 0.992 | 41.2 | 0.074 | R | 0.002 | < 0.001 | 1.15 (0.96, 1.39) |
| Asian | 9 | 0.90 (0.75, 1.09) | 0.284 | 11.0 | 0.343 | F | 0.006 | 0.006 | 0.99 (0.83, 1.18) | ||
| Chinese | 8 | 0.87 (0.71, 1.08) | 0.214 | 18.5 | 0.283 | F | 0.0141 | 0.006 | 0.98 (0.81, 1.19) | ||
SNP single nucleotide polymorphism, M model for meta-analysis, F Fixed-effects model, R Random-effects model, PHP-value for heterogeneity test, PORP-value for OR test, PBeggP-value for Begg’s test, PEggerP-value for Egger’s test, ORS OR derived from trim and fill sensitive analysis
Association of IL28B polymorphism rs12979860 with HBV-related HCC
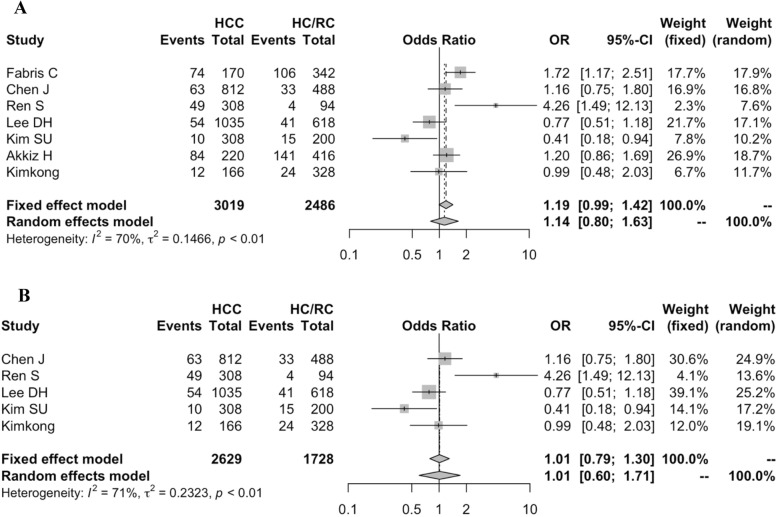
HBV-related HCC is the most serious consequence of persistent HBV infection [35]. In order to explore the disease outcome-specific association with genetic factors, we further investigated the association of IL28B polymorphisms with the risk of HBV-related HCC. However, few studies provided sufficient information on that, hence We only assess the association of rs12979860 C/T with the development of HBV-related HCC in the allelic model. As shown in Fig. 3, 7 studies comprising 3019 cases and 2486 controls were included in this meta-analysis. A random-effects model was used to estimate the association of the rs12979860 polymorphism with the risk of HCC in HCC patients and healthy controls (HC) or recovered controls (RC). No significant association was observed of the SNP rs12979860 with HBV-related HCC susceptibility (T vs C: OR = 1.14, 95% CI = 0.80–1.63). Subgroup analysis was performed for Asians population, but rs12979860 had no significant association with the development of HBV-related HCC (T vs C: OR = 1.01, 95% CI = 0.60–1.71).
Fig. 3.
Forest plot for association between IL28B rs12979860 polymorphism and the development of HBV-related HCC (T vs C). a Overall analysis; b Subgroup analysis
Sensitivity analysis
Association of IL28B polymorphisms with HBV persistence
Heterogeneity of the associations between IL28B polymorphisms and persistent HBV infection varied from 0 to 44.7%. We found low heterogeneity for the SNP rs12979860 in the allelic and recessive models (T vs C: I2 = 24.6, PH = 0.21; TT vs CT + CC: I2 = 0, PH = 0.50). However, heterogeneity in the dominant model for IL28B polymorphisms (rs12979860, rs12980275 and rs8099917) was significant. Subgroup analyses were carried out to identify sources of heterogeneity. After stratifying by Asian population, there was no heterogeneity for rs12980275 and rs8099917 in the dominant model (rs12980275 in T vs G: I2 = 0, PH = 0.80; rs8099917 in A vs G: I2 = 11%, PH = 0.34). However, heterogeneity in CC vs CT + TT for rs12979860 was still higher than 25%. We further performed the subgroup analysis stratified by Chinese population, but heterogeneity remained for CC vs CT + TT (I2 = 40.8%, PH = 0.095). Sequential omission of individual studies (Additional file 1), after excluding the study of Kim SU [13], the significant pooled OR was no longer significant for CC vs CT + TT (OR = 0.88, 95% CI = 0.75–1.01), but heterogeneity still existed (I2 = 40%). In addition, there was a significant decline in heterogeneity (I2 = 27%), with excluding the study of Ren S [19]. However, it was still higher than 25%.
Association of IL28B polymorphism rs12979860 with HBV-related HCC
Heterogeneity of these 7 studies was 70%. Sensitivity analysis was performed using the sequential omission approach. However, the heterogeneity remained high, and a random-effects model was used to estimate the pooled ORs. The overall result was not influenced by the sequential omission approach. The 7 pooled ORs and corresponding 95% CIs were presented in Additional file 1.
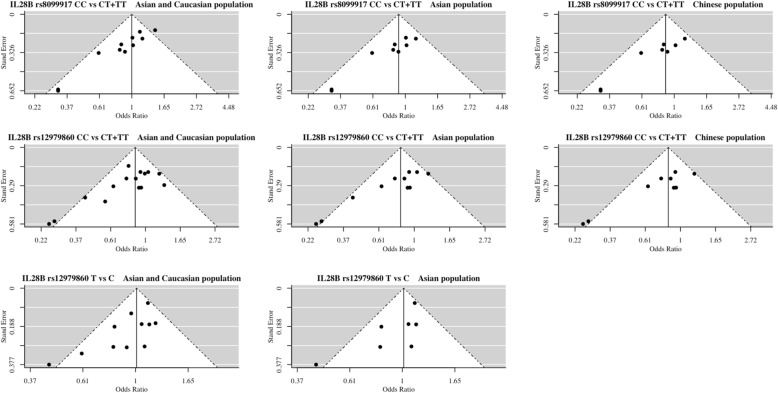
Publication Bias
Association of IL28B polymorphisms with HBV persistence
In overall and subgroup analysis, publication bias occurred in the allelic and dominant model for rs12979860 and occurred in the dominant model for rs8099917. Begg’s rank correlation, Egger’s weighted regression and trim and fill method were used as the sensitivity analysis to examine the publication bias (Table 2). The presence of publication bias was shown in Fig. 4.
Fig. 4.
Funnel plot analysis to examine publication bias
Association of IL28B polymorphism rs12979860 with HBV-related HCC
There was no obvious asymmetry in the funnel plot for the total population or the Asian population. No publication bias was observed for the association of SNP rs12979860 with HBV-related HCC susceptibility (Total population: PBegg = 0.773, PEgger = 0.700; Asian population: PBegg = 0.817, PEgger = 0.400).
Discussion
In this meta-analysis, we examined the association of IL28B polymorphisms with the outcome of HBV infection. We performed a meta-analysis of the associations of IL28B polymorphisms (rs12979860, rs12980275 and rs8099917) with HBV persistence. Then we assessed the association of the SNP rs12979860 with the risk of HBV-related HCC. Overall, there was no strong evidence of an association of IL28B polymorphisms with HBV infection. As such, these 3 SNPs do not appear to be risk factors for hepatitis B progression. This meta-analysis provides a reliable and comprehensive assessment of the association of IL28B polymorphisms with HBV infection. This study is the first systematic review that included all relevant eligible English publications in the recent 8 years. Meanwhile, it had a larger sample size comprising 5587 cases and 4295 than previous studies [36, 37]. Previous studies investigating these associations may have included low-quality publications [38, 39].
Firstly, rs12979860, rs12980275 and rs8099917 had no association with HBV persistence, overall or by subgroup although the heterogeneity was still higher than 25% but was not explained by any individual study. This meta-analysis is consistent with the study of DH Lee [26] showing that IL28B polymorphisms are unrelated to HBV infection, assessed from HBsAg seroclearance, rather than the risk of persistent HBV infection and progression of HBV-related HCC. Moreover, according to the strict inclusion criteria we excluded some studies [38, 39] included by Lee. In addition, we included totally 18 recent publications in this meta-analysis, and the quantity was greater than in the 11 publications included in Lee’s study. Similarly, this study is consistent with the study of Tang Sd. et al. [40]. However, there is a difference between that study and this study. In their study, only 9 publications were included and only the rs12979860 C/T polymorphism was investigated to evaluate the relationship. In our study, 18 publications were included to investigate the association of 3 SNPs with HBV persistence. For subgroup analysis, results differed from that in Chen J’s study [24]. They found a significant association of rs12979680 with the risk of persistent HBV infection among Chinese population under an additive model. However, we found no such association in either an allelic, dominant or recessive model. This difference might have occurred because of sample size. There were 406 cases and 244 controls included in Chen J’s study. Smaller studies may have a higher chance of false positives. A published study with small sample size and significant result is more likely to be a false positive because of the low sensitivity [41]. Meanwhile, there was no general principle of gene model selection [42]. However, it is easier to apply logistic regression analysis to dominant and recessive model, which would be more precise to estimate the pooled OR [42].
IFN-λ3 is coded by the IL28B gene and has an antiviral effect on chronic HCV [43]. Many studies [44–47] have shown an association of IL28B polymorphisms with HCV infection. However, the nucleotide analogs may have an additional pharmacological effect by inducing IFN-λ3 production, which facilitates interferon-stimulated genes and decreases the production of HBsAg [48]. Moreover, HBV and HCV share a similar natural history, pathogenesis and transmission modality. In order to provide novel insights for HBV treatment, focus has been on the association of IL28B polymorphisms with HBV infection [49]. Although several studies [24, 27, 32] have demonstrated IL28B polymorphisms predict HBV infection. There is a possibility of producing a false positive result. In the study of Martin MP [20], despite they revealed that the SNP rs12979860 was associated with spontaneous clearance of HBV, this significant association was merely found among one-fifth of patients who were also chronic HCV carriers and 69% patients with HIV coinfection. Therefore, we excluded participants testing positive for anti-HCV or anti-HIV, because the correlation of these diseases with IL28B genes would potentially increase the likelihood of false positive in our estimate.
Secondly, rs12979860 had no association with the risk of HBV-related HCC, overall or by subgroup. No publication bias was found by either Begg’s rank correlation method or Egger’s weighted regression method. Nevertheless, this estimate might be underpowered due to the limited number of studies. We obtained consistent findings with a few studies [15, 17, 26]. However, this meta-analysis failed to reach a consensus with the studies of Fabris and Ren S [19, 23]. In the study of Fabris, the researchers found rs12979860 polymorphism contributed to the risk of HCC among chronic HCV patients. However, they did not observe the association of the rs12979860 polymorphism with HBV-related HCC. That might explain the discrepancy. GWAS have identified the IL28B gene as a major determinant of the course of HCV infection, and experimental evidence indicates that IFN-λ3 coded by IL28B gene has an anti-tumor effect [50, 51]. The SNP rs12979860 C/T might only have significant association with the risk of HCV-related HCC. In the study of Ren S, they found a significant association between rs12979860 and HBV-related HCC in the Chinese population. However, we obtained an inconsistent result based on the combined evidence. As HBV involves a complicated interplay between host and virus, no single molecular analysis was expected to fully unravel the disease mechanism. Multiple molecular levels could interact and also show plasticity in different physiological conditions and disease stages [52]. Hence there is a great requirement for additional novel approaches that could combine data from different molecular levels and could help to determine the causal path from genotype to phenotype.
Another thing we would highlight about the HBV-related HCC, is that it is thought to involve the interaction between environmental factors and inherited susceptibility [53]. For instance, recent case-control studies found a positive association between African dietary iron overload and HBV/HCV-related HCC [54–56]. The underlying mechanism could be excessive free irons generating harmful “reactive oxygen species” (ROS) under aerobic conditions in the human body. ROS could damage the balance between oxidative and reductive events, and consequently destroy DNA in hepatocytes and induce HCC [57]. This finding requires further investigation due to the limitation of observational studies. However, dietary iron overload might increase the risk of developing HBV-relate HCC in Africans and might disguise the potential association of the IL28B gene with the disease.
This study is a candidate gene association study which is different from GWAS. Candidate gene studies are relatively cheap and quick to perform, and are concentrated on the selection of a putative candidate genetic variants according to its relevance in the mechanism of the disease investigated. However, candidate gene studies are susceptible to publication bias, and they are underpowered due to high rates of false positive findings and low rates of replication [58]. In contrast, GWAS are more reliable and comprehensive approach for identifying loci in the genome that might be relevant to the interest disease. Millions of SNPs in the genome are assessed for genotype-phenotype association. However, the cost of genotyping for a GWAS is at least an order of magnitude higher than the cost to genotype several candidate genes [59]. Hence, candidate gene studies are a cost-effective approach to identify genetic risk factors in the absence of GWAS for a specific disease. Currently, no GWAS has assessed the association of IL28B with HBV infection. The unknown history of HBV exposure in the controls or relatively small sample size of the study reduces power to identify genetic variants with modest effect [60].
Conventionally, the association between genotype variants and disease in candidate gene studies was discovered from case-control studies. However, there might be a spurious correlation despite the observed statistical significance. Candidate gene studies are prone to false positive findings for the following reasons. Firstly, the allele may be in linkage disequilibrium with an allele at another locus that directly affects the expression of the phenotype. Secondly, the allele itself is functional and directly affects the expression of the phenotype [61]. Thirdly, differences in the frequency of genetic variants within the population might lead to false positive results. In this study, the minor allele frequencies of these 3 SNPs in Asians were significantly lower than those of the Caucasians, suggesting the presence of population specificity [62]. Lastly, some systemic technical bias would be generated due to different DNA samples from cases and controls, and eventually induced a false positive relationship [63].
There were also several advantages that would further strengthen this study compared to previous similar studies. Above all, the publications we included in this meta-analysis were high-quality based on the NOS score, which increased the internal validity. Moreover, we followed the strict inclusion criteria such as no-coinfection with HCV or HIV, which decreased the bias generated by correlations of these diseases with IL28B polymorphisms. Thirdly, this meta-analysis included a total of 5587 cases and 4295, comprising 4913 cases and 3865 controls for rs12979860 C/T, 3765 cases and 2506 controls for rs12980275 A/G, and 3912 cases and 2900 controls for rs8099917 T/G, which were even more than twice the subjects included in previous studies. In addition, we performed a more comprehensive subgroup analysis by ethnicity, because of potential differences between populations. Lastly, this study is the first systematic review that included all relevant eligible English publications in the recent 8 years. Systematic reviews provide a synthesis of evidence for clinical decisions. Hence having an up-to-date and comprehensive review is imperative.
The findings of this systematic review and meta-analysis were limited by several factors. Firstly, the study design included only case-control studies. However, genetic case-control studies are susceptible to selection bias. The key requirement for a case-control study to avoid selection bias is that the cases and controls are selected from the same underlying population, which is difficult to ascertain. Thirdly, the genotype distribution deviated from HWE in 1 study, which might be attributed to population stratification or potential selection bias. Fourthly, we did not have original data for all studies to account for potential confounding however, genetic case control studies are not open to confounding, because few factors are determinants of genetic make-up. Fifthly, the assay methods used to characterize the polymorphisms differed in these studies, i.e. the sensitivity and specificity of identifying IL28B polymorphisms were different, which might reduce the reliability. Lastly, although we included 18 studies in this meta-analysis, the presence of heterogeneity and publication bias indicated potential issue. There was still a great need for large, high methodological quality publications from a genome wide association study in both Caucasian and Asian populations to verify our findings.
Conclusion
Genetic epidemiology is a vital composition of public health research. However, spurious associations might also generate inefficient treatments and waste medical resources. This study is a cost-effective approach to investigating the association of IL28B with HBV infection, in the absence of GWAS for this specific association. However, there were still several limitations that might influence the effect of genetic factors on disease outcome. Further well-designed large-scale studies, especially related to the gene-ethnic group and phenotype-environmental interaction are warranted to confirm the real contribution of these polymorphisms to the outcome of HBV infection.
Supplementary information
Additional file 1: Supplement 1. Minor allele frequencies of IL28B polymorphisms in persistent HBV infection patients. Supplement 2. Heterogeneity analysis of each IL28B polymorphism.
Acknowledgements
We would like to express appreciation to Prof. Schooling, Prof. Cowling, Dr. Maggie Kwok and Dr. Eric Lau (The University of Hong Kong) for their time and constructive suggestions on this study.
Abbreviations
- IL 28B
Interleukin (IL) 28B
- HBV
Hepatitis B virus
- SNP
Single nucleotide polymorphism
- HIV
Human immunodeficiency virus
- HCV
Hepatitis C virus
- HHC
Hepatitis hepatocellular carcinoma
- HCC
Hepatocellular carcinoma
- DALYs
Disability-adjusted life-years
- IFN-λ3
Interferon-λ3
- GWAS
Genome-wide association studies
- PEG-IFN-α
Pegylated interferon-α
- OR
Odds ratio
- CI
Confidence interval
- HBsAg
Hepatitis B surface antigen
- anti-HBc
Hepatitis B core antibody
- anti-HBs
Hepatitis B surface antibody
- HWE
Hardy-Weinberg equilibrium
- NOS
Newcastle-Ottawa Scale
- HC
Healthy control
- RC
Recovered control
Authors’ contributions
JS designed the study and served as principal investigators. JYZ and LWF carried out the research, enrolled patients, included the initial data, and reviewed the manuscript. JYZ, XYZ, LWF and HP collected, analyzed, and interpreted the data. JYZ carried out statistical analysis. JYZ and XYZ wrote the manuscript, and all authors contributed to the final draft. All authors read and approved the final manuscript.
Funding
This work was supported by CAMS Innovation Fund for Medical Sciences (2017-I2M-3-018), the Tianjin Municipal Science and Technology Commission Major Project (18ZXDBSY00070).
The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12881-020-01026-w.
References
- 1.Hepatitis B. https://www.who.int/news-room/fact-sheets/detail/hepatitis-b. Accessed 10 May 2019..
- 2.Bagheri Amiri F, Mostafavi E, Mirzazadeh A. Hiv, Hbv And Hcv coinfection prevalence in Iran. A systematic review and meta-analysis. PloS one. 2016;11(3):e0151946. doi: 10.1371/journal.pone.0151946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muruka C, Muruka A. Guidelines for environmental health Management in Children’s homes in sub-Sahara Africa. Int J Environ Res Public Health. 2007;4(4):319–331. doi: 10.3390/ijerph200704040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.HIV/HBV and HIV/HCV Co-infection in Sub-Saharan Africa: Transmission, Disease Outcomes, and Treatment Options. https://www.worldgastroenterology.org/publications/e-wgn/e-wgn-expert-point-of-view-articles-collection/hiv-hbv-and-hiv-hcv-co-infection-in-sub-saharan-africa. Accessed 11 May 2019.
- 5.Clements CJ, Coghlan B, Creati M, et al. Global control of hepatitis B virus: does treatment-induced antigenic change affect immunization? Bull World Health Organ. 2010;88(1):66–73. doi: 10.2471/BLT.08.065722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zidan A, Scheuerlein H, Schüle S, et al. Epidemiological pattern of hepatitis B and hepatitis C as etiological agents for hepatocellular carcinoma in iran and worldwide. Hepat Mon. 2012;12(10 HCC):e6894. doi: 10.5812/hepatmon.6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das S, Ramakrishnan K, Behera SK, et al. Hepatitis B vaccine and immunoglobulin: key concepts. J Clin Transl Hepatol. 2019;7(2):165–171. doi: 10.14218/JCTH.2018.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shariff MIF, Cox IJ, Gomaa AI, et al. Hepatocellular carcinoma: current trends in worldwide epidemiology, risk factors, diagnosis and therapeutics. Expert Rev Gastroenterol Hepatol. 2009;3(4):353–367. doi: 10.1586/egh.09.35. [DOI] [PubMed] [Google Scholar]
- 9.Croagh CMN, Lubel JS. Natural history of chronic hepatitis B: phases in a complex relationship. World J Gastroenterol. 2014;20(30):10395–10404. doi: 10.3748/wjg.v20.i30.10395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Indolfi LG, Mangone LG, Calvo LP, et al. Interleukin 28B rs12979860 single-nucleotide polymorphism predicts spontaneous clearance of hepatitis C virus in children. J Pediatr Gastroenterol Nutr. 2014;58(5):666–668. doi: 10.1097/MPG.0000000000000275. [DOI] [PubMed] [Google Scholar]
- 11.Barathiraja S, Gangadhara PAV, Umapathi V, et al. Expression and purification of biologically active bovine interferon λ3 (IL28B) in Pichia pastoris. Protein Expr Purif. 2018;145:14–18. doi: 10.1016/j.pep.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Sonneveld MJ, Wong VWS, Woltman AM, et al. Polymorphisms Near IL28B and Serologic Response to Peginterferon in HBeAg-Positive Patients With Chronic Hepatitis B. Gastroenterology. 2012;142(3):513–20.e1. doi: 10.1053/j.gastro.2011.11.025. [DOI] [PubMed] [Google Scholar]
- 13.Kim SU, Song KJ, Chang HY, et al. Association between IL28B polymorphisms and spontaneous clearance of hepatitis B virus infection. PLoS One. 2013;8(7):e69166. doi: 10.1371/journal.pone.0069166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song Y, Shen Y, Xia X, et al. Association between genetic polymorphisms of the gene and leukomonocyte in Chinese hepatitis B virus-infected individuals. PeerJ. 2017;5(12):e4149. doi: 10.7717/peerj.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He J, Yu G, Li Z, et al. Influence of interleukin-28B polymorphism on progression to hepatitis virus-induced hepatocellular carcinoma. Tumor Biol. 2014;35(9):8757–8763. doi: 10.1007/s13277-014-2142-3. [DOI] [PubMed] [Google Scholar]
- 16.Suo GJ, Zhao ZX. Association of the interleukin-28B gene polymorphism with development of hepatitis virus-related hepatocellular carcinoma and liver cirrhosis: a meta-analysis. Genet Mol Res. 2013;12(3):3708. doi: 10.4238/2013.September.19.1. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Zhu S-L, Chen J, et al. Meta-analysis of associations of interleukin-28B polymorphisms rs8099917 and rs12979860 with development of hepatitis virus-related hepatocellular carcinoma. Onco Targets Ther. 2016;9:3249. doi: 10.2147/OTT.S104904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snast I, Reiter O, Atzmony L, et al. Psychological stress and psoriasis: a systematic review and meta-analysis. Br J Dermatol. 2018;178(5):1044–1055. doi: 10.1111/bjd.16116. [DOI] [PubMed] [Google Scholar]
- 19.Ren S, Lu J, Du X, et al. Genetic variation in IL28B is associated with the development of hepatitis B-related hepatocellular carcinoma. Cancer Immunol Immunother. 2012;61(9):1433–1439. doi: 10.1007/s00262-012-1203-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin MP, Qi Y, Goedert JJ, et al. IL28B polymorphism does not determine outcomes of hepatitis B virus or HIV infection. J Infect Dis. 2010;202(11):1749. doi: 10.1086/657146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng LJ, Guo JS, Zhang Z, et al. IL28B rs12979860 polymorphism does not influence outcomes of hepatitis B virus infection. Tissue Antigens. 2012;79(4):302–305. doi: 10.1111/j.1399-0039.2011.01835.x. [DOI] [PubMed] [Google Scholar]
- 22.Li W, Jiang Y, Jin Q, et al. Expression and gene polymorphisms of interleukin 28B and hepatitis B virus infection in a Chinese Han population. Liver Int. 2011;31(8):1118–1126. doi: 10.1111/j.1478-3231.2011.02507.x. [DOI] [PubMed] [Google Scholar]
- 23.Fabris C, Falleti E, Cussigh A, et al. IL-28B rs12979860 C/T allele distribution in patients with liver cirrhosis: role in the course of chronic viral hepatitis and the development of HCC. J Hepatol. 2011;54(4):716–722. doi: 10.1016/j.jhep.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Wang L, Li Y, et al. Association Analysis between SNPs in IL-28B Gene and the Progress of Hepatitis B Infection in Han Chinese. PLoS One. 2012;7(12):e50787. doi: 10.1371/journal.pone.0050787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martín-Carbonero L, Rallón NI, Benito JM, et al. Short communication: does interleukin-28B single nucleotide polymorphisms influence the natural history of hepatitis B? AIDS Res Hum Retrovir. 2012;28(10):1262–1264. doi: 10.1089/AID.2011.0365. [DOI] [PubMed] [Google Scholar]
- 26.Lee DH, Cho Y, Seo JY, et al. Polymorphisms near interleukin 28B gene are not associated with hepatitis B virus clearance, hepatitis B e antigen clearance and hepatocellular carcinoma occurrence. Intervirology. 2013;56(2):84–90. doi: 10.1159/000342526. [DOI] [PubMed] [Google Scholar]
- 27.Al-Qahtani AA, Al-Anazi MR, Abdo AA, et al. Genetic variation in interleukin 28B and correlation with chronic hepatitis B virus infection in Saudi Arabian patients. Liver Int. 2014;34(7):e208–ee16. doi: 10.1111/liv.12347. [DOI] [PubMed] [Google Scholar]
- 28.Seto W-K, Wong D, Kopaniszen M, et al. HLA-DP and IL28B polymorphisms: influence of host genome on hepatitis B surface antigen Seroclearance in chronic hepatitis B. Clin Infect Dis. 2013;56(12):1695. doi: 10.1093/cid/cit121. [DOI] [PubMed] [Google Scholar]
- 29.Karatayli SC, Bozdayi M, Karatayli E, et al. Interleukin-28 gene polymorphisms may contribute to HBsAg persistence and the development of HBeAg-negative chronic hepatitis B. Liver Int. 2015;35(3):846–853. doi: 10.1111/liv.12595. [DOI] [PubMed] [Google Scholar]
- 30.Akkiz H, Kuran S, Akgollu E, et al. The role of interleukin 28B gene polymorphism in Turkish patients with hepatocellular carcinoma. Ann Hepatol. 2014;13(6):788–795. [PubMed] [Google Scholar]
- 31.Liao Y, Cai B, Li Y, et al. Association of HLA-DP/DQ, STAT4 and IL-28B variants with HBV viral clearance in Tibetans and Uygurs in China. Liver Int. 2015;35(3):886–896. doi: 10.1111/liv.12643. [DOI] [PubMed] [Google Scholar]
- 32.Shi X, Chi X, Pan Y, et al. IL28B is associated with outcomes of chronic HBV infection. Yonsei Med J. 2015;56(3):625–633. doi: 10.3349/ymj.2015.56.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kimkong I, Chankaew J, Kunanopparat A, et al. Gene polymorphisms of interleukin 28B and the risk to chronic hepatitis B virus infection in Thai. Tissue Antigens. 2015;85(3):177–181. doi: 10.1111/tan.12517. [DOI] [PubMed] [Google Scholar]
- 34.Gerami S, Ghojazadeh M. Interleukin 28B Genetic Polymorphism and Spontaneous Recovery from Hepatitis B Virus Infection in an Iranian Azeri Population. Hepat Mon. 2017;17:9. [Google Scholar]
- 35.Ni YH, Chang MH, Hsu HY, et al. Different hepatitis B virus core gene mutations in children with chronic infection and hepatocellular carcinoma. Gut. 2003;52(1):122–125. doi: 10.1136/gut.52.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee DH, Lee JH, Kim YJ, et al. Relationship between polymorphisms near the IL28B gene and spontaneous HBsAg seroclearance: a systematic review and meta-analysis. J Viral Hepat. 2014;21(3):163–170. doi: 10.1111/jvh.12193. [DOI] [PubMed] [Google Scholar]
- 37.Xia P, Zhou M, Dong D, et al. Association of polymorphisms in interleukin-18 and interleukin-28B genes with outcomes of hepatitis B virus infections: a meta-analysis. Tumor Biol. 2014;35(2):1129–1137. doi: 10.1007/s13277-013-1151-y. [DOI] [PubMed] [Google Scholar]
- 38.Yapali S, Cetintas A, Salman E, et al. 492 IL28B genotype has no impact on spontaneous hepatitis B surface antigen (HBsAg) clearance in inactive carries: a long term, case-matched retrospective cohort study. J Hepatol. 2012;56:S193. [Google Scholar]
- 39.Linda D, Michael JC, Jonathan D, et al. A multicentre molecular analysis of hepatitis B and blood-borne virus coinfections in Viet Nam. PLoS One. 2012;7(6):e39027. doi: 10.1371/journal.pone.0039027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang S, Yue M, Wang J, et al. Associations of IFN-γ rs2430561 T/a, IL28B rs12979860 C/T and ERα rs2077647 T/C polymorphisms with outcomes of hepatitis B virus infection: a meta-analysis. J Biomed Res. 2014;28(6):484–493. doi: 10.7555/JBR.28.20130162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ioannidis JPA. Why Most published research findings are false (essay) PLoS Med. 2005;2(8):e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horita N, Kaneko T. Genetic model selection for a case–control study and a meta-analysis. Meta Gene. 2015;5:1–8. doi: 10.1016/j.mgene.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rallón NI, Soriano V, Naggie S, et al. Impact of IL28B gene polymorphisms on interferon-λ3 plasma levels during pegylated interferon-α/ribavirin therapy for chronic hepatitis C in patients coinfected with HIV. J Antimicrob Chemother. 2012;67(5):1246–1249. doi: 10.1093/jac/dkr598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayes CN, Michio I, Hiroshi A, Kazuaki C. Genetics of IL28B and HCV—response to infection and treatment. Nat Rev Gastroenterol Hepatol. 2012;9(7):406. doi: 10.1038/nrgastro.2012.101. [DOI] [PubMed] [Google Scholar]
- 45.Bibert S, Roger T, Calandra T, et al. IL28B expression depends on a novel TT/−G polymorphism which improves HCV clearance prediction. J Exp Med. 2013;210(6):1109. doi: 10.1084/jem.20130012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y, Xu HX, Wang LJ, et al. Meta-analysis: IL28B polymorphisms predict sustained viral response in HCV patients treated with pegylated interferon-α and ribavirin. Aliment Pharmacol Ther. 2012;36(2):91–103. doi: 10.1111/j.1365-2036.2012.05131.x. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka Y, Nishida N, Sugiyama M, et al. Genome-wide association of IL28B with response to pegylated interferon-α and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 48.Murata K, Asano M, Matsumoto A, et al. Induction of IFN-λ3 as an additional effect of nucleotide, not nucleoside, analogues: a new potential target for HBV infection. Gut. 2018;67(2):362–371. doi: 10.1136/gutjnl-2016-312653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singhatiraj E. HIV Co-Infections with Hepatitis B and C. J Aids Clin Res. 2013;01:S3. [Google Scholar]
- 50.Li Q, Kawamura K, Ma G, et al. Interferon-λ induces G1 phase arrest or apoptosis in oesophageal carcinoma cells and produces anti-tumour effects in combination with anti-cancer agents. Eur J Cancer. 2010;46(1):180–190. doi: 10.1016/j.ejca.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 51.Zitzmann K, Brand S, Baehs S, et al. Novel interferon-λs induce antiproliferative effects in neuroendocrine tumor cells. Biochem Biophys Res Commun. 2006;344(4):1334–1341. doi: 10.1016/j.bbrc.2006.04.043. [DOI] [PubMed] [Google Scholar]
- 52.Riva E, Scagnolari C, Turriziani O, et al. Hepatitis C virus and interferon type III (interferon-λ3/interleukin-28B and interferon-λ4): genetic basis of susceptibility to infection and response to antiviral treatment. Clin Microbiol Infect. 2014;20(12):1237–1245. doi: 10.1111/1469-0691.12797. [DOI] [PubMed] [Google Scholar]
- 53.David JH. Gene–environment interactions in human diseases. Nat Rev Genet. 2005;6(4):287. doi: 10.1038/nrg1578. [DOI] [PubMed] [Google Scholar]
- 54.Hagen TM, Huang S, Curnutte J, et al. Extensive oxidative DNA damage in hepatocytes of transgenic mice with chronic active hepatitis destined to develop hepatocellular carcinoma. Proc Natl Acad Sci U S A. 1994;91(26):12808–12812. doi: 10.1073/pnas.91.26.12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han X, Liehr JG. 8-hydroxylation of guanine bases in kidney and liver DNA of hamsters treated with estradiol: role of free radicals in estrogen-induced carcinogenesis. Cancer Res. 1994;54(21):5515–5517. [PubMed] [Google Scholar]
- 56.Asare GA, Mossanda KS, Kew MC, et al. Hepatocellular carcinoma caused by iron overload: a possible mechanism of direct hepatocarcinogenicity. Toxicology. 2006;219(1):41–52. doi: 10.1016/j.tox.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 57.Deodutta R, Dorak MT. Environmental factors, genes, and the development of human cancers. New York: Springer; 2010. [Google Scholar]
- 58.Ioannidis PAJ, Tarone KR, McLaughlin KJ. The false-positive to false-negative ratio in epidemiologic studies. Epidemiology. 2011;22(4):450–456. doi: 10.1097/EDE.0b013e31821b506e. [DOI] [PubMed] [Google Scholar]
- 59.Wilkening S, Chen B, Bermejo JL, et al. Is there still a need for candidate gene approaches in the era of genome-wide association studies? Genomics. 2009;93(5):415–419. doi: 10.1016/j.ygeno.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 60.Li Y, Si L, Zhai Y, et al. Genome-wide association study identifies 8p21.3 associated with persistent hepatitis B virus infection among Chinese. Nat Commun. 2016;7:11664. doi: 10.1038/ncomms11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siliverman E, Palmer L. Case-control association studies for the genetics of complex respiratory diseases. Am J Respir Cell Mol Biol. 2000;22(6):645–648. doi: 10.1165/ajrcmb.22.6.f191. [DOI] [PubMed] [Google Scholar]
- 62.Fraser HB, Lam LL, Neumann SM, Kobor MS. Population-specificity of human DNA methylation. Genome Biol. 2012;13(2):R8. doi: 10.1186/gb-2012-13-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat Genet. 2007;39(7):S37–S42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplement 1. Minor allele frequencies of IL28B polymorphisms in persistent HBV infection patients. Supplement 2. Heterogeneity analysis of each IL28B polymorphism.
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.