Abstract
Despite the serious public health impact of Crimean-Congo hemorrhagic fever (CCHF), the efficacy of antivirals targeting the causative agent, CCHF virus (CCHFV), remains debatable. Neutralizing monoclonal antibodies (MAbs) targeting the CCHFV glycoprotein Gc have been reported to protect mice against challenge with the prototype CCHFV strain, IbAr10200. However, due to extensive sequence diversity of CCHFV glycoproteins, it is unknown whether these MAbs neutralize other CCHFV strains. We initially used a CCHF virus-like particle (VLP) system to generate 11 VLP moieties, each possessing a glycoprotein from a genetically diverse CCHFV strain isolated in either Africa, Asia, the Middle East, or southeastern Europe. We used these VLPs in biosafety level 2 conditions to efficiently screen MAb cross-neutralization potency. Of the 16 MAbs tested, 3 (8A1, 11E7, and 30F7) demonstrated cross-neutralization activity with most CCHF VLPs, with 8A1 neutralizing all VLPs tested. Although binding studies suggest that none of the MAbs compete for the same epitope, combining 11E7, 30F7, or both 11E7 and 30F7 with 8A1 had no additive effect on increasing neutralization in this system. To confirm our findings from the VLP system, the 3 MAbs capable of strain cross-neutralization were confirmed to effectively neutralize 5 diverse CCHFV strains in vitro. Passaging CCHFV strains in the presence of sub-neutralizing concentrations of MAbs did not generate escape mutants resistant to subsequent neutralization. This study demonstrates the utility of the VLP system for screening neutralizing MAbs against multiple CCHFV strains, and provides the first evidence that a single MAb can effectively neutralize a number of diverse CCHFV strains in vitro, which may lead to development of future CCHF therapeutics.
Keywords: Crimean-Congo hemorrhagic fever virus, Virus-like particles, Monoclonal antibodies, Neutralization assay
1. Introduction
Crimean-Congo hemorrhagic fever virus (CCHFV) is the causative agent of an acute severe viral hemorrhagic fever (CCHF) in humans. Geographically, CCHFV or CCHFV reactive antibodies have been detected throughout Southern Eurasia and Africa (Bente et al., 2013). Case fatality rates of CCHF vary from ~5% to 30% (Bente et al., 2013; Ergonul, 2012; Papa et al., 2016). Due to the broad and expanding geographic distribution of Hyalomma ticks, the primary vector and reservoir of CCHFV, CCHF outbreaks may increase in frequency and spread to new areas (Estrada-Pen~a et al., 2015). Currently, prophylactic and therapeutic options available for treating CCHF patients are limited to administration of the antiviral drug ribavirin, which has not shown clear clinical benefit in historic meta analyses (Duygu et al., 2012; Koksal et al., 2010; Soares-Weiser et al., 2010). Therefore, development of novel CCHFV therapeutics is of utmost importance in combating the increasing public health burden of CCHF.
In recent years, several groups have shown that polyclonal or monoclonal antibodies (MAbs), or combinations of MAbs, can prevent fatal disease when administered to animals experimentally infected with hemorrhagic fever viruses (Cross et al., 2016; Qiu et al., 2014; Zeitlin et al., 2016). Antibody therapy has been attempted in several instances of human CCHF (Kubar et al., 2011; Van Eeden et al., 1985) and has shown modest success in small studies, but its efficacy has not been assessed in large or randomized clinical trials. Furthermore, mouse studies have suggested an important role for antibodies in protection from CCHF (Canakoglu et al., 2015; Dowall et al., 2016). Together, these data suggest that antibody treatment may be an effective therapy against CCHFV, but a number of questions remain.
Mouse studies have shown that immune responses against the CCHFV glycoproteins expressed as the complete glycoprotein precursor, GPC, are important for protection (Buttigieg et al., 2014; Hinkula et al., 2017), while immune responses against partial GPC subunits delay the time to death (Kortekaas et al., 2015). However, the GPC gene is more genetically variable than the other viral proteins such as the nucleocapsid protein (NP) and the viral RNA-dependent RNA-polymerase (L); the encoded surface glycoproteins may vary by over 25% at the amino acid level among strains co-circulating in the same territory (Goedhals et al., 2014; Papa et al., 2014). Thus, GPC may be a more difficult target for the immune system or antibody therapy, especially when heterologous strains of the virus are in concurrent circulation (Flyak et al., 2016; Wec et al., 2016).
Here, we studied the cross-strain neutralization potential of a panel of previously reported MAbs raised against the IbAr10200 prototype CCHFV strain (Bertolotti-Ciarlet et al., 2005). Potent virus neutralization activity has been reported as essential for several successful antibody therapies (Geisbert et al., 2014; Qiu et al., 2014; Wu et al., 2008). We assessed here under biosafety level 2 (BSL-2) conditions the ability of MAbs to cross-neutralize CCHFV strains using the transcription and entry-competent virus-like particle (tecVLP) system. tecVLPs consist of viral proteins and a minigenome that together generate particles that are morphologically consistent with CCHFV. Therefore, they mimic the viral replication cycle and subsequent cell entry and transcription without generating infectious virus. We then confirmed our findings with tecVLPs using CCHFV strains under BSL-4 conditions.
2. Materials and methods
2.1. Biosafety statement
Procedures involving infectious CCHFV were conducted in a BSL-4 facility according to institutionally approved standard operating procedures. Other procedures were performed under BSL-2 conditions.
2.2. Cell lines and antibodies
BSR-T7 cells were obtained from K.K. Conzelmann (Ludwig-Maximilians-Universität) and propagated in DMEM supplemented with 10% FBS, media additives (1% sodium pyruvate, 1% non-essential amino acids, and 1% penicillin/streptomycin), and 400 ng/mL gentamycin. SW-13 cells were obtained from P. Leyssen (Rega Instituut KU) and propagated in DMEM supplemented with 10% FBS and media additives. All cells were grown in a humidified 37 °C, 5% CO2 incubator.
Murine MAbs targeting CCHFV strain IbAr10200 PreGn (5A5, 6B12, 7F5, 8F10, 10E11, 11F6, and 13G8), PreGc (1H6, 3E3, 11E7, 12A9, 13G5, and 30F7), and NP (2B11 and 9D5) were obtained from the Joel M. Dalrymple-Clarence J. Peters USAMRIID Antibody Collection through BEI Resources. Antibodies were purified by protein G affinity chromatography to >95% purity (purity range 97.5 – 99.8%) as determined by Experion Pro260 analysis. The PreGc MAb 8A1 was produced from hybridoma cells (from G. Ludwig, USAMRIID) by GenScript Inc. 8A1 was purified by protein A affinity chromatography, and had 95% purity as determined by SDS-PAGE. CCHFV hyperimmune mouse ascetic fluid (HMAF) used was the same as previously reported (Bergeron et al., 2015).
2.3. Viruses and tecVLPs
The ORFs of the GPC of CCHFV isolates ArD15786 (DQ211627), Baghdad-12 (AJ538197), Kosova Hoti (EU037902), NIV112143 (JN572085), SPU18/88 (KJ682810), Sudan Al-Fulah 3–2008 (HQ378185), and YL04057 (FJ562094) were codon-optimized and synthesized by GenScript Inc, and cloned into the previously described expression vector pCAGGS (pC-GPC). Including the previously described IbAr10200, Oman199809166, Turkey200406546, and Afg09 tecVLPs, 11 tecVLP moieties were generated (Fig. 1) (Zivcec et al., 2015).
Fig. 1.
Phylogenetic analysis of amino acid sequences of Crimean-Congo hemorrhagic fever virus (CCHFV) glycoprotein precursors. Maximum likelihood analysis was used to compare complete amino acid sequences of the glycoprotein precursor proteins from the indicated CCHFV strains. Accession numbers are listed and bootstrap support values are indicated at the nodes. Highlighted in blue are strains that were sequenced during the course of these experiments (813040 UAE and 813042 UAE) and only used as infectious CCHFV. Highlighted in red are sequences obtained from GenBank that were used to generate transcription and entry competent virus-like particles (tecVLPs). Purple frames outline sequences used as both tecVLPs and infectious CCHFV strains.
The previously described CCHFV helper plasmids encoding the strain IbAr10200 NP (pC-NP), the codon-optimized L (pCLCK-L, possessing an R substitution at position 16) helper plasmids, and the pL-Luc minigenome plasmids were used in all experiments. BSR-T7 cells were transfected with pC-NP, pCLCK-L, pL-Luc, and a pC-GPC plasmid using TransIT-LT1 Transfection Reagent according to manufacturer’s recommendations (Mirus Bio LLC). Media were replaced with fresh culture medium the following day, and tecVLP-containing cell supernatants were collected and concentrated as previously described (Zivcec et al., 2015).
CCHFV isolates Turkey-812955 (KY362515, KY362517, and KY362519), Oman-812956 (KY362514, KY362516, and KY362518), UAE-813040 (MF289414, MF289415, and MF289416), and UAE-813042 (MF289417, MF289418, and MF289419) were propagated in SW-13 cells. CCHFV strain IbAr10200 was generated by reverse genetic approaches as previously described, and propagated in SW-13 cells (recIbAr10200) (Bergeron et al., 2015). CCHFV identity and sequence, and exclusion of contaminants in the virus stocks, were confirmed by next-generation sequencing using a MiniSeq System according to manufacturer’s instructions (Illumina, Inc).
2.4. CCHFV and tecVLP neutralization assay
tecVLP neutralization assays were conducted as previously described (Zivcec et al., 2015). For the CCHFV plaque reduction/ neutralization test (PRNT), MAbs were diluted in SW-13 media to equal starting concentrations (10 αg/mL), and further diluted in a 2-fold dilution series (concentration range 101 to 8 10−2 αg/mL, or ~1:100- to 1:12800-fold dilutions). MAb dilutions were mixed with equal volumes of CCHFV isolates diluted to ~100 TCID50/well in SW-13 media, and incubated for 1 h at 37 °C. The mixture was then applied to confluent monolayers of SW-13 cells in 6-well plates and incubated for 1 h at 37 ○C. Following incubation, the inocula were removed, and the cells were washed with DMEM, overlaid with 1–1.2% Avicel (FMC Health and Nutrition), and incubated at 37 °C for 3 – 4 days. The overlays were removed, fixed with formalin, and stained with crystal violet. Plaques were counted visually, and reductions in the number of plaques were reported as the titer reduction percentage.
2.5. Serial passaging of CCHFV in the presence of neutralizing MAb
CCHFV strains recIbAr10200 and Turkey-812955 were passaged in the presence of sub-neutralizing doses of 8A1 (0.625 or 0.3125 αg/mL) or 30F7 (1.25 or 0.625 αg/mL), or in SW-13 media. Both strains were incubated with MAbs and used to infect SW-13 cells in 6-well plates as described in 2.4. Following infection, the inocula were replaced with their respective MAb-containing or plain SW-13 media and incubated for 2 days at 37 °C. Following incubation, supernatants were aliquoted and frozen at 80 ○C. CCHFV RNA levels were monitored by quantitative RT-PCR assays as previously described (Bergeron et al., 2015; Spengler et al., 2017). Five CCHFV passages were performed; efforts to passage the virus for longer than 5 passages in the presence of sub-neutralizing level of MAbs all resulted in reduction of CCHFV RNA to undetectable levels. Changes in GPC sequences were determined by next-generation sequencing. Briefly, the GPC region of Turkey-812955 and recIbAr10200 was amplified from the extracted RNA using the SuperScript® III One-Step RT-PCR System with Platinum Taq DNA Polymerase (Thermo Scientific) (Supplemental Table 1). GPC DNA fragments were purified and sequenced using a MiniSeq System according to manufacturer’s instructions (Illumina, Inc).
2.6. CCHFV competitive ELISA
Horseradish peroxidase (HRP) was conjugated to MAbs 8A1 and 11E7 using the HRP Conjugation Kit (Abcam Plc) to create 8A1-HRP and 11E7-HRP. HRP did not effectively conjugate to MAb 30F7 or diminished 30F7’s binding to its substrate, so the binding of this MAb could not be assessed. 96-well ELISA plates were coated with monoclonal antibodies 12A9 and 13G5 (1:1000 dilution in PBS) overnight at 4 °C. Plates were washed with 0.1% Tween-20 PBS. Slurries of BSR-T7 cells transfected with IbAr10200 tecVLP or mock-transfected were diluted 1:20 in SuperBlock blocking buffer (SB, Thermo Scientific) with 0.5% Tween-20 and 0.5% Triton X-100 (Sigma-Aldrich), added to the coated wells, and incubated at 37 °C for 1–2 h. ELISA plates were washed at least 3 times and incubated with MAbs (8A1, 11E7, 30F7, or 9D5 [negative control]; 1 αg/mL in SB-0.1% Tween-20) for 1–2 h at 37 °C. The plates were washed and incubated with 1 αg/mL 8A1-HRP or 11E7-HRP for 1–2 h at 37 °C. HRP signal was developed using the ABTS 2-Component Microwell Peroxidase Substrate kit (SeraCare Life Sciences). Competition was determined as the percentage of signal of a conjugated MAb (8A1-HRP or 11E7-HRP) following incubation with a competing MAb (8A1, 11E7, or 30F7) compared to control MAb (9D5).
2.7. MAb peptide screening
MAbs 1H6, 3E3, 8A1, 11E7, 12A9, 13G5, and 30F7, and purified HMAF (positive control) were sent to JPT Peptide Technologies GmbH for seromarker discovery. Briefly, peptides (15 aa linear epitopes with 11 aa overlap) of the entire Gc region of CCHFV IbAr10200 were synthesized (Supplementary Table 2) and chemoselectively immobilized onto microarrays via N-terminus using a tag. MAb binding to these peptides was determined by a peptide ELISA. Data were normalized to the control slide and reported as fold increase over signal generated by normal purified mouse IgG:
2.8. Statistical analyses
One- or Two-way analyses of variance (ANOVA) with Dunnett’s or Sidak’s multiple comparisons tests were performed using GraphPad Prism version 6.0d for Mac OS X (GraphPad Software, Inc). For MAb inhibitory dilutions, GraphPad Prism was used to fita 4-parameter equation to semilog plots of the concentration-response data. The plot was used to interpolate the concentration of MAb that inhibited 50% of the NanoLuc signal or resulted in 50% reduction in plaque counts in target cells (EC50).
3. Results
3.1. 8A1, 11E7, and 30F7 broadly cross-neutralize CCHF tecVLPs
To expand the tecVLP panel across additional CCHFV GPC clades (Fig. 1), we generated 7 more tecVLP moieties possessing glycoproteins from diverse CCHFV strains, and tested the collection of 11 tecVLP moieties against a panel of 16 MAbs targeting CCHFV NP (negative control) or the mature glycoproteins Gn and Gc. The GPCs were selected to have approximately 2 strains from each of the 6 GPC clades represented, including both recently circulating and historical strains. Luciferase signal measured in target cells incubated with neutralized tecVLP mixtures was compared to luciferase signal of tecVLPs neutralized by the negative control. When diluted to 5 αg/mL, MAbs targeting NP or the PreGn region of GPC did not neutralize tecVLP signal appreciably, while most MAbs targeting the PreGc region neutralized one or more tecVLP moieties (Fig. 2). Of the 7 PreGc-targeting MAbs, 8A1, 11E7, and 30F7 cross-neutralized >50% of the signal in at least 6 of the 11 tecVLPs tested, and were therefore selected for additional characterization of dosing and combinations. No evidence of cell toxicity was observed following treatment with any tecVLP or MAb (data not shown).
Fig. 2.
Assessing cross-neutralizing activity of monoclonal antibodies (MAbs) against CCHFV tecVLPs. tecVLPs possessing glycoproteins from strains IbAr10200, Sudan Al-Fulah 2009–03, Turkey 200406546, Kosova Hoti, Oman199809166, SPU18/88, ArD15786, YL04057, Afg09, NIV112143, and Baghdad-12 were incubated with MAbs diluted to a final concentration of 5 αg/mL. Percent inhibition was calculated by comparing the luciferase signal to that of tecVLPs incubated with the monoclonal 9D5, which targets the CCHFV nucleoprotein and does not neutralize viral entry. Data are shown as standard error of the a mean (n = 4).
3.2. 8A1 is the most potent and most broadly cross-neutralizing MAb
While 8A1, 11E7, and 30F7 all neutralized the IbAr10200 VLP signal at ≤1 αg/mL (with EC50 values of 0.046, 0.499, and 0.777 mg/ mL, respectively), the EC50 values of these MAbs for neutralizing other tecVLP moieties varied significantly (Table 1, Supplemental Fig. 1). 8A1 consistently neutralized the signal of every tecVLP moiety more strongly and at lower concentrations than either 11E7 or 30F7. 8A1 neutralized 87.9 ± 0.4% to 99.7 ± 0.0% of tecVLP signals at EC50 values of 2 αg/mL (Table 1, Supplemental Fig. 1A). In our experiments, 30F7 could neutralize most tecVLPs at a lower concentration and higher maximal inhibition percentage than 11E7 (Supplemental Fig. 1B and C), and 11E7 generally performed the most poorly of the 3 cross-neutralizing MAbs (Supplemental Fig. 1C).
Table 1.
MAb-mediated neutralization of tecVLPs in SW-13 cells.
| MAb |
8A1 |
11E7 |
30F7 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| tecVLP | EC50 (μg/mL) | 95% Confidence interval | Maximal RLU inhibition (%) | EC50 (μg/mL) | 95% Confidence interval | Maximal RLU inhibition (%) | EC50 (μg/mL) | 95% Confidence interval | Maximal RLU inhibition (%) |
| IbAr10200 | 0.046 | 0.031–0.070 | 99.639 ± 0.033 | 0.499 | 0.089–2.798 | 83.978 ± 0.329 | 0.777 | 0.451–1.34 | 98.682 ± 0.224 |
| Sudan Al-Fulah | 1.303 | 0.653–2.601 | 94.452 ± 0.184 | 0.572 | 2.080e(−4)–1575 | 53.108 ± 3.329 | 1.358 | 0.362–5.096 | 66.772 ± 4.409 |
| Turky | 1.412 | 0.948–2.104 | 92.766 ± 0.110 | >1000 | Ambiguous | 71.948 ± 0.603 | 0.897 | 0.284–2.834 | 79.023 ± 1.372 |
| Hoti | 1.043 | 0.4473–2.433 | 96.343 ± 0.143 | >1000 | Ambiguous | 52.368 ± 2.003 | 0.629 | 0.299–1.321 | 72.346 ± 3.639 |
| Oman | 0.780 | 0.595–1.022 | 87.925 ± 0.421 | >1000 | Ambiguous | 43.052 ± 1.720 | 1.320 | 0.611–2.855 | 72.368 ± 1.604 |
| SPU18/88 | 1.082 | 0.449–2.606 | 99.735 ± 0.029 | 0.137 | 0.06593–0.2858 | 95.563 ± 0.183 | 1.124 | 0.574–2.203 | 95.559 ± 0.700 |
| ArD15786 | 2.057 | 0.187–22.67 | 99.670 ± 0.060 | 1.285 | 0.1707–9.668 | 80.706 ± 1.148 | 1.055 | 0.336–3.316 | 95.361 ± 0.280 |
| YL04057 | 0.124 | 0.092–0.168 | 99.774 ± 0.022 | >1000 | Ambiguous | 71.599 ± 1.688 | 0.403 | 0.273–0.595 | 99.368 ± 0.089 |
| Afg09 | 0.668 | 0.521–0.857 | 94.711 ± 0.140 | >1000 | Ambiguous | 69.726 ± 0.264 | 1.785 | 0.547–5.828 | 81.615 ± 2.091 |
| NIV112143 | 1.410 | 0.607–3.280 | 92.205 ± 0.508 | >1000 | Ambiguous | 41.826 ± 1.838 | >1000 | Ambiguous | 52.170 ± 1.685 |
| Baghdad-12 | 0.534 | 0.175–1.628 | 94.734 ± 1.603 | 0.036 | 0.020–0.063 | 99.657 ± 0.164 | 1.267 | 0.328–4.889 | 97.114 ± 1.631 |
MAbs 8A1, 11E7, and 30F7 were diluted in a 2-fold dilution series from 101 to 8 × 10−2 αg/mL and incubated with tecVLPs. Percent inhibition was calculated by comparing signal from tecVLP incubated with MAbs to mock-treated tecVLP signal. The data were used to fit a 4-paramenter equation to semilog plots of the concentration-response data and obtain an effective concentration 50% (EC50) for each monoclonal antibody against each tecVLP moiety. The EC50 values and 95% confidence intervals are shown as well as maximal inhibition. Maximal inhibition is shown as standard error of the a mean (n = 4).
3.3. 11E7 and/or 30F7 do not enhance 8A1 neutralizing potency
Combining any 2 of 8A1, 11E7, and 30F7 did not significantly affect EC50 values or maximal inhibition of tecVLP signal; indeed, the 8A1/11E7 combination may have had lower neutralization potency than 8A1 alone (Table 2, Supplemental Fig. 2). Combining all 3 MAbs also did not enhance the neutralizing potency or maximal inhibition of 8A1 alone.
Table 2.
Neutralizing potency of MAb combinations against tecVLPs in SW-13 cells.
| MAb | 8A1/11E7 | 8A1/30F7 | ||||
| tecVLP | EC50 (μg/mL) | 95% Confidence interval | Maximal RLU inhibition (%) | EC50 (μg/mL) | 95% Confidence interval | Maximal RLU inhibition (%) |
| IbAr10200 | 0.701 | 0.545–0.901 | 99.250 ± 0.107 | 0.300 | 0.245–0.368 | 99.596 ± 0.035 |
| Sudan Al-Fulah | 2.137 | 0.811–5.630 | 77.166 ± 1.854 | 0.623 | 0.300–1.295 | 92.492 ± 0.783 |
| Turkey | 2.521 | 1.538–4.133 | 75.7838 ± 1.196 | 1.485 | 0.478–4.611 | 91.150 ± 0.748 |
| Hoti | 1.936 | 0.841–4.458 | 82.022 ± 0.590 | 0.518 | 0.373–0.721 | 93.128 ± 0.267 |
| Oman | 1.431 | 0.885–2.316 | 78.350 ± 0.757 | 0.416 | 0.322–0.544 | 90.723 ± 1.155 |
| SPU18/88 | 0.481 | 0.357–0.647 | 98.732 ± 0.923 | 0.378 | 0.122–1.170 | 99.864 ± 0.060 |
| ArD15786 | 3.502 | 0.607–20.200 | 97.857 ± 0.361 | 0.610 | 0.153–2.441 | 99.628 ± 0.028 |
| YL04057 | 0.22 | 0.148–0.325 | 99.269 ± 0.052 | 0.117 | 0.075–0.181 | 99.609 ± 0.071 |
| Afg09 | 1.501 | 1.100–2.049 | 92.285 ± 0.574 | 0.374 | 0.288–0.486 | 93.741 ± 0.388 |
| NIV112143 | 3.836 | 0.103–142.9 | 80.730 ± 0.987 | 0.911 | 0.273–3.042 | 89.616 ± 0.776 |
| Baghdad-12 | 0.084 | 0.054–0.130 | 92.747 ± 4.152 | 0.146 | 0.033–0.646 | 99.973 ± 0.350 |
| MAb | 11E7/30F7 | 8A1/11E7/30F7 | ||||
| tecVLP | EC50 (μg/mL) | 95% Confidence interval | Maximal RLU inhibition (%) | EC50 (μg/mL) | 95% Confidence interval | Maximal RLU inhibition (%) |
| IbAr10200 | 0.197 | 0.124–0.311 | 99.407 ± 0.154 | 0.094 | 0.074–0.119 | 99.761 ± 0.034 |
| Sudan Al-Fulah | 1.346 | 0.114–15.840 | 76.376 ± 3.099 | 0.393 | 0.294–0.524 | 91.814 ± 1.071 |
| Turkey | 0.526 | 0.314–0.882 | 92.349 ± 1.845 | 0.264 | 0.198–0.352 | 96.095 ± 0.906 |
| Hoti | 0.951 | 0.552–1.639 | 80.918 ± 2.995 | 0.238 | 0.181–0.313 | 94.643 ± 0.460 |
| Oman | 0.532 | 0.292–0.971 | 86.627 ± 2.426 | 0.162 | 0.121–0.216 | 90.647 ± 1.437 |
| SPU18/88 | 0.156 | 0.091–0.269 | 99.498 ± 0.066 | 0.119 | 0.092–0.152 | 99.437 ± 0.063 |
| ArD15786 | 1.919 | 0.053–69.22 | 96.787 ± 0.398 | 0.169 | 0.130–0.219 | 99.593 ± 0.156 |
| YL04057 | 0.374 | 0.203–0.694 | 99.446 ± 0.103 | 0.052 | 0.040–0.066 | 99.485 ± 0.117 |
| Afg09 | 0.425 | 0.017–373 | 94.520 ± 1.103 | 0.129 | 0.103–0.161 | 96.643 ± 0.358 |
| NIV112143 | 0.672 | 0.190–2.372 | 63.796 ± 6.478 | 0.400 | 0.211–0.757 | 86.476 ± 2.854 |
| Baghdad-12 | 0.065 | 0.024–0.176 | 90.436 ± 2.905 | 0.045 | 0.029–0.070 | 98.243 ± 0.710 |
Indicated combinations of 8A1, 11E7, and 30F7 were diluted in 2-fold series from 101 to 8× 10−2 αg/mL and incubated with tecVLPs. Percent inhibition was calculated by comparing the signal from tecVLPs incubated with MAbs to signal from mock-treated tecVLPs. EC50 values and 95% confidence intervals are shown, as well as maximal inhibition for each combination of antibodies. Maximal inhibition is shown as standard error of the a mean (n = 4).
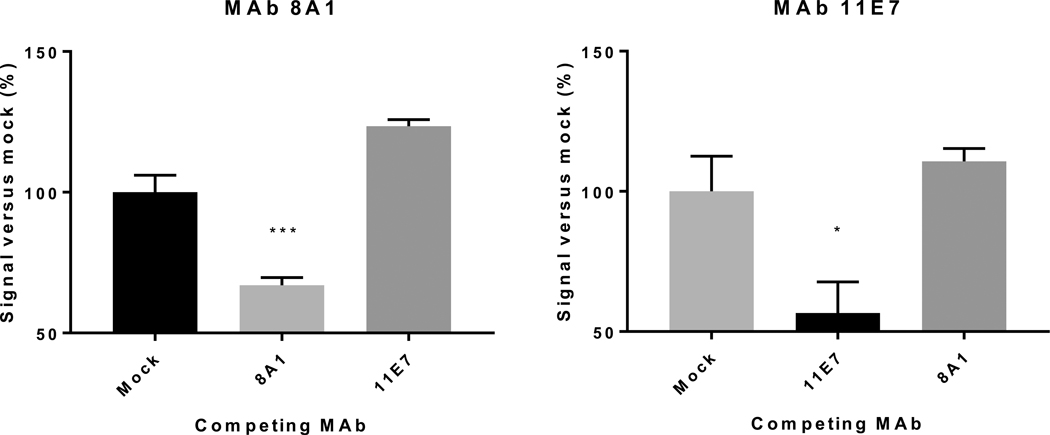
3.4. 8A1, 11E7, and 30F7 do not block binding of heterologous MAbs
To determine whether cross-neutralizing MAbs compete for binding, we conjugated HRP to MAb and performed a competition ELISA (Fig. 3). MAb 30F7 did not successfully couple with HRP, but did not interfere with 8A1 and 11E7 binding (data not shown). Adding 8A1 did not inhibit 11E7-HRP signal, nor did adding 11E7 inhibit 8A1-HRP signal. However, using 8A1 together with 8A1-HRP, and 11E7 with 11E7-HRP resulted in maximal HRP signal reduction of ~25% compared to the negative control.
Fig. 3.
Competition ELISA of cross-neutralizing MAbs. Monoclonal antibodies 8A1, 11E7, or 9D5 (negative control; this antibody targets the nucleoprotein) were used at a final concentration of 1 αg/mL to block the attachment of horseradish peroxidase (HRP)-conjugated 8A1 or 11E7 (1 αg/mL concentration). Percent HRP signal was calculated by comparing to HRP signal of samples with 9D5. Data were analyzed by one-way ANOVA, compared to mock signal using Dunnett’s multiple comparison test and are shown as standard error of the a mean (n = 8). * = p < 0.05, ***p < 0.01.
3.5. Peptide scan ELISA shows weak binding of anti-Gc antibodies
To assess peptide binding in Gc-targeting MAbs, we performed a peptide scan-based ELISA. Ultimately, all MAbs gave low binding signal indicative of low reactivity with linear peptides. In the assay, of all the cross-neutralizing MAbs, 11E7 reacted most strongly and had highest reactivity to peptides 77 (55.2-fold increase; NTSWMSWDGCDLDYY1359), 58 (34.7-fold increase; NIQQKLP-PEIITLHP1283), and 87 (33.7-fold increase; LNIETDYTKNFHFHS1399) (Supplemental Table 2). Of the 3 cross-neutralizing MAbs, 8A1 had intermediate reactivity to peptides and reacted to peptides 149 (20.6-fold increase; FKYRHLKDDEETGYR1647), 44 (20.5-fold increase; KDLFTDYMFVKWKVE1227), and 76 (19.5-fold increase; EPHFNTSWMSWDGCD1355). 30F7 had the weakest interaction of the cross-neutralizing MAbs, and most strongly interacted with peptides 48 (15.2-fold; IKTEAIVCVELTSQE1243), 44 (11.1-fold; KDLFTDYMFVKWKVE1227), and 5 (9.7-fold; SLETSLSIEAPWGAI1071). HMAF, which was previously shown to bind linear Gc epitopes (Erickson et al., 2007), was used as positive control, and bound to Gc peptides more potently than any of the MAbs (up to 613.7-fold).
3.6. tecVLP assays effectively predict the neutralizing potency of MAbs against CCHFV strains
To confirm that the neutralization potency of MAbs against tecVLPs reflected their neutralizing potency against infectious CCHFV strains (Fig. 1), we used the MAbs in a PRNT assay. In line with the tecVLP data, 8A1 was able to neutralize all the CCHFV strains used, and demonstrated significantly higher neutralization potency, both in terms of its 50% PRNT (PRNT50) dose and the maximal inhibition, than 11E7 or 30F7 for all CCHFV strains tested (Table 3, Supplemental Fig. 3). In contrast to the tecVLP data, however, 11E7 neutralized all strains by >50%, with <1 αg/mL PRNT50 values against all strains, which were not statistically different from those of 30F7. However, similarly to the tecVLP data, 30F7 showed higher maximal neutralization than 11E7.
Table 3.
MAb-mediated neutralization of CCHFV strains in SW-13 cells.
| Strain | 8A1 |
11E7 |
30F7 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| PRNT50 (μg/mL) | 95% Confidence interval | Maximal inhibition (%) | PRNT50 (μg/mL) | 95% Confidence interval | Maximal inhibition (%) | PRNT50 (μg/mL) | 95% Confidence interval | Maximal inhibition (%) | |
| IbAr10200 | 0.180 | 0.159–0.203 | 98.565 ± 0.359 | 0.332 | 0.242–0.454 | 68.062 ± 1.435 | 0.310 | 0.253–0.380 | 96.411 ± 0.949 |
| Turkey | 0.186 | 0.167–0.206 | 98.848 ± 0.230 | 0.284 | 0.235–0.344 | 86.636 ± 1.004 | 0.384 | 0.337–0.436 | 93.548 ± 0.610 |
| Oman | 0.165 | 0.135–0.202 | 98.463 ± 0.307 | 0.367 | 0.260–0.518 | 75.102 ± 1.844 | 0.230 | 0.191–0.276 | 84.016 ± 1.108 |
| 813040 UAE | 0.570 | 0.486–0.668 | 95.101 ± 1.256 | 0.607 | 0.500–0.736 | 81.268 ± 1.525 | 3.931 | 1.003–15.400 | 78.0978 ± 1.605 |
| 813042 UAE | 0.598 | 0.395–0.906 | 94.052 ± 0.372 | 0.593 | 0.404–0.871 | 69.888 ± 1.115 | 0.311 | 0.231–0.421 | 71.163 ± 4.581 |
MAbs 8A1, 11E7, and 30F7 were used in 2-fold dilution series from 101 to 1.6 × 10−1 αg/mL in a plaque reduction/neutralization 50% (PRNT50) assay with CCHFV strains IbAr10200, Turkey-812955, Oman-812956, 813040 UAE, and 813042 UAE. Plaques were counted visually, and percent inhibition was calculated by comparing the number of plaques formed by antibody-incubated CCHFV to those formed by mock-treated CCHFV. PRNT50 values, 95% confidence intervals, and maximal inhibition for each antibody are shown. Maximal inhibition is shown as standard error about a mean (n = 3).
3.7. No escape CCHFV mutants were detected after growing the virus in the presence of MAbs
To examine the likelihood of developing escape mutations in response to MAb neutralization, strains recIbAr10200 and Turkey-812955 were passaged in the presence 8A1 or 30F7 (using a dose of ~EC90 and ~EC85, respectively). In each passage, CCHFV RNA levels were monitored by qRT-PCR (data not shown). However, by passage 4, cell supernatants contained no detectable CCHFV RNA. Thus we halved the amount of MAbs used in order to continue the study for passages 4 and 5 (a dose of ~1 EC70 of both 8A1 and 30F7). During passages 4 and 5, the amount of CCHFV RNA increased steadily, so at passage 6, starting MAb concentrations were again used. This resulted in the disappearance of CCHFV RNA in 3 separate attempts to propagate the virus past passage 6. Next-generation sequencing of the CCHFV GPC Gc region of the strains at passage 5 did not show any consensus sequence changes or sub-stantial changes in polymorphism frequency compared to mock-treated viruses (Supplemental Table 3).
4. Discussion
Over the past 2 decades, the range of CCHFV circulation and the total number of identified CCHF cases have steadily increased (Bente et al., 2013; Ergonul, 2012). This increase has been accompanied by reports of multiple strains co-circulating within a geographic region, highlighting the need to develop molecular diagnostics and therapeutics that target multiple CCHFV strains (Gargili et al., 2011; Goedhals et al., 2014; Papa et al., 2014). MAbs have demonstrated protective efficacy and safety in other animal viral disease models and in patients; however, MAb effects are usually specific against an individual virus or specific strain (Flyak et al., 2016; Wec et al., 2016).
Using tecVLPs in BSL-2, we identified 3 Gc-targeting MAbs that neutralized the luciferase signal of multiple tecVLP moieties. The 3 MAbs were also able to neutralize wild-type CCHFV strains in BSL-4 at even lower effective concentrations, and to neutralize strains absent from the tecVLP panel, showing that the tecVLP system is highly predictive for screening neutralizing MAbs (Tables 1 and 3). The increased EC50 values compared to PRNT50 values observed are likely due to the concentration of tecVLP versus CCHFV used. The total number of infectious units used was relatively low (~100 TCID50/well) while the total amount of tecVLP used was higher (~104 units/well) to obtain a broader dynamic range. In addition, followin transfection of cells by GPC plasmids, glycoproteins are secreted into the supernatant (data not shown) and may have interfered with antibody neutralization. This interference is likely to have had the greatest impact on 11E7 as a weaker neutralizing MAb compared to other MAbs. As 8A1 neutralized all tested CCHFV strains and tecVLPs moieties, it likely targets a conserved epitope essential for viral entry. 8A1 did not bind strongly to any Gc linear peptides, and the identified peptide binding regions were not highly conserved between the tecVLP moieties or CCHFV strains (data not shown), indicating that this MAb likely detects conformational epitopes. 30F7 and 11E7 also bound linear epitopes weakly, but showed lower neutralizing potency against CCHFV strains. In contrast to neutralizing MAbs, polyclonal purified HMAF, which was developed following infection of wild-type mice with CCHFV and contains antibodies directed against both linear and conformational epitopes of CCHFV NP and glycoproteins (Erickson et al., 2007), strongly bound to linear epitopes throughout the middle and C-terminal end of Gc (Supplemental Table 2). As we were unable to definitively determine that the MAbs bound linear peptides, we conclude that their neutralizing potency does not correlate to recognition of linear epitopes.
Despite several attempts to generate escape mutants to antibodies 8A1 and 30F7, we could not produce a virus that was phenotypically different from the parent strain or resistant to MAb neutralization. After 5 passages in the presence of either MAb, both CCHFV strains recIbAr10200 and Turkey-812955 remained susceptible to neutralization. Furthermore, next-generation sequencing did not reveal large differences in Gc consensus sequences or frequency of polymorphisms in virus from the final passage (Supplemental Table 3). These data suggest that these MAbs target conserved Gc regions that are resistant to mutations.
Individually 8A1, 11E7, and 30F7 have shown partial protection following lethal strain IbAr10200 CCHFV infection in the suckling mouse model (Bertolotti-Ciarlet et al., 2005). Multiple animal disease models, including those for Ebola, Lassa, and Chikungunya viruses, have shown that MAb cocktails ( 2) are frequently required for robust protective effects following infection. Use of redundant MAbs in cocktail therapies can often prevent the emergence of viable escape mutants, increase the treatment time frame, and engage additional methods of adaptive immunity (e.g., antibody-dependent cell cytotoxicity) (Cross et al., 2016; Pal et al., 2013; Qiu et al., 2012). While 8A1, 11E7, and 30F7 varied in neutralizing potencies against different CCHFV strains and tecVLPs, they did not interfere with each other’s binding or neutralization potency (Table 2), and 8A1 and 30F7 treatment did not give rise to viable escape mutants. Together, these MAbs may act more potently than single MAbs, against a number of current CCHFV strains and possibly newly emerging ones.
In conclusion, we have screened a panel of murine MAbs at BSL-2 and BSL-4 for their ability to neutralize multiple CCHFV strains. Our data highlight the utility of the tecVLP system in both low- and medium-throughput (6- to 96-well) screening. We identified 3 cross-neutralizing MAbs that effectively neutralize tecVLPs and CCHFV strains with high potency. MAb 8A1 was identified as highest in neutralizing potency and broadest cross-neutralizing ability. The MAbs do not compete for the same Gc epitopes but are not synergistic. Our studies lay the groundwork for testing a rational selection of MAb-based therapies in murine CCHFV disease models, and further confirm the Gc region of CCHFV as a conserved therapeutic target.
Supplementary Material
Acknowledgements
The authors would like to thank Tatyana Klimova for critical editing of the manuscript, Karl-Klaus Conzelmann (Ludwig-Maximilians-Universität, Munich, Germany) for providing BSR-T7 cells, Pieter Leyssen (Rega Instituut KU, Leuven, Netherlands) for providing SW-13 cells, USAMRIID for providing hybridomas, and the rest of Viral Special Pathogens Branch for input.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.antiviral.2017.08.014.
References
- Bente DA, Forrester NL, Watts DM, McAuley AJ, Whitehouse CA, Bray M, 2013. Crimean-Congo hemorrhagic fever: history, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antivir. Res 100, 159–189. 10.1016/j.antiviral.2013.07.006. [DOI] [PubMed] [Google Scholar]
- Bergeron E´., Zivcec M, Chakrabarti AK, Nichol ST, Albarinõ CG, Spiropoulou CF, 2015. Recovery of recombinant Crimean Congo hemorrhagic fever virus reveals a function for non-structural glycoproteins cleavage by furin. PLoS Pathog. 11, e1004879 10.1371/journal.ppat.1004879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti-Ciarlet A, Smith J, Strecker K, Paragas J, Altamura LA, Mcfalls JM, Frias-sta N, Schmaljohn CS, Doms RW, 2005. Cellular localization and antigenic characterization of Crimean-Congo hemorrhagic fever virus glycoproteins. J. Virol 79, 6152–6161. 10.1128/JVI.79.10.6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttigieg KR, Dowall SD, Findlay-Wilson S, Miloszewska A, Rayner E, Hewson R, Carroll MW, 2014. A novel vaccine against Crimean-Congo haemorrhagic fever protects 100% of animals against lethal challenge in a mouse model. PLoS One 9 10.1371/journal.pone.0091516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canakoglu N, Berber E, Tonbak S, Ertek M, Sozdutmaz I, Aktas M, Kalkan A, Ozdarendeli A, 2015. Immunization of knock-out a/b interferon receptor mice against high lethal dose of Crimean-Congo hemorrhagic fever virus with a cell culture based vaccine. PLoS Negl. Trop. Dis 1–14. 10.1371/journal.pntd.0003579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross RW, Mire CE, Branco LM, Geisbert JB, Rowland MM, Heinrich ML, Goba A, Momoh M, Grant DS, Fullah M, Khan SH, Robinson JE, Geisbert TW, Garry RF, 2016. Treatment of Lassa virus infection in outbred Guinea pigs with first-in-class human monoclonal antibodies. Antivir. Res 133, 218–222. 10.1016/j.antiviral.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowall SD, Graham VA, Rayner E, Hunter L, Watson R, Taylor I, Rule A, Carroll MW, 2016. Protective effects of a modified Vaccinia Ankara-based vaccine candidate against Crimean-Congo Haemorrhagic Fever virus require both cellular and humoral responses. PLoS One 11, 1–13. 10.1371/journal.pone.0156637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duygu F, Kaya T, Baysan P, 2012. Re-evaluation of 400 Crimean-Congo hemorrhagic fever cases in an endemic area: is ribavirin treatment suitable? Vector Borne Zoonotic Dis. 12, 812–816. 10.1089/vbz.2011.0694. [DOI] [PubMed] [Google Scholar]
- Ergonul O, 2012. Crimean-Congo hemorrhagic fever virus: new outbreaks, new discoveries. Curr. Opin. Virol 2, 215–220. http://dx.doi.org/101016/j.coviro.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Erickson BR, Deyde V, Sanchez AJ, Vincent MJ, Nichol ST, 2007. N-linked glycosylation of Gn (but not Gc) is important for Crimean Congo hemorrhagic fever virus glycoprotein localization and transport. Virology 361, 348–355. 10.1016/j.virol.2006.11.023. [DOI] [PubMed] [Google Scholar]
- Estrada-Pen~a A, De La Fuente J, Latapia T, Ortega C, 2015. The impact of climate trends on a tick affecting public health: a retrospective modeling approach for Hyalomma marginatum (ixodidae). 10, 1–16. 10.1371/journal.pone.0125760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyak AI, Shen X, Murin CD, Turner HL, David JA, Fusco ML, Lampley R, Kose N, Ilinykh PA, Kuzmina N, Branchizio A, King H, Brown L, Bryan C, Davidson E, Doranz BJ, Slaughter JC, Sapparapu G, Klages C, Ksiazek TG, Saphire EO, Ward AB, Bukreyev A, Crowe JE, 2016. Cross-reactive and potent neutralizing antibody responses in human survivors of natural Ebola-virus infection. Cell 164, 392–405. 10.1016/j.cell.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargili A, Midilli K, Ergonul O, Ergin S, Alp HG, Vatansever Z, Iyisan S, Cerit C, Yilmaz G, Altas K, Estrada-Pen~a A, 2011. Crimean-Congo hemorrhagic fever in European part of Turkey: genetic analysis of the virus strains from ticks and a seroepidemiological study in humans. Vector Borne Zoonotic Dis. 11, 747–752. 10.1089/vbz.2010.0030. [DOI] [PubMed] [Google Scholar]
- Geisbert TW, Mire CE, Geisbert JB, Chan Y-P, Agans KN, Feldmann F, Fenton K. a, Zhu Z, Dimitrov DS, Scott DP, Bossart KN, Feldmann H, Broder CC, 2014. Therapeutic treatment of nipah virus infection in nonhuman primates with a neutralizing human monoclonal antibody. Sci. Transl. Med 6 10.1126/scitranslmed.3008929,242ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedhals D, Bester PA, Paweska JT, Swanepoel R, Burt FJ, 2014. Next-generation sequencing of southern African Crimean-Congo haemorrhagic fever virus isolates reveals a high frequency of M segment reassortment. Epidemiol. Infect 142, 1952–1962. 10.1017/S0950268814000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinkula J, Devignot S, Åkerstroäm S, Karlberg H, Wattrang E, Bereczky S, Mousavi-Jazi M, Risinger C, Lindegren G, Vernersson C, Paweska J, Jansen van Vuren P, Blixt O, Brun A, Weber F, Mirazimi A, 2017. Immunization with DNA plasmids coding for Crimean-Congo hemorrhagic fever virus capsid and envelope proteins and/or virus-like particles induces protection and survival in challenged mice. J. Virol 10.1128/JVI.02076-16.JVI.02076-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koksal I, Yilmaz G, Aksoy F, Aydin H, Yavuz I, Iskender S, Akcay K, Erensoy S, Caylan R, Aydin K, 2010. The efficacy of ribavirin in the treatment of Crimean-Congo hemorrhagic fever in Eastern Black Sea region in Turkey. J. Clin. Virol 47, 65–68. 10.1016/j.jcv.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Kortekaas J, Vloet RPM, McAuley AJ, Shen X, Bosch BJ, de Vries L, Moormann RJM, Bente DA, 2015. Crimean-Congo hemorrhagic fever virus subunit vaccines induce high levels of neutralizing antibodies but no protection in STAT1 knockout mice. Vector-Borne Zoonotic Dis. 15, 759–764. 10.1089/vbz.2015.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubar A, Haciomeroglu M, Ozkul A, Bagriacik U, Akinci E, Sener K, Bodur H, 2011. Prompt administration of Crimean-Congo hemorrhagic fever ( CCHF ) virus hyperimmunoglobulin in patients diagnosed with CCHF and viral load monitorization by Reverse Transcriptase-PCR. Jpn. J. Infect. Dis 64, 439–443. [PubMed] [Google Scholar]
- Pal P, Dowd KA, Brien JD, Edeling MA, Gorlatov S, Johnson S, Lee I, Akahata W, Nabel GJ, Richter MKS, Smit JM, Fremont DH, Pierson TC, Heise MT, Diamond MS, 2013. Development of a highly protective combination monoclonal antibody therapy against Chikungunya virus. PLoS Pathog. 9 10.1371/journal.ppat.1003312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa A, Sidira P, Larichev V, Gavrilova L, Kuzmina K, Mousavi-Jazi M, Mirazimi A, Stro€her U, Nichol S, 2014. Crimean-Congo hemorrhagic fever virus, Greece. Emerg. Infect. Dis 20, 288–290. 10.3201/eid2002.130690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa A, Tsergouli K, Çağlayık D, Bino S, Como N, Uyar Y, Korukluoglu G, 2016. Cytokines as biomarkers of Crimean-Congo hemorrhagic fever. J. Med. Virol 88, 21–27. 10.1002/jmv.24312. [DOI] [PubMed] [Google Scholar]
- Qiu X, Fernando L, Melito PL, Audet J, Feldmann H, Kobinger G, Alimonti JB, Jones SM, 2012. Ebola GP-specific monoclonal antibodies protect mice and Guinea pigs from lethal Ebola virus infection. PLoS Negl. Trop. Dis 6, 31–34. 10.1371/journal.pntd.0001575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, Fausther-Bovendo H, Wei H, Aviles J, Hiatt E, Johnson A, Morton J, Swope K, Bohorov O, Bohorova N, Goodman C, Kim D, Pauly MH, Velasco J, Pettitt J, Olinger GG, Whaley K, Xu B, Strong JE, Zeitlin L, Kobinger GP, 2014. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 514, 47–53. 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares-Weiser K, Thomas S, Thomson G, Garner P, 2010. Ribavirin for Crimean-Congo hemorrhagic fever: systematic review and meta-analysis. BMC Infect. Dis 10, 207 10.1186/1471-2334-10-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler JR, Keating MK, Mcelroy AK, Zivcec M, Coleman JD, Zaki SR, Nichol ST, Spiropoulou CF, 2017. Crimean-Congo hemorrhagic fever in humanized mice reveals Glial cells as primary targets of neurological infection. J. Infect. Dis 10.1093/infdis/jix215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eeden PJ, Van Eeden SF, Joubert JR, King JB, Van de Wal BW, Michell WL, Joubert JR, Van Eeden PJ, King JB, 1985. A nosocomial outbreak of Crimean-Congo haemorrhagic fever at Tygerberg Hospital Part II. Management of patients. South Afr. Med. J 68, 718–721. [PubMed] [Google Scholar]
- Wec AZ, Nyakatura EK, Herbert AS, Howell KA, Shulenin S, Jangra RK, Bharrhan S, Kuehne AI, 2016. A “Trojan horse” bispecific-antibody strategy for broad protection against ebolaviruses. Science 354, 12598–12603, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Pfarr DS, Losonsky GA, Kiener PA, 2008. Immunoprophylaxis of RSV infection: advancing from RSV-IGIV to palivizumab and motavizumab. Curr. Top. Microbiol. Immunol 317, 103–123. 10.1007/978-3-540-72146-8_4. [DOI] [PubMed] [Google Scholar]
- Zeitlin L, Geisbert JB, Deer DJ, Fenton KA, Bohorov O, Bohorova N, Goodman C, Kim D, Hiatt A, Pauly MH, Velasco J, Whaley KJ, Altmann F, Gruber C, Steinkellner H, Honko AN, Kuehne AI, Aman MJ, Sahandi S, Enterlein S, Zhan X, Enria D, Geisbert TW, 2016. Monoclonal antibody therapy for Junin virus infection. Proc. Natl. Acad. Sci. U. S. A 113, 4458–4463. 10.1073/pnas.1600996113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivcec M, Metcalfe MG, Albarinõ CG, Guerrero LW, Pegan SD, Spiropoulou CF, Bergeron É, 2015. Assessment of inhibitors of pathogenic Crimean-Congo hemorrhagic fever virus strains using virus-like particles. PLoS Negl. Trop. Dis 9, e0004259 10.1371/journal.pntd.0004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.