Abstract
Bacteria undergoing anaerobic fermentation must maintain redox balance. In vivo metabolic evolution schemes based on this principle have been limited to targeting NADH-dependent reactions. Here, we developed a facile, specific, and high-throughput growth-based selection platform for NADPH-consuming reactions in vivo, based on an engineered NADPH-producing glycolytic pathway in Escherichia coli. We used the selection system in the directed evolution of a NADH-dependent d-lactate dehydrogenase from Lactobacillus delbrueckii toward utilization of NADPH. Through one round of selection, we obtained multiple enzyme variants with superior NADPH-dependent activities and protein expression levels; these mutants may serve as important tools in biomanufacturing d-lactate as a renewable polymer building block. Importantly, sequence analysis and computational protein modeling revealed that diverging evolutionary paths during the selection resulted in two distinct cofactor binding modes, which suggests that the high throughput of our selection system allowed deep searching of protein sequence space to discover diverse candidates en masse.
Keywords: redox balance, high-throughput selection, NADPH-dependent pathways, d-lactate dehydrogenase, metabolic engineering
Graphical Abstract

NADPH-dependent metabolic reactions are indispensable tools in biomanufacturing. A vast number of important targets including most natural products, amino acids, and lipids are synthesized via NADPH-dependent pathways. Furthermore, NADPH serves as the preferred electron donor for some of the largest and most versatile classes of enzymes such as the cytochromes P4501,2 and enoate reductases,3,4 which find broad applications in in vitro biotransformation and in vivo metabolic engineering.3,5,6 The importance of NADPH-dependent reactions was also highlighted in the recent efforts on production of biorenewables directly from CO2 in photosynthetic organisms, which exhibit much greater availability of NADPH than NADH.7–10
In the era of next-generation sequencing and genome mining, new NADPH-dependent enzymes are being predicted with unprecedented rate. However, validation and characterization of their catalytic activities remains a time-consuming and labor-intensive task, which often involves enzyme-specific assays in vitro. NADPH-dependent enzymes have also been obtained by altering the cofactor specificity of NADH-dependent enzymes in directed evolution,11,12 but similar challenges exist: screening large libraries of protein variants can be experimentally intractable with 96-well plate-based enzymatic assays.11 More importantly, in vitro assays may not accurately reflect the enzymes’ functionality in vivo because the assay conditions differ from the environment inside the cells, it is difficult to recapitulate in vitro the interfering and/or regulatory effects of other intracellular metabolites on the enzymes. Additionally, protein solubility may be an issue in vivo, especially for heterologous or engineered enzymes.
Recently, a fluorescence-activated cell sorting (FACS)-based method was established for selecting NADPH-consuming reactions in vivo in E. coli.13 In this system, the presence of NADPH-consuming reactions lowered the intracellular NADPH level, which in turn activated the expression of a fluorescent protein reporter through the transcriptional regulator SoxR. This method conferred much higher throughput (~106 variants per screen) compared to the 96-well plate-based methods. However, this approach required extensive expertise in FACS, and suffered from relatively high false positive rate, presumably due to cell-to-cell heterogeneity in gene expression. Moreover, the throughput was still not ideal to accommodate large protein libraries generated from random or multiple-site-saturated mutagenesis.14
In this work, we developed a growth-based selection platform in E. coli to address the above-mentioned limitations. We utilize growth as the readout, which allows for substantially higher throughput (~108 variants per round of selection). The selection platform operates based on fermentative redox balance principles, which have been explored to optimize NADH-dependent metabolic pathways (Figure 1). Briefly, glycolysis serves as the essential source of ATP in E. coli under anaerobic conditions. When the native mixed-acid fermentation pathways are disrupted, cell growth will depend on the heterologously introduced NADH-oxidizing reactions, with the latter forming a closed redox cycle with the native glycolysis (Figure 1A). Using this strategy, fermentative pathways for succinate,15 C4–C8 linear alcohols,16,17 L-alanine,18 and 2,3-butanediol19 have been optimized. In contrast, NADPH-consuming reactions do not restore redox balance due to cofactor mismatch (Figure 1B).19 Therefore, we hypothesized that if glycolysis can be engineered to produce NADPH, the aforementioned selection scheme might be adapted to exclusively respond to NADPH-dependent reactions (Figure 1C, D).
Figure 1.

Schematics of the redox balance-based selection platforms. (A) Native glycolysis forms a closed redox loop with NADH-dependent reactions, which enables growth of E. coli in anaerobic conditions. (B) Redox balance cannot be achieved with native glycolysis and NADPH-dependent reactions. (C) and (D) In contrast, engineered NADPH-generating glycolysis only pairs with NADPH-dependent reactions to support anaerobic growth.
Here, we successfully built and characterized the proposed selection system, which features the highly active and specific NADPH-generating glyceraldehyde-3-phosphate dehydrogenase (GapDH) GapN from Streptococcus mutans in place of E. coli’s native GapA. We also employed this selection platform as a tool to evolve the exclusively NADH-dependent L. delbrueckii d-lactate dehydrogenase (d-Ldh) to become highly active for NADPH (kcat/KM around 2.5 × 104 mM−1 s−1, a more than 470-fold increase compared to wild type).
RESULTS AND DISCUSSION
The design of our selection system involves a drastic change in E. coli’s central metabolism: the cofactor preference of glycolysis is switched. While NADPH-dependent glycolytic pathways have been engineered in E. coli,20–23 it is unknown whether the cofactor swapped pathways can provide sufficient flux to sustain anaerobic growth. A greater rate of glycolysis is required in anaerobic than in aerobic conditions,24 and NAD(P)H has a much smaller pool size compared to NAD(H),25 which may limit the redox cycling rate.
Hence, we first sought to identify NADPH-generating GapDHs with high in vivo activity. We designed a growth complementation system where E. coli’s native NADH-generating GapA in glycolysis was disrupted (Figure 2A). As a result, the ΔgapA strain W3CG26 was unable to grow in minimal medium using glucose as the sole substrate (Figure 2B), since all glucose-utilization pathways including the Embden–Meyerhof–Parnas (EMP) pathway, Entner–Dou-doroff (ED) pathway, and Pentose phosphate pathway (PPP) were blocked (Figure 2A, B). Next, we introduced four NADPH-generating GapDHs on a multiple-copy plasmid: GapN from Streptococcus mutans20,27,28 (on plasmid pLZ2, Table S1), GapN and GapC from Clostridium acetobutylicum21,29 (on pLZ3 and pLZ4), and GapB from Bacillus subtilis23 (on pLZ5). A plasmid pEK28 carrying E. coli gapA served as a control. Under these conditions, only S. mutans GapN and C. acetobutylicum GapC restored cell growth with comparable rate to E. coli GapA, suggesting their high activity in vivo. Interestingly, B. subtilis GapB, which was previously shown to complement W3CG in rich medium, failed to do so in minimal medium.23 By using minimal medium, we imposed higher demands on glycolytic activity, presumably because all cellular components need to be synthesized de novo from glucose.
Figure 2.
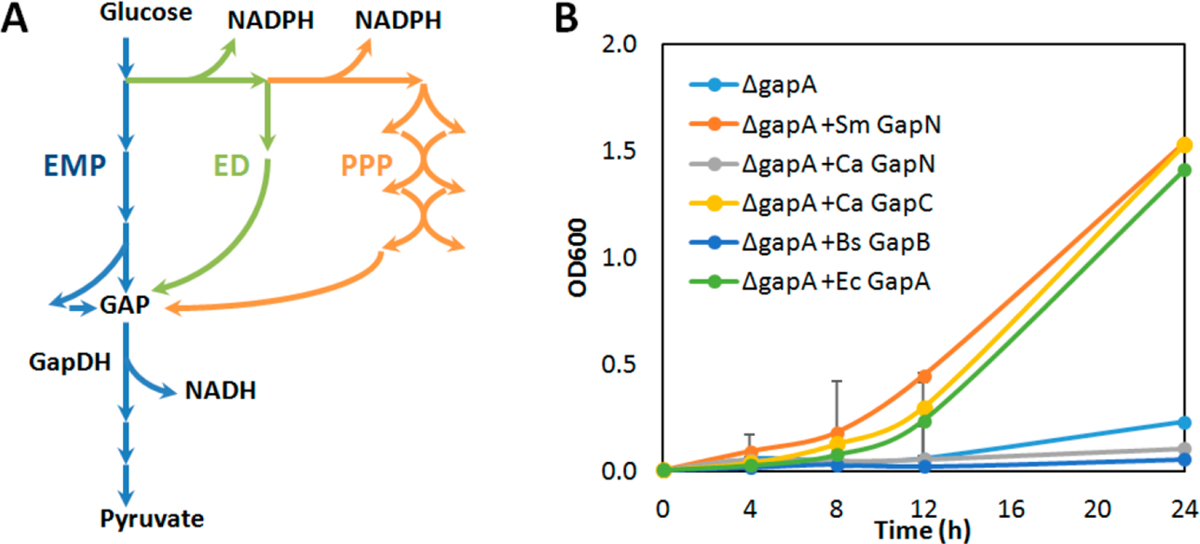
Complementation of heterologous NADPH-generating GapDHs to GapA deficiency in E. coli. (A) Overview of glycolytic pathways in E. coli. (B) Growth restoration of heterologous GapDHs in ΔgapA strain W3CG in minimal medium with glucose as the sole substrate. Error bars represent standard deviations of three replicates. n = 3. Ec, Escherichia coli; Sm, Streptococcus mutans; Ca, Clostridium acetobutylicum; Bs, Bacillus subtilis.
Next, we tested if the engineered NADPH-generating glycolysis mediated by S. mutans GapN or C. acetobutylicum GapC could support growth under anaerobic conditions where an artificial fermentative pathway to recycle NADPH was also introduced (Figure 1D). The model fermentative reaction we used is the reduction of acetoin to 2,3-butanediol.19,30 Depending on the choice of enzymes, this reaction can either recycle NADH (catalyzed by Klebsiella pneumoniae BudC) or NADPH (catalyzed by Clostridium beijerinckii Adh). Consistent with previous reports,19 when the mixed-acid fermentation-deficient E. coli strain JCL16631 was cultured in anaerobic conditions with glucose and the substrate acetoin, only the NADH-dependent K. pneumoniae BudC (on pLZ6) enabled growth (Figure 3A, B). In contrast, when E. coli GapA was replaced by S. mutans GapN in strain W3CG carrying pLZ2, only the NADPH-dependent C. beijerinckii Adh (on pLZ7) enabled growth in the same condition (Figure 3C, D), indicating the establishment of NADPH-specific redox balance in vivo. The high specificity toward NADPH-consuming reactions may be attributed to the high cofactor specificity of S. mutans GapN (Figure S1). Similar growth behavior was observed on plates (Figure 3E, F), where colonies only formed under anaerobic conditions when C. beijerinckii Adh, but not K. pneumoniae BudC, was introduced into W3CG strain harboring pLZ2. NADP(H)-and NAD(H)-dependent reactions can cross-talk in vivo through the transhydrogenases PntAB and UdhA.19 In our system, the fact that no growth was observed with the NADH-consuming pathways indicated that the native expression levels of these endogenous transhydrogenases may not be sufficient to cause significant leakage. On the other hand, glycolysis mediated by C. acetobutylicum GapC, in strain W3CG carrying pLZ4, supported anaerobic growth with both NADH-and NADPH-consuming enzymes (Figure S2), which is possibly due to the less stringent cofactor specificity of C. acetobutylicum GapC.29
Figure 3.
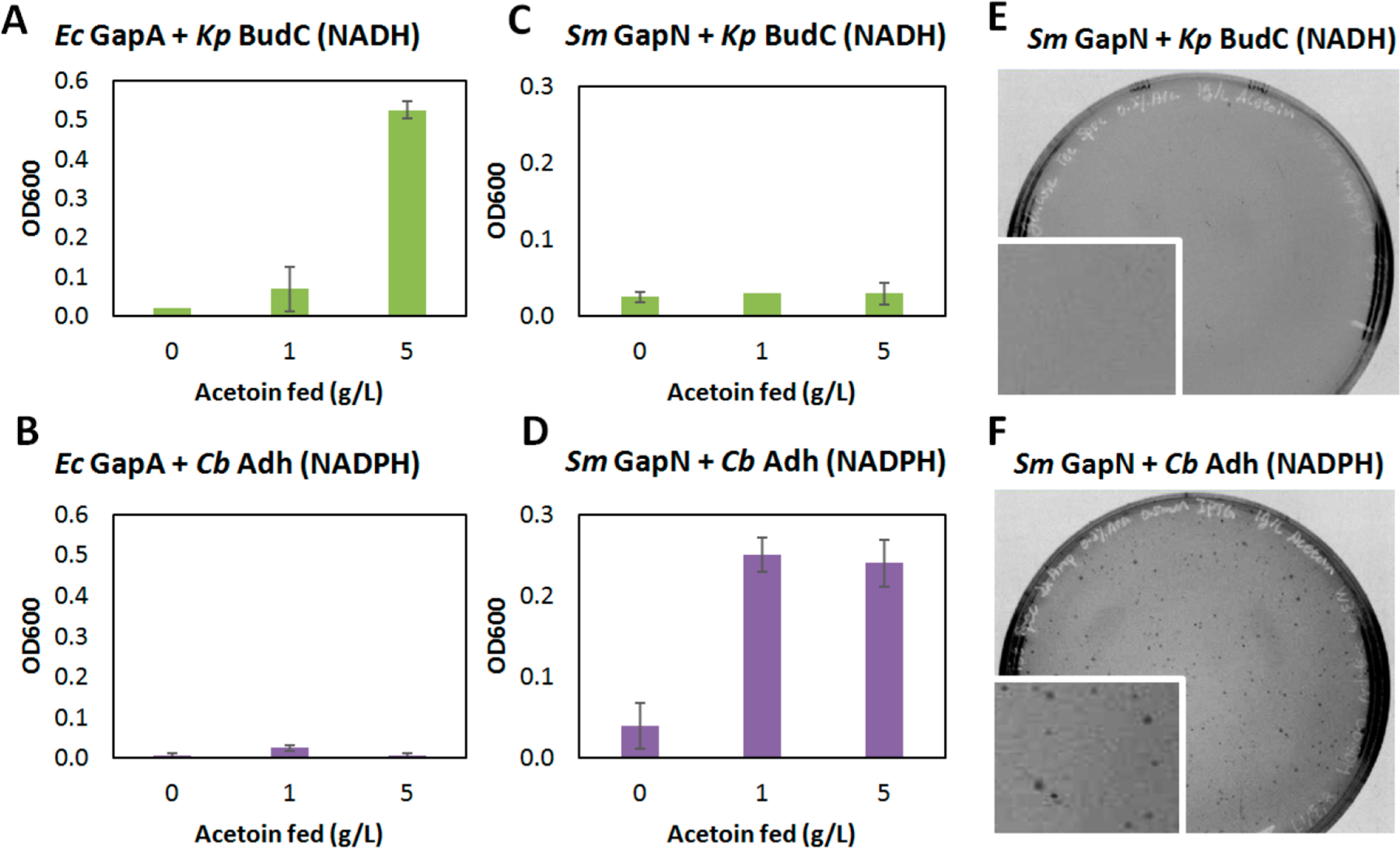
Anaerobic growth behavior with native and engineered glycolysis. (A) and (B) Native glycolysis mediated by Ec GapA only supports anaerobic growth in the presence of NADH-dependent Kp BudC. (C) and (D) Engineered glycolysis mediated by Sm GapN only supports anaerobic growth in the presence of NADPH-dependent Cb Adh. (E) and (F) Colonies formed on plates grown anaerobically when Cb Adh, but not Kp BudC, was introduced in the strain harboring Sm GapN-mediated glycolysis. Error bars represent standard deviations of three replicates. n = 3. Ec, Escherichia coli; Kp, Klebsiella pneumoniae; Cb, Clostridium beijerinckii; Sm, Streptococcus mutans.
These results suggested that the strain W3CG/pLZ2 could potentially be used as the host for the anaerobic growth rescue-based selection for NADPH-consuming reactions of interest. The transformation efficiency of W3CG/pLZ2 was determined to be ~2 × 108 colony forming units per 1 μg of plasmid DNA (Supporting Information). Therefore, the throughput of the selection system was estimated to be in the order of magnitude of 108 per round, which was substantially higher than the 96-well plate-based method (~104, assuming 100 plates are assayed per round) or the FACS-based method (~106).13
As a proof-of-concept, we then applied the selection method in directed protein evolution aimed at engineering NADH-dependent enzymes to utilize NADPH, where both high throughput and high specificity to NADPH are required. d-Lactate is an essential building block of high thermostability polylactate (PLA), a renewable and biodegradable polymer of broad applications. Recent interest in producing d-lactate from CO in cyanobacteria7,10,24 has motivated the development of an efficient NADPH-dependent d-Ldh.32 Specifically, the NADH-dependent L. delbrueckii d-Ldh has been engineered to utilize NADPH through bioinformatics-aided rational design.32 Now with the in vivo selection system in hand, we sought to ask these questions: first, whether more active variants could be discovered through searching a larger sequence space, and second, whether enzymes more compatible with in vivo applications could be discovered. The previously reported variant SRT (D176S, I177R, F178T) showed a ~ 6-fold lower protein expression yield compared to the wild type (Table S2), which is commonly seen in engineered enzymes harboring multiple mutations.11 We aimed to discover better folded enzyme variants with higher expression levels.
We performed site-saturated mutagenesis using NNK degenerate codons at the D176, I177 and F178 positions of d-Ldh (Figure 4A). The resulting DNA library (pLZ82) was transformed into the selection strain W3CG/pLZ2, generating a library of ~6.2 × 107 in size (Supporting Information). A fraction of the library (~1.5 × 105 independent transformants, which was sufficient to cover 5-fold of the theoretical library size of 20 × 20 × 20 = 8 × 103) was plated on LB agar containing 10g/L glucose and 5g/L pyruvate, and incubated anaerobically at 37 °C for 74 h. At the end of the incubation, ~0.8 × 103 colonies formed. We randomly picked 20 relatively large colonies, extracted the mutated d-Ldh plasmids, and retransformed the plasmids into the selection strain. All retransformants were verified to have anaerobic growth capability in liquid culture, with the wild type L. delbrueckii d-Ldh (on pLZ75) serving as a negative control (Figure 4B). The growth restoration was augmented by supplementation of pyruvate, the substrate of d-Ldh (Figure 4B). Sequences of the 20 variants were characterized and summarized in Table S3. We chose 8 variants of high (variant 1, 2, 3), medium (variant 8, 11, 13), and low (variant 18) anaerobic growth rescuing capabilities to further characterize using purified proteins. As shown in Figure 4C, all 8 variants gained significant NADPH-dependent activities as measured in enzyme assays in vitro, suggesting that the false positive rate of selection was very low. Additionally, enzyme assay results suggested that no selection pressure was imposed on NADH-dependent activities (Figure S3). While a weak trend exists, the specific activities measured in vitro do not strictly correlate to the variants’ growth enabling capabilities in vivo. One cause of this discrepancy might be that the specific activities measured in vitro do not accurately reflect the in vivo activities, as mentioned above. Another cause could be that the variants had different levels of active protein after purification (Table S2), indicating different expression inside the cells. Since NADPH provides the reducing power used by many essential anabolic pathways, it is also possible that extremely active NADPH-consuming enzymes could negatively affect the growth. To test if this is a potential source of bias, future work is needed to compare selection outcomes where the library is expressed with different copy number plasmids or promoter strengths.
Figure 4.
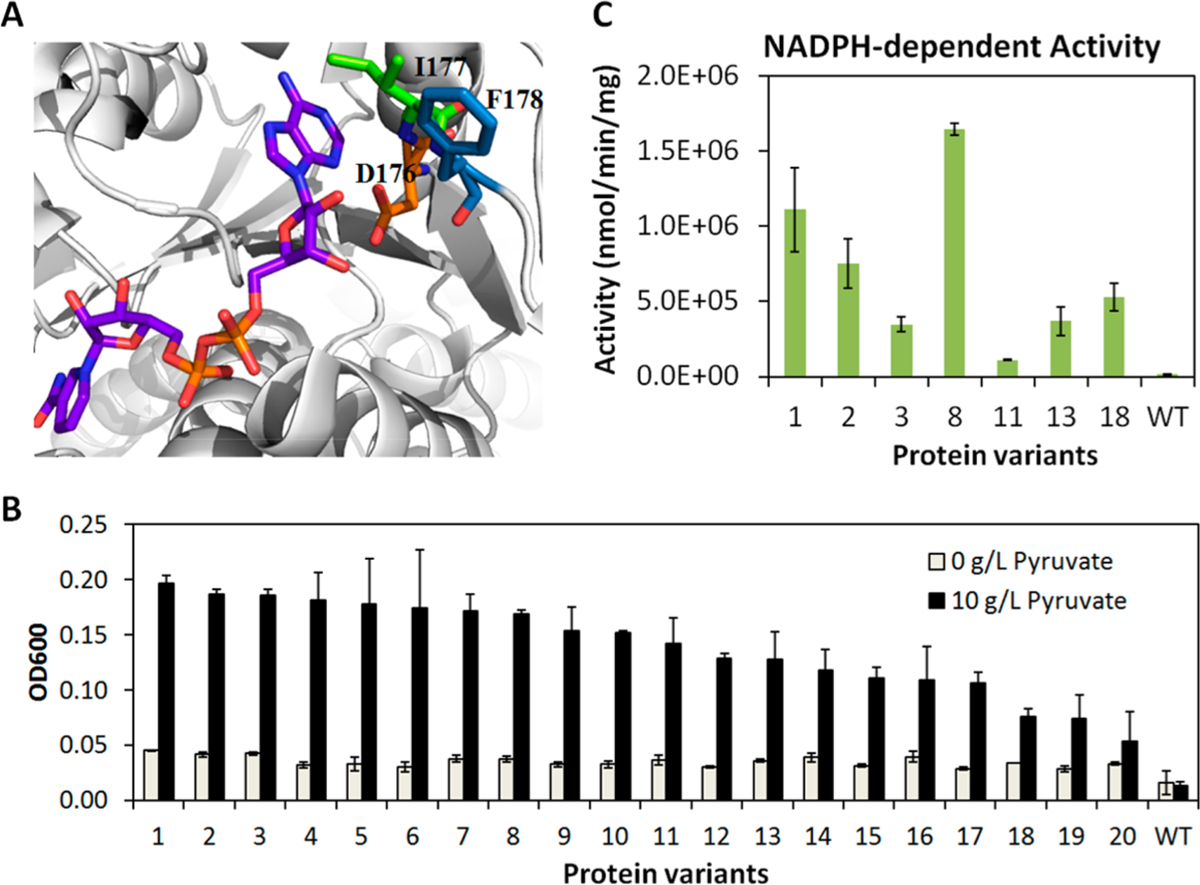
Selection of d-Ldh library and characterization of selected enzyme variants. (A) Structure of L. delbrueckii d-Ldh (PDB: 1J49), showing D176, I177, and F178 residues’ role in NADH binding. (B) Validation of anaerobic growth capabilities of 20 selected variants in liquid culture. (C) NADPH-dependent activities of 8 selected variants, as well as the wild type, as measured using purified protein in enzyme assays in vitro. Error bars represent standard deviations of three replicates. n = 3.
We further characterized the kinetic parameters of the most active hit, variant 8, GLW (D176G, I177L, F178W) in comparison to SRT and wild type d-Ldh (Table 1). The catalytic efficiency kcat/KM of GLW for the NADPH-dependent reaction is around 2.5 × 104 mM−1 s−1, which is ~472-fold higher than that of wild type. GLW is also ~60% more active than the previously identified variant SRT. To our knowledge, GLW represents the most active NADPH-dependent d-Ldh enzyme reported so far.32 Noticeably, GLW also has ~2-fold higher expression levels compared to SRT as measured by soluble protein yield in E. coli (Table S2).
Table 1.
Kinetic Parameters of d-Ldh Variants
| enzyme | NADH | NADPH | |
|---|---|---|---|
| d-Ldh | KM [mM] | 0.07 ± 0.01 | 0.59 ± 0.17 |
| kcat [s−1] | 303.1 ± 61.1 | 32.0 ± 16.1 | |
| kcat/KM [mM−1 s−1] | 4359.2 ± 324.8 | 54.1 ± 12.2 | |
| d-Ldh SRT | KM [mM] | 0.32 ± 0.01 | 0.11 ± 0.01 |
| kcat [s−1] | 896.1 ± 17.9 | 1801.1 ± 173.6 | |
| kcat/KM [mM−1 s−1] | 2767.7 ± 161.2 | 16091.1 ± 648.9 | |
| d-Ldh GLW | KM [mM] | 0.11 ± 0.01 | 0.12 ± 0.01 |
| kcat [s−1] | 1727.3 ± 184.0 | 3062.90 ± 15.10 | |
| kcat/KM [mM−1 s−1] | 14992.1 ± 377.2 | 25526.6 ± 2216.5 |
The mutation pattern of GLW revealed that strengthened hydrophobic interactions might be the main contributing factor for NADP(H) binding. To explore this hypothesis, we performed molecular modeling of the NADPH bound SRT and GLW with Rosetta. NADPH bound structures of SRT and GLW were generated through remodeling of the wild type with mutated sequences, followed by docking of the cofactor (Figure 5A, B). Distinct NADPH binding modes were observed, with the SRT model showing side-chain electrostatic interactions with the cofactor (Figure 5A) and the GLW model binding through a combination of hydrophobic packing and backbone hydrogen bonding (Figure 5B). Specifically, L177 enables hydrophobic packing of the loop against the adenosine ring of the cofactor; the structural flexibility and minimal steric interactions of G176 allows optimal positioning of L177; and the bulky W178 may serve to enhance loop rigidity to maintain the productive binding mode and avoid steric clash at the 2′ phosphate group. The models suggest that the function of GLW requires the concerted action of all three mutations, which would not be easily designed without a library-based approach.
Figure 5.
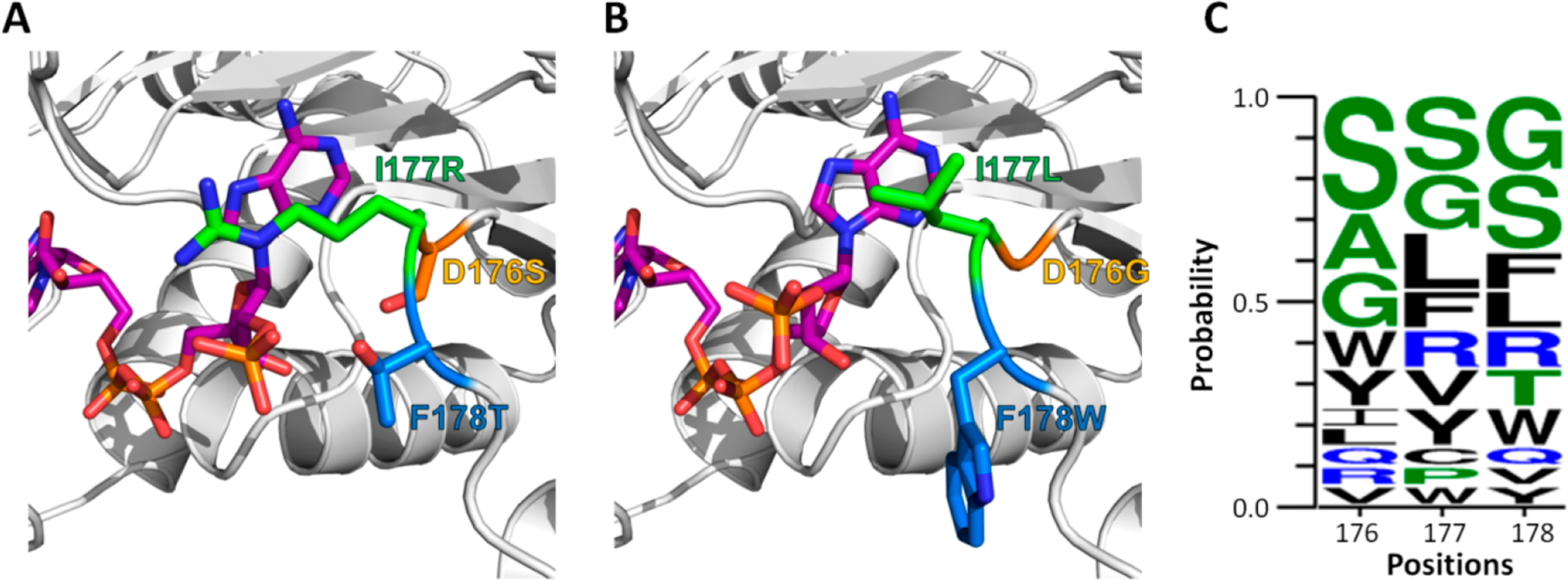
Two distinct cofactor binding modes in selected enzyme variants. (A) and (B) Rosetta models of SRT and GLW variants with NADPH bound, respectively. (C) Logo plot summarizing mutation patterns of the 20 variants at the three randomized positions.
The logo plot in Figure 5C shows the prevalence of these two distinct cofactor binding modes in the selected enzyme population. Specifically, the negatively charged D176 in the wild type enzyme was eliminated during selection. Instead, the positively charged residue R was selected, mediating the proposed cofactor binding mode through electrostatic interactions.32 The selected variants potentially using this cofactor binding mode include the previously reported SRT variant (variant 3), as well as its close homologue SRS (D176S, I177R, F178S, variant 1). On the other hand, bulky and hydrophobic residues such as W, Y, I, L, V, and F were also enriched, consistent with the presence of a second cofactor binding mode through hydrophobic interactions. These results suggested that with a selection system of sufficient throughput, we can begin to harness the power of divergent evolution and explore diverse mechanisms of enzyme functions.
In summary, we have demonstrated that the in vivo selection system for NADPH-dependent reactions established in this study has very high throughput (potentially ~108 per round of selection), stringent specificity for NADPH, low false positive rate, and easy readout using cell growth. As a proof-of-concept, we successfully applied the selection system toward the directed evolution of L. delbrueckii d-Ldh and obtained diverse variants with NADPH-dependent activities. One of the variant, GLW, represents the most active NADPH-dependent d-Ldh reported so far, and is suggested to interact with the cofactor with a different binding mode than what has been explored previously in rational design. We envision that this selection platform may be applicable to the discovery and optimization of individual enzymes and multienzyme pathways that consume NADPH in vivo. More fundamentally, this study also demonstrated that NADP(H) can function as a catabolic redox cofactor in anaerobic fermentation in E. coli.
MATERIALS AND METHODS
Media and Growth Conditions.
Cloning was performed with E. coli XL-1 blue, and protein expression was completed with E. coli BL21 (DE3). E. coli W3CG was cultured in M9 minimal medium supplemented with 15 g/L glycerol, 4 g/L malate, and 10 μg/mL of tetracycline. Concentrations utilized for antibiotic selection were 200 μg/mL for ampicillin and 50 μg/mL for spectinomycin. All strains were cultured at 37 °C with 250 rpm agitation. Induction was initiated with final concentrations of 0.2% arabinose for strains with PBAD promoter, and 0.5 mM IPTG for strains with Plac promoter.
Strains and Plasmids.
The strains and plasmids used in this study are summarized in Table S1. The construction of the plasmids, including the d-Ldh library pLZ82, is detailed in the Supporting Information.
Aerobic and Anaerobic Growth Rescue.
The culture conditions used to generate growth data in Figure 2 and Figure 3 are detailed in the Supporting Information.
Selection of d-Ldh Library.
E. coli W3CG harboring plasmid pLZ2 was transformed with the d-Ldh library pLZ82 by electroporation (see electroporation method in the Supporting Information). After rescuing in SOC medium aerobically for 45 min, transformants were plated on LB agar plates containing 10 g/L glucose, 0.2% arabinose, 0.5 mM IPTG, 5 g/L pyruvate and appropriate antibiotics, then placed into a BD GasPak EZ anaerobic bag and incubated at 37 °C for 74 h. The colonies were picked and restreaked onto the same type of plates and again incubated in anaerobic conditions to obtain single colonies. Subsequently, single colonies were cultured in liquid medium to extract plasmids using QIAprep Spin Miniprep kit (Qiagen, Maryland U.S.A).
Purification and Characterization of d-Ldh Variants.
The conditions used to purify d-Ldh variants are detailed in the Supporting Information. The d-Ldh activity was measured using previously reported methods.32 The NADPH-or NADH-dependent reaction was started by adding the purified protein to the assay mixture (200 μL) containing 150 mM sodium phosphate (pH 6.5), 300 μM NADH or NADPH, 2.5 mM MgCl2 and 30 mM sodium pyruvate. The initial reaction rate as measured by the decrease of absorbance at 340 nm was monitored by a spectrophotometer at 30 °C.
Rosetta Modeling.
Models were generated through Rosetta.33 Mutations were introduced onto the crystal structure for lactate dehydrogenase (PDB: 1J49), followed by optimization of the NADPH binding pose.34 A conformer library for NADPH was produced with OpenBabel.35 Docking trials including loop remodeling, side chain rotamer sampling, ligand and backbone torsion flexibilities were carried out resulting in a total of 500 decoys per variant. Final models were selected based on examination of Rosetta interface energies (Supporting Information).
Supplementary Material
ACKNOWLEDGMENTS
The work is funded by the start-up fund of UC Irvine to H.L.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acssyn-bio.8b00179.
Plasmids and strains (Table S1), Plasmid and d-Ldh library construction, enzyme assays of Streptococcus mutans GapN (Figure S1), anaerobic growth mediated by Clostridium acetobutylicum gapC (Figure S2), determination of the transformation efficiency of W3CG/pLZ2, protein expression and purification of d-Ldh variants, protein expression levels of selected d-Ldh variants (Table S2), mutations in the 20 d-Ldh variants obtained from anaerobic selection (Table S3), NADH-dependent activities of selected d-Ldh variants (Figure S3), aerobic and anaerobic growth rescue conditions, Rosetta modeling of d-Ldh variants (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Dohr O, Paine MJ, Friedberg T, Roberts GC, and Wolf CR (2001) Engineering of a functional human NADH-dependent cytochrome P450 system. Proc. Natl. Acad. Sci. U. S. A 98, 81–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Maurer SC, Kühnel K, Kaysser LA, Eiben S, Schmid RD, and Urlacher VB (2005) Catalytic Hydroxylation in Biphasic Systems using CYP102A1Mutants. Adv. Synth. Catal 347, 1090–1098. [Google Scholar]
- (3).Chaparro-Riggers JF, Rogers TA, Vazquez-Figueroa E, Polizzi KM, and Bommarius AS (2007) Comparison of Three Enoate Reductases and their Potential Use for Biotransformations. Adv. Synth. Catal 349, 1521–1531. [Google Scholar]
- (4).Knaus T, Paul CE, Levy CW, de Vries S, Mutti FG, Hollmann F, and Scrutton NS (2016) Better than Nature: Nicotinamide Biomimetics That Outperform Natural Coenzymes. J. Am. Chem. Soc 138, 1033–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Biggs BW, Lim CG, Sagliani K, Shankar S, Stephanopoulos G, De Mey M, and Ajikumar PK (2016) Overcoming heterologous protein interdependency to optimize P450-mediated Taxol precursor synthesis in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 113, 3209–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Jung ST, Lauchli R, and Arnold FH (2011) Cytochrome P450: taming a wild type enzyme. Curr. Opin. Biotechnol 22, 809–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Angermayr SA, van der Woude AD, Correddu D, Vreugdenhil A, Verrone V, and Hellingwerf KJ (2014) Exploring metabolic engineering design principles for the photosynthetic production of lactic acid by Synechocystis sp. PCC6803. Biotechnol. Biofuels 7, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Atsumi S, Higashide W, and Liao JC (2009) Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol 27, 1177–1180. [DOI] [PubMed] [Google Scholar]
- (9).Lan EI, and Liao JC (2012) ATP drives direct photosynthetic production of 1-butanol in cyanobacteria. Proc. Natl. Acad. Sci. U. S. A 109, 6018–6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Li C, Tao F, Ni J, Wang Y, Yao F, and Xu P (2015) Enhancing the light-driven production of d-lactate by engineering cyanobacterium using a combinational strategy. Sci. Rep 5, 9777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Cahn JKB, Werlang CA, Baumschlager A, Brinkmann-Chen S, Mayo SL, and Arnold FH (2017) A General Tool for Engineering the NAD/NADP Cofactor Preference of Oxidoreductases. ACS Synth. Biol 6, 326–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).You C, Huang R, Wei X, Zhu Z, and Zhang Y-HP (2017) Protein engineering of oxidoreductases utilizing nicotinamide-based coenzymes, with applications in synthetic biology. Synthetic and Systems Biotechnology 2, 208–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Siedler S, Schendzielorz G, Binder S, Eggeling L, Bringer S, and Bott M (2014) SoxR as a Single-Cell Biosensor for NADPH-Consuming Enzymes in Escherichia coli. ACS Synth. Biol 3, 41–47. [DOI] [PubMed] [Google Scholar]
- (14).Brinkmann-Chen S, Flock T, Cahn JK, Snow CD, Brustad EM, McIntosh JA, Meinhold P, Zhang L, and Arnold FH (2013) General approach to reversing ketol-acid reductoisomerase cofactor dependence from NADPH to NADH. Proc. Natl. Acad. Sci. U. S. A 110, 10946–10951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Jantama K, Haupt MJ, Svoronos SA, Zhang X, Moore JC, Shanmugam KT, and Ingram LO (2008) Combining metabolic engineering and metabolic evolution to develop non-recombinant strains of Escherichia coli C that produce succinate and malate. Biotechnol. Bioeng 99, 1140–1153. [DOI] [PubMed] [Google Scholar]
- (16).Machado HB, Dekishima Y, Luo H, Lan EI, and Liao JC (2012) A selection platform for carbon chain elongation using the CoA-dependent pathway to produce linear higher alcohols. Metab. Eng 14, 504–511. [DOI] [PubMed] [Google Scholar]
- (17).Shen CR, Lan EI, Dekishima Y, Baez A, Cho KM, and Liao JC (2011) Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl. Environ. Microbiol 77, 2905–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Zhang X, Jantama K, Moore JC, Shanmugam KT, and Ingram LO (2007) Production of L -alanine by metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol 77, 355–366. [DOI] [PubMed] [Google Scholar]
- (19).Liang K, and Shen CR (2017) Selection of an endogenous 2,3-butanediol pathway in Escherichia coli by fermentative redox balance. Metab. Eng 39, 181–191. [DOI] [PubMed] [Google Scholar]
- (20).Centeno-Leija S, Utrilla J, Flores N, Rodriguez A, Gosset G, and Martinez A (2013) Metabolic and transcriptional response of Escherichia coli with a NADP(+)-dependent glyceraldehyde 3-phosphate dehydrogenase from Streptococcus mutans. Antonie van Leeuwenhoek 104, 913–924. [DOI] [PubMed] [Google Scholar]
- (21).Martinez I, Zhu J, Lin H, Bennett GN, and San KY (2008) Replacing Escherichia coli NAD-dependent glyceraldehyde 3-phosphate dehydrogenase (GAPDH) with a NADP-dependent enzyme from Clostridium acetobutylicum facilitates NADPH dependent pathways. Metab. Eng 10, 352–359. [DOI] [PubMed] [Google Scholar]
- (22).Valverde F, Losada M, and Serrano A (1999) Engineering a central metabolic pathway: glycolysis with no net phosphorylation in an Escherichia coli gap mutant complemented with a plant GapN gene. FEBS Lett 449, 153–158. [DOI] [PubMed] [Google Scholar]
- (23).Wang Y, San KY, and Bennett GN (2013) Improvement of NADPH bioavailability in Escherichia coli by replacing NAD(+)-dependent glyceraldehyde-3-phosphate dehydrogenase GapA with NADP (+)-dependent GapB from Bacillus subtilis and addition of NAD kinase. J. Ind. Microbiol. Biotechnol 40, 1449–1460. [DOI] [PubMed] [Google Scholar]
- (24).Varman AM, Yu Y, You L, and Tang YJ (2013) Photoautotrophic production of d-lactic acid in an engineered cyanobacterium. Microb. Cell Fact 12, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, and Rabinowitz JD (2009) Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol 5, 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Ganter C, and Pluckthun A (1990) Glycine to alanine substitutions in helices of glyceraldehyde-3-phosphate dehydrogenase: effects on stability. Biochemistry 29, 9395–9402. [DOI] [PubMed] [Google Scholar]
- (27).Komati Reddy G, Lindner SN, and Wendisch VF (2015) Metabolic engineering of an ATP-neutral Embden-Meyerhof-Parnas pathway in Corynebacterium glutamicum: growth restoration by an adaptive point mutation in NADH dehydrogenase. Appl. Environ. Microbiol 81, 1996–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Takeno S, Hori K, Ohtani S, Mimura A, Mitsuhashi S, and Ikeda M (2016) l-Lysine production independent of the oxidative pentose phosphate pathway by Corynebacterium glutamicum with the Streptococcus mutans gapN gene. Metab. Eng 37, 1–10. [DOI] [PubMed] [Google Scholar]
- (29).Liu J, Qi H, Wang C, and Wen J (2015) Model-driven intracellular redox status modulation for increasing isobutanol production in Escherichia coli. Biotechnol. Biofuels 8, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Yan Y, Lee CC, and Liao JC (2009) Enantioselective synthesis of pure (R,R)-2,3-butanediol in Escherichia coli with stereospecific secondary alcohol dehydrogenases. Org. Biomol. Chem 7, 3914–3917. [DOI] [PubMed] [Google Scholar]
- (31).Atsumi S, Cann AF, Connor MR, Shen CR, Smith KM, Brynildsen MP, Chou KJ, Hanai T, and Liao JC (2008) Metabolic engineering of Escherichia coli for 1-butanol production. Metab. Eng 10, 305–311. [DOI] [PubMed] [Google Scholar]
- (32).Meng H, Liu P, Sun H, Cai Z, Zhou J, Lin J, and Li Y (2016) Engineering a d-lactate dehydrogenase that can super-efficiently utilize NADPH and NADH as cofactors. Sci. Rep 6, 24887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Fleishman SJ, Leaver-Fay A, Corn JE, Strauch EM, Khare SD, Koga N, Ashworth J, Murphy P, Richter F, Lemmon G, Meiler J, and Baker D (2011) RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite. PLoS One 6, e20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Razeto A, Kochhar S, Hottinger H, Dauter M, Wilson KS, and Lamzin VS (2002) Domain closure, substrate specificity and catalysis of d-lactate dehydrogenase from Lactobacillus bulgaricus. J. Mol. Biol 318, 109–119. [DOI] [PubMed] [Google Scholar]
- (35).O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, and Hutchison GR (2011) Open Babel: An open chemical toolbox. J. Cheminf 3, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


