Abstract
Background
In Gram-negative bacteria, Type Va and Vc autotransporters are proteins that contain both a secreted virulence factor (the “passenger” domain) and a β-barrel that aids its export. While it is known that the folding and insertion of the β-barrel domain utilizes the β-barrel assembly machinery (BAM) complex, how the passenger domain is secreted and folded across the membrane remains to be determined. The hairpin model states that passenger domain secretion occurs independently through the fully-formed and membrane-inserted β-barrel domain via a hairpin folding intermediate. In contrast, the BamA-assisted model states that the passenger domain is secreted through a hybrid of BamA, the essential subunit of the BAM complex, and the β-barrel domain of the autotransporter.
Methods
To ascertain the models’ plausibility, we have used molecular dynamics to simulate passenger domain secretion for two autotransporters, EspP and YadA.
Results
We observed that each protein’s β-barrel is unable to accommodate the secreting passenger domain in a hairpin configuration without major structural distortions. Additionally, the force required for secretion through EspP’s β-barrel is more than that through the BamA β-barrel.
Conclusions
Secretion of autotransporters most likely occurs through an incompletely formed β-barrel domain of the autotransporter in conjunction with BamA.
General significance
Secretion of virulence factors is a process used by practically all pathogenic Gram-negative bacteria. Understanding this process is a necessary step towards limiting their infectious capacity.
Keywords: autotransporters, secretion systems, membrane proteins, molecular dynamics
Introduction
Many, if not all, pathogenic Gram-negative and Gram-positive bacteria use surface-localized virulence factors to infect and/or undermine the host immune system. One of the ways that Gram-negative bacteria export these virulence factors is via the type V secretion system, also known as autotransporters [1]. Two sub-types, type Va and Vc, are composed of an N-terminal passenger domain (which usually contains the virulence factor [2]) and a C-terminal β-barrel translocon [3-8]. Some autotransporters contain an α-helical linker which connects the β-barrel to its passenger domain [3, 5, 6, 9]. The name autotransporter originally comes from the understanding that they require no accessory factors nor external energy to secrete across the outer membrane [10, 11], although more recently it has been recognized that insertion of the β-barrel translocon is mediated by the β-barrel assembly machinery (BAM) complex [12, 13].
After secretion across the inner membrane in the unfolded form [14], autotransporters are guided by chaperone proteins [15, 16] in the periplasm to the BAM complex, which assists their insertion into the outer membrane [17-21]. How the passenger domain is exported, however, is still not well understood, mainly due to the lack of traditional energy sources, i.e. ATP and ion gradients, in the periplasm [22]. One hypothesis is that passenger domain folding on the extracellular side provides the energy for secretion in a manner akin to a “Brownian ratchet” [23, 24]. Based on this evidence, there are two leading models for passenger domain secretion: the hairpin model and the BamA-assisted model [25]. The hairpin model states that the passenger domain is exported through its own β-barrel domain via a hairpin intermediate without assistance from other proteins. In contrast, the BamA-assisted model states that the β-barrel domain stays in conjunction with the BAM complex while the passenger domain is secreted through a hybrid pore composed of BamA and its own β-barrel domain [26, 27]. Experimental evidence supporting the latter model includes the observed association of BamA with stalled secretion intermediates [20, 28].
Here, we test the validity of the two models using molecular dynamics simulations for two types of autotransporters, type Va (classical autotransporters) and type Vc (trimeric autotransporters). We use EspP as an example of a type Va classical autotransporter [12, 29], and YadA as an example of a type Vc trimeric autotransporter adhesin [30]. EspP is a member of the serine protease autotransporter of Enterobacteriaceae (SPATE) family [31]. As a classical autotransporter, it is composed of a monomeric N-terminal passenger domain, a 12-stranded C-terminal β-barrel translocon, and an α-helical linker that gets cleaved after secretion, releasing the passenger domain from the cell surface via an autoproteolytic reaction [32]. Its cleavable α-linker region connecting the passenger domain and translocon has been found to be necessary for folding and stability of the β-barrel translocon [3, 33, 34]. EspP’s known virulence functions include biofilm formation and cell adhesion [35]. It has been implicated in several diseases and symptoms, including diarrhea and inflammation [29, 36]. Its possible use as a vaccine against Shiga toxins has also been explored [37, 38].
YadA is a trimeric autotransporter adhesin (TAA) from the family of type Vc autotransporters [30]. It is expressed by both Yersinia enterocolitica and Yersinia pseudotuberculosis [39] and is present in Yersiniapestis with a frameshift mutation [40]. It mainly functions as an adhesin, although it also has been reported to act as an invasin in Y. pseudotuberculosis [41]. The passenger domain is an N-terminal trimeric β-roll, connected to the TM domain via a trimeric coiled-coil stalk [42, 43] and its transmembrane β-barrel domain is 12-stranded, with each YadA monomer contributing four β-strands. The structure of a shortened construct of YadA containing the β-barrel and part of the stalk, called YadA-M, has been resolved using solid-state NMR [44-46]. In this structure, a region in the coiled-coiled domain consisting of small residues, termed the ASSA region, was identified that exhibited low structural propensity and predicted increased dynamics. Later functional studies showed that mutating the first alanine of this linker region to a proline stalls passenger domain secretion [47]. This finding is similar to earlier studies, which showed that secretion can be stalled or inhibited by mutation of a conserved glycine residue [48], and the corresponding residue in Hia forms an stabilizing interaction between the α-helix and β-sheet [49]. Additionally, an arginine residue in the C-terminal β-barrel plays a role in preventing the misfolding of the passenger domain [50, 51].
We used previous experimental evidence for the formation of a passenger domain hairpin intermediate to first address whether this intermediate could be accommodated by the fully-formed β-barrel domains of these two autotransporters. Our simulation results indicate that while this is possible, major structural distortions of the β-barrel domain would need to occur for passenger domain secretion. We next tested passenger domain transport through the BamA β-barrel via steered molecular dynamics, where we found the forces required for transport are lower than those required for transport through the autotransporter β-barrel domains alone. We therefore propose that passenger domain secretion likely occurs before the autotransporter β-barrel is fully folded and passes through an asymmetric hybrid barrel as recently proposed [52].
Methods
Molecular Dynamics (MD)
All-atom MD simulations were performed using NAMD 2.11 [53] and the CHARMM36m [54, 55] force field parameters for proteins and lipids with the TIP3P-CHARMM water model [56]. All simulations were performed under periodic boundary conditions with a cutoff at 12 Å for short-range electrostatic and Lennard-Jones interactions and a force-based switching function starting at 10 Å. The particle-mesh Ewald method [57] with a grid spacing of 1 Å was used for long-range electrostatic interaction calculations. Bonds between a heavy atom and a hydrogen atom were maintained to be rigid, while all other bonds remain flexible. Unless otherwise stated, each system was equilibrated for 250 ns under an isothermal-isobaric ensemble (NPT) at 310 K and 1 bar, with a timestep of 2 fs. A Langevin thermostat with a damping coefficient of 1 ps −1 was used for temperature control and a Langevin piston with a period of 0.1 ps and decay of 0.05 ps was used for pressure control. The production run of each system was run three times, in order to check the reproducibility of our simulations [58]. VMD was used for all visualization [59]. The total simulation time was around 6 μs.
System Generation
All systems were generated using CHARMM-GUI [60, 61], with the lipid compositions as listed in Supplementary Tables S1 and S2. For the YadA systems, in order to simulate in a native-like environment, the lipid composition was chosen based on previous studies of Y. pestis [62, 63]. For EspP, the system was constructed using an asymmetric outer membrane with LPS from E.coli K12 and 15:4:1 ratio of DPPE, PVPG and PVCL2 as used in previous studies [64, 65]. The systems generated by CHARMM-GUI were prepared for equilibration using the following steps for 1 ns each: 1) minimization for 10,000 steps, 2) melting of lipid tails, 3) restraining only the protein, and 4) restraining the protein backbone. For the EspP hairpin system, an additional step was used to restrain the backbone of the β-barrel after restraining only the protein backbone for 1 ns. For the YadA hairpin system, the backbone was restrained for 10 ns instead of 1 ns. Any mutations were performed before removing the restraint in protein backbone. For the elongated hairpin system, post sidechain or hairpin relaxation systems were used as a starting point. The hairpin and elongated hairpin systems were constructed by creating a linear amino acid chain based on their respective sequences using the Molefacture plugin of VMD, followed by additional solvation and ionization. For BamA-EspP system, the passenger domain of EspP was stretched and positioned through an opening in the lumen of the BamA β-barrel.
Steered Molecular Dynamics (SMD)
Steered molecular dynamics simulations were run using the collective variables module (colvars) [66]. In order to simulate the secretion of the hairpin using the elongated hairpin systems, a C α atom of the passenger domain was pulled up wards (+z direction) along the membrane normal in a piece-wise fashion. Specifically, for hairpin systems, the C α atom of the passenger domain closest to the top of the β-barrel along the membrane normal was pulled until it became the highest C α atom of the hairpin. For BamA systems, the C α atom of the passenger domain was pulled through the lumen of the BamA β-barrel. The pulling rate was set to be 1 Å /ns, in accordance with a similar study [67]. The force constant was 10 kcal/mol/Å2. To hold the other side of the hairpin within the β-barrel, two atoms of the hairpin were restrained with respect to the position of the β-barrel, the C α atom of the hairpin residues closest to the top of the β-barrel and to the bottom of the β-barrel. For the YadA system, which is a trimeric structure, all three hairpins were restrained. Each SMD simulation was run three times.
Results
The YadA β-barrel domain is unable to accommodate the passenger domain as a hairpin intermediate
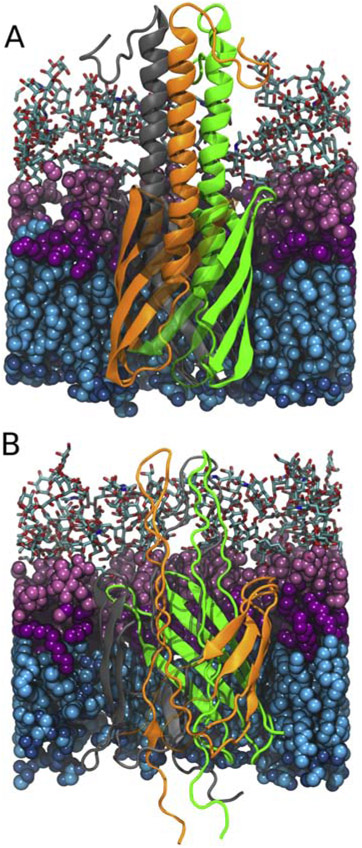
To test if the YadA β-barrel is large enough to accommodate the trimeric passenger domain in a hairpin conformation, we constructed a total of five YadA-M structures based on its NMR structure [44, 45]. which is a truncated construct composed of the β-barrel domain and a partial segment of the α-helical passenger domain (PDB ID:2LME). These five structures are (1) the wildtype structure (WT), (2) the linker region proline mutant from Chauhan et al. [47] (A354P), (3-4) the hairpin structures based on the WT and mutant structures (WThairpin and A354P hairpin, respectively), and (5) a β-barrel-only structure (WTbarrel). WT and WThairpin structures are shown in Fig. 1. For hairpin systems, the hairpin was constructed by stretching the linker and the passenger domain and placing them as a hairpin structure inside the β-barrel. The membrane of all YadA systems has a lipid composition based on Y. pestis with specific esters from Tornabene et al. [62] and fatty acids from Whittaker et al. [63] (Supplementary Table S1). After equilibration, all systems were simulated for the durations given in Table 1.
Figure 1:
Comparison between the original YadA-M (PDB: 2LME, [45]) structure (A) and the generated hairpin intermediate (B). The hairpin structure is constructed by stretching the linker and the passenger domain. Lipopolysaccharides in the outer leaflet are shown in pink (with sugar groups in licorice, polar headgroups in light pink and lipid tails in magenta). Phospholipids in the inner leaflet are shown in blue (with lipid tails in cyan and polar heads in dark blue). YadA β-strands are semi-transparent to clearly show the α-helical domain in panel A and the hairpin folding intermediate in panel B. The individual YadA monomers are shown in green, orange, and grey.
Table 1:
Simulation Details for YadA. All systems were constructed using the YadA-M NMR structure PDB 2LME [45].
| Structure | Source | System Size | Runs |
|---|---|---|---|
| WT | 2LME | 73K atoms | 250 ns x 3 |
| A354P | Based on PDB 2LME, ASSA to PSSA | 74K atoms | 250 ns x 3 |
| WT hairpn | Based on PDB 2LME, Hairpin | 84K atoms | 100 ns x 3 |
| A354P hairpin | Based on Hairpin, ASSA to PSSA | 84K atoms | 100 ns x 3 |
| WTbarrel | Based on PDB 2LME, β-barrel Only | 65K atoms | 100 ns x 3 |
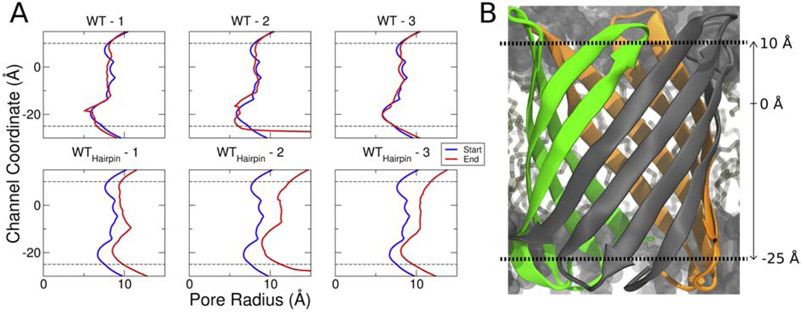
For both the WThairpin and A354P hairpin systems, the β-barrel expands dramatically at the beginning of the production simulations compared to their non-hairpin counterparts (Fig. 1). To quantify this, we measured the pore radius over the channel coordinate using the program HOLE [68] (Fig. 2). While the pore radii of the WT replicas do not change significantly over the duration of the simulation, those of the WThairpin replicas expand by ~ 3 −4 Å. Similarly, when the pore radii for the mutant systems were measured, only the hairpin structures exhibited β-barrel expansion (Fig. S1). The simulation snapshots show that the YadA β-barrel with three hairpins inside cannot be fully folded. This result agrees with previous experimental evidence, which indicated that the β-barrel domain of TAAs may not be fully folded during secretion [47, 69]. Our results suggest that the β-barrel maintains its partially folded form either during or after BAM assembly until the entire passenger domain is secreted.
Figure 2:
Passenger domain hairpin folding intermediates result in β-barrel expansion during equilibration. (A) Channel coordinate vs. pore radius for the original structures and the hairpin structures in three different replicas. The dotted lines on the top and the bottom indicate the top and bottom positions of the β-barrel. The blue line is a channel profile for the crystal structure, and the red line is at the end of the simulation. (B) Channel coordinate position relative to the β-barrel of YadA. Throughout the 250 ns simulation, the pore radii of the WT replicas remain relatively unchanged, while those of the WT hairpin replicas expand by 3 to 4 Å.
In an attempt to provide experimental evidence on whether YadA β-barrel expansion is required for passenger domain transport, we constructed four double-cysteine mutants that could form either intra- or inter-monomer disulfide bonds to prevent potential expansion during passenger domain export. Unfortunately, these double-cysteine mutants did not present in the bacterial membrane, despite the addition of TCEP to the growth media, or the use of a DegP deficient E. coli strain for expression (Fig. S3 [70]).
A proline mutation in the YadA linker region drives the α-helices apart
Chauhan et al. recently demonstrated that the mutation of Ala354 to a proline in a conserved ASSA motif mostly inhibits passenger domain secretion [47]. In order to investigate the possible structural effect of the A354P mutation on YadA, we first used steered molecular dynamics (SMD) to pull one of the three passenger domain hairpins through the β-barrel (Table 2). The force required to pull the hairpin through the β-barrel over time was then compared between the WT hairpin systems and A354Phairpin systems over three replicas. While the force was slightly lower for the A354P hairpin system compared to that of the WT hairpin system, it was not statistically significant over most of the pulled distance (Fig. S4).
Table 2:
Simulation Details for Hairpin Secretions
| Structure | Source | System Size | Runs |
|---|---|---|---|
| WT hairpin | PDB ID: 2LME [45] | 134K atoms | 274 ns, 275 ns, 296 ns |
| A354P hairpin | Based on WThairpin, A354P | 135K atoms | 278 ns, 335 ns, 290 ns |
| Linker-stretched | PDB ID: 3SLJ [9]. Linker and passenger domain stretched | 179K atoms | 493 ns, 378 ns, 237 ns |
| Linker-helix | PDB ID: 3SLJ [9]. Only passenger domain stretched | 177K atoms | 369 ns, 297 ns, 241 ns |
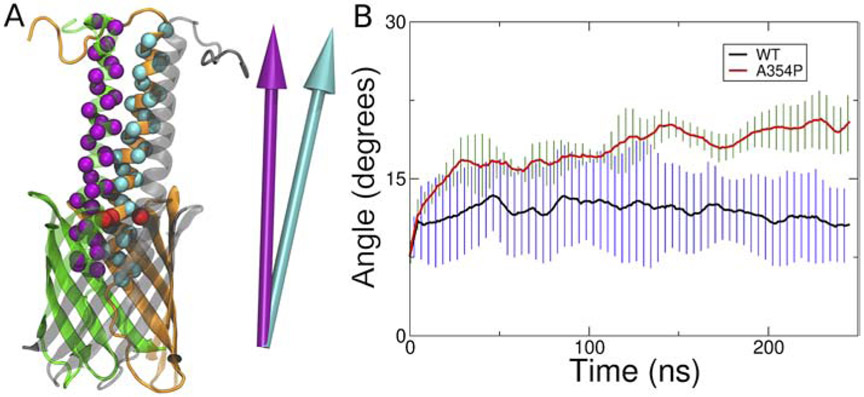
To determine if the mutation alters the structure of YadA, we ran equilibrium simulations of the WT and A354P forms of the YadA-M structure (PDB 2LME [45]). YadA-M is a construct in which the passenger domain is removed beyond three ±-helical domains extending above the β-barrel, into the extracellular space. We measured the relative motion of these three ±-helical domains, quantified by the relative angles between their principal axes (Fig. 3A). The helices remain relatively stable over the course of the simulation for the WT systems (Fig. 3B and Fig. S2). However, all A354P system replicas show an increase in the relative angle over time, indicating that the helical regions are driven apart. Specifically, the helices of WT systems maintain relative helical angles under 15°, while the helices of mutant systems go over 15°. The averages of the relative helical angles for the last 50 ns for both WT and A354P systems make the difference more apparent. For WT systems, the average is 11° while for A354P systems, it is 20°. Combined with the result from the secretion and YadA-M simulations, we conclude that although the secretion of a single passenger domain is easier with the proline mutation, it actually hinders the formation of the trimeric structure of the passenger domain, effectively stalling the full secretion. This is also supported by the result of Chauhan et al., showing that the majority of WT secretes all three helices, while A354P secretion is mostly blocked; when helices do secrete, it is typically only one or two [47].
Figure 3:
The A354P mutation drives the YadA helices apart. (A) How the relative angles were measured. First, the principal axes of the C α atoms of the helices are calculated (as represented by purple and cyan arrows). Then, the angle between the principal axes is measured. A larger angle indicates a greater separation of the helices. The C α atoms of A354 are indicated in red. (B) Average of relative angles formed between the principal axes of each helix pair. The data shown are running averages taken over 10 ns. Black and red lines represent the WT and A354P, and the error bars are represented in blue and green, respectively.
EspP β-barrel is not large enough to secrete the passenger domain in a hairpin conformation
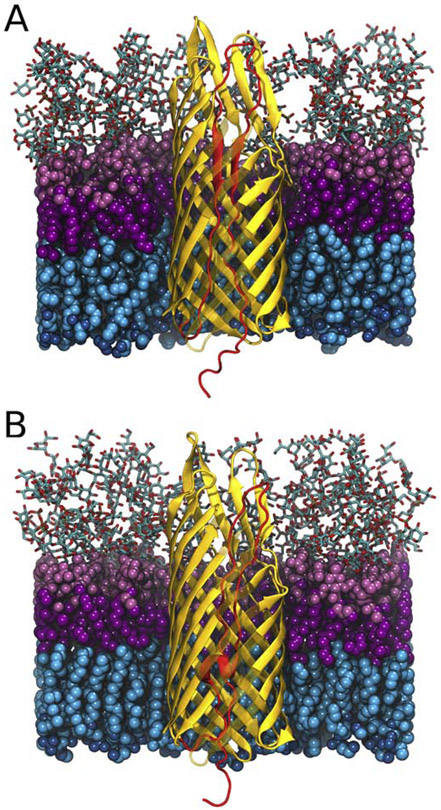
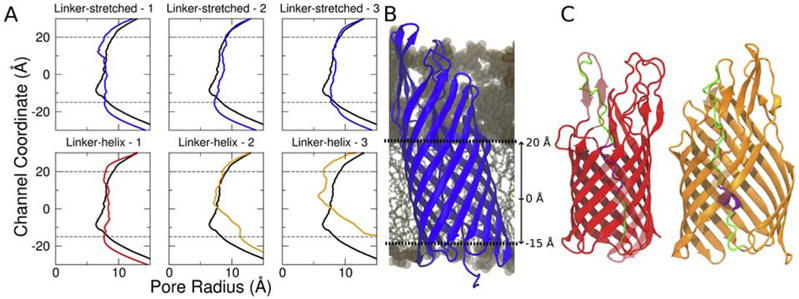
For TAAs, we showed that the β-barrel would need to expand/distort to the point of rupturing the β-barrel in order for all three ±-helices of the passenger domain to be exported in a hairpin conformation simultaneously [47, 71] (Figs. 1 and 2). However, monomeric autotransporters only need to export one hairpin through the β-barrel during secretion. Therefore, we examined whether the EspP β-barrel is large enough to accommodate the hairpin structure during secretion. Another difference between YadA and EspP is that the linker region of EspP is folded into an α-helical structure in its fully secreted state. To investigate the role of this short α-helical linker region (residue 1024 to 1030) of EspP, we have constructed two different hairpin structures of EspP (based on PDB ID: 3SLJ [9]): 1)a hairpin structure with the linker and passenger domain stretched (linker-stretched), 2) that with only passenger domain stretched (linker-helix); both systems are shown in Fig. 4. SMD was used to induce secretion for three runs over 250-500 ns each (Table 2). The force used to pull the passenger domain out of the β-barrel is comparable for linker-stretched and linker-helix structures (Fig. S5). However, when the pore size of the C-terminal β-barrel region was measured (Fig. 5 A and B), a significant increase was observed for two out of three replicas of the linker-helix structure, while the pore size of the linker-stretched structure remained constant. We examined the trajectories and discovered that, in the initial 50 ns of the simulation, the pre-folded linker region actually unfolds on its own in the replica in which the β-barrel did not expand, while the β-barrel is disrupted in the other two replicas. Specifically, the β-barrel opens between the β5 strand and β6 strand for both of these replicas (Fig. 5C). Once disrupted, the two β-strands do not come back together for the rest of the simulations. These differences in β-barrel structure or the initial unfolding of the pre-folded linker region among three replicas imply that the C-terminal β-barrel for EspP does not have enough space for both the folded linker region and the hairpin, although an unfolded hairpin can be accommodated.
Figure 4:
Comparison between the linker-stretched and linker-helix structures. (A) EspP structure [9] with the hairpin (red) made by stretching the linker and passenger domain. (B) The same structure but with only the passenger domain stretched to make a hairpin (red). Lipopolysaccharides in the outer leaflet are shown in pink (with sugar groups in licorice, polar headgroups in light pink and lipid tails in magenta), and phospholipids in the inner leaflet are shown in blue (with lipid tails in cyan and polar heads in dark blue). Some β-strands are semi-transparent to clearly show the differences between the hairpin domains.
Figure 5:
The linker-helix EspP structure either unfolds the linker region or disrupts its β-barrel to secrete its passenger domain. (A) The EspP β-barrel pore sizes for both the linker-stretched and linker-helix EspP structure for three different replicas. The black line represents the initial pore size. The blue, red and orange lines represent the final pore size for each simulation. While both the linker-stretched replicas and one linker-helix do not demonstrate a significant pore size change over time, the other linker-helix replicas show pore expansion over time. (B) The channel coordinate position relative to the EspP β-barrel. (C) The mid-secretion structures of linker-native - 1 and linker-native - 2 (in red and in orange, respectively). In the initial 50 ns of simulation, linker-native - 1 unfolds the folded linker region and preserves its β-barrel structure (red). Linker-native - 2 and linker-native - 3 keep its linker region folded while disrupting the β-barrel structure (orange).
The EspP passenger domain requires less force to secrete through the BamA β-barrel than through the EspP β-barrel
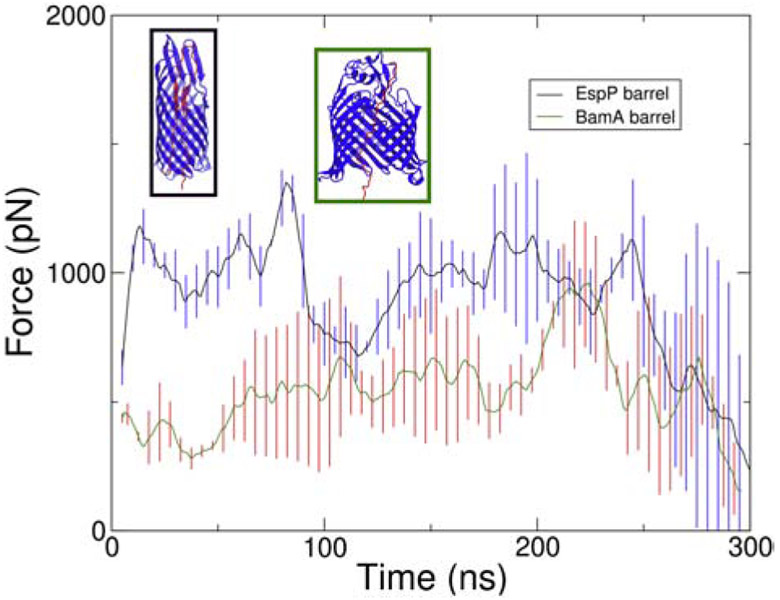
Our results indicate that the EspP β-barrel is not large enough for passenger domain secretion via a folded hairpin intermediate. We therefore wondered whether the secretion through BamA is favored over secretion via an unfolded hairpin conformation in EspP alone. We constructed and simulated a BamA-assisted model by placing the passenger domain from inside the BamA β-barrel in an extended state, and pulling them through the opening of the BamA β-barrel (PDB ID: 5D0O) [72]. The SMD simulations were run for durations indicated in Table S3. The forces required to secrete the passenger domain through the EspP β-barrel and the BamA β-barrel were compared. The force required to pull the EspP passenger domain through the BamA β-barrel was lower than that for pulling it through the EspP β-barrel over most of the pulled distance (Fig. 6, all replicas in Fig. S6). To gain further insight into the causes of the large forces, we computed the electrostatic and van der Waals interaction energies between the β-barrel and the passenger domain over time. The results show that the passenger domain is less attracted to the BamA β-barrel when compared to its native β-barrel, suggesting secretion through the BamA β-barrel is easier (Fig. S7).
Figure 6:
Comparison of force vs. time of linker-stretched hairpin structure and BamA β-barrel with EspP passenger domain. The black line shows the average force vs. time of the secretion of linker stretched hairpin structure from three replicas and the green line shows that of the BamA β-barrel with the EspP passenger domain from three replicas. Both lines are running average of the raw data taken over 10 ns. The error bars for linker-stretched hairpin structure and BamA β-barrel with EspP passenger domain are shown in blue and red, respectively.
Discussion
In this study, we explored the plausibility of the hairpin model of passenger domain secretion in autotransporters. MD simulations of two model autotransporters from the type Va (EspP) and type Vc (YadA) families were performed. For each autotransporter, a hairpin model was constructed and simulated at equilibrium to judge their stability and with steered MD to mimic secretion. Results for YadA (Figs. 1 and 2) show that to accommodate the passenger domain in the hairpin conformation, the β-barrel needs to expand, partially disrupting it. While a disrupted β-barrel in the membrane seems unlikely, an incomplete trimeric structure has been observed in the periplasm for other trimeric autotransporters [69], suggesting that the partial β-barrel formation observed in this study may be relevant in some contexts.
In experiments, the A354P mutation in YadA prevents secretion [47]. Chauhanet al. [47] showed that A354P Yada-M has the same β-sheet content as YadA-M, is stable under semi-native SDS-page conditions, and can be extracted and purified from the membrane in the folded form. These results suggest that even without complete passenger domain secretion, the YadA-M β-barrel is released from the BAM complex into the membrane. Our equilibrium simulations of YadA-M structure (lacking the passenger domain) show that the A354P mutation results in separation between the α-helical linkers (Fig. 3). This separation would limit association of the three passenger domain monomers at the extracellular surface, which we hypothesize is necessary to enforce secretion, similar to the folding of passenger domains in monomeric autotransporters [24]. While we carried out experiments attempting to capture intermediate states of the protein via cross-linking of the β-strands, multiple YadA cysteine-constructs designed based on the simulations were not found in the membrane, despite the use of reducing agents and a DegP-deletion strain (Fig. S3). Thus, we believe that plasticity of the β-barrel is necessary for its membrane insertion and likely for passenger-domain secretion as well.
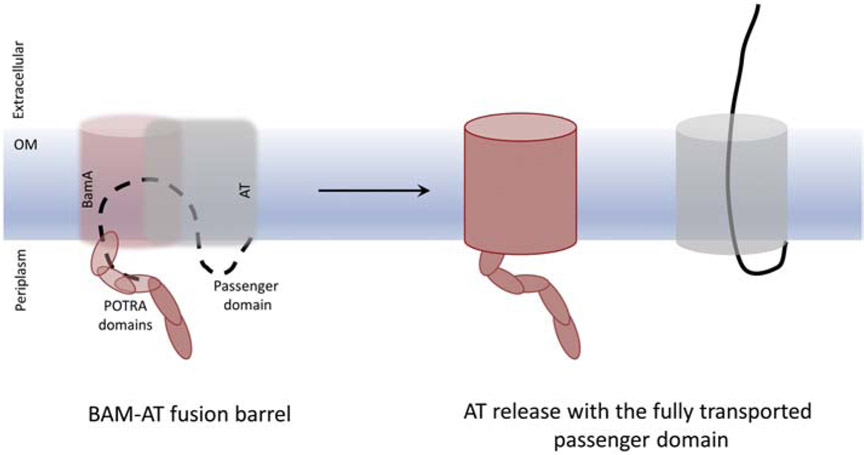
We also tested the plausibility of the BamA-assisted model for EspP. Although we did not model a hybrid EspP-BamA β-barrel, we did examine secretion of the passenger domain through BamA alone. When the passenger domain was secreted through BamA using the same method as for the hairpin model through EspP, the force required is much lower (Fig. 6). However, in some of the simulations of secretion through BamA alone, the lateral gate [73] of the BamA β-barrel also separated (data not shown). This suggests that a larger β-barrel, such as a hybrid barrel formed between BamA’s and EspP’s β-barrels during the latter’s insertion, would be more amenable to secretion than either β-barrel alone [1, 25]. Indeed, recently, Doyle and Bernstein [52] proposed that BamA integrates EspP into the membrane via an asymmetric ’swing’ mechanism, where the β-barrel is folded in the periplasm with the assistance of BamA, and then ’swung’ into the membrane where it closes to form a mature β-barrel. It is tempting to speculate that membrane insertion of the β-barrel and passenger domain secretion are tightly coupled, with secretion occurring through the immature β-barrel or its hybrid with BamA (Fig. 7). This modified BamA-assisted model would satisfy both our findings, as well as previous experimental findings on passenger domain secretion. However, it still leaves some uncertainty about trimeric autotransporters, which would either need to be inserted sequentially by BamA or form a proto-barrel in the periplasm prior to engagement with BamA. The latter could occur via interactions with a “periplasmic funnel” through other BAM-complex proteins leading to BamA [13]
Figure 7:
Preferred model for passenger domain transport. A passenger domain hairpin intermediate forms in a hybrida BamA-AT β-barrel (left panel), and secretion of the passenger domain occurs before the release and closure of the AT β-barrel domain to form the fully folded AT (right panel). The BamA and AT β-barrels are semi-transparent to clearly show the AT passenger domain.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health (R01-GM123169) and by the National Science Foundation Physics of Living Systems Student Research Network (PHY-1806833). Support from the Norwegian Center for Molecular Medicine (NCMM - to DL) is gratefully acknowledged. MOR acknowledges the Research Council of Norway for funding. Computational resources were provided through the Extreme Science and Engineering Discovery Environment (XSEDE; TG-MCB130173), which is supported by NSF Grant ACI-1548562. Additional resources were provided by the Partnership for an Advanced Computing Environment (PACE) at the Georgia Institute of Technology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Bernstein HD, Type V Secretion in Gram-Negative Bacteria, EcoSal Plus 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Henderson IR, Nataro JP, Virulence functions of autotransporter proteins, Infect. Immun 69 (2001) 1231–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Oomen CJ, van Ulsen P, van Gelder P, Feijen M, Tommassen J, Gros P, Structure of the translocator domain of a bacterial autotransporter, EMBO J. 23 (2004) 1257–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Barnard TJ, Dautin N, Lukacik P, Bernstein HD, Buchanan SK, Autotransporter structure reveals intra-barrel cleavage followed by conformational changes, Nat. Struct. Mol. Biol 14 (2007) 1214–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].van den Berg B, Crystal structure of a full-length autotransporter, J. Mol. Biol 396 (2010) 627–633. [DOI] [PubMed] [Google Scholar]
- [6].Tajima N, Kawai F, Park SY, Tame JR, A novel intein-like autoproteolytic mechanism in autotransporter proteins, J. Mol. Biol 402 (2010) 645–656. [DOI] [PubMed] [Google Scholar]
- [7].Zhai Y, Zhang K, Huo Y, Zhu Y, Zhou Q, Lu J, Black I, Pang X, Roszak AW, Zhang X, Isaacs NW, Sun F, Autotransporter passenger domain secretion requires a hydrophobic cavity at the extracellular entrance of the β-domain pore, Biochem. J 435 (2011) 577–587. [DOI] [PubMed] [Google Scholar]
- [8].Gawarzewski I, DiMaio F, Winterer E, Tschapek B, Smits SHJ, Jose J, Schmitt L, Crystal structure of the transport unit of the autotransporter adhesin involved in diffuse adherence from Escherichia coli, J. Struct. Biol 187 (2014) 20–29. [DOI] [PubMed] [Google Scholar]
- [9].Barnard TJ, Gumbart J, Peterson JH, Noinaj N, Easley NC, Dautin N, Kuszak AJ, Tajkhorshid E, Bernstein HD, Buchanan SK, Molecular basis for the activation of a catalytic asparagine residue in a self-cleaving bacterial autotransporter, J. Mol. Biol 415 (2012)128–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Klauser T, Pohlner J, Meyer TF, The secretion pathway of IgA protease-type proteins in gram-negative bacteria, Bioessays 15 (1993) 799–805. [DOI] [PubMed] [Google Scholar]
- [11].Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala’Aldeen D, Type V protein secretion pathway: the autotransporter story, Microbiol. Mol. Biol. Rev 68 (2004) 692–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Leo JC, Grin I, Linke D, Type V secretion: mechanism(s) of autotransport through the bacterial outer membrane, Philos. Trans. R. Soc. Lond. B, Biol. Sci 367 (2012) 1088–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Leo JC, Linke D, A unified model for BAM function that takes into account type Vc secretion and species differences in BAM composition, AIMS Microbiol. 4 (2018) 455–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Natale P, Bruser T, Driessen AJ, Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane-distinct translocases and mechanisms, Biochim. Biophys. Acta 1778 (2008) 1735–1756. [DOI] [PubMed] [Google Scholar]
- [15].Ruiz-Perez F, Henderson IR, Leyton DL, Rossiter AE, Zhang Y, Nataro JP, Roles of Periplasmic Chaperone Proteins in the Biogenesis of Serine Protease Autotransporters of Enterobacteriaceae, J. Bacteriol 191 (2009) 6571–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ruiz-Perez F, Henderson IR, Nataro JP, Interaction of FkpA, a peptidyl-prolyl cis/trans isomerase with EspP autotransporter protein, Gut Microbes 1 (2010) 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Plummer AM, Fleming KG, From chaperones to the membrane with a BAM!, Trends Biochem. Sci 41 (2016) 872–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bakelar I, Buchanan SK, Noinaj N, The structure of the β -barrel assembly machinery complex, Science 351 (2016) 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sauri A, Soprova Z, WickstrÖm D, de Gier J-W, Vander Schors RC, Smit AB, Jong WS, Luirink J, The Bam (Omp85) complex is involved in secretion of the autotransporter haemoglobin protease, Microbiology 155 (2009) 3982–3991. [DOI] [PubMed] [Google Scholar]
- [20].leva R, Bernstein HD, Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane, Proc. Natl. Acad. Sci. USA 106 (2009) 19120–19125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lehr U, Schutz M, Oberhettinger P, Ruiz-Perez F, Donald JW, Palmer T, Linke D, Henderson IR, Autenrieth IB, C-terminal amino acid residues of the trimeric autotransporter adhesin YadA of Yersinia enterocolitica are decisive for its recognition and assembly by BamA, Mol. Microbiol 78 (2010) 932–946. [DOI] [PubMed] [Google Scholar]
- [22].Thanassi DG, Stathopoulos C, Karkal A, Li H, Protein secretion in the absence of ATP: the autotransporter, two-partner secretion and chaperone/usher pathways of gram-negative bacteria (review), Mol. Memb. Biol 22 (2005) 63–72. [DOI] [PubMed] [Google Scholar]
- [23].lunker M, Besingi RN, Clark PL, Vectorial transport and folding of an autotransporter virulence protein during outer membrane secretion, Mol. Microbiol 71 (2009) 1323–1332. [DOI] [PubMed] [Google Scholar]
- [24].Braselmann E, Clark PL, Autotransporters: The Cellular Environment Reshapes a Folding Mechanism to Promote Protein Transport, J. Phys. Chem. Lett 3 (2012) 1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bernstein HD, Looks can be deceiving: recent insights into the mechanism of protein secretion by the autotransporter pathway, Mol. Microbiol 97 (2015) 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ieva R, Tian P, Peterson JH, Bernstein HD, Sequential and spatially restricted interactions of assembly factors with an autotransporter βdomain, Proc. Natl. Acad. Sci. USA 108 (2011) E383–E391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pavlova O, Peterson JH, leva R, Bernstein HD, Mechanistic link between barrel assembly and the initiation of autotransporter secretion, Proc. Natl. Acad. Sci. USA 110 (2013) E938–E947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Roman-Hernandez G, Peterson JH, Bernstein HD, Reconstitution of bacterial autotransporter assembly using purified components, eLife 3 (2014) e04234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Brunder W, Schmidt H, Karch H, EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V, Mol. Microbiol 24 (1997) 767–778. [DOI] [PubMed] [Google Scholar]
- [30].Linke D, Riess T, Autenrieth IB, Lupas A, Kempt VA; Trimeric autotransporter adhesins: variable structure, common function, Trends Microbiol. 14(2006) 264–270. [DOI] [PubMed] [Google Scholar]
- [31].Weiss A, Brockmeyer J, Prevalence, biogenesis, and functionality of the serine protease autotransporter EspP, Toxins (Basel) 5 (2012)25–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Dautin N, Barnard TJ, Anderson DE, Bernstein HD, Cleavage of a bacterial autotransporter by an evolutionarily convergent autocatalytic mechanism, EMBO J. 26 (2007) 1942–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ieva R, Skillman KM, Bernstein HD, Incorporation of a polypeptide segment into the β-domain pore during the assembly of a bacterial autotransporter, Mol. Microbiol 67 (2008) 188–201. [DOI] [PubMed] [Google Scholar]
- [34].Dé E, Saint N, Glinel K, Meli AC, Lévy D, Jacob-Dubuisson F, Influence of the passenger domain of a model autotransporter on the properties of its translocator domain, Mol. Membr. Biol 25 (2008) 192–202. [DOI] [PubMed] [Google Scholar]
- [35].Xicohtencatl-Cortes J, Saldana Z, Deng W, Castaneda E, Freer E, Tarr PI, Finlay BB, Puente JL, Giron JA, Bacterial macroscopic rope-like fibers with cytopathic and adhesive properties, J. Biol. Chem 285 (2010) 32336–32342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Djafari S, Ebel F, Deibel C, Kramer S, Hudel M, Chakraborty T, Characterizat ion of an exported protease from Shiga toxin-producing Escherichia coli, Mol. Microbiol 25 (1997) 771–784. [DOI] [PubMed] [Google Scholar]
- [37].In J, Lukyanenko V, Foulke-Abel J, Hubbard AL, Delannoy M, Hansen A-M, Kaper JB, Boisen N, Nataro JP, Zhu C, Boedeker EC, Giron JA, Kovbasnjuk O, Serine protease EspP from enterohemorrhagic Escherichia coli is sufficient to induce shiga toxin macropinocytosis in intestinal epithelium, PLoS One 8 (2013) e69196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Byrd W, Ruiz-Perez F, Setty P, Zhu C, Boedeker EC, Secretion of the Shiga toxin B subunit (Stx1B) via an autotransporter protein optimizes the protective immune response to the antigen expressed in an attenuated E. coli (rEPEC E22 Δ ler) vaccine strain, Vet. Microbiol 211 (2017) 180–188. [DOI] [PubMed] [Google Scholar]
- [39].Leo JC, Skurnik M, Adhesins of human pathogens from the genus Yersinia, Adv. Exp. Med. Biol 715(2011)1–15. [DOI] [PubMed] [Google Scholar]
- [40].Rosqvist R, Skurnik M, Wolf-Watz H, Increased virulence of Yersinia pseudotuberculosis by two independent mutations, Nature 334 (1988) 522–525. [DOI] [PubMed] [Google Scholar]
- [41].Eitel J, Dersch P, The YadA protein of Yersinia pseudotuberculosis mediates high-efficiency uptake into human cells under environmental conditions in which invasin is repressed, Infect. Immun 70 (2002)4880–4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Nummelin H, Merckel MC, Leo JC, Lankinen H, Skurnik M, Goldman A, The Yersinia adhesin YadA collagen-binding domain structure is a novel left-handed parallel β-roll, EMBO J. 23 (2004) 701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Koretke KK, Szezesny P, Gruber M, Lupas AN, Model structure of the prototypical non-fimbrial adhesin YadA of Yersinia enterocolitica, J. Struct. Biol 155 (2006) 154–161. [DOI] [PubMed] [Google Scholar]
- [44].Wollmann P, Zeth K, Lupas AN, Linke D, Purification of the YadA membrane anchor for secondary structure analysis and crystallization, Int. J. Biol. Macromol 39 (2006) 3–9. [DOI] [PubMed] [Google Scholar]
- [45].Shahid SA, Bardiaux B, Franks WT, Krabben L, Habeck M, van Rossum BJ, Linke D, Membrane-protein structure determination by solid-state NMR spectroscopy of microcrystals, Nat. Methods 9 (2012) 1212–1217. [DOI] [PubMed] [Google Scholar]
- [46].Shahid SA, Markovic S, Linke D, van Rossum B-J, Assignment and secondary structure of the YadA membrane protein by solid-state MAS NMR, Sci. Rep 2 (2012)803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Chauhan N, Hatlem D, Orwick-Rydmark M, Schneider K, Floetenmeyer M, van Rossum B, Leo JC, Linke D, Insights into the autotransport process of a trimeric autotransporter, Yersinia Adhesin A(YadA), Mol. Microbiol 111 (2019)844–862. [DOI] [PubMed] [Google Scholar]
- [48].Grosskinsky U, Schütz M, Fritz M, Schmid Y, Lamparter MC, Szczesny P, Lupas AN, Autenrieth IB, Linke D, A Conserved Glycine Residue of Trimeric Autotransporter Domains Plays a Key Role in Yersinia Adhesin A Autotransport, J. Bacteriol 189(2007)9011–9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Holdbrook DA, Piggot TJ, Sansom MS, Khalid S, Stability and membrane interactions of an autotransport protein: MD simulations of the Hia translocator domain in a complex membrane environment, Biochim. Biophys. Acta - Biomembr 1828 (2013)715–723. [DOI] [PubMed] [Google Scholar]
- [50].Aoki E, Sato D, Fujiwara K, Ikeguchi M, Electrostatic repulsion between unique arginine residues is essential for the efficient in vitro assembly of the transmembrane domain of a trimeric autotransporter, Biochemistry 56 (2017) 2139–2148. [DOI] [PubMed] [Google Scholar]
- [51].Aoki E, Ikeguchi M, In vitro assembly of Haemophilus influenzae adhesin transmembrane domain and studies on the electrostatic repulsion at the interface, Biophys. Rev 11 (2019)303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Doyle MT, Bernstein HD, Bacterial outer membrane proteins assemble via asymmetric interactions with the BamA β-barrel, Nat. Commun 10 (2019) 3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K, Scalable molecular dynamics with NAMD, J. Comput. Chem 26(2005)1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Best RB, Zhu X, Shim J, Lopes PEM, Mittal J, Feig M, MacKerell AD, Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone φ , ψ and Side-Chain χ 1 and χ 2 Dihedral Angles, J. Chem. Theory Comput 8 (2012)3257–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Huang J, Rauscher S, Nawrocki G, Ran T, Feig M, de Groot BL, Grubmuller H, MacKerell AD, CHARMM36m: an improved force field for folded and intrinsically disordered proteins, Nat. Methods 14 (2017)71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML, Comparison of simple potential functions for simulating liquid water, J. Chem. Phys 79 (1983) 926–935. [Google Scholar]
- [57].Darden T, Perera L, Li L, Pedersen L, New tricks for modelers from the crystallography toolkit: the particle mesh Ewald algorithm and its use in nucleic acid simulations, Structure 7 (1999) 55–60. [DOI] [PubMed] [Google Scholar]
- [58].Knapp B, Ospina L, Deane CM, Avoiding False Positive Conclusions in Molecular Simulation: The Importance of Replicas, J. Chem. Theory Comput 14 (2018) 6127–6138. [DOI] [PubMed] [Google Scholar]
- [59].Humphrey W, Dalke A, Schulten K, VMD: Visual Molecular Dynamics, J. Mol. Graphics 14 (1996)33–38. [DOI] [PubMed] [Google Scholar]
- [60].Jo S, Kim T, Iyer VG, Im W, CHARMM-GUI: A web-based graphical user interface for CHARMM, J. Comput. Chem 29 (2008) 1859–1865. [DOI] [PubMed] [Google Scholar]
- [61].Lee J, Patel DS, Sthle J, Park S-J, Kern NR, Kim S, Lee J, Cheng X, Valvano MA, Holst O, Knirel YA, Qi Y, Jo S, Klauda JB, Widmalm G, Im W, CHARMM-GUI Membrane Builder for Complex Biological Membrane Simulations with Glycolipids and Lipoglycans, J. Chem. Theory Comput 15 (2019) 775–786. [DOI] [PubMed] [Google Scholar]
- [62].Tornabene TG, Lipid composition of selected strains of Yersinia pestis and Yersinia pseudotuberculosis, Biochim. Biophys. Acta - Lipids Lipid Metab. 306 (1973) 173–185. [DOI] [PubMed] [Google Scholar]
- [63].Whittaker P, Comparison of Yersinia pestis to other closely related Yersinia species using fatty acid profiles, Food Chem. 116(2009) 629–632. [Google Scholar]
- [64].Wu EL, Fleming PJ, Yeom MS, Widmalm G, Klauda JB, Fleming KG, Im W, E. coli outer membrane and interactions with OmpLA, Biophys. J 106 (2014)2493–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Hwang H, Paracini N, Parks JM, Lakey JH, Gumbart JC, Distribution of mechanical stress in the Escherichia coli cell envelope, Biochim. Biophys. Acta 1860 (2018) 2566–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Fiorin G, Klein ML, Hénin J, Using collective variables to drive molecular dynamics simulations, Mol. Phys 111 (2013)3345–3362. [Google Scholar]
- [67].Powers RE, Gaudet R, Sotomayor M, A partial calcium-free linker confers flexibility to inner-ear protocadherin-15, Structure 25 (2017)482–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Smart OS, Neduvelil JG, Wang X, Wallace B, Sansom MS, Hole: A program for the analysis of the pore dimensions of ion channel structural models, J. Mol. Graphics 14 (1996) 354–360. [DOI] [PubMed] [Google Scholar]
- [69].Sikdar R, Peterson JH, Anderson DE, Bernstein HD, Folding of a bacterial integral outer membrane protein is initiated in the periplasm, Nat. Commun 8 (2017) 1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Meuskens I, Michalik M, Chauhan N, Linke D, Leo JC, A New Strain Collection for Improved Expression of Outer Membrane Proteins, Front. Cell. Infect. Microbiol 7 (2017) 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Bassler J, Alvarez BH, Hartmann MD, Lupas AN, A domain dictionary of trimeric autotransporter adhesins, Int. J. Med. Microbiol 305 (2015) 265–275. [DOI] [PubMed] [Google Scholar]
- [72].Gu Y, Li H, Dong H, Zeng Y, Zhang Z, Paterson NG, Stansfeld PJ, Wang Z, Zhang Y, Wang W, Dong C, Structural basis of outer membrane protein insertion by the BAM complex, Nature 531 (2016) 64–69. [DOI] [PubMed] [Google Scholar]
- [73].Lundquist K, Bakelar J, Noinaj N, Gumbart JC, C-terminal kink formation is required for lateral gating in BamA, Proc. Natl. Acad. Sci. USA 115 (2018) E7942–E7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.