Abstract
We investigated the cytolytic and mechanistic activity of anti-CD19 chimeric antigen receptor natural killer (CD19.CAR.NK92) therapy in lymphoma cell lines (diffuse large B-cell, follicular, and Burkitt lymphoma), including rituximab- and obinutuzumab-resistant cells, patient-derived cells, and a human xenograft model. Altogether, CD19.CAR.NK92 therapy significantly increased cytolytic activity at E:T ratios (1:1–10:1) via LDH release and prominent induction of apoptosis in all cell lines, including in anti-CD20 resistant lymphoma cells. The kinetics of CD19.CAR.NK92 cell death measured via droplet-based single cell microfluidics analysis showed that most lymphoma cells were killed by single contact, with anti-CD20 resistant cell lines requiring significantly longer contact duration with NK cells. Additionally, systems biology transcriptomic analyses of flow-sorted lymphoma cells co-cultured with CD19.CAR.NK92 revealed conserved activation of IFNγ signaling, execution of apoptosis, ligand binding, and immunoregulatory and chemokine signaling pathways. Furthermore, a 92-plex cytokine panel analysis showed increased secretion of granzymes, increased secretion of FASL, CCL3 and IL10 in anti-CD20 resistant SUDHL-4 cells with induction of genes relevant to mTOR and G2/M checkpoint activation were noted in all anti-CD20 resistant cells co-cultured with CD19.CAR.NK92 cells. Collectively, CD19.CAR.NK92 was associated with potent anti-lymphoma activity across a host of sensitive and resistant lymphoma cells that involved distinct immuno-biologic mechanisms.
INTRODUCTION
B-cell non-Hodgkin lymphomas (bNHL) are the most common form of lymphoma in the Western World. bNHLs are generally treatable, however the vast majority of indolent bNHL patients are incurable and a significant minority of patients with aggressive bNHL die from the disease. Improved therapeutics for NHLs are desired, especially ‘targeted’ immunologic agents with favorable side effect panels.
The human natural killer (NK) cell line, NK-92, isolated from a patient with NK cell lymphoma, is fully characterized, expandable with maintained cytotoxicity, and available as clinical grade, “off the shelf” cellular product [1–8]. Notably, NK-92 cells lack most killer-cell immunoglobulin-like receptors (KIRs) with few exceptions (e.g., KIR2DL4). Several studies have demonstrated that NK-92 kills cancer cells [5–7, 9–11]. In vitro cytotoxicity assays demonstrated that NK-92 cells maintain high degrees of cytotoxicity at effector:target ratios (10:1) vs an array of human cancer lines[9]. NK-92 was also shown to be effective in myeloma and chronic lymphocytic leukemia animal/primary models [10, 11].
To enhance target specificity, NK-92 cells were bioengineered to express chimeric antigen receptors (CARs) against target antigens expressed on tumor cells (e.g., CD19). CARs are composed of an extra-cellular domain consisting monoclonal antibody derived from single chain variable fragment (scFv) fused with CD8 transmembrane domain and intracytoplasmic signal transduction domain derived from CD3 (zeta) [1, 2, 12]. Although peripheral blood derived NK cells are utilized for generation of CAR-NK cells, improvements to increasing the gene transfer efficiency, overcoming limitations related to in vitro expansion, in vivo persistence following the infusion, and reducing lag time delays associated with manufacturing of CAR-NK cells are apparent [13]. Similar disadvantages also are relevant to CAR-T manufacturing process resulting in treatment delays that may not be tenable for patients with clinically aggressive disease [14]. Thus, availability of “off the shelf” engineered versions of continuously expanding NK92-CAR cells provides a potential novel targeted product for urgent or immediate therapeutic need. In vivo studies using CD19.CAR.NK92 have shown efficient drug distribution and cell kill in leukemia murine models [2, 12].
CD19 is a cell surface protein ubiquitously expressed through all stages of B cell development and consistently present in all malignant B cells, including in bNHL [15]. Targeting CD19 is an attractive strategy for the treatment of bNHL with CAR modified T or NK cells. Patients with bNHL are traditionally treated with anti-CD20 antibody therapy (i.e., rituximab or obinutuzumab), either alone or in combination with chemotherapy platforms [16]. However, many bNHL patients treated with anti-CD20 antibody therapy develop disease relapse or become refractory, which continues to be a major unmet need. Potential factors involved in resistance to anti-CD20 antibody therapy include loss of CD20 expression on the cell surface of B lymphocytes and deficiencies related to host immune factors, such as FC receptor polymorphism, immune suppression that impede NK, T or macrophage dependent antibody directed cell mediated cytotoxicity[16]. Targeting CD19 is rationale and the availability of “off the shelf” CD19.CAR.NK92 may provide a viable option for bNHL patients with CD20 antibody resistant aggressive disease and/or for patients either unfit or unable to wait for manufactured CAR-T or CAR-NK therapies.
Thus, our goal in this study was to establish the mechanistic rationale for NK-based therapy in bNHL and to determine the therapeutic potency and kinetics of the targeted “off the shelf” therapy, CD19-CAR-NK, in a host of in vitro bNHL cell lines (including anti-CD20 resistant cells), primary patient derived cells, and an in vivo xenograft model. Furthermore, we utilized unbiased systems biology approach to characterize the biological pathways and genes responsive to CD19.CAR.NK92 activity both in anti-CD20 sensitive and resistant bNHL models.
MATERIALS AND METHODS
Cell culture
DLBCL cell lines SUDHL4 and SUDHL10, Burkitt’s cell line Raji, were purchased from American Type Culture Collection (Manassas, VA, USA), STR profiling authenticated were maintained in RPMI-1640 medium supplemented with 10% Fetal Bovine Serum and 1% antibiotic-antimycotic solution (Corning Cellgro, Manassas, VA, USA). Rituximab (RR) and obinutuzumab (OR) resistant cells were established by continuously culturing SUDHL4 and SUDHL10 with increasing concentration of rituximab and obinutuzumab (5–20μg/mL) for over a period of 8 weeks and characterized resistance as decreased CD20 expression (Figure S1), using anti-CD20-Cy5 staining (#347201, BD Bioscience, San Jose, CA, USA) and flow cytometry. CD19 antigen density were determined by flow cytometry, using Quantibrite anti-CD19-PE (Clone SJ25C1)#340364, and Quantibrite-PE calibration beads (BD Bioscience, San Jose, CA, USA). Primary human lymphoma cells (EL-2 and EL-5) were isolated from IRB exempt discarded tissue specimens of patients with relapsed/refractory diffuse large B-cell lymphoma (DLBCL) obtained through Tufts Tumor Repository, as described previously [17]. Activated NK-92 (aNK) and CD19.CAR.NK92 cells were provided as gift from Nantkwest, Inc. (Woburn, MA, USA) and the details of CAR construct used in design and development of CD19.CAR.NK92 cells is outlined in [1, 2], and (Supplemental Methods). CD19.CAR.NK92 maintained in MyeloCult H5100 (STEMCELL Technologies, Cambridge, MA, USA) and supplemented with recombinant human IL-2 (500IU/ml) (STEMCELL Technologies). aNK cells engineered to express endogenously retained IL2 [18] were maintained in X-Vivo 10 media (Lonza, Walkersville, MD, USA) supplemented with 5% human serum. Primary human CD56+ NK cells (≥90% purity) used in microfluidic single cell analyses and peripheral blood mononuclear cells (PBMC) purchased from Stemcell Technologies (Cambridge, MA, USA), cultured in RPMI-1640 media consisting 10% FBS and 50 ng/mL IL-2 (Peprotech, Rocky Hill, NJ, USA).
Transcriptomic analyses
SUDHL4, SUDHL4-RR, SUDHL4-OR, SUDHL10, SUDHL10-RR, SUDHL10-OR, Raji, EL-2 and EL-5 cell lines were co-cultured with CD19.CAR.NK92 for two hours at effector:target (E:T) ratio 1:1 with each consisting 5X106 cells maintained at 37°C and 5% CO2. Following co-culture, flow sorting of CD19 positive target bNHL cells preloaded with cell-tracking dye QDOT655 (Molecular Probes, Eugene, OR), was performed by gating out GFP+ CD19.CAR.NK92 cells. Isolated bNHL cells were used for isolation of total mRNA using RNeasy Minikit (Qiagen, Germantown, MD), and microarray experiments were performed using human HT-12 bead array chips (Illumina, San Diego, CA, USA) at Yale Center for Genomic Analysis. All experiments were performed in triplicates. Background correction, data normalization, statistical analysis, Ingenuity Pathway Analysis (IPA), Gene Set Enrichment Analysis (GSEA), Cytoscape network analysis, and determination of key significant genes were performed as previously reported [19–21] and outlined in Supplemental Methods. Genes associated with NK ligands were compiled through a comprehensive search from literature. The raw expression data from these experiments are available at NCBI Gene Expression Omnibus (GEO) database, with following identifiers: GSE120315 and GSE120316.
Cell-mediated cytotoxicity and apoptosis
NK cells (both CD19.CAR.NK92 and aNK), anti-CD3/CD28, IL-2 activated PBMC and lymphoma cells were co-cultured at E:T ratios ranging 1:1–10:1 in Lymphocyte Growth Medium LGM3 (Lonza) for 4 hours in U bottom 96-well cell culture plates. Supernatants were collected to determine percent target cell lysis based on GAPDH release, as outlined in the instructions supplied with AcellaTox-Glo assay kit (Cell Technology, Fremont, CA, USA) or LDH release assay (Promega, Madison, WI, USA). CD19 staining and apoptosis assay based on Annexin V-PE / 7-AAD staining by flow cytometry were performed following the protocol as provided by the manufacturer, BD Biosciences. All experiments were performed in experimental triplicates, with error bars represented in data indicating standard deviations of mean, statistical significant differences determined by 2-tailed, t-test, comparing control with experimental conditions.
Microfluidic analysis
Target cells labeled with 2μM Calcein AM (Thermo Fisher, Waltham, MA; excitation/emission: 495nm/515nm) at 37°C for 20 minutes, were washed once with media and loaded in a syringe at 1.5 million/mL concentration. Both target cells and unlabeled CD19.CAR.NK92 were introduced in the microfluidic device, oil droplets were generated and image acquisition were performed as outlined before [17]. Target cell death was indicated by significant (>80%) loss of Calcein AM fluorescence, number of contacts made and duration of contacts between cell pairs were determined as described previously [17, 22]. Death Index calculated for each target cell as the ratio of death time to total contact time with the NK cell and represented as averaged value for cells undergoing single contact or multiple (2–9) contacts with NK cells. All statistical analysis was performed using non-parametric two-sided Mann Whitney U test, and p value <0.05 was considered statistically significant.
Cytokine release assay
For the determination of immune factors secreted during CD19.CAR.NK92 mediated cytolytic activity of bNHL cells, 5000 cells were co-cultured at 1:1 (E:T) for 24 hours in 96 well U bottom plate in replicates. Supernatant were collected and analyzed using proximity expression assay analyzing predesigned 92-plex cytokines immuno-oncology panel and service available from Olink Proteomics Inc. (Watertown, MA, USA). Cytokine panel analyzed is listed in Table S1, results were reported as normalized protein expression Log2 NPx values, as reported [24].
RESULTS
CD19 selective efficacy and cell death with CD19.CAR.NK92 in bNHL
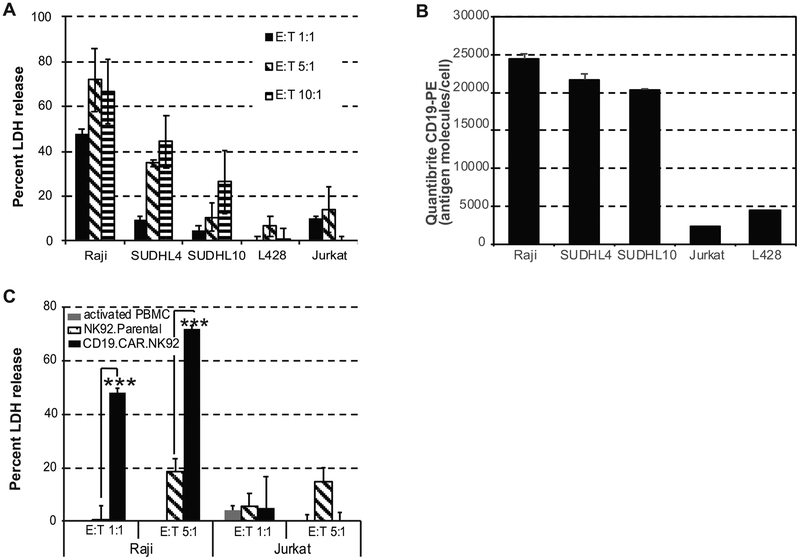
We examined the cytolytic activity of CD19.CAR.NK92 against various target cells including CD19+ bNHL cells (Raji, SUDHL4 and SUDHL10), CD19 negative Jurkat T cell lymphoma (TCL), and L428 Hodgkin lymphoma (HL) cells. CD19.CAR.NK92 (effector) cells were co-cultured with target cells at effector to target (E:T) ratios 1:1–10:1, for 4 hours followed by assessment of LDH release from the cell culture supernatants, as a measure of cytolysis resulting from cytolytic activity mediated by CD19-CAR-NK. There was significant cytolytic activity in all CD19+ bNHL cells (Raji, SUDHL4 and SUDHL10) at E:T ratios (1:1–10:1), while minimal activity was observed in CD19 negative Jurkat or L428 cell lines (Figure 1A). CD19 expression, in terms of antigen molecules/cell in these target cells evaluated based on calibrated Quantibrite anti-CD19-PE immunostaining and flow cytometry assay confirmed lack of CD19 staining in Jurkat and L428, (Figure 1B), which corresponded to poor CD19.CAR.NK92 activity observed in these cells, while highly expressed in the bNHL cells (Figure 1B).
Figure 1. CD19.CAR.NK92 mediated cytotoxicity in B cell non-Hodgkin lymphoma (bNHL).
A) Bar graph representing percent LDH release (y-axis) from target B and T lymphoma cells, with increasing Effector (CD19.CAR.NK92) to Target (E:T) ratio represented in x-axis from 4-hour co-culture experiments indicate increased cytotoxicity occurring only in B lymphoma cells (Raji, SUDHL4, and SUDHL10). B) Bar graph representing average CD19 antigen density/cell determined based on calibrated Quantibrite CD19-PE immunostaining and flow cytometry analysis of B and T lymphoma cells. C) Bar graph representing percent LDH release (y-axis) comparing the cytotoxic effects of (PBMC, activated NK92 and CD19.CAR.NK92 in target Raji or Jurkat (TCL) at indicated E:T ratios, show significant CD19+ selective cytotoxic activity of CD19.CAR.NK92 against Raji cells compared to parental NK92 cells (*** denotes P<0.005). All experiments were performed in triplicates, error bars represent standard deviations.
Next, we compared cytotoxicity of CD19.CAR.NK92 with mixed lymphocyte reaction (MLR) consisting anti-CD3/CD28 and IL-2 activated peripheral blood mononuclear cells (PBMC), IL-2 secreting; activated NK-92 (aNK) cells in Raji; or Jurkat cells via LDH release assay. Results from these experiments showed minimal cytotoxicity with aNK (<5%) compared with 45% with CD19.CAR.NK92 at E:T ratio 1:1 (P<0.001), and 20% with aNK compared with 70% using CD19.CAR.NK92 (P<0.001) at E:T ratio 5:1 (Figure 1C). In Jurkat cells at E:T ratio of 5:1, aNK exhibited cytolytic activity comparable with Raji cells (Figure 1C).
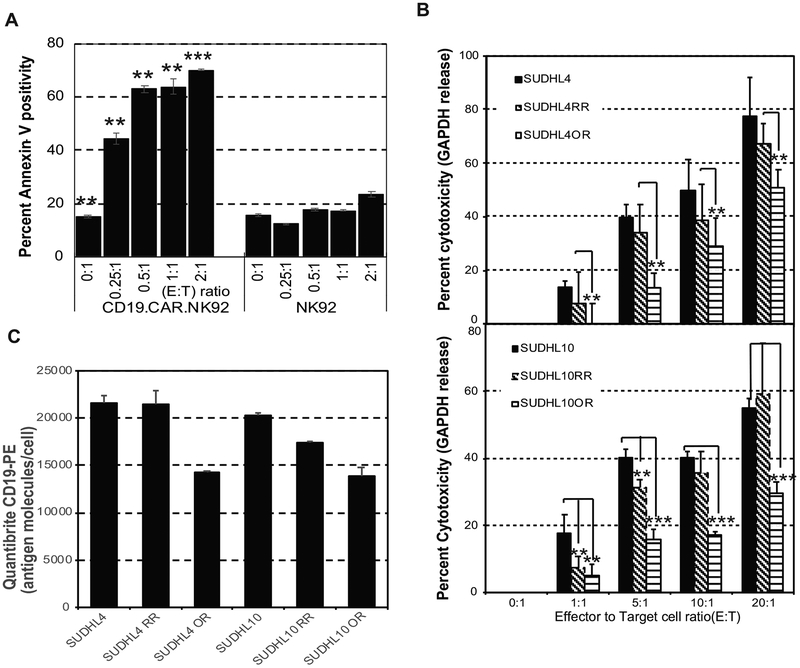
While granzymes and perforin triggered cytolytic activity occur rapidly (<4 hours), induction of apoptosis by secreted factors occur at 24 hours. We then compared the ability of CD19.CAR.NK92 vs. aNK to induce apoptosis in target Raji cells by co-culturing for 24 hours, followed by Annexin-V-PE/7-AAD staining and flow cytometry analysis gating out CD19.CAR.NK92 GFP positive cells. CD19.CAR.NK92 induced potent apoptotic cell death at E:T ratio as low as 0.25:1 (45%) and >60% E:T ratios 0.5:1 – 2:1 (Figure 2A), while Annexin-V/7AAD positivity in Raji cells were barely detectable in the presence of aNK at E:T 2:1 (Figure 2A). These results suggested NK-92 engineered to express CD19 as CD19.CAR.NK92 was selectively active against CD19+ Raji and other bNHL cells.
Figure 2. CD19.CAR.NK92 mediated cytotoxicity in CD20 sensitive and resistant B cell non-Hodgkin lymphoma (bNHL).
A) Bar graph representing percent Annexin-V positivity determined based on flow cytometry comparing the apoptotic effects of activated NK92 and CD19.CAR.NK92 in target Raji cells at indicated E:T ratios, from 24-hour co-culture indicate that co-culture of Raji cells with CD19.CAR.NK92 resulted in significant apoptotic activity compared to parental NK92. (** denotes P<0.05, *** denotes P<0.005). B) Bar graph represents percent GAPDH release (y-axis) from target anti-CD20 sensitive or resistant (RR and OR) SU-DHL4 or SU-DHL10 cells, with increasing Effector (CD19.CAR.NK92) to Target (E:T) ratio represented in x-axis from 4 hour co-culture experiments, indicate no significant differences in the cytotoxic responses in RR cells compared to sensitive cells, with significant lower cytotoxic responses in OR cells compared to sensitive cells, with CD19.CAR.NK92. (** denotes P<0.05, *** denotes P<0.005). C) Bar graph represents average CD19 antigen density/cell determined based on calibrated Quantibrite CD19-PE immunostaining and flow cytometry analysis indicate generalized reduction in the antigen density of CD19 expression in OR cells compared anti-CD20 sensitive or RR, SU-DHL4 or SU-DHL10 cells. All experiments were performed as experimental triplicates, with error bars representing standard deviations of mean and statistical significant differences (P values)comparing control with experimental conditions were determined by 2-tailed, t-test.
CD19.CAR.NK92 activity in anti-CD20 sensitive and resistant bNHL cells.
Here, 103 target cells of SUDHL4 or SUDHL10 sensitive and anti-CD20-resistant cells (SUDHL4-RR, SUDHL4-OR, SUDHL10-RR, SUDHL10-OR) were co-cultured with CD19.CAR.NK92 at E:T of 1:1– 20:1 for 4 hours. We observed E:T ratio dependent increase in cytolytic activity by CD19.CAR.NK92 on all target cells (Figure 2B), including in RR SUDHL4 and SUDHL10, while relatively lower cytolytic activity was seen in OR cells in both bNHL models (Figure 2B). Flow cytometric assessments with calibrated Quantibrite anti-CD19-PE immunostaining revealed lowered expression of CD19 antigen molecules/cell, in SUDHL4-OR (14330±69) and SUDHL10-OR (13847±1004) compared with SUDHL4 (21626+772) and SUDHL10 (20342+176) cells (Figure 2C). In addition, the in vivo efficacy of CD19.CAR.NK92 investigated using human tumor xenografts derived from SUDHL10 or OR cells, revealed considerable lag time delay in the tumor growth with six doses of CD19.CAR.NK92 treatment compared with untreated SCID mice (Figure S2 & Supplemental Methods).
Dynamic efficacy of CD19.CAR.NK92 vis-à-vis microfluidic analyses
Next, we determined the heterogeneity in dynamic cellular interaction between CD19.CAR.NK92 cells and bNHL cells, since NK cells have been shown to interact with and kill other target cell types with variable kinetics[22]. Rapid target cell recognition and stable conjugation may lead to robust target killing activity, as observed in our prior studies [17]. The dynamics of target cell (NHL cells) interaction with CD19.CAR.NK92, unmodified NK92 cells, or primary human CD56+ NK cells encapsulated in fluid-droplets of size 90 ± 10μm diameter at 1:1 ratio were evaluated (Figure S3.A). We observed a higher rate of target cell (SUDHL10) conjugations made by CD19.CAR.NK92 cells (75 ± 7%) compared with NK92 (58 ± 2%) or primary NK cells (33 ± 12%) (Figure S3.A). Next, we observed that 55% of CD19.CAR.NK92 cells were able to establish initial contacts with target SUDHL10 cells rapidly (<5 minutes) compared with 45% and 30%, respectively, by NK92 and primary NK cells (Figure S3.B). Furthermore, we noted dynamic variations in the nature of interactions that occurred between CD19.CAR.NK92 cells and target cells in anti-CD20 sensitive (SUDHL10) cells vs. resistant RR and OR cells (Figure 2).
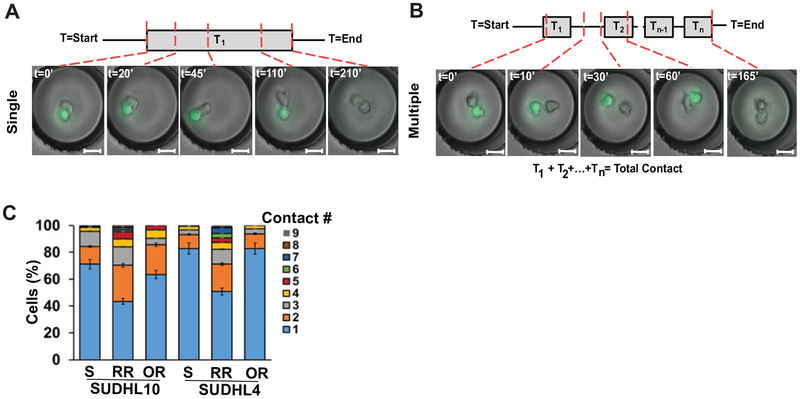
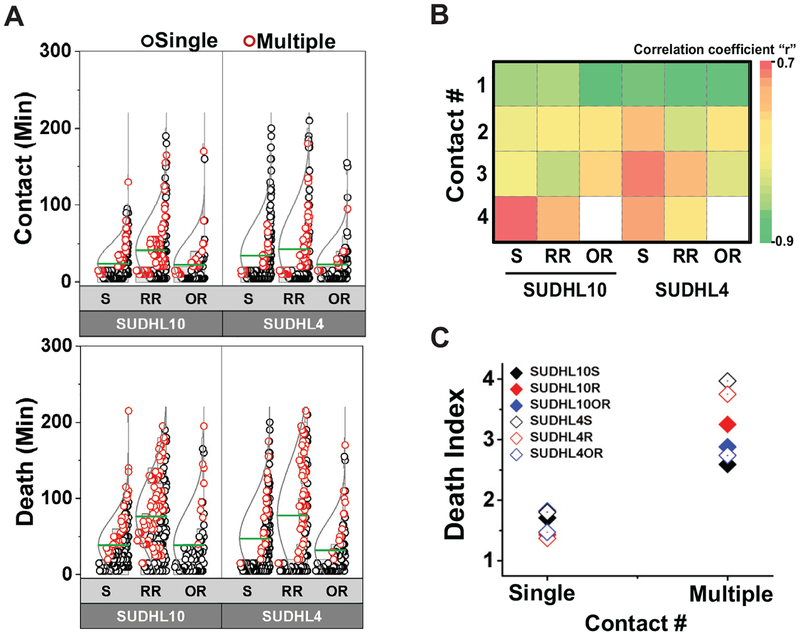
Discrete E-T cell pairs formed either single long-lasting contacts (Figure 3A) or multiple short contacts (Figure 3B). While the parental and OR target cells primarily formed single contacts with CD19.CAR.NK92 cells (>60% for SUDHL10, >80% for SUDHL4), the RR cells in both target lines formed more multiple (2–9) contacts with NK cells (Figure 3C). CD19.CAR.NK92 cells compared to the parental SUDHL10 cells (Figure 4A, mean: 24 min, P<0.0001). Consequently, there was a time delay in cell death in SUDHL10-RR (mean: 76 minutes) compared to SUDHL10 (mean: 38 minutes, P<0.0001) (Figure 4A). In contrast, SUDHL10-OR cells had contact profiles and death times similar to parental cells. However, CD19.CAR.NK92 cells mediated death of OR cells faster than RR cells for both cell lines (mean values for SUDHL10-OR/RR: 38/76 minutes, respectively, P<0.01; SUDHL4-OR/RR: 32/78 minutes respectively, P<0.01).
Figure 3. Microfluidic real-time single cell interaction kinetics of CD19.CAR.NK92 treated bNHL cells.
A-B) Interaction between CD19.CAR.NK92 cells and target cells in microfluidic droplets depicting single (A) or multiple (B) contacts between distinct cell pairs. T = 0–210’ indicates minutes elapsed. C) Frequency of contacts between CD19.CAR.NK92 - SUDHL4/10 cell pairs.
Figure 4. Differential single cell interaction kinetics between CD19.CAR.NK92 and anti-CD20 senstive and resistant bNHL cells.
A) Distribution of total contact duration and death times of SUDHL4/10 cell pairs treated with CD19.CAR.NK92. - Cell pairs undergoing single contacts are indicated by black circles and cell pairs demonstrating multiple (2–9) contacts are indicated by red circles. Mean of the distributions are represented by green lines. *P< 0.05, **P< 0.01, ***P<0.0001. (B) Matrix of correlation between total contact duration and time of death for effector-target cell pairs. (C) Averaged efficiency of cell death depicted by Death Index, the ratio between time of death to contact time TD/TC for single cell pairs. All data obtained from n=181, 216, and 103 cells for SUDHL10, SUDHL10 RR and SUDHL10 OR respectively; n=170, 139, and 111 cells for SUDHL4, SUDHL4 RR and SUDHL4 OR respectively (mean values for SUDHL10 sensitive, RR, OR, respectively: 28 vs. 66 minutes, 59 vs 90 minutes, 28 vs. 70 minutes, P<0.0001 for all; SUDHL4 sensitive, RR, OR respectively: 41 vs. 80 minutes, 47 vs 111 minutes, 25 vs. 69 minutes, P<0.0001 for all). Mean ± SEM from two independent replicates.
We then categorized target cell death resulting from single (1) vs. multiple (2–9) NK cell contacts to determine the efficacy of NK-mediated lysis and observed inverse correlation between contact numbers with total contact duration and time of irrespective of CD20 resistance (range: 0.9–0.7 at single contact, −0.7–0.4 at 4 contacts, Figure 4B). We further normalized death of individual cell by total contact duration with NK cell defined as Death Index (DI) (Figure 4C). As shown in Figure 4C, DI of cells undergoing multiple contacts were higher compared with DI of single-contacts in all bNHL target cells. Taken together, these findings implied that multiple interrupted contacts between CD19.CAR.NK92 cells and target cells delineated inefficient bNHL cell cytolysis and therefore target cell death resulting from single contact is a key determinant of effective NK cell-mediated cell kill.
Transcriptomic analyses
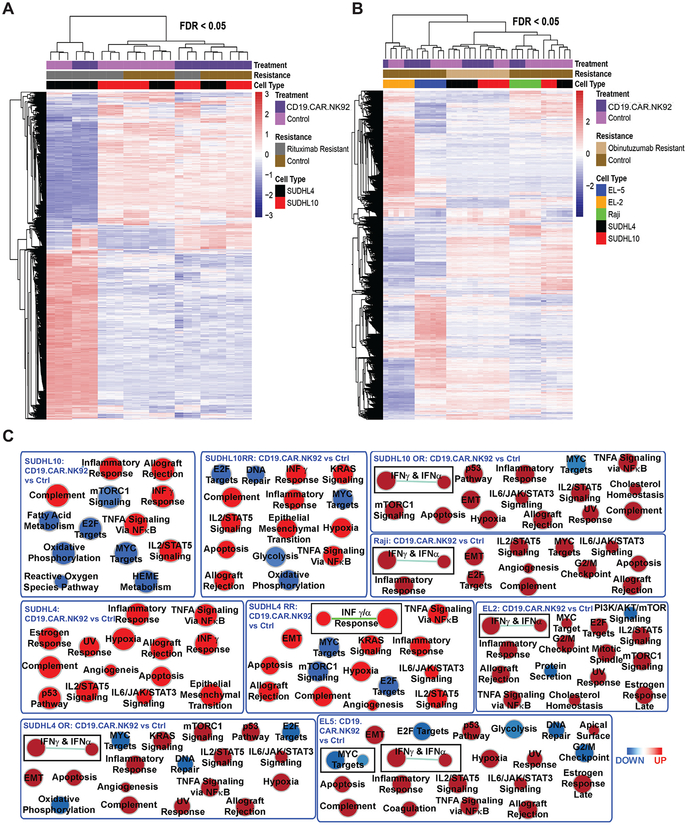
Microarray analysis of mRNA isolated from flow sorted bNHL and primary cells following 2-hour co-culture with CD19.CAR.NK92 at 1:1 E:T ratio indicated that anti-CD20 sensitive (SUDHL4 or SUDHL10) and resistant cells (SUDHL4 or SUDHL10 RR cells; Figure 5A) and (SUDHL4 or SUDHL10 OR cells; primary NHL cells Figure 5A–B) had distinct patterns of gene expression profiling (GEP) by PCA and hierarchical clustering analysis.
Figure 5. Transcriptomics and GSEA analysis of global biological responses to CD19.CAR.NK92 in bNHL cells. A-B).
Hierarchical clustering and heatmap analysis of differentially genes from the transcriptome of flow sorted bNHL cells co-cultured with CD19.CAR.NK92. comparing with triplicates of (A) untreated SUDHL4, SUDHL10 and corresponding RR cells and (B) comparing with untreated SUDHL4, SUDHL10, corresponding OR cells, Raji and primary lymphoma cells (EL-2 and EL-5), determined by ANOVA with FDR<0.05. The identified differentially expressed genes were utilized for subsequent GSEA analysis. C) Hallmark analysis of significant biological functions by GSEA represented as nodes, with size reflecting the amounts of genes involved in each biological process, that were either activated (Red) or inhibited (blue) in the bNHL cells co-cultured with CD19.CAR.NK92 compared to untreated SUDHL4, SUDHL10, anti-CD20 resistant (RR and OR) cells, Raji and primary lymphoma cells (EL-2 and EL-5).
PCA plots show the GEPs from SUDHL10, SUDHL10 RR and SUDHL4 RR clustered together along the same axis principal component PC2, while SUDHL4 RR and SUDHL4 were clustered on same axis on PC1 (Figure S4A) with CD19.CAR.NK treatment. Similarly, SUDHL4 OR or SUDHL10 OR cells were observed to cluster together on PC2 following CD19.CAR.NK92 exposure (Figure S4B) and the clustering of CD19-CAR-NK-treated primary cells or Raji cells appeared to co-localize with untreated control cells (Figure S4).
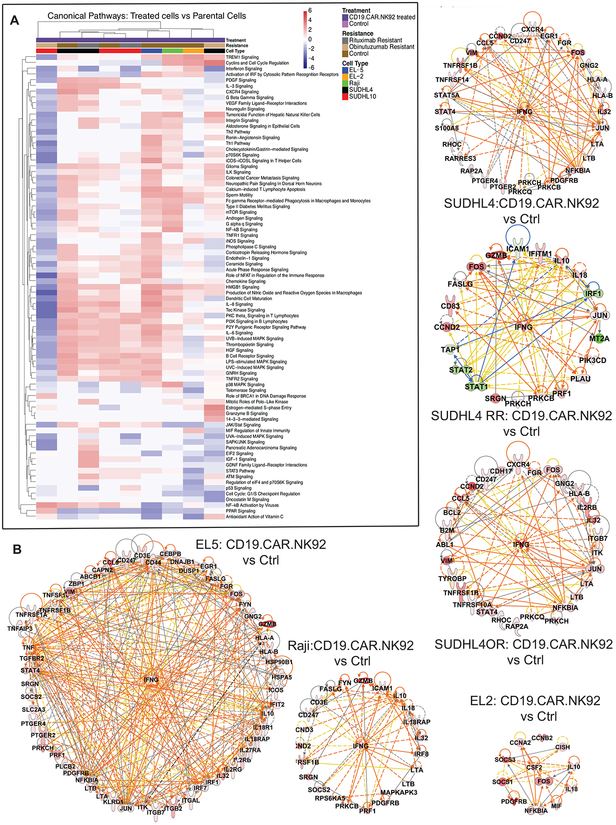
GSEA utilizing C2 canonical pathway database and network analysis revealed conserved upregulation of apoptosis execution, interferon signaling, meiotic recombination and immunoregulatory interactions among anti-CD20 sensitive and resistant (SUDHL4 and SUDHL10 cells), Raji and primary lymphoma cells (EL2 and EL5 cells) following CD19.CAR.NK92 exposure (Figure S5). While topographical networks shown in Figure S5 represent predictable interactions, GSEA of “hallmark signatures” based on well-defined ‘founder gene sets’ and dependent on coherent gene expression pattern from the experiment is the preferred method for identifying discriminative biological properties from transcriptomic evaluations [25]. Hallmark biological analysis indicated upregulation of hypoxia, inflammatory and complement responses, allograft rejection, IFN and TNF signaling, IL-2/STAT5 or IL-6/JAK/STAT3 signaling events, as key biological responses to CD19.CAR.NK92 exposure in bNHL cells. Furthermore, variably conserved downregulation of MYC and E2F target genes, oxidative phosphorylation, mTOR signaling were also noted in most bNHL cells, except Raji and SU-DHL4 cells (Figure 5C and S5). Canonical pathways determined using differentially expressed gene sets from CD19.CAR.NK92 treated bNHL cells based on IPA also determined upregulation (in Red) of several immune signaling including, IFN, TNF, CXCR4, Th1, Th2, IL-3, IL-6, IL-8 and Granzyme B signaling pathways associated with NK mediated cytolytic activity (Figure 6A).
Figure 6. Canonical Pathway and key gene analysis of bNHL cells treated with CD19.CAR.NK92.
A) Heatmap represents results indicate general consensus in the canonical pathways that were either activated or inhibited by CD19.CAR.NK92 treatment versus controls bNHL cells, determined using differentially expressed gene sets and IPA with z-score > 2 indicating activation (red) and z-score < −2 indicating inhibition (blue).
B) The interactions of the common key significant genes represented using IPA, with key genes determined based on gene sets with overlapping functionalities and from multi-platform analysis (GSEA, IPA), with colors denoting the observed up (Red) or down (Green) regulation in gene expression, in the bNHL cells that were co-cultured with CD19.CAR.NK92 compared to untreated SUDHL4, SUDHL10, anti-CD20 resistant (RR and OR) cells, Raji and primary lymphoma cells (EL-2 and EL-5), identified that IFNγ (as central hub) and interactive network of genes related to cytokine functions are upregulated. Data represented in these figure were derived from results provided in Figure 3, consisting experimental triplicates of CD19.CAR.NK92 treated bNHL cells.
Analysis of key gene functions determined based on overlapping gene sets represented from GSEA gene sets (Figure 5C & S5, Table S2) and IPA predicted upstream regulators and bio-functions (Figure 6A), as previously described [20] identified network of interactive key genes with IFNγ as the most highly connected gene, observed among primary bNHL cells, Raji, SUDHL4, SUDHL4 RR and SUDHL4 OR cells, following CD19.CAR.NK92 (Figure 6B). Interestingly, upregulated expression of IL-10 and IL-18 were observed in CD19.CAR.NK92 treated primary bNHL cells, Raji and SUDHL4-RR cells (Figure 6B).
Previous studies have demonstrated that IFNγ transactivates and induces the gene expression of IL10 in human B cells [26]. Furthermore, IL-18 is known act upon NK cells promoting IFNγ secretion, stimulation and expansion of NK cells with potentiation of its cytotoxicity against tumor cells [27], which suggests these cytokines may be related to the pro-inflammatory activity in response to CD19.CAR.NK92 treatment. Additionally, several immune response mechanisms including TNF, IL2, IL5, IL6, STAT5 signaling, inflammatory and complement pathway responses were predicted as activated based on the differentially expressed gene set determined from GSEA analysis (Figure 5C). For further validation, we focused our analysis on CD19.CAR.NK92 activated immune or inflammatory responses via investigating ‘secreted’ cytokine and immune factors responses from bNHL treated with CD19-CAR-NK.
Differential secretomic and transcriptomic responses
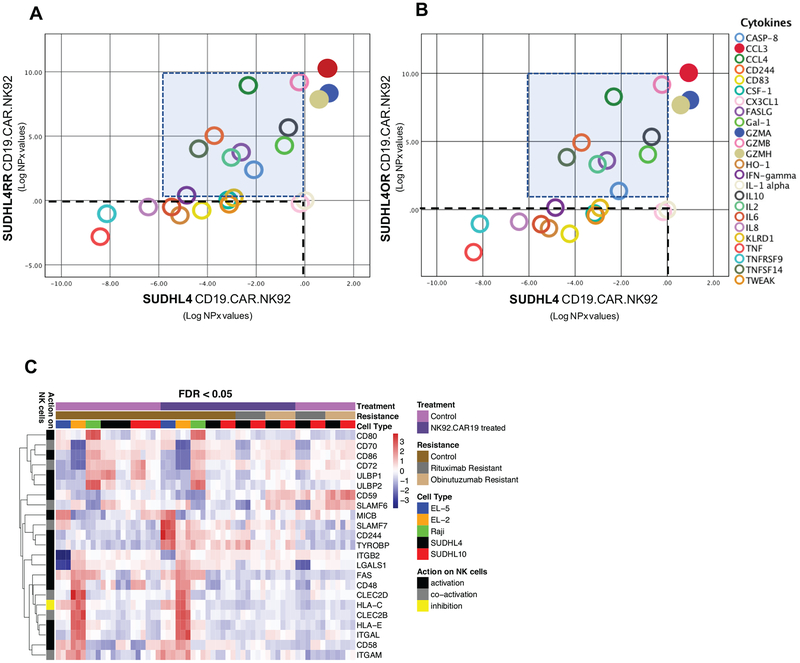
We utilized the OLink 92-plex cytokine panel for an unbiased quantification of secreted factors using the cell culture supernatants (to maximize the detectability of late responding cytokines) from bNHL cells with CD19-CAR-NK and co-cultured bNHL/CD19.CAR.NK92 cells. Based on log2 normalized protein expression (NPX values), we observed >3 to 10.7-fold increase in FASLG, Granzymes (GZMA, GZMH), TNFSF14, Gal-1, IL2, IL10, CCL3, CCL4, CD244 and 1.3–2.4-fold increase in Caspase-8 secretion, all associated with cell death or inflammatory responses in CD19.CAR.NK92 SUDHL4 RR and OR cells compared to SUDHL-4 cells (Figure 7A–B). Further, analysis of NK regulatory ligand expression of from the transcriptomic data indicated upregulation of NK cell activatory CD59 and SLAMF6 ligands and downregulation of HLA-C, associated with inhibition of NK lytic activity in anti-CD20 resistant SUDHL4 (Figure 7C), suggesting that such differential expression of NK regulatory ligands in anti-CD20 resistant SUDHL4 could be responsible for robust induction of cytokine secretions in the presence of CD19.CAR.NK92 cells.
Figure 7. Secretome and NK ligand expression of CD19.CAR.NK92 treated anti-CD20 sensitive and resistant bNHL cells.
A-B) Scatter plots showing results of cytokines detected from 92-plex cytokine panel proximity extension assay represented as log2 normalized protein expression (NPX) comparing CD19.CAR.NK92 treated anti-CD20 sensitive (in x-axis) and resistant SUDHL4 (in y-axis), following subtraction of background and basal cytokine secretions from co-cultured cells. Cytokine secretion with CD19.CAR.NK92 treatment remaining at or below basal levels are indicated within dotted lines (connecting x and y axis), cytokine secretions selectively induced in experimental replciations of anti-CD20 resistant SUDHL4 cells treated with CD19.CAR.NK92 indicated within shaded box, which includes FASLG, Granzymes (GZMA, GZMH), TNFSF14, Gal-1, IL2, IL10, CCL3, CCL4, CD244. C) Heatmap representation comparing the changes in the differential expression of NK cell interaction ligands before and after treatment, from experimental triplicates of CD19.CAR.NK92 treated bNHL cells, and the effect of these ligands on NK cell activity in NHL cells, such as, upregulation of NK cell activatory CD59 and SLAMF6 ligands with CD19.CAR.NK92 is observed in anti-CD20 resistant SUDHL4.
Comparing the differential GEP from CD19.CAR.NK92 treated anti-CD20 sensitive vsresistant bNHL transcriptome, by GSEA revealed enrichment of gene sets associated with hypoxia, mTOR, E2F, mitotic spindle and G2/M checkpoint mechanisms in rituximab or obinutuzumab resistant SUDHL-4 and SUDHL10 cells compared to anti-CD20 sensitive parental cells, with FDR (q<0.0005) (Table 1), outlining the explicit impact of CD19.CAR.NK92 among all anti-CD20 resistant bNHL cells. These pathways are likely targets for potential drug combination investigation with CD19-CAR-NK, such as mTOR or cell cycle checkpoint activation could antagonize non-cytolytic apoptotic mode of cell killing functions by NK cells, [28, 29] (ie., FASL mediated killing by CD19-CAR-NK).
Table 1. Gene set enrichment analysis of differentially expressed genes from CD19.CAR.NK92 treatment in anti-CD20 resistant cells vs parental anti-CD20 sensitive cells.
List of hallmark mechanisms identified as “activated” and based numbers of differentially expressed participating genes and q-values, were determined from GSEA of transcriptomic data consisting experimental triplicates of CD19.CAR.NK92 treated bNHL cells, identified biological mechanisms with predicted co-targeting potential for the enhancement of CD19.CAR.NK92 activity in anti-CD20 resistant RR or OR cells.
| Rituximab Resistant Cells | Obinutuzumab Resistant Cells | |||||||
|---|---|---|---|---|---|---|---|---|
| CD19.CAR.NK92 treated - SUDHL10RR VS Parental | CD19.CAR.NK92 treated - SUDHL4RR VS Parental | CD19.CAR.NK92 treated -SUDHL10OR VS Parental | CD19.CAR.NK92 treated - SUDHL4OR VS Parental | |||||
| HALLMARK PATHWAYS | q-Value | Genes | q-Value | Genes | q-Value | Genes | q-Value | Genes |
| TNFA_SIGNALING_VIA_NFKB | 0.000281 | 4 | 4.43E-05 | 23 | 1.69E-06 | 4 | 1.40E-07 | 10 |
| INTERFERON_GAMMA_RESPONSE | 3.22E-17 | 13.5 | 2.91E-19 | 2 | 9.24E-26 | 2.5 | 4.40E-19 | 26 |
| INTERFERON_ALPHA_RESPONSE | 7.31E-13 | 26 | 1.60E-15 | 2 | 9.65E-24 | 5.5 | 5.05E-13 | 24 |
| ALLOGRAFT_REJECTION | 0.022409 | 11.5 | 6.48E-05 | 11 | 1.64E-05 | 15 | 3.21E-05 | 21 |
| IL2_STAT5_SIGNALING | 0.009847 | 26 | 6.09E-05 | 5 | 2.82E-05 | 10 | 0.000154 | 25 |
| MTORC1_SIGNALING | 1.20E-05 | 28 | 0.000103 | 20.5 | 1.64E-05 | 20 | 4.12E-06 | 6 |
| APOPTOSIS | 0.022409 | 22.5 | 3.51E-08 | 9 | 1.34E-07 | 14 | 1.20E-05 | 35 |
| P53_PATHWAY | 0.022409 | 17 | 1.56E-06 | 34.5 | 8.41E-05 | 10.5 | 0.000154 | 18.5 |
| HYPOXIA | 0.039886 | 23.5 | 0.000204 | 26 | 0.011481 | 17 | 0.006607 | 26 |
| MITOTIC_SPINDLE | 0.031408 | 30.5 | 1.56E-06 | 12 | 0.004035 | 36 | 2.03E-06 | 20.5 |
| E2F_TARGETS | 0.000887 | 18 | 0.000204 | 41 | 1.54E-07 | 35.5 | 6.27E-07 | 31.5 |
| G2M_CHECKPOINT | 0.000534 | 30 | 0.000495 | 30.5 | 1.50E-07 | 44.5 | 1.41E-08 | 25.5 |
DISCUSSION
We revealed here that CD19.CAR.NK92 induced prominent cytolytic activity in rituximab- and obinutuzumab-resistant cells, patient-derived primary cells, and a human lymphoma xenograft model. Most importantly, we assessed the biological responses of target bNHL cells and interactive behaviors with CD19.CAR.NK92. We identified mechanisms of activity including dynamic assessment of single cell interaction kinetics via as well as conserved activation of IFNγ signaling, execution of apoptosis, ligand binding, immunoregulatory or chemokine signaling pathways via transcriptomic analyses of bNHL cells following the encounter with CD19.CAR.NK92 cells. In interpreting these observations, several factors should be considered.
The NK-92 cell line is a continuously growing cell line, that lacks all the inhibitory receptors except KIR2DL4 and constitutively express NKG2D activation receptor [30, 31]. The highly cytotoxic nature of the NK-92 cell line is in part accounted by the lack of the KIRs and constitutive expression of the activation receptors [6, 32]. Safety of NK-92 was evaluated in phase I clinical studies found NK-92 to be well tolerated and without presence of graft versus host disease (GVHD) [31, 33, 34][35]. Several engineered versions of NK-92 have been developed targeting various hematologic and solid malignancies, and CD19.CAR.NK92 specificity is engineered to target malignant B-cells. In vivo studies using CD19.NK.CAR92 have shown efficient drug distribution and cell kill in chronic lymphocytic leukemia and acute lymphoblastic leukemia murine models [2, 12]. CD19 selective CD19.CAR.NK92 mediated cell killing of Raji cells has been reported previously, demonstrating that CD19.CAR.NK92 but not parental NK92 had retained both in vitro and in vivo cytolytic activity against bNHL cells [36]. Further cord blood derived NK cells engineered to express IL15, anti-CD19 and iC9 suicidal gene evaluated against lymphoma tumor xenografts derived using Raji cells, also showed comparable efficacy, delineating that both NK92 or cord blood derived NK cells are suited for CD19 targeted therapy against bNHL [37].
Although, previous studies with either NK92 or cord blood derived NK cells expressing CD19.CAR.NK92 reported anti-lymphoma activity, the biological activity of bNHL cells and the context of rituximab and/or obinutuzumab CD20 sensitivity and resistance in the presence of CD19.CAR.NK92 has not been reported before. Results from our experiments in bNHL including anti-CD20 sensitive and resistant bNHL cells indicated that CD19.CAR.NK92 induced potent cytolytic activity in all bNHL cells regardless of anti-CD20 sensitivity or resistance. Microfluidics based dynamic assessment of single cell interaction kinetics revealed mechanistic commonalities across all bNHL target lines, including significantly delayed death for cells that required multiple contacts with CD19.CAR.NK92 cells. Further investigation of the heterogeneity in bNHL target cell subpopulations may shed light on the specific mechanisms of death at a single-cell level, such as the physiological relevance of multiple contacts between the NK and RR cells, extended contact duration and delayed death for RR cells. Interestingly, upregulation of CD59 and SLAMF6 ligands are associated with NK cell activation [38, 39] and downregulation of HLA-C, inhibitor of NK lytic activity [40] were noted in anti-CD20 resistant RR and OR cells (i.e., SUDHL4 and SUDHL10), indicating that further characterization of differential NK receptor-ligand expersion and interactions could possibly explain heterogeneity in RR or OR cells responses to CD19.CAR.NK92s, as observed in the microfluidics assays. Interestingly, increased expression of CD59 is reported to be associated with poor outcome and response to rituximab based therapies in bNHL [41, 42], while CD59 is known to promote innate cytotoxic activity NK cells against target K562 erythroleukemic cells [38], suggesting that differential expression of such NK interactory ligands could play an important role for CD19.CAR.NK92 mediated cytolytic activity in anti-CD20 resistant cells, which warrants further investigations. In validating the response of cytokine factors predicted among key gene events, we observed partially conserved validations including IFNγ, FASLG, IL-10 and TNFSF14 from the secretomic analysis, while other predicted cytokine activity were undetected potentially due to factors such as differential expression and secretion kinetics, or stability of these cytokines. In addition, we observed increased secretion of CCL3 along with cytolytic granzymes with CD19.CAR.NK92 in anti-CD20 sensitive and resistant cells. CCL3 serves as an important chemotactic mediator essential for recruitment of NK, T, macrophage and dendritic cells to the tumor microenvironment, and promotion of immune mediated tumor rejection function [43, 44]. Taken together, based on cell interaction kinetics we observed that CD19.CAR.NK92 could kill both anti-CD20 sensitive and resistant cells; kill RR and sensitive cells by equal extent through functional cytotoxicity; and kill OR and sensitive cells rapidly through single contact interactions.
In conclusion, we demonstrated that CD19.CAR.NK92 has potent anti-lymphoma activity across a host of lymphoma cells including RR and OR bNHL cells Further pre-clinical analyses to delineate mechanisms of resistance as well as potential biomarkers for clinical application of this novel “off the shelf” immunotherapeutic agent is needed.
Supplementary Material
ACKNOWLEDGEMENTS
A.M.E, T.K, R.D supported by TUFTS NIH CTSI pilot funding UL1TR002544, NIH grants 1R33CA223908-01 and R01 GM127714-01A1.
Footnotes
CONFLICT OF INTERESTS
Funding in part of this study for provided by Nantkwest to A.M.E, other roles for A.M.E advisory board (with honorarium): Bayer, Seattle Genetics, Affimed, Verastem, Pharmacyclics, Research to Practice, and Physician Education Resource; Research support: Takeda, Seattle Genetics, Merck, NIH/NCI, Leukemia and Lymphoma Society, and ORIEN. Y.C founder and consultant with financial interests in Oncomics LLC.
REFERENCES
- 1.Boissel L, Betancur M, Lu W, Wels WS, Marino T, Van Etten RA, et al. Comparison of mRNA and lentiviral based transfection of natural killer cells with chimeric antigen receptors recognizing lymphoid antigens. Leukemia & lymphoma. 2012;53(5):958–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boissel L, Betancur M, Wels WS, Tuncer H, Klingemann H. Transfection with mRNA for CD19 specific chimeric antigen receptor restores NK cell mediated killing of CLL cells. Leukemia research. 2009;33(9):1255–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gong JH, Maki G, Klingemann HG. Characterization of a human cell line (NK-92) with phenotypical and functional characteristics of activated natural killer cells. Leukemia. 1994;8(4):652–8. [PubMed] [Google Scholar]
- 4.Klingemann H. Challenges of cancer therapy with natural killer cells. Cytotherapy. 2015;17(3):245–9. [DOI] [PubMed] [Google Scholar]
- 5.Klingemann HG. Natural killer cell-based immunotherapeutic strategies. Cytotherapy. 2005;7(1):16–22. [DOI] [PubMed] [Google Scholar]
- 6.Maki G, Klingemann HG, Martinson JA, Tam YK. Factors regulating the cytotoxic activity of the human natural killer cell line, NK-92. J Hematother Stem Cell Res. 2001;10(3):369–83. [DOI] [PubMed] [Google Scholar]
- 7.Suck G, Odendahl M, Nowakowska P, Seidl C, Wels WS, Klingemann HG, et al. NK-92: an ‘off-the-shelf therapeutic’ for adoptive natural killer cell-based cancer immunotherapy. Cancer Immunol Immunother. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tam YK, Martinson JA, Doligosa K, Klingemann HG. Ex vivo expansion of the highly cytotoxic human natural killer-92 cell-line under current good manufacturing practice conditions for clinical adoptive cellular immunotherapy. Cytotherapy. 2003;5(3):259–72. [DOI] [PubMed] [Google Scholar]
- 9.Yan Y, Steinherz P, Klingemann HG, Dennig D, Childs BH, McGuirk J, et al. Antileukemia activity of a natural killer cell line against human leukemias. Clin Cancer Res. 1998;4(11):2859–68. [PubMed] [Google Scholar]
- 10.Swift BE, Williams BA, Kosaka Y, Wang XH, Medin JA, Viswanathan S, et al. Natural killer cell lines preferentially kill clonogenic multiple myeloma cells and decrease myeloma engraftment in a bioluminescent xenograft mouse model. Haematologica. 2012;97(7):1020–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weitzman J, Betancur M, Boissel L, Rabinowitz AP, Klein A, Klingemann H. Variable contribution of monoclonal antibodies to ADCC in patients with chronic lymphocytic leukemia. Leukemia & lymphoma. 2009;50(8):1361–8. [DOI] [PubMed] [Google Scholar]
- 12.Boissel L, Betancur-Boissel M, Lu W, Krause DS, Van Etten RA, Wels WS, et al. Retargeting NK-92 cells by means of CD19- and CD20-specific chimeric antigen receptors compares favorably with antibody-dependent cellular cytotoxicity. Oncoimmunology. 2013;2(10):e26527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C, Oberoi P, Oelsner S, Waldmann A, Lindner A, Tonn T, et al. Chimeric Antigen Receptor-Engineered NK-92 Cells: An Off-the-Shelf Cellular Therapeutic for Targeted Elimination of Cancer Cells and Induction of Protective Antitumor Immunity. Front Immunol. 2017;8:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chavez JC, Bachmeier C, Kharfan-Dabaja MA. CAR T-cell therapy for B-cell lymphomas: clinical trial results of available products. Ther Adv Hematol. 2019;10:2040620719841581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maude SL, Teachey DT, Porter DL, Grupp SA. CD19-targeted chimeric antigen receptor T-cell therapy for acute lymphoblastic leukemia. Blood. 2015;125(26):4017–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salles G, Barrett M, Foa R, Maurer J, O’Brien S, Valente N, et al. Rituximab in B-Cell Hematologic Malignancies: A Review of 20 Years of Clinical Experience. Adv Ther. 2017;34(10):2232–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarkar S, Sabhachandani P, Ravi D, Potdar S, Purvey S, Beheshti A, et al. Dynamic Analysis of Human Natural Killer Cell Response at Single-Cell Resolution in B-Cell Non-Hodgkin Lymphoma. Front Immunol. 2017;8:1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jochems C, Hodge JW, Fantini M, Fujii R, Morillon YM 2nd, Greiner JW, et al. An NK cell line (haNK) expressing high levels of granzyme and engineered to express the high affinity CD16 allele. Oncotarget. 2016;7(52):86359–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beheshti A, Benzekry S, McDonald JT, Ma L, Peluso M, Hahnfeldt P, et al. Host age is a systemic regulator of gene expression impacting cancer progression. Cancer Res. 2015;75(6):1134–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ravi D, Beheshti A, Abermil N, Passero F, Sharma J, Coyle M, et al. Proteasomal Inhibition by Ixazomib Induces CHK1 and MYC-Dependent Cell Death in T-cell and Hodgkin Lymphoma. Cancer Res. 2016;76(11):3319–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beheshti A, Neuberg D, McDonald JT, Vanderburg CR, Evens AM. The Impact of Age and Sex in DLBCL: Systems Biology Analyses Identify Distinct Molecular Changes and Signaling Networks. Cancer Inform. 2015;14:141–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vanherberghen B, Olofsson PE, Forslund E, Sternberg-Simon M, Khorshidi MA, Pacouret S, et al. Classification of human natural killer cells based on migration behavior and cytotoxic response. Blood. 2013;121(8):1326–34. [DOI] [PubMed] [Google Scholar]
- 23.Boissel LK H,; Khan J,; Soon-Shiong P. Intra-Tumor Injection of CAR-Engineered NK Cells Induces Tumor Regression and Protection Against Tumor Re-Challenge. Blood Suppl. 2016;128(22):466. [Google Scholar]
- 24.Assarsson E, Lundberg M, Holmquist G, Bjorkesten J, Thorsen SB, Ekman D, et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 2014;9(4):e95192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ziegler-Heitbrock L, Lotzerich M, Schaefer A, Werner T, Frankenberger M, Benkhart E. IFN-alpha induces the human IL-10 gene by recruiting both IFN regulatory factor 1 and Stat3. J Immunol. 2003;171(1):285–90. [DOI] [PubMed] [Google Scholar]
- 27.Son YI, Dallal RM, Mailliard RB, Egawa S, Jonak ZL, Lotze MT. Interleukin-18 (IL-18) synergizes with IL-2 to enhance cytotoxicity, interferon-gamma production, and expansion of natural killer cells. Cancer Res. 2001;61(3):884–8. [PubMed] [Google Scholar]
- 28.Carnevale G, Carpino G, Cardinale V, Pisciotta A, Riccio M, Bertoni L, et al. Activation of Fas/FasL pathway and the role of c-FLIP in primary culture of human cholangiocarcinoma cells. Sci Rep. 2017;7(1):14419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weichhart T, Hengstschlager M, Linke M. Regulation of innate immune cell function by mTOR. Nat Rev Immunol. 2015;15(10):599–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klingemann HG, Wong E, Maki G. A cytotoxic NK-cell line (NK-92) for ex vivo purging of leukemia from blood. Biol Blood Marrow Transplant. 1996;2(2):68–75. [PubMed] [Google Scholar]
- 31.Tonn T, Schwabe D, Klingemann HG, Becker S, Esser R, Koehl U, et al. Treatment of patients with advanced cancer with the natural killer cell line NK-92. Cytotherapy. 2013;15(12):1563–70. [DOI] [PubMed] [Google Scholar]
- 32.Tonn T, Becker S, Esser R, Schwabe D, Seifried E. Cellular immunotherapy of malignancies using the clonal natural killer cell line NK-92. J Hematother Stem Cell Res. 2001;10(4):535–44. [DOI] [PubMed] [Google Scholar]
- 33.Arai S, Meagher R, Swearingen M, Myint H, Rich E, Martinson J, et al. Infusion of the allogeneic cell line NK-92 in patients with advanced renal cell cancer or melanoma: a phase I trial. Cytotherapy. 2008;10(6):625–32. [DOI] [PubMed] [Google Scholar]
- 34.Boyiadzis M, Agha M, Redner RL, Sehgal A, Im A, Hou JZ, et al. Phase 1 clinical trial of adoptive immunotherapy using “off-the-shelf” activated natural killer cells in patients with refractory and relapsed acute myeloid leukemia. Cytotherapy. 2017;19(10):1225–32. [DOI] [PubMed] [Google Scholar]
- 35.Williams BA, Law AD, Routy B, denHollander N, Gupta V, Wang XH, et al. A phase I trial of NK-92 cells for refractory hematological malignancies relapsing after autologous hematopoietic cell transplantation shows safety and evidence of efficacy. Oncotarget. 2017;8(51):89256–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oelsner S, Friede ME, Zhang C, Wagner J, Badura S, Bader P, et al. Continuously expanding CAR NK-92 cells display selective cytotoxicity against B-cell leukemia and lymphoma. Cytotherapy. 2017;19(2):235–49. [DOI] [PubMed] [Google Scholar]
- 37.Liu E, Tong Y, Dotti G, Shaim H, Savoldo B, Mukherjee M, et al. Cord blood NK cells engineered to express IL-15 and a CD19-targeted CAR show long-term persistence and potent antitumor activity. Leukemia. 2018;32(2):520–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Omidvar N, Wang EC, Brennan P, Longhi MP, Smith RA, Morgan BP. Expression of glycosylphosphatidylinositol-anchored CD59 on target cells enhances human NK cell-mediated cytotoxicity. J Immunol. 2006;176(5):2915–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu N, Zhong MC, Roncagalli R, Perez-Quintero LA, Guo H, Zhang Z, et al. A hematopoietic cell-driven mechanism involving SLAMF6 receptor, SAP adaptors and SHP-1 phosphatase regulates NK cell education. Nat Immunol. 2016;17(4):387–96. [DOI] [PubMed] [Google Scholar]
- 40.Winter CC, Gumperz JE, Parham P, Long EO, Wagtmann N. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J Immunol. 1998;161(2):571–7. [PubMed] [Google Scholar]
- 41.Song G, Cho WC, Gu L, He B, Pan Y, Wang S. Increased CD59 protein expression is associated with the outcome of patients with diffuse large B-cell lymphoma treated with R-CHOP. Med Oncol. 2014;31(7):56. [DOI] [PubMed] [Google Scholar]
- 42.Treon SP, Mitsiades C, Mitsiades N, Young G, Doss D, Schlossman R, et al. Tumor cell expression of CD59 is associated with resistance to CD20 serotherapy in patients with B-cell malignancies. J Immunother. 2001;24(3):263–71. [PubMed] [Google Scholar]
- 43.Woo J, Iyer S, Cornejo MC, Mori N, Gao L, Sipos I, et al. Stress protein-induced immunosuppression: inhibition of cellular immune effector functions following overexpression of haem oxygenase (HSP 32). Transpl Immunol. 1998;6(2):84–93. [DOI] [PubMed] [Google Scholar]
- 44.Allen F, Bobanga ID, Rauhe P, Barkauskas D, Teich N, Tong C, et al. CCL3 augments tumor rejection and enhances CD8(+) T cell infiltration through NK and CD103(+) dendritic cell recruitment via IFNgamma. Oncoimmunology. 2018;7(3):e1393598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.