Abstract
MYB transcription factors are one of the most important mediators for the survival of plants under multiple stress responses. In the present study, EaMYB18, encoding a single R3 repeat MYB DNA binding domain was isolated from stress-tolerant wild relative species of sugarcane Erianthus arundinaceus. In silico analysis of 948 bp coding mRNA sequence of EaMYB18 exhibited the presence of four exons and three introns. Further, the EaMYB18 gene was transformed in tobacco and its stable inheritance was confirmed through antibiotic resistance screening, PCR amplification and Southern hybridization blotting. Results of the estimation of MDA, proline, total chlorophyll and antioxidant activities of EaMYB18 transgenic tobacco lines exhibited least oxidative damage under drought and cold stress over the untransformed ones, the over-expression of EaMYB18 has improved drought and cold stress tolerance ability in tobacco. The comparative physiological and biochemical analysis of transgenic tobacco plants overexpressing SoMYB18, SsMYB18 and EaMYB18, revealed that the EaMYB18 and SsMYB18 transgenic plants demonstrated effective tolerance to drought and cold stresses, while SoMYB18 showed improved tolerance to salt stress alone. Amongst these three genes, EaMYB18 displayed the highest potential for drought and cold stress tolerances as compared to SoMYB18 and SsMYB18 genes.
Electronic supplementary material
The online version of this article (10.1007/s13205-020-02197-2) contains supplementary material, which is available to authorized users.
Keywords: Abiotic stress, MYB, Sugarcane, Transcription factor, Tobacco
Introduction
Abiotic stress affects plants at different levels by reducing the CO2 assimilation rate, hampering stomatal opening, water potential, transpiration rate, plant growth rate, and leaf cell size (Zhu 2002). Abiotic stresses (drought, soil salinity, and temperature extremes) that cause the depletion of cellular water content, are the major factors limiting the productivity of plants (Hare et al. 1998). Upon exposure to different stresses, many plant species protect themselves through dynamic physiological, biochemical, and molecular response mechanisms. This comprises of expression of stress-specific genes, enabling their survival under erratic environmental circumstances (Hare et al. 1998; Prabu et al. 2011; Pagariya et al. 2012; Agarwal et al. 2013; Chen et al. 2015). Numerous studies have reported the overproduction of reactive oxygen species (ROS) and hydrogen peroxide in response to drought and salt stress (Pagariya et al. 2012; Shingote et al. 2015; Mahajan and Yadav 2014; Movahedi et al. 2014). To prevent the cell injuries induced by such ROS, cells are equipped with enzymatic scavenging systems such as ascorbate peroxidase (APX), glutathione reductase (GR), glutathione-S transferase (GST) and catalase (CAT) (Mahajan and Yadav 2014; Movahedi et al. 2014; Chen et al. 2015). A close relationship between the rate and extent of antioxidant activity concerning plant stress tolerance has been reported in many plant species (Sairam et al. 2005). Therefore, acquisitive stress tolerance in plants can be achieved through the improvement of its antioxidant system. The plant genotype plays a crucial role in these responses to environmental cues. The key regulatory genes, particularly transcription factors act as a nodal point in interweaving a complicated signal transduction network in plant adaptation to abiotic stresses (Rahaie et al. 2010; Sairam et al. 2005; Baldoni et al. 2013; Ambawat et al. 2013; Agarwal et al. 2013; Li et al. 2014a, b).
Previously, abiotic stress response studies in different plant systems have not only testified the expression of transcription factor and stress-responsive genes but also ingrained their role in protecting the plant cells from stress responses (Seo et al. 2009; Zhang et al. 2011; Ambawat et al. 2013; Agarwal et al. 2013; Li et al. 2014a, b; Chen et al. 2015).
Erianthus arundinaceus (E. arundinaceus) is a sugarcane wild relative species with excellent vigour and adaptability to water deficits and other environmental aberrations (Piperidis et al. 2000; Augustine et al. 2014). For accomplishing sustainable plant stress tolerance through genetic engineering, the identification, isolation and characterization of the novel stress-responsive genes from the wild gene pool are necessary. The MYB proteins belong to a large family of transcription factors and are functionally diverse in all eukaryotes. Copious MYB proteins have been previously reported in the sugarcane, Arabidopsis, rice, maize and cotton genomes (Rahaie et al. 2010; Baldoni et al. 2013; Agarwal et al. 2013; Chen et al. 2015), and most of these functions as transcription factors with varying numbers of MYB-DNA binding domain repeats (Dubos et al. 2010).
Drought and salt are the primary causes of lower cane yield in rain-fed and saline soil confined areas of India as well as other countries across the globe. Erianthus sp. has the mechanisms to tolerate extreme drought conditions and they continue to their normal growth during the summer season, where the growth of commercial sugarcane cultivars ceases. In our previous studies, we have successfully isolated and functionally characterized the transgenic tobacco plants over-expressing orthologous MYB18 genes from sugarcane variety Co740 and Saccharum spontaneum, a wild relative species of sugarcane. Physiological and biochemical estimations of stable transgenic tobacco plants over-expressing SoMYB18 and SsMYB18 genes have exhibited high tolerance to salt and drought stress respectively (Shingote et al. 2015, 2016). In expectancy, to find a more effective multiple stress tolerance gene from the C4 plant species, an ortholog MYB18 transcription factor gene of E. arundinaceus was isolated and functionally characterized in transgenic tobacco plants accompanied by its comparative physio-biochemical analysis with formerly reported MYB18 genes.
Materials and methods
Sugarcane wild relative species E. arundinaceus was collected from Vasantdada Sugar Institute, Manjari (Bk), Pune, Maharashtra, India for the experimental work, and used to isolate and clone the genomic EaMYB18 gene. Tobacco (Nicotiana tabacum) was used to generate EaMYB18-transgenic plants. All the reagents and chemicals used in the study were of high purity, analytical grade and procured from reliable sources.
Cloning and sequence analysis of EaMYB18 family member
The genomic DNA from sugarcane species E. arundinaceus was isolated by the CTAB method (Kale et al. 2017). A pair of primers reported in our previous study MYB18 FP and MYB18 RP were used to amplify full-length MYB18 gene (Shingote et al. 2015; Table S1). The obtained EaMYB18 sequence was analyzed using different in silico tools to explore conserved regions in the gene sequence. The open reading frame (ORF) was defined by FGENISH software and amino acid sequence alignment was obtained with the Clustal W2 program, then based on homologies the consensus R2R3 MYB DNA-binding domain were orderly grouped. The phylogeny was obtained using the Clustal W program and the neighbour-joining method (1000 bootstraps).
Generation of EaMYB18 over-expressing transgenic tobacco plants
The full-length genomic sequence encoding EaMYB18 gene was cloned into binary vector pBinAR between the restriction sites KpnI and BamHI under the control of CaMV 35S promoter with OCS terminator. The MYB18pBINKpnI_FP and MYB18pBINBamHI_RP primers were used for EaMYB18 amplification (Table S1). In vitro grown tobacco plants (1-month-old) were used for transformation using Agrobacterium tumefaciens (LBA4404) containing pBinAR::EaMYB18 as per standard procedure (Horsch et al. 1985).
Screening of EaMYB18 putative transformed plants
The transformed plants were selected on MS (Murashige and Skoog 1962) medium containing 50 mg L−1 of Kanamycin. After rooting and acclimatization, the regenerated plants were grown in a greenhouse to set seeds by self-pollination. The seeds of transgenic T1 plant lines were selected on MS medium containing 50 mg L−1 Kanamycin. Resistant plants were transferred to a close greenhouse, allowed to produce seeds and used in further analyses. For polymerase chain reaction (PCR) studies, genomic DNA was isolated from in vitro developed plantlets of both control (untransformed) as well as EaMYB18 transgenic lines. Integration of the transgene was screened by PCR, and Southern blotting using EaMYB18 gene-specific primers (Table S1). The transgene expression of Southern positive EaMYB18 transgenic lines was confirmed through reverse transcription PCR (RT-PCR) analysis as per the method given by Shingote et al. (2016). The EaMYB18 primers were designed according to the FGENESH predicted mRNA sequences of the EaMYB18 gene to encompass the ORF from start to stop codon. The PCR products were separated on 1% (w/v) agarose gel.
Abiotic stress treatments of untransformed and EaMYB18 transgenic tobacco
Seeds of EaMYB18 expressing transgenic and untransformed tobacco plants of T2 generation were sterilized and germinated on medium containing 50 mg L−1 kanamycin. For the salt and drought treatments 2–3-week-old plants were treated with 200 mM NaCl and 4% polyethylene glycol (PEG-8000) incubated at 25 ± 2 °C under 16 h light/8 h dark, respectively. Leaf tissues were collected at 0, 6, 12, 24 and 48 h sampling points. For the cold treatment, transferred 2–3-week-old plants from 22 °C to 4 °C and harvested the samples at 0, 6, 12, 24 and 48 h sampling points. Untransformed plants were kept under the ambient conditions. All the treatment experiments were performed in triplicates.
Measurement of the malondialdehyde (MDA), proline and total chlorophyll contents
Following the treatment, the seedlings were harvested for the measurements of physiological parameters. All the measurements were repeated three times. The proline and MDA contents were estimated as described by Shingote et al. (2015). Chlorophyll content was measured spectrophotometrically according to the method of Liu et al. (2011).
Enzyme extraction
Enzyme extraction was carried out with few modifications in Mahajan and Yadav’s (2014) protocol. Total protein concentrations were measured by the spectrophotometric method previously described by Bradford (1976) using BSA as standard. The crude supernatant was then immediately used for CAT, peroxidase (POX) and superoxide dismutase (SOD) assays according to the method given by Mahajan and Yadav (2014) and Liu et al. (2011). The CAT, POX and SOD activities were determined spectrophotometrically and were expressed in U mg−1 protein min−1. All the measurements were made in triplicate until and unless stated, and values are expressed as a mean ± SEM.
Results and discussion
PCR amplification, cloning, and sequencing of EaMYB18
The genomic EaMYB18 gene was PCR amplified from the genomic DNA of E. arundinaceus using MYB18 primers (Table S1). The reaction conditions were optimized to obtain a single band of 1652 bp length (Fig. S1). The amplified fragment was then cloned in pGEM™T-Easy vector (Promega) and sequenced. The full-length EaMYB18 sequence was subjected to different analyses using bioinformatics tools to explore conserved regions within the gene sequence.
Phylogenetic and in silico characteristics of EaMYB18
BLAST results indicated that the isolated sequence belongs to MYB genes showing homologies with the existing MYB18 sequences. The full-length genomic EaMYB18 (Accession no. MN907120) was 1466 bp in length from start to stop codon with 62 bp 5′UTR and 124 bp 3′UTR region, respectively. This genomic sequence of 1466 bp consisted of four exons which were further separated by three introns (Fig. 1). SWI3, ADA2, N-CoR, and TFIIIB DNA binding domains spanned exons 1, 2 and 3, comprised a single MYB repeat domain. The FGENISH predicted mRNA sequence of 948 bp encoded 315 amino acid proteins (Fig. 1b). The predicted amino acid sequence BLAST has showed 93% similarity with the available full-length MYB sequences from Oryza brachyantha [XP_006644076.1], O. sativa [NP_001054597.1], Zea mays [NP_001132070.1, ACF79741.1] and Sorghum bicolour [XP_002440559.1] (Fig. 2). EaMYB18 gene has the characteristics of transcriptional activators; implying transcription, RNA processing, modification, cell division and chromosome partitioning. The known and predicted DNA binding residues and the SANT domains are important for the DNA binding activity of the EaMYB18 protein. The highly conserved tryptophan (W) residues involved in the folding of the DNA binding domain, are also signified in the EaMYB18 sequence (Fig. 3). An alignment analysis of conserved SANT domain using Clustal W program and neighbour-joining method exhibited the highest percent similarity coverage within the Saccharum complex MYB18 group.
Fig. 1.
Depiction of mRNA transcript architecture of EaMYB18 gene. (a Genomic DNA containing three introns (I) and four exons (E). b FGENESH predicted mRNA transcript)
Fig. 2.
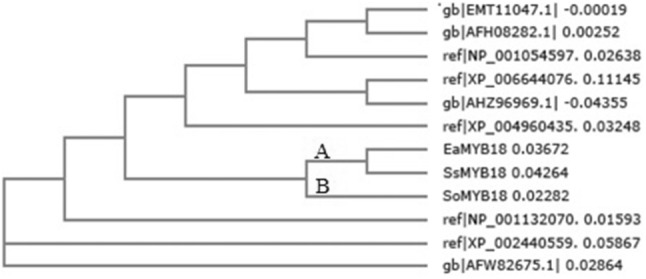
Neighbour–joining tree of predicted MYB proteins. (The phylogeny was based on an alignment derived using the Clustal W program and the Neighbour-joining method (1000 bootstraps). The phylogeny with respect to the conserved SANT could be segregate the sugarcane MYB18 genes into 2 groups viz; A and B
Fig. 3.
Bioinformatics analysis of EaMYB18 from Erianthus spp. (Amino acid sequence alignment was obtained with ClustalW2, the SANT/MYB DNA binding region indicated by black line and the single MYB domain is covered by box, star indicates identical amino acid residues and dots are the conserved or semi-conserved substitutions observed and dashes indicate gaps)
In silico analysis of the EaMYB18 sequence revealed a phylogenetic tree branched into a distinct monocot clade, which advocated that the sequence of EaMYB18 belongs to monocot MYB transcription factors. The sequence information decodes that SoMYB18 has ‘type 2’ while SsMYB18 and EaMYB18 have ‘type1’ MYB DNA binding domain. Being prime wild species of the Saccharum complex, SsMYB18 and EaMYB18 are evolutionary close to each other and are hence placed in the same clade. However, SoMYB18 is placed slightly distant as it is a commercial sugarcane hybrid developed through the conventional breeding process and recombination events that might have also occurred during the breeding process (Figs. 1and 3). The single-repeat MYB domain of EaMYB18 gene is comprised of 42 amino acids and is similar as reported in SsMYB18. Furthermore, single repeat MYB proteins were also reported from other plant species signifying their importance in cellular processes (Baranowskijet al. 1994; Ganesan et al. 2012). The highest percent similarity coverage was illustrated amongst the EaMYB18, SsMYB18, and SoMYB18 gene sequences, which revealed that they might have evolved from the same MYB family ancestor.
Development of transgenic tobacco lines over-expressing the EaMYB18 gene
EaMYB18 gene was cloned in the pBinAR binary vector and the construct was confirmed by PCR, restriction digestion and sequencing (Fig. S2). The EaMYB18 recombinant pBinAR vector was transformed through Agrobacterium-mediated method and the positive plants were selected on kanamycin (50 mg L−1) selection medium (Fig. S3). Out of the 48 putative transgenic tobacco plants, 45 plants were found to be PCR positive (Fig. S4). The Southern analysis confirmed the integration of the EaMYB18 gene into the tobacco genome, where, T0-6, T0-12 and T0-32 showed single gene integration while T0-36 possessed double integration (Fig. S5). Three transgenic tobacco lines (T0-6, T0-12 and T0-32) were hardened under greenhouse conditions. Based on kanamycin resistance, homozygous transgenic lines were selected for further analysis (Fig. S6). The T2 generation EaMYB18 expressing transgenic plants of T0-6, T0-12 and T0-32 derived lines were used further for the functional analyses.
Performance of EaMYB18 transgenic tobacco plants under drought, salt, and cold stresses
To confirm the role of EaMYB18 in the regulation of stress tolerance, transgenic lines over-expressing EaMYB18 were obtained in T2 generation. Three single event transgenic tobacco lines T2-6, T2-12, and T2-32 were further studied for the physiological and biochemical parameters under the stress conditions. The effect of EaMYB18 transgenic plants was correlated with the changes in proline, MDA, and chlorophyll contents with respective untransformed plants under the stress conditions (Fig. 4, Table S2). Further, the activation of specific enzymes (SOD, POX and CAT) was determined for ROS. Amongst the three events, T2-6 event has shown significant results and therefore comparative results of this event with untransformed plants were described and discussed in detail (Table S2, S3, and S5). The transgene expression of southern positive transgenic lines (T2-6) was confirmed with the RT-PCR analysis. All the three lines showed expression of transgene (EaMYB18) under control, drought, salt, and cold stress conditions, whereas the expression was not detected in non-transformed tobacco plants (Fig. S7).
Fig. 4.
Stress induction experiments of T2 generation single copy EaMYB18 transgenic tobacco plants (TT) and respective untransformed tobacco plants (UT). 4% PEG stress, 200 mM NaCl stress and 4 °C cold stress at 6 h, 12 h, 24 h and 48 h intervals
Determination of lipid peroxidation (MDA)
Along the inclining PEG stress period, MDA content increased gradually in the untransformed tobacco plants but it remained steady in the transgenic tobacco plants. Under PEG stress, the relative difference in MDA content of transgenic tobacco plants over the respective untransformed controls was up by 21%, 104% and 108% at 12 h, 24 h and 48 h, respectively (Fig. 5). During the NaCl stress, there was a decrease in MDA content by 19%, 9%, 61% and 55% at 6 h, 12 h, 24 h, and 48 h, respectively as compared to the respective control untransformed tobacco plants (Table S2). The MDA content in untransformed tobacco plants was higher in all the cold treatments than the transformed tobacco plants by 78%, 14%, 42% and 71% at 6 h, 12 h, 24 h, and 48 h, respectively (Table S2). MDA is a peroxidized product of fatty acid lipids in the cell wall, and this serves as an indicator of membrane integrity. The results demonstrated that an increase in stress period was proportionate with the accumulation of MDA in untransformed plants but it was steady in EaMYB18 over-expressing tobacco plants. Thus, the lower amount of MDA content suggested that transgenic lines over-expressing EaMYB18 showed less oxidative damage under a prolonged duration of the induced drought, salt, and cold stress, respectively.
Fig. 5.
MDA content of un-transformed tobacco (UT) verses EaMYB18 over-expressing transgenic tobacco
Determination of proline accumulation
Accumulation of proline to counteract the effects of osmotic stress is a common adaptive mechanism for the plant under stress conditions (Yang et al. 2012). It was observed that during the PEG stress, the relative difference in the accumulation of proline in transgenic and untransformed tobacco was negligible at 0 h, 6 h, 12 h and further declined at 24 h (3.1-fold), and then an increase of almost 3 folds was seen at 48 h (Fig. 6a). Initially, the proline content increased under the NaCl stress in both transgenic and untransformed plants. However, it was almost one-fold higher in transgenic plants (6 h and 12 h) as compared to untransformed ones (Fig. 6b). The proline content got abruptly increased in the untransformed tobacco at 2 4 h followed by a decline at 48 h than the transformed one, while the proline accumulation remained steady at 48 h under the NaCl stress. Upon exposure to cold stress, the proline content was higher in transgenic tobacco plants over the untransformed ones except at 12 h (decreased by 0.8 folds; Fig. 6c). In transgenic tobacco plants, the relative proline content showed an increment by 10 log folds at 6 h.
Fig. 6.
Proline content of un-transformed tobacco (UT) verses EaMYB18 over-expressing transgenic tobacco
Total chlorophyll of EaMYB18 over-expressing transgenic tobacco plants
During the initial exposure of PEG stress, the total chlorophyll content of transformed tobacco plants at 0 h and 6 h was 2% and 52% higher than that of respective untransformed plants, respectively. The relative difference in total chlorophyll content of EaMYB18 transformed against the untransformed plants was not noteworthy for the rest PEG stress period (Fig. 7a). Upon exposure to salt stress, the total chlorophyll content of both transformed and untransformed plants increased at 6 h (1.74 and 1.66 mg g−1 fresh weight) and further declined by 18% at 12 h in transformed plants over the untransformed plants. Further, the chlorophyll content got reduced in untransformed tobacco plants at 24 h and 48 h, while it was 40% and 27% higher in EaMYB18 overexpressing plants, respectively (Fig. 7b). Alike the salt stress, a similar trend was visualized under the cold stress. The total chlorophyll content increased in untransformed tobacco. The total chlorophyll content was higher at 0 h, 12 h, 24 h and 48 h under cold stress, while a 27% decrease was observed at 6 h with respective untransformed plants (Fig. 7c).
Fig. 7.
Total chlorophyll content of un-transformed tobacco (UT) verses EaMYB18 over-expressing transgenic tobacco. Bars represent mean values ± SE (n = 9). Bars represent mean values ± SE (n = 9). Note: blue colour indicates untransformed tobacco, while red indicates transformed tobacco plants over-expressing EaMYB18 gene
Over-expression of the MYB transcription factor gene in the heterologous plant system induces stress tolerance. Hence, a few MYB TFs are being used to transform drought and salt-sensitive crops to improve these traits (Agarwal et al. 2013; Li et al. 2014a, b). As indicative, biochemical determinants were analyzed in EaMYB18 transgenic tobacco plants for examining the level of tolerance to drought, salt and cold stresses. MDA is widely recognized as a key biochemical component for lipid peroxidation (Muley et al. 2019) and hence a key major to gauge plant responses to abiotic stress (RoyChoudhury et al. 2007). The results of our study displayed a steady increase in the MDA content of untransformed tobacco plants, while it was comparatively steady in the transformed tobacco plants for the management of all three stresses. Moreover, analogous results have been reported in transgenic plants expressing stress tolerant genes in tobacco under abiotic stress (RoyChudhury et al. 2007; Wang et al. 2014; Mahajan and Yadav 2014). Initially, the MDA content increased in both EaMYB18 transgenic as well as untransformed tobacco (6 h and 12 h) but it was remarkably stable in transgenic for further stress treatments (24 h and 48 h). The reason for this might be the plants were suddenly exposed to stress but over-expression of EaMYB18 might have helped the transgenic tobacco in tolerating the auxiliary stress. In conclusion, less MDA accumulation under prolonged stress in EaMYB18 transgenic tobacco plants exhibited less oxidative damage under the severe drought, salt, and cold stress conditions.
Accumulation of proline to counteract the effects of osmotic stress is one of the adaptive mechanisms for plant responses to stress conditions (Yang et al. 2012). In our studies, under PEG stress initially, the proline content was relatively less (6 h, 12 h, and 24 h) while the proline accumulation reached the highest peak at 48 h to 701.1 µmole g−1 fresh weight (Table S2). Thus, supporting the osmotic adjustments of EaMYB18 transformed tobacco plants at the utmost stress treatment. Other researchers have also reported a similar trend under different abiotic stresses by over-expressing AtMYB2A in Arabidopsis (Mao et al. 2011), OsMYB3R2, OsMYB2 and OsMYB4 in rice (Ma et al. 2009; El-Kereamy et al. 2012; Xiong et al. 2014). Under NaCl stress conditions the proline content was initially higher 116% and 17% in EaMYB18 transgenic plants (6 h and 12 h) as compared to the untransformed. It was lesser in transgenic at 24 h and almost equal at 48 h (Table S2). Upon exposure to cold stress, the proline content was higher in transgenic tobacco plants than the untransformed plants except at 12 h (decreased by onefold; Table S2). A sudden increase in logarithmic proline content was observed in transgenic tobacco plants (1029%) at 6 h, and this might be an instantaneous response of the plant to unexpected cold shock. Nonetheless, it got declined upon further exposure to cold stress (81%). The highest proline accumulation was reported in transgenic plants at 48 h under PEG stress, and this entails that EaMYB18 might have played a crucial role in regulating proline biosynthesis genes under drought stress. However, the decreased osmotic potential in stressed EaMYB18 transgenic was not completely consistent with their free proline contents. It may be due to (1) proline may act as an initial (salt) and late (PEG) signaling/regulatory molecule able to activate multiple responses, through the components of the adaptation process (Pagariya et al. 2012), (2) EaMYB18 could be partially held accountable for the osmotic potential reduction through proline accumulation (Mao et al. 2011), (3) The EaMYB18 over-expression might have incited the cross-talk between different gene cascades of tolerance, resulting in less proline accumulation at early stress treatments.
Photosynthesis is a key energy source for plant metabolism is the primary processes to be challenged by biotic and abiotic stresses in plants (Liu et al. 2011; Muley et al. 2019). Analysis of photosynthetic performance suggested a crucial role of EaMYB18 in scaling up pigment synthesis at the initial stress period but incapable to do so under prolonged drought stress (Fig. 7a). Upon exposure to salt and cold stress, the total chlorophyll content of transformed plants was higher at 24 h and 48 h whereas a relative decline was reported in untransformed plants (Fig. 7b). Several researchers have previously reported a clear effect on leaf pigment contents, normal physiology and entire metabolic balance of plant under the stress conditions which were pertinent to our findings as well (Liu et al. 2011; Mao et al. 2011; Ganesan et al. 2012). The EaMYB18could have contributed significantly towards the inhibition of the chlorophyll degradation under salt and cold but not under the drought stress.
Effect of EaMYB18 over-expression on the antioxidant system of transgenic tobacco plants
It has been reported that plants have an array of antioxidant defence enzymes to scavenge excess ROS and maintain cellular ROS homeostasis (Muley et al. 2019). We have evaluated CAT, POX and SOD activities of transgenic tobacco plants exposed to PEG, NaCl and cold stress to ascertain the role of EaMYB18 in escalating the activities of antioxidant enzymes.
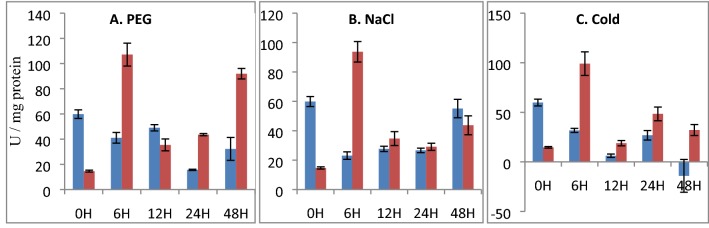
Under normal conditions, the activity of CAT was lower in transformed plants than the untransformed tobacco plants (3 folds). As shown in Fig. 8a, the activity of CAT increased profoundly in EaMYB18 transgenic tobacco plants (160% higher) at 6 h PEG stress, while it slightly decreased to 38% at 12 h further followed by an increment of 177% and 185% at 24 h and 48 h than the respective untransformed plants (Table S3). Similar to PEG stress, under the NaCl stress, the CAT activity increased radically at 6 h (306%) in EaMYB18 transformed tobacco, and it was higher at 12 h and 24 h by 25% and 9% than the untransformed tobacco, respectively, but it decreased slightly at 48 h (Fig. 8b). Under cold stress, the CAT activity in EaMYB18 transformed tobacco plants were consistently higher than the respective untransformed plants by 212%, 205%, 80%, and 228% at 6 h, 12 h, 24 h, and 48 h, respectively (Table S3; Fig. 8c). Under all three stresses, the activity of CAT in the transgenic tobacco plants was highest at the initial stress (6 h) period proposing that EaMYB18 could have played a central role in mitigating the stress by regulating the ROS scavenging mechanism of the plant.
Fig. 8.
CAT activity of un-transformed tobacco (UT) verses EaMYB18 over-expressing transgenic tobacco
A noticeable elevation in the activity of POX was recorded in all the stress samples with a slight decline at 24 h, under all the PEG stress treatments (Fig. 9a). The increase was 415% at 6 h, 152% at 12 h and 147% at 48 h, while 26% reduction at 24 h was witnessed in EaMYB18 transformants concerning untransformed plants (Table S3). Under NaCl stress, the POX activity was 69% higher at 12 h as compared with the untransformed plants, but it got diminished in other samples (Fig. 9b). An identical trend was recorded for POX activity during the cold stress. The relative POX activity was 1.2 folds higher at 12 h, while it got reduced by almost 30% in all other transgenic tobacco plants (Fig. 9c).
Fig. 9.
POX activity of un-transformed tobacco (UT) verses EaMYB18 over-expressing transgenic tobacco
Initially, the SOD activity was considerably higher (18 folds) in transgenic tobacco plants as compared to the untransformed ones (Fig. 10). At 6 h PEG stress, the SOD activity of transgenic plants was 4.3 folds higher whilst a trivial relative difference was observed during the further sampling points (0.7, 0.08 and 0.6 folds). Upon exposure to the salt stress, the percent relative SOD activity was significantly elevated by 620%, 83%, and 308% at 6 h, 12 h and 48 h in transformed than the untransformed plants, respectively, however a decline of 17% was observed at 24 h (Table S3, Fig. 10b). SOD activity during cold stress followed an identical trend like drought stress. The general trend inferred that EaMYB18 over-expressing plants possessed relatively higher SOD activity (121%) at 6 h, while a sharp decline by 248%, 226%, and 9% at 12 h, 24 h, and 48 h, respectively (Fig. 10c) over the untransformed plants.
Fig. 10.
SOD activity of un-transformed tobacco (UT) verses EaMYB18 over-expressing transgenic tobacco. Bars represent mean values ± SE (n = 9). Note: blue colour indicates untransformed tobacco, while red indicates transformed tobacco plants over-expressing EaMYB18 gene
In consensus to our studies, tobacco plants over-expressing mangrove species R3-MYB transcription factor exhibited enhanced tolerance to abiotic stress intermediated through the elevated levels of ROS-scavenging enzyme activities was reported by previous studies (Ganesan et al. 2012). An impulsive increase in CAT activity at 6 h under drought conditions in our studies can be allocated to the plant’s spontaneous confront to unexpected stress, while a further decline at 12 h can be ascribed to the failure of first-line defence of plants system to cope up with the stress. Further, an increase in the CAT activity at later stages of drought stress (24 h and 48 h) and higher CAT activity at all the stages except at 48 h under salt stress perhaps was the consequence of EaMYB18 mediated activation of second-line of defence. In our studies, the CAT activity of EaMYB18 transgenic plants was significantly higher than the respective untransformed plants under drought, salt and cold stresses (Fig. 8). Similar findings were reported by Yang et al. (2012) and Movahedi et al. (2014) in rice and poplar, respectively. Besides, a few recent studies also reported enhanced CAT activity in transgenic tobacco expressing rice Rab16A and Zea maize ZmMPK5 genes (RoyChoudhury et al. 2007; Zhang et al. 2014a, b).
POX plays an essential role in scavenging ROS and protecting cells in higher plants, by consuming H2O2 (Gill and Tuteja, 2010). The higher POX activity in PEG stressed transgenic lines could be due to the over-expression of EaMYB18 triggered up-regulation of defence responsive genes. Akin results were reported by Chen et al. (2015) and Li et al. (2015) while expressing GbMYB5 and SpWRKY1 genes, respectively, in tobacco plants under the drought stress. The POX activity was higher in transgenic lines under both salt and cold stress at 12 h only delineating that EaMYB18 may not have a significant role in regulating the POX activity at other stress intervals. The SOD activity in both drought and cold stress was 18 folds higher at 0 h and 6 h in EaMYB18 transgenic tobacco plants while the relative difference in further sampling points was nonsignificant (Fig. 10). Upon exposure to salt stress, the relative percent SOD activity of transformed plants was significantly elevated in all sampling points except at 24 h (Fig. 10B). Thus, hinting that EaMYB18 has a role in upregulating the SOD activity under prolonged salt stress, whilst it has an initial contribution only at early stress regulation in drought and cold stresses.
POX and CAT functions in the elimination of H2O2 and converts it to water, while SOD is an important enzyme in catalyzing the ROS (Muley et al. 2019). In our results, it was found that the EaMYB18 over-expressing plants showed tolerance to all three stresses through the collective activities of these enzymes. Although a relative percentage of CAT and POX activities of EaMYB18 transgenic were noted, a variable decline concerning an increase of stress period, while a notable increase was observed in the SOD activity in comparison with the respective control plants (Table S3).
In conformity, the results concluded that the joint activity and inter-correlation between these enzymes might have an important role in preventing the formation of ROS and achieving stress tolerance. The balanced regulation in the activities of these enzymes can be induced by ROS directly or indirectly (Pagariya et al. 2012). In contrast, few researchers have stated that ROS enzyme activities were closely related to stress tolerance of many transgenic tobacco and Arabidopsis plants (Ramegowda et al. 2012; Mahajan and Yadav 2014; Zhang et al. 2014a, b). These results anticipated that EaMYB18 over-expression has played an important role in balancing the combined effects of CAT, POX, and SOD to combat or alleviate the stress damage in plants.
Taken together, our results suggested that over-expression of EaMYB18 in tobacco enhances the osmotic tolerance by decreasing the lipid peroxidation under PEG, salt and cold stress conditions. The EaMYB18 over expression might have triggered the stress tolerance through a different cascade of genes that resulted in relatively less proline accumulation at initial stress treatments while higher accumulation upon extended drought and cold stress (48 h). The findings suggested the participatory role of EaMYB18 through increased synthesis of pigments at the initial drought stress period. This hinted the effective role of MYB TFs in the maintenance of plant metabolism thereby preventing chloroplast deformation due to salt and cold stress, respectively. Enzyme assay results projected that the over-expression of EaMYB18 could have collectively regulated the CAT, POX and SOD activities and thus resulted in attaining stress tolerance of transgenic tobacco plants. Chen et al. (2015) reported akin results in transgenic tobacco over-expressing GbMYB5 and reported enhanced accumulations of CAT, POX and CAT enzymes with reduced production of MDA in transgenic tobacco under drought stress.
Comparative effect of EaMYB18, SsMYB18 and SoMYB18 during salt, drought and cold stresses
A detailed characterization of SoMYB18 and SsMYB18 transcription factors were reported in our previous findings (Shingote et al. 2015, 2016). For evaluation of the best potential MYB18 ortholog, we have compared the performance of these three transcription factors under different stress conditions. The sequence analysis showed that SoMYB18 was 1557 bp member of the R2R3-MYB subfamily containing two MYB DNA binding domains (R2, R3) and a SANT/MYB DNA binding domain, whereas SsMYB18 and EaMYB18 contain single-repeat R3 MYB domain with 1470 and 1466 bp in length, respectively (Table S4). The EaMYB18 and SsMYB18 configuration were similar to that of other R3 MYBs formerly reported by Baranowskij et al. (1994) and Ganesan et al. (2012). Besides, all three genes have variable numbers and sizes of introns and exons with a significant difference at the mRNA level. The DNA binding domains of all three transcription factors were as: SoMYB18 had two SANT (R2-R3), CARD, ARM and CLECT; SsMYB18comprised of single SANT (R3), REB1 and PLN03091; and EaMYB18 contained a single SANT (R3) and PLN03091, respectively (Table S4). Thus, there was a predictable difference in all three MYB18 gene sequences at genomic, mRNA and protein/domain level isolated from sugarcane and its wild relative species. This intimated that there could be a difference in their DNA binding ability to different regulatory elements entailing a diverse ability to combat drought, salt, and cold stresses, respectively.
We evaluated the best-performing transgenic events each of EaMYB18, SsMYB18 and SoMYB18 through a relative percentage change in the biochemical parameter of transgenic over the respective transformed tobacco plants as represented. The improved stress tolerance of all three MYB18 transgenic plants was correlated with changes in the MDA, proline, and total chlorophyll contents (Figs. S8–S10). Although these changes are not the only way through which plants cope with the environmental abrasions but these physiological changes are considered as important indicators of plant tolerances to stress.
MDA is an important parameter related to responses of plants to abiotic stress and widely recognized as a parameter for membrane stability index (RoyChoudhury et al. 2007). Under drought stress, the relative percentage of MDA content was trivial but with an increase in stress period, there was a slight increase in EaMYB18 and SoMYB18 transgenic plants at 6 h followed by a significant decline at 12 h (Fig. S8). The comparative decrease in the relative percent of MDA content was most significant in EaMYB18 (51% and 52%) at 24 h and 48 h, respectively (Fig. S8). The relative percentage of MDA got reduced in the SsMYB18 transgenic and found to be stable throughout the sampling period. Under salt stress regime relative MDA percentage in all the three MYB18 transgenics got significantly declined till 24 h of the stress period. It was almost the same in all three transgenics, and an approximate decline of around 37% whilst the highest decline of 51% was noted at 48 h in SsMYB18 transgenic lines than the un-transformed plants, respectively. Upon exposure to cold stress, the relative percent MDA of all three transgenic lines got decreased and it was most significant in EaMYB18 transgenic (− 41%) at 48 h (Fig. S8). These results inferred that EaMYB18 and SsMYB18 transgenic lines exhibited the least oxidative damage under the severe drought and cold stress conditions than the SoMYB18 transgenic lines. Whereas, SoMYB18 has shown significantly less oxidative damage under only salt stress and moderate under drought and cold stress.
Proline is one of the most common osmolytes in plants and its accumulation is associated with responses of plants to the external stress conditions (Zhang et al. 2014a, b). When the plants were exposed to PEG stress, at the initial period (6 h) the relative percentage of proline content was significantly higher in the SoMYB18 transgenic plants. Under the salt and cold stresses, it was considerably higher in EaMYB18 transgenic plants (Fig. S9). During the extended stress regime, it was higher in EaMYB18 transgenic under PEG and cold stress whilst it was highly significant (almost 600%) in SoMYB18 under salt stress. Consequently, EaMYB18 transgenic showed an accumulation of more amount of proline under the prolonged drought and cold stress, while SoMYB18 in salt stress only (Fig. S9).
Adaptations in total chlorophyll content are also reported as important cues of plant response to its growth and stress situations (Li et al. 2014a, b; Muley et al. 2019). After exposure to drought stress the EaMYB18 transgenic demonstrated a lesser amount relative percentage of chlorophyll content than the SsMYB18 and SoMYB18, respectively (Fig. S10). The relative percent of chlorophyll content was most significant in SsMYB18 transgenic under salt and cold stress, whereas for SoMYB18 it was significant under the salt stress only. These results endorsed the vital role of EaMYB18 and SoMYB18 in up-regulating the synthesis of pigments only at the initial period but unable to do so under prolonged drought stress. On the other hand, SoMYB18 regulated it effectively throughout the regime, EaMYB18 regulated it moderately higher upon prolonged stress exposure (24 h and 48 h). Thus, SsMYB18 most predominantly prevented the degradation of chlorophyll under drought and cold stress while SoMYB18 under salt stress only (Fig. S10).
The relative percentage of CAT activity under all three stresses increased noticeably in EaMYB18 and SsMYB18 lines. Particularly at extreme drought stress period, it was highest in EaMYB18, while it was most significant in SoMYB18 under salt and SsMYB18 under cold stress, respectively. These results suggested that EaMYB18 has a significant role in the upregulation of CAT activity under drought, while SoMYB18 and SsMYB18 regulate CAT activity under cold stress, respectively (Fig. S11). Thus, it can be said that EaMYB18 and SsMYB18 could have probably played a role in developing drought and cold stress tolerance, and a moderate salt stresses tolerance by enhancing the CAT activity. Furthermore, EaMYB18 was also seemed to be more effective in managing extended drought stress. SoMYB18 upregulated the CAT activity under salt stress only and appeared as a promising candidate to manage salt stress (Fig. S11). The relative percentage of POX activity under drought stress was higher in SsMYB18 and EaMYB18 transgenic at all sampling points (except EaMYB18 at 24 h). It was more significant in SsMYB18 transgenic, and moderately higher in EaMYB18 at the initial sampling point, while it was non-significant during all sampling points in SoMYB18 transgenic plants. These results inferred that EaMYB18 could have an involvement in the upregulation of the POX activity, but lesser as compared with SsMYB18 under drought, salt, and cold stress, respectively, while under the same circumstances SoMYB18 might not have significant role in up-regulating the POX activity (Fig. S12). Upon exposure to PEG, salt and cold stress, the relative percentage of SOD activity was higher in SoMYB18 transgenic plants when compared with the SsMYB18 and EaMYB18 plants (Fig. S13). SsMYB18 and EaMYB18 transgenic plants have significant SOD activity only under salt stress while it was non-significant under drought and cold stresses (Fig. S13).
Conclusion
Taken together, overexpression of EaMYB18 transcription factors might lead the changes in CAT, POX and SOD activities as revealed from the enzyme assay data. These metabolic changes can be assigned to over-expression of EaMYB18 transcription factor which might have allowed plants for effective osmoregulation, less oxidative damage and combined balanced activity of ROS scavenging enzymes under stress conditions. The comparative physiological and biochemical analysis of transgenic tobacco plants overexpressing SoMYB18, SsMYB18 and EaMYB18, revealed that EaMYB18 and SsMYB18 transgenic plants demonstrated effective tolerance to drought and cold stresses, while SoMYB18 shown improved tolerance to salt stress alone. Amongst these three genes, EaMYB18 seems to have more potential for drought and cold stress tolerances than SoMYB18 and SsMYB18 genes. Further identification of target pathway genes and unraveling signaling networks through which EaMYB18interacts to alleviate or combat the environmental stresses. It will provide further cues to improve the EaMYB18 based stress tolerance of C3 and C4 crop species.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors appreciatively recognize the Director General, Vasantdada Sugar Institute, Pune, India for financial and research support, and Dr. S. Anandhan, Directorate of Onion and Garlic Research, Rajgurunagar, India and Dr. Som Dutt, Central Potato Research Institute, Shimla, India for their precious suggestions during this research work and for critically proof reading the manuscript. The financial grant in terms of CREST fellowship to PGK by DBT, New Delhi is also thankfully accredited.
Author contributions
The overall concept was conceived by PGK and PRS, Experiments were designed by PRS, PGK, KHB and MCP, Experiments were performed by PRS and ABM.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Agarwal PK, Shukla PS, Gupta K, Jha B. Bioengineering for salinity tolerance in plants: state of the art. Mol Biotechnol. 2013;54:102–123. doi: 10.1007/s12033-012-9538-3. [DOI] [PubMed] [Google Scholar]
- Ambawat S, Sharma P, Yadav NR, Yadav RC. MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants. 2013;19:307–321. doi: 10.1007/s12298-013-0179-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine SM, Ashwin NJ, Syamaladevi DP, Appunu C, Chakravarthi M, Ravichandran V, Tuteja N, Subramonian N. Overexpression of EaDREB2 and pyramiding of EaDREB2 with the pea DNA helicase gene (PDH45) enhance drought and salinity tolerance in sugarcane (Saccharum spp. hybrid) Plant Cell Rep. 2014;34:247–263. doi: 10.1007/s00299-014-1704-6. [DOI] [PubMed] [Google Scholar]
- Baldoni E, Genga A, Medici A, Coraggio I, Locatelli F. The OsMyb4 gene family: stress response and transcriptional auto-regulation mechanisms. Biol Plant. 2013;57:691–700. doi: 10.1007/s10535-013-0331-3. [DOI] [Google Scholar]
- Baranowskij N, Frohberg C, Prat S, Willmitzer L. A novel DNA binding protein with homology to Myb oncoproteins containing only one repeat can function as a transcriptional activator. EMBO J. 1994;13:5383–5392. doi: 10.1002/j.1460-2075.1994.tb06873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chen T, Li W, Hu X, Guo J, Liu A, Zhang B. A cotton MYB transcription factor, GbMYB5, is positively involved in plant adaptive response to drought stress. Plant Cell Physiol. 2015;56:917–929. doi: 10.1093/pcp/pcv019. [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- El-kereamy A, Bi YM, Ranathunge K, Beatty PH, Good AG, Rothstein SJ. The rice R2R3-MYB transcription factor OsMYB55 Is involved in the tolerance to high temperature and modulates amino acid metabolism. PLoS ONE. 2012 doi: 10.1371/journal.pone.0052030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan G, Sankararamasubramanian HM, Harikrishnan M, Ganpudi A, Ashwin G, Parida A. A MYB transcription factor from the grey mangrove is induced by stress and confers NaCl tolerance in tobacco. J Exp Bot. 2012;63:4549–4561. doi: 10.1093/jxb/ers135. [DOI] [PubMed] [Google Scholar]
- Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem. 2010;48:909–930. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Hare PD, Cress WA, Van SJ. Dissecting the roles of osmolyte accumulation during stress. Plant Cell Environ. 1998;21:535–553. doi: 10.1046/j.1365-3040.1998.00309.x. [DOI] [Google Scholar]
- Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT. A simple and general method for hybridization revealed the expected. Science. 1985;227:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Kale PB, Mirajkar SJ, Shingote PR. A practical manual for basic techniques in molecular biology. Germany: Lambert Academic Publishing; 2017. [Google Scholar]
- Li J, Bin-Luan YS, Liu Z. Overexpression of SpWRKY1 promotes resistance to Phytophthora nicotianae and tolerance to salt and drought stress in transgenic tobacco. Physiol. Plant. 2015;155:248–266. doi: 10.1111/ppl.12315. [DOI] [PubMed] [Google Scholar]
- Li C, Ng CKY, Fan LM. MYB transcription factors, active players in abiotic stress signaling. Environ Exp Bot. 2014 doi: 10.1016/j.envexpbot.2014.06.014. [DOI] [Google Scholar]
- Li HL, Guo D, Peng SQ. Molecular and functional characterization of the JcMYB1, encoding a putative R2R3-MYB transcription factor in Jatropha curcas. Plant Growth Regul. 2014;75:45–53. doi: 10.1007/s10725-014-9930-z. [DOI] [Google Scholar]
- Liu H, Zhou X, Dong N, Liu X, Zhang H, Zhang Z. Expression of a wheat MYB gene in transgenic tobacco enhances resistance to Ralstonia solanacearum, and to drought and salt stresses. Funct Integr Genomics. 2011;11:431–443. doi: 10.1007/s10142-011-0228-1. [DOI] [PubMed] [Google Scholar]
- Ma Q, Dai X, Xu Y, Guo J, Liu Y, Chen N, Xiao J, Zhang D, Xu Z, Zhang X, Chong K. Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol. 2009;150:244–256. doi: 10.1104/pp.108.133454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan M, Yadav SK. Overexpression of a tea flavanone 3-hydroxylase gene confers tolerance to salt stress and Alternaria solani in transgenic tobacco. Plant Mol Biol. 2014;85:551–573. doi: 10.1007/s11103-014-0203-z. [DOI] [PubMed] [Google Scholar]
- Mao X, Jia D, Li A, Zhang H, Tian S, Zhang X, Jia J, Jing R. Transgenic expression of TaMYB2A confers enhanced tolerance to multiple abiotic stresses in Arabidopsis. Funct Integr Genomics. 2011;11:445–465. doi: 10.1007/s10142-011-0218-3. [DOI] [PubMed] [Google Scholar]
- Movahedi A, Zhang J, Gao P, Yang Y, Wang L, Yin T, Kadkhodaei S, Ebrahimi M, Zhuge Q. Expression of the chickpea CarNAC3 gene enhances salinity and drought tolerance in transgenic poplars. Plant Cell Tissue Organ Cult. 2014;120:141–154. doi: 10.1007/s11240-014-0588-z. [DOI] [Google Scholar]
- Muley AB, Shingote PR, Patil AP, Dalvi SG, Suprasanna P. Gamma radiation degradation of chitosan for application in growth promotion and induction of stress tolerance in potato (Solanum tuberosum L.) Carbohydr Polym. 2019;210:289–301. doi: 10.1016/j.carbpol.2019.01.056. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Planta. 1962;15(3):473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Pagariya MC, Devarumath RM, Kawar PG. Biochemical characterization and identification of differentially expressed candidate genes in salt stressed sugarcane. Plant Sci. 2012;184:1–13. doi: 10.1016/j.plantsci.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Piperidis G, Christopher MJ, Carroll BJ, Berding N, D’Hont A. Molecular contribution to selection of intergeneric hybrids between sugarcane and the wild species Erianthus arundinaceus. Genome. 2000;43:1033–1037. doi: 10.1139/g00-059. [DOI] [PubMed] [Google Scholar]
- Prabu G, Kawar PG, Pagariya MC, Prasad DT. Identification of water deficit stress upregulated genes in sugarcane. Plant Mol Biol Report. 2011;29:291–304. doi: 10.1007/s11105-010-0230-0. [DOI] [Google Scholar]
- Rahaie M, Xue GP, Naghavi MR, Alizadeh H, Schenk PM. A MYB gene from wheat (Triticum aestivum L.) is up-regulated during salt and drought stresses and differentially regulated between salt-tolerant and sensitive genotypes. Plant Cell Rep. 2010;29:835–844. doi: 10.1007/s00299-010-0868-y. [DOI] [PubMed] [Google Scholar]
- Ramegowda V, Senthil-Kumar M, Nataraja KN, Reddy MK, Mysore KS, Udayakumar M. Expression of a finger millet transcription factor, ECNAC1, in tobacco confers abiotic stress-tolerance. PLoS ONE. 2012 doi: 10.1371/journal.pone.0040397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RoyChoudhury A, Roy C, Sengupta DN. Transgenic tobacco plants overexpressing the heterologous lea gene Rab16A from rice during high salt and water deficit display enhanced tolerance to salinity stress. Plant Cell Rep. 2007;26:1839–1859. doi: 10.1007/s00299-007-0371-2. [DOI] [PubMed] [Google Scholar]
- Sairam RK, Srivastava GC, Agarwal S, Meena RC. Differences in antioxidant activity in response to salinity stress in tolerant and susceptible wheat genotypes. Biol Plant. 2005;49:85–91. doi: 10.1007/s10535-005-5091-2. [DOI] [Google Scholar]
- Seo PJ, Xiang F, Qiao M, Park JY, Lee YN, Kim SG, Lee YH, Park WJ, Park CM. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009;151:275–289. doi: 10.1104/pp.109.144220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingote PR, Kawar PG, Pagariya MC, Kuhikar RS, Thorat AS, Babu KH. SoMYB18, a sugarcane MYB transcription factor improves salt and dehydration tolerance in tobacco. Acta Physiol Plant. 2015;37:217. doi: 10.1007/s11738-015-1961-1. [DOI] [Google Scholar]
- Shingote PR, Kawar PG, Pagariya MC, Rathod PR, Kharte SB. Ectopic Expression of SsMYB18, a novel MYB transcription factor from Saccharum spontaneum augments salt and cold tolerance in tobacco. Sugar tech. 2016 doi: 10.1007/s12355-016-0466-6. [DOI] [Google Scholar]
- Wang RK, Cao ZH, Hao YJ. Over-expression of a R2R3 MYB gene MdSIMYB1 increases tolerance to multiple stresses in transgenic tobacco and apples. Physiol Plant. 2014;150:76–87. doi: 10.1111/ppl.12069. [DOI] [PubMed] [Google Scholar]
- Xiong H, Li J, Liu P, Duan J, Zhao Y, Guo X, Li Y, Zhang H, Ali J, Li Z. Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice. PLoS ONE. 2014;9:1–13. doi: 10.1371/journal.pone.0092913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Dai X, Zhang WH. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot. 2012;63:2541–2556. doi: 10.1093/jxb/err431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ju HW, Chung MS, Huang P, Ahn SJ, Kim CS. The R-R-type MYB-like transcription factor, AtMYBL, is involved in promoting leaf senescence and modulates an abiotic stress response in Arabidopsis. Plant Cell Physiol. 2011;52:138–148. doi: 10.1093/pcp/pcq180. [DOI] [PubMed] [Google Scholar]
- Zhang D, Jiang S, Pan J, Kong X, Zhou Y, Liu Y, Li D. The over-expression of a maize mitogen-activated protein kinase gene (ZmMPK5) confers salt stress tolerance and induces defence responses in tobacco. Plant Biol. 2014;16:558–570. doi: 10.1111/plb.12084. [DOI] [PubMed] [Google Scholar]
- Zhang L, Liu G, Zhao G, Xia C, Jia J, Liu X, Kong X. Characterization of a wheat R2R3-MYB transcription factor gene, TaMYB19, involved in enhanced abiotic stresses in Arabidopsis. Plant Cell Physiol. 2014;55:1802–1812. doi: 10.1093/pcp/pcu109. [DOI] [PubMed] [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.