Dear Editor,
Cellular communication is crucial during basic development and in the maintenance of homeostasis in adult organisms, and such communication involves the connexin and pannexin (PANX) families of large-pore channels. The PANX family was identified in 20001 and consists of PANX1, PANX2, and PANX3.2 Among the three subtypes, the most studied is PANX1 because of its potential effects on multiple physiological and pathological functions,3 including microbial infection, cancer progression, ischemia, and neurological disorders. Very recently, we also identified mutations in PANX1 causing familial female infertility characterized by oocyte death.4 PANX1 is a major ATP release and nucleotide permeation channel, which is N-glycosylated and forms three species, GLY0, GLY1, and GLY2.5 Cleavage of the C-terminus by Caspase 3/7 activates the channel and causes the release of ATP and UTP, which functions as a “find-me” signal during apoptosis.6 It is also known that membrane distortion or increased intracellular calcium or extracellular potassium levels can activate the channel.7 In addition, PANX1 channel activity can be regulated by various receptors to maintain cellular electrochemical and metabolic homeostasis.8
It has been widely believed that PANX1 assembles into a hexamer and functions as a hemichannel to allow the passage of molecules < 1.5 kDa.9 The PANX1 protomer is predicted to contain four transmembrane helices with both N- and C-termini inside the cytoplasm.5 In 2017, Chiu et al. used negative staining to produce low-resolution 2D class-averaged images illustrating a hexameric assembly of the PANX1 channel.10 However, applying six-fold symmetry during data processing is likely to have caused artifacts. To determine the overall architecture and assembly of the channel, we resolved the structure of human wild-type (WT) PANX1 by single-particle cryo-electron microscopy (cryo-EM) at near-atomic resolution. This revealed a heptameric assembly and the constriction sites that regulate molecule permeation through the channel.
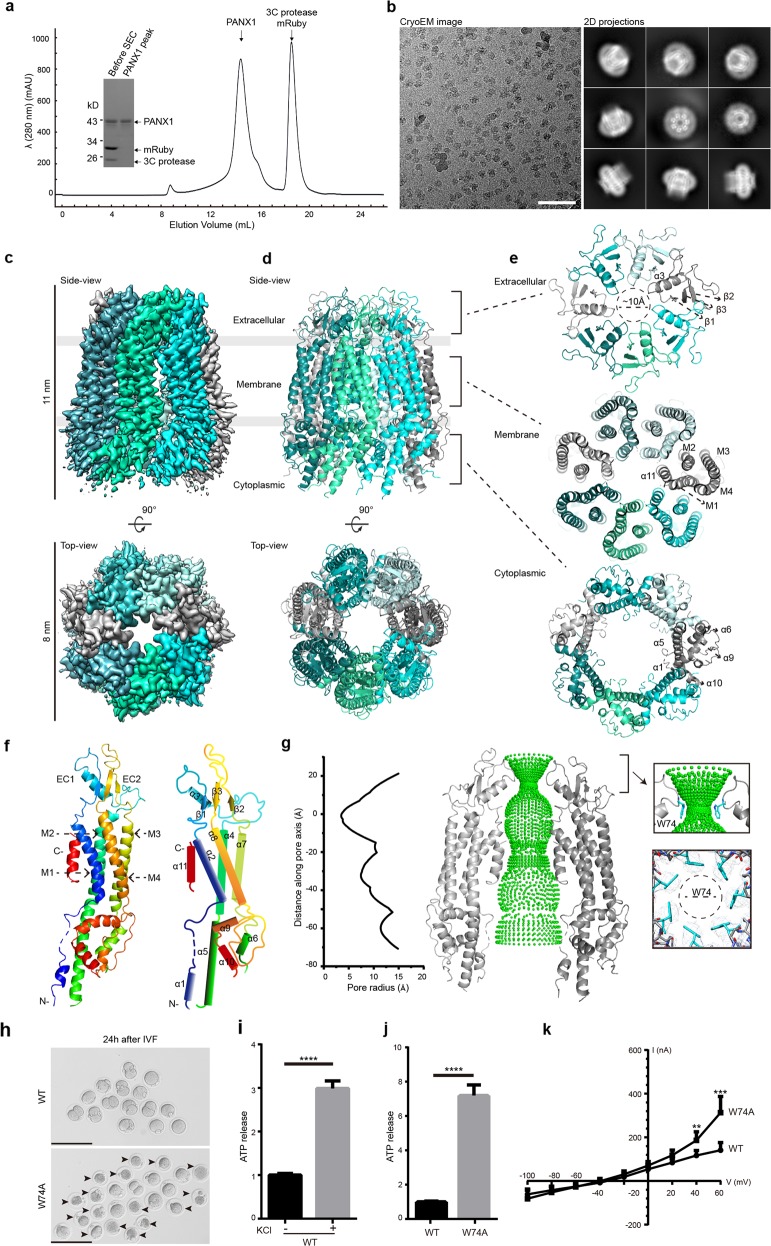
We expressed the full-length WT human PANX1 protein fused at its C-terminus with a 3C protease cleavage site and red fluorescent protein mRuby in HEK293S GnTI− cells, and we performed the buffer screen using fluorescence-detection size-exclusion chromatography (FSEC). The polymeric PANX1 protein displayed decent monodispersity and stability in digitonin-solubilized buffer, and neither pH nor salt concentration altered the overall FSEC profiles in our experiments (Supplementary information, Fig. S1). For large-scale purification, the fused protein was first purified by affinity purification and then digested with 3C protease. The PANX1 protein complex was subsequently separated from mRuby by size-exclusion chromatography. Protein purity was further verified by SDS-PAGE with Coomassie blue staining, indicating a single monomer band (Fig. 1a).
Fig. 1. Cryo-EM structure of the human PANX1 channel.
a A representative size-exclusion chromatogram profile of the purified polymeric PANX1 protein. The inset panel shows the affinity-purified PANX1 protein digested by 3C protease and the peak fraction on an SDS-PAGE gel stained with Coomassie blue. b A representative cryo-EM micrograph and 2D class-averaged images of the PANX1 protein. Scale bar, 60 nm. c, d 3D reconstruction map and model of the PANX1 channel viewed from the side and top with each subunit individually colored. The extracellular, transmembrane, and cytoplasmic regions are indicated. e Three top-down views of the PANX1 extracellular, transmembrane, and cytoplasmic regions corresponding to the brackets in d. Secondary structures are marked. f Protomer structure of PANX1 colored by spectrum and a schematic representation of the secondary structures. g Sphere representation of the pore pathway of the PANX1 channel and the pore radius plot produced by the HOLE program. For clarity, only two subunits are shown. The Cα position of W74 is set to zero. The restriction site controlled by W74 and the local density map are shown in the right panel. h GV oocytes injected with WT or W74A cRNA collected at 6 h after in vitro fertilization. The arrowheads mark the dead zygotes and bars represent 200 µm. i The ATP content measured in HEK293S GnTI− cells transfected with WT PANX1 plasmid, with or without 100 mM KCl stimulation. j The ATP contents measured in HEK293S GnTI− cells transfected with WT or W74A PANX1 plasmid. Bars indicate means ± SEM (n = 4). The data were normalized to the WT group. k Average I-V curves from X. laevis oocytes expressing WT or W74A PANX1. Data are shown as means ± SD (n = 4-8). **P < 0.01, ***P < 0.001 and ****P < 0.0001.
Cryo-EM images were initially analyzed using the cisTEM. Without applying any symmetry, 2D class-averaged images of top-down views revealed that the PANX1 channel assembles as a heptamer (Fig. 1b). To improve the resolution of the PANX1 complex, RELION 3.0 was used and C7 symmetry was applied during 3D classification and refinement. The whole work-flow for image processing is illustrated in Supplementary information, Fig. S2a. By combining a total of 73,421 well-defined particles, we obtained a final reconstruction at an overall resolution of 3.2 Å (Fig. 1c and Supplementary information, Fig. S2). The density map showed prominent side-chain features in the transmembrane helix region that allowed us to unambiguously register the residues for the four transmembrane helices (Supplementary information, Fig. S3). The N- and C-termini showed relatively poor density. Based on the cryo-EM density map, we built a de novo atomic model of the PANX1 complex (Fig. 1d). Our model had good geometry as evidenced by the validation statistics (Supplementary information, Table S1). Loops consisting of amino acids 20–24, 163–186, and 356–409 were presumably disordered and thus were not modeled.
Our structure of PANX1 clearly shows that the seven protomers form an inverted pail-shaped assembly, with external dimensions of ~11 nm in length and ~8 nm in width (Fig. 1c). Each protomer comprises an N-terminal helix (NTH), a transmembrane domain (TMD), an extracellular domain (ECD), and a C-terminal domain (CTD) (Fig. 1e, f). Through the measurement of ATP release, we confirmed that ATP could pass through our WT PANX1 channel, and adding 100 mM K+ in the extracellular medium could further increase ATP release (Fig. 1i). Because no K+ was present in our purification system, we presume that our structure is trapped in an inactive or closed state.
Our structure shows that the NTH is located intracellularly, in line with previous predictions.5 The TMD contains four α-helices (M1–M4), with M1 and M2 proximal, and M3 and M4 distal along the central axis (Fig. 1e). The ECD consists of two segments (EC1 and EC2) that connect M1–M2 and M3–M4, respectively. EC1 has a short α-helix, while EC2 is made up of anti-parallel β-sheets, and two disulfide crosslinks between EC1 and EC2 are formed by the Cys66–Cys265 and Cys84–Cys246 pairs. These cysteines are highly conserved in the PANX family (Supplementary information, Fig. S4), and mutations of any of the four cysteines result in non-functional PANX1 channels.11 Four α-helices (α5, α6, α9 and α10) are clearly visible in the CTD, while two large loops (163–186 and 356–409) are missing. There was notable additional electron density present at the channel vestibule within the TMD region, and we built the α11 helix of CTD to fit this density. However, the general poor density of the CTD implies its intrinsic flexibility and involvement in channel gating.12
Atom distance measurements using HOLE suggest that the restriction ring for molecular permeation is mainly controlled by residue W74 (located in EC1), with a radius of less than 5 Å (Fig. 1e, g). To validate the molecular mechanism in the restriction site, we made an alanine substitution at the W74 position. Approximately 60% mouse oocytes injected with W74A cRNA died 24 h after in vitro fertilization, and HEK293S GnTI− cells overexpressing W74A channels showed increased release of ATP (Fig. 1h, j). We also investigated the effect of the W74A mutation on channel activity by performing electrophysiological recordings in Xenopus laevis oocytes. The I-V curve showed that the W74A receptors had increased activity compared to WT receptors at positive holding potentials of both 40 mV and 60 mV (Fig. 1k). Thus, we provide functional validation that W74 forms the extracellular restriction site for molecular permeation.
Cellular communication through gap junction channels is important in regulation of organism homeostasis. Innexin6 (INX-6), calcium homeostasis modulator family member 2 (CALHM2), and Connexin46 (Cx46) are gap junction proteins with resolved structures. By structural comparison, similarities and differences between PANX1, INX-6, CALHM2, and Cx46 are clearly visible. INX-6, CALHM2, and Cx46 form octameric, undecameric, and hexameric gap junctions, respectively, while PANX1 forms a heptameric hemichannel. Due to different assembly forms and sizes, these molecules occupy unequal spaces in the membrane (Supplementary information, Fig. S5a, b). In terms of individual protomer, PANX1, INX-6, and Cx46 share topological similarities in their TMD and ECD regions (Supplementary information, Fig. S5c), but they also exhibit differences in some local regions. For example, both PANX1 and INX-6 have their EC2 domain in-between the inner and outer lobes of EC1, while the outer lobe of EC1 is absent in Cx46. PANX1, INX-6, and CALHM2 all have two disulfide bonds, whereas Cx46 has three (Supplementary information, Fig. S5d). In addition, the CTDs display less conserved structural homology among these four large-pore channels.
In summary, we present here the cryo-EM structure of the human PANX1 channel and elaborate on its architecture and symmetry. The structure shows an unexpected heptameric assembly, which is completely different from existing evidence, suggesting that it forms a hexamer, and the molecular permeation through the channel seems to be controlled by residue W74 in the extracellular region. The determination of the PANX1 channel structure will lay foundation for mechanistic understanding of human disease-causing mutations in PANX1 in our previous study of channelopathy.4
The cryo-EM density map for human pannexin 1 channel has been deposited in EM Database under the accession code EMD-30028, and the coordinate for the structure has been deposited in Protein Data Bank under accession code 6M02.
Supplementary information
Supplementary information, Figs, Table and Materials
Acknowledgements
We are grateful to Dr Qingxia Wang (Cryo-Electron Microscopy Research Center at Shanghai Institute of Materia Medica) and Dr Yu Kong (EM facility at the Institute of Neuroscience) for facility support, to Dr Xiang Zhang and Hong Wu (Shanghai YueXin Life-science Tech) for computational support, to Dr Shanshuang Chen (Shanghai Jiao Tong University) for data analysis, and to Dr Qing Sang (Fudan University) for mouse oocyte microinjection. This work was financially supported by the National Key R&D Program of China (2017YFA0505700, 2016YFC1000600, 2017YFC1001500, 2018YFC1003800), the National Natural Science Foundation of China (31771115, 81725006, 81822019, 81771581, 81571501, 81971450, 81971382), the Strategic Priority Research Program of the Chinese Academy of Science (XDB32020000, XDA12010317), the Shanghai Municipal Science and Technology Major Project (2018SHZDZX05, 2017SHZDZX01), the CAS Key Laboratory of Receptor Research (SIMM1804YKF-03), the Thousand Young Talents Program, the 100 Talents Program of the Chinese Academy of Sciences, the Natural Science Foundation of Shanghai (18ZR1447700), and Shanghai Rising-Star Program (grant 17QA1404800).
Author contributions
R.Q., L.W., and S.Z. designed the project; R.Q. performed the protein purification and sample preparation; R.Q., L.D., and J.Z. collected the data; L.D. and X.Y. processed the cryo-EM images and built the model; and R.Q. and S.Z. analyzed the data and wrote the manuscript with inputs from all co-authors.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Ronggui Qu, Lili Dong
Contributor Information
Xuekui Yu, Email: xkyu@simm.ac.cn.
Lei Wang, Email: wangleiwanglei@fudan.edu.cn.
Shujia Zhu, Email: shujiazhu@ion.ac.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-020-0298-5.
References
- 1.Panchin Y, et al. Curr. Biol. 2000;10:R473–R474. doi: 10.1016/S0960-9822(00)00576-5. [DOI] [PubMed] [Google Scholar]
- 2.Bruzzone R, Hormuzdi SG, Barbe MT, Herb A, Monyer H. Proc. Natl. Acad. Sci. USA. 2003;100:13644–13649. doi: 10.1073/pnas.2233464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Penuela S, Harland L, Simek J, Laird DW. Biochem. J. 2014;461:371–381. doi: 10.1042/BJ20140447. [DOI] [PubMed] [Google Scholar]
- 4.Sang Q, et al. Sci. Transl. Med. 2019;11:eaav8731. doi: 10.1126/scitranslmed.aav8731. [DOI] [PubMed] [Google Scholar]
- 5.Boassa D, et al. J. Biol. Chem. 2007;282:31733–31743. doi: 10.1074/jbc.M702422200. [DOI] [PubMed] [Google Scholar]
- 6.Chekeni FB, et al. Nature. 2010;467:863–867. doi: 10.1038/nature09413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahl G. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2015;370:20140191. doi: 10.1098/rstb.2014.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isakson BE, Thompson RJ. Channels. 2014;8:118–123. doi: 10.4161/chan.27978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Dahl G. Am. J. Physiol. Cell Physiol. 2018;315:C290–C299. doi: 10.1152/ajpcell.00302.2017. [DOI] [PubMed] [Google Scholar]
- 10.Chiu YH, et al. Nat. Commun. 2017;8:14324. doi: 10.1038/ncomms14324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bunse S, et al. J. Membr. Biol. 2011;244:21–33. doi: 10.1007/s00232-011-9393-3. [DOI] [PubMed] [Google Scholar]
- 12.Sandilos JK, et al. J. Biol. Chem. 2012;287:11303–11311. doi: 10.1074/jbc.M111.323378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information, Figs, Table and Materials