Abstract
Rett syndrome (RTT), a severe postnatal neurodevelopmental disorder, is caused by mutations in the X-linked gene encoding methyl-CpG-binding protein 2 (MeCP2). MeCP2 is a chromatin organizer regulating gene expression. RTT-causing mutations have been shown to affect this function. However, the mechanism by which MeCP2 organizes chromatin is unclear. In this study, we found that MeCP2 can induce compaction and liquid–liquid phase separation of nucleosomal arrays in vitro, and DNA methylation further enhances formation of chromatin condensates by MeCP2. Interestingly, RTT-causing mutations compromise MeCP2-mediated chromatin phase separation, while benign variants have little effect on this process. Moreover, MeCP2 competes with linker histone H1 to form mutually exclusive chromatin condensates in vitro and distinct heterochromatin foci in vivo. RTT-causing mutations reduce or even abolish the ability of MeCP2 to compete with histone H1 and to form chromatin condensates. Together, our results identify a novel mechanism by which phase separation underlies MeCP2-mediated heterochromatin formation and reveal the potential link between this process and the pathology of RTT.
Subject terms: Nucleosomes, Mechanisms of disease
Introduction
Rett syndrome (RTT) is a severe postnatal neurodevelopmental disorder characterized by the loss of acquired motor and language skills, autistic features, and unusual stereotyped movements.1–3 RTT is caused by mutations in the X-linked gene encoding methyl-CpG-binding protein 2 (MeCP2).4 MeCP2 was initially identified as a transcriptional repressor that can selectively bind to methylated CpG dinucleotides flanked by AT-rich segments.5–8 MeCP2 is a chromatin organizer ubiquitous to all cell types; however, it is most highly expressed in neurons.9,10 MeCP2 has been proposed to play an important role in transcriptional repression via interaction of its transcriptional repression domain (TRD) with the histone deacetylase (HADC)-containing co-repressor complexes Sin3A and NCoR/SMRT, which creates a locally hypo-acetylated repressive chromatin state.11,12 In addition, it is also well documented that MeCP2 is able to repress transcription by mechanisms independent of histone deacetylation and DNA methylation.13,14 Numerous in vitro experiments showed that MeCP2 can modulate higher order chromatin structures and architecture by competing with linker histone H1 and compacting nucleosomal arrays.8,15–17 Furthermore, in vivo studies showed that MeCP2 deficiency results in a doubling of histone H1 levels and global changes of chromatin organization in neurons, which suggests that MeCP2 serves as an alternative chromatin architecture protein.10
Recently, a number of studies have shown that liquid–liquid phase separation (LLPS) drives the formation of heterochromatin mediated by HP1, thus providing a new perspective on the mechanisms that regulate heterochromatin assembly and functions.18–22 In line with this, recent in vivo studies showed that MeCP2 is also able to reorganize and cluster global heterochromatin architecture into punctate structures called chromocenters.10,16,23–25 This suggests that MeCP2 may also mediate formation of heterochromatin by LLPS. Indeed, in this study, we found that MeCP2 can induce LLPS of nucleosomal arrays (NA) in vitro, and almost all RTT-causing mutations weaken MeCP2-mediated chromatin LLPS. We further showed that MeCP2 competes with linker histone H1 to form mutually exclusive phase-separated chromatin compartments, and that MeCP2 variants harboring RTT-associated mutations fail to form chromatin condensates and hence lose the ability to compete with H1. Our results provide a new perspective on the mechanisms by which LLPS drives formation of distinct heterochromatin foci mediated by MeCP2 and linker histone H1. These mechanisms may be associated with RTT pathogenesis.
Results
MeCP2 induces compaction and LLPS of nucleosomal arrays in vitro
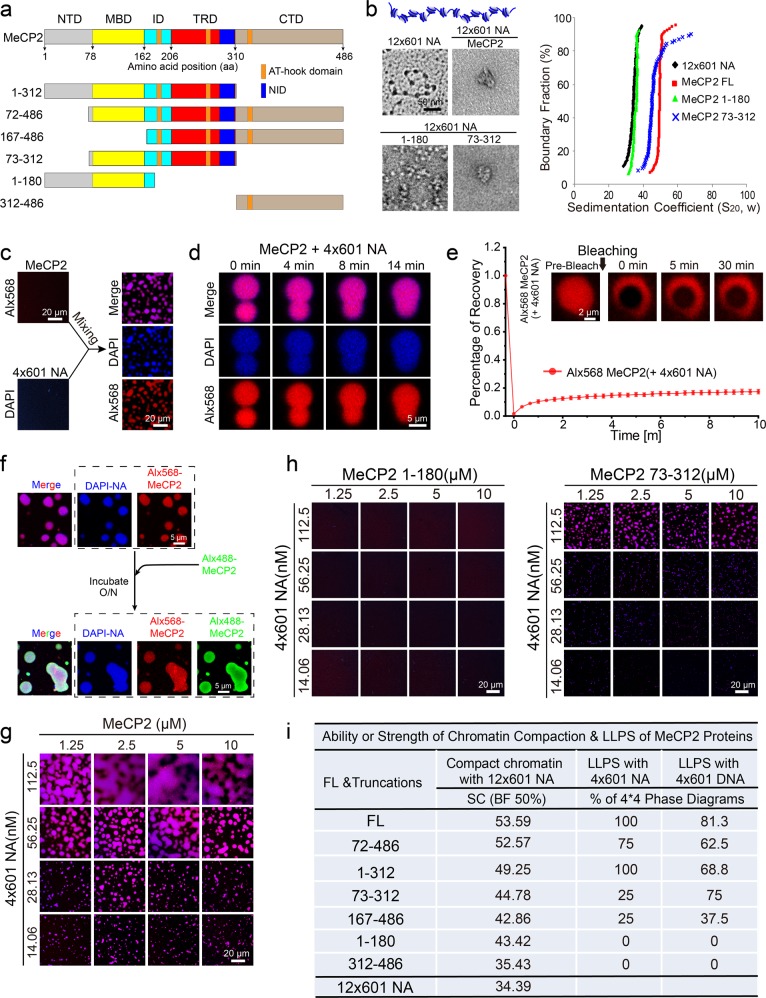
MeCP2 contains a disordered N-terminal domain (NTD), a methyl-CpG-binding domain (MBD), an intervening domain (ID), a transcription repression domain (TRD) and a disordered C-terminal domain (CTD)26 (Fig. 1a). In addition, the C-terminal fragment of MeCP2 contains three AT-hook motifs (Fig. 1a), which preferentially bind AT-rich DNA sequences.27 Similar to previous reports,15,16 we showed by electron microscopy (EM) analysis (see Materials and Methods) that full-length (FL) MeCP2 compacts beads-on-a-string nucleosomal arrays (12 repeats of 177-bp “601 nucleosome positioning sequence”, 12×601-NA) into condensed structures (Fig. 1b). Accordingly, analytical ultracentrifugation (AUC) assays (see Materials and Methods) showed that MeCP2-compacted 12×601-NA has a much larger sedimentation coefficient (~50S) than 12×601-NA alone (~35S) (Fig. 1b). Using EM and AUC analysis, we next dissected which domain(s) of MeCP2 is responsible for its chromatin compaction function using a variety of truncated proteins (Fig. 1a, b; Supplementary information, Figs. S1a and S2a). The 72–486 aa truncation of MeCP2 shows a similar sedimentation coefficient profile and compaction behavior as the FL protein (Supplementary information, Fig. S2a), which suggests that the NTD of MeCP2 contributes little to chromatin compaction. All other five MeCP2 truncations compact 12×601-NA to varying degrees (Fig. 1b; Supplementary information, Fig. S2a). Together, our results reveal that the remaining three major domains — MBD, TRD, and CTD — contribute to the chromatin compaction function of MeCP2.
Fig. 1. MeCP2 drives chromatin compaction and chromatin liquid–liquid phase separation in vitro.
a Diagram of the structural domains of FL MeCP2. Truncated proteins for functional analysis of domain are shown underneath. b EM images (left panels) and sedimentation coefficient distribution plots (right panel) for the reconstituted 12×177-bp 601-nucleosomal arrays (12×601-NA), and chromatin fiber compacted by FL MeCP2 and two truncations, 1–180 and 73–312 aa. Scale bar for EM images, 50 nm. c Left panels, in vitro phase separation assays for MeCP2 protein or reconstituted 4×601-NA alone. MeCP2 protein was partially (5%) labeled with Alexa Fluor 568. DNA in NA was stained using DAPI. Right panels, puncta formed by MeCP2 protein with 4×601-NA in vitro. Scale bar, 20 µm. d Fusion upon contact of droplets formed by MeCP2 protein with 4×601-NA. Scale bar, 5 µm. e In vivo FRAP analysis of puncta formed by MeCP2 protein with 4×601-NA. Top, snapshots of a punctum before and after bleaching. Scale bar, 2 µm. Bottom, average fluorescence recovery traces of Alx568-labeled MeCP2 in puncta (n = 6). All data are presented as mean ± SD. f Alx488-labeled MeCP2 (Alx488-MeCP2) penetrates into pre-formed Alx568-MeCP2-4×601-NA droplets. Scale bar, 5 µm. g Phase diagram of MeCP2 with 4×601-NA. Only the merge channel is shown here. Scale bars, 20 µm. h Phase diagrams of MeCP2 domain truncations (1–180, 73–312 aa) with 4×601-NA. Scale bars, 20 µm. i Summary of the ability or strength of chromatin compaction and phase separation of FL MeCP2 and its domain truncations with nucleosome arrays or DNA. The ability or strength of chromatin compaction is indicated by the sedimentation coefficient of AUC when the boundary fraction is 50% in Supplementary information, Fig. S2b. The ability or strength of phase separation is indicated by the percentage of 4 × 4 phase diagrams when MeCP2 proteins were mixed with 4×601-NA or DNA in Supplementary information, Fig. S2e, f, for example, 75% indicated that 12 of the 16 (4 × 4) conditions in the phase diagram underwent phase separation.
Most recently, we and other groups have shown that heterochromatin domains are assembled, at least in part, via LLPS.18–22 Therefore, we next investigated whether MeCP2 induces LLPS of NA to form droplet-like chromatin condensates in vitro. MeCP2 alone does not undergo LLPS under physiological salt condition (Fig. 1c; Supplementary information, Fig. S1b) unless a crowding agent, such as PEG8000, is added (Supplementary information, Fig. S1c, d). In contrast, mixing MeCP2 with 4×601-NA at physiological salt condition causes striking punctum formation (Fig. 1c). Of note, MeCP2 also forms numerous puncta when mixed with 4×601 DNA (Supplementary information, Fig. S1b, e). Interestingly, these in vitro chromatin puncta are reminiscent of MeCP2-associated heterochromatin domains observed in cells (see later sections and also23). Thus, we hypothesized that the puncta derived from mixing MeCP2 and NA or DNA were also driven by LLPS.
Several lines of evidence are consistent with this hypothesis. We found that pairs of puncta slowly fused and relaxed into larger ones upon contact (Fig. 1d; Supplementary information, Fig. S1f). Although fluorescence recovery after photobleaching (FRAP) assays showed that MeCP2 only marginally recovered within ten minutes after photobleaching (Fig. 1e; Supplementary information, Fig. S1g), MeCP2 was able to penetrate into pre-formed MeCP2/4×601-NA puncta over night (Fig. 1f). When mono-nucleosome (Mono-N) was used, MeCP2/Mono-N phase separation was much weaker than with 4×601-NA (Fig. 1g; Supplementary information, Fig. S1h, i), indicating that the punctum formation ability of MeCP2 with its binding partners is valency-dependent. In addition, MeCP2 protein was more dynamic in droplets with Mono-N than those with 4×601-NA (Fig. 1e; Supplementary information, Fig. S1j). These data implied that 4×601-NA or 4×601 DNA samples, but not Mono-N, contained sites that were able to sequester MeCP2 tightly in the droplets. Furthermore, MeCP2 can induce the formation of chromatin or DNA condensates when mixed with 4×601-NA, Mono-N, or 4×601 DNA over a large concentration range with optimal concentration ratios reminiscent of the phase diagram of two interacting multivalent components (Fig. 1g; Supplementary information, Fig. S1e, h, i). Taking all these data together, we concluded that MeCP2 can indeed undergo LLPS when bound to NA or DNA.
As shown above, distinct functional domains of MeCP2 contribute differently to chromatin compaction (Fig. 1b; Supplementary information, Fig. S2a, b). We next assessed which domain(s) of MeCP2 harbors the ability to undergo LLPS when mixed with 4×601-NA or 4×601 DNA. Our results showed that neither the NTD-MBD protein (1–180 aa) nor the CTD protein (312–486 aa) could undergo LLPS when mixed with 4×601-NA (Fig. 1h; Supplementary information, Fig. S2c). The truncations TRD-CTD (167–486 aa) and MBD-TRD (73–312 aa) underwent weak LLPS with 4×601-NA (Fig. 1h; Supplementary information, Fig. S2c). Finally, the truncations NTD-MBD-TRD (1–312 aa) and MBD-TRD-CTD (72–486 aa) showed a stronger LLPS ability than the other truncations but were still a little weaker than the FL MeCP2 (Supplementary information, Fig. S2c). These MeCP2 truncations were of similar ranking in terms of LLPS ability when mixed with 4×601 DNA (Supplementary information, Fig. S2d). In addition, the 72–486 truncation has a similar ability for LLPS with 4×601 DNA as FL protein. Interestingly, as summarized in Fig. 1i, the LLPS ability of these MeCP2 truncations correspond well with their chromatin compaction abilities (Fig. 1i; Supplementary information, Fig. S2b, e, f). This suggests that chromatin compaction may be correlated with LLPS in heterochromatin formation.
DNA methylation regulates MeCP2-driven chromatin LLPS in vitro
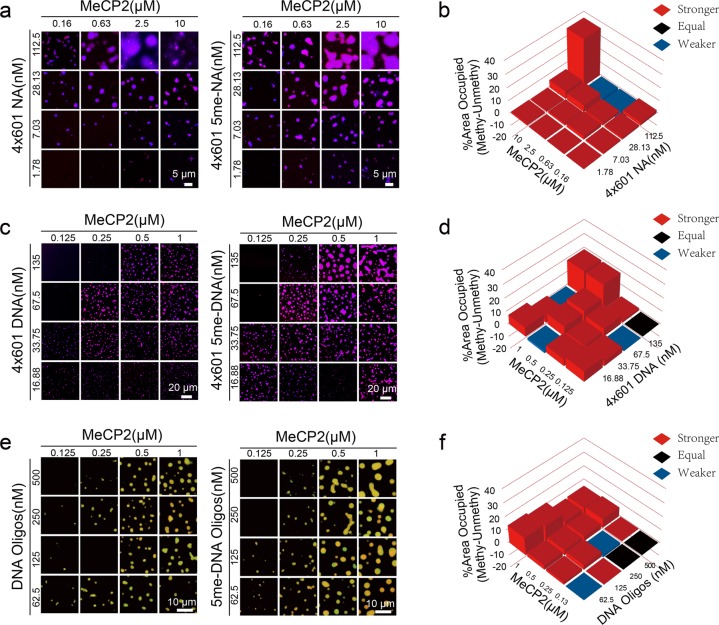
MeCP2 was first identified as a methylated DNA-binding protein.5–8 Further studies showed that MeCP2 not only binds to methylated DNA but also interacts with unmethylated and hydroxymethylated DNA.16,28–30 Therefore, we examined whether DNA methylation affects the phase separation ability of MeCP2 with DNA or NA. To generate substrates for this assay, we methylated 4×601 DNA with the methyltransferase M.SssI in vitro (4×601 5me-DNA) (Supplementary information, Fig. S3a, b). We also used methylated 4×601-NA (4×601 5me-NA) assembled from 4×601 5me-DNA. To ensure a sample containing 100% methylation at designated sites, we also synthesized a methylated DNA oligo (5me-oligo) (Supplementary information, Fig. S3c). Similar to unmethylated substrates, these methylated DNAs or nucleosomal arrays did not undergo LLPS by themselves under physiological salt condition (Supplementary information, Fig. S3d), whereas they readily underwent LLPS after mixing with MeCP2 (Fig. 2a, c, e). The LLPS-inducing ability of MeCP2 was moderately enhanced with methylated substrates (NA, DNA or Oligo) compared to the unmethylated ones (Fig. 2b, d, e).
Fig. 2. Effect of DNA methylation on LLPS of MeCP2 with 4×601 DNA or 4×601-NA in vitro.
a Phase diagrams of MeCP2 with 4×601-NA or 4×601 5me-NA. Scale bars, 5 µm. b Comparison of phase diagrams of MeCP2 in a. Value percentage = [Area occupied percentage of Methylation (4×601 5me-NA with MeCP2)] – [Area occupied percentage of Unmethylation (4×601-NA with MeCP2)]. Red squares indicate DNA methylation enhanced the LLPS of MeCP2; black squares indicate no LLPS or methylated DNA acted the same with unmodified DNA the LLPS of MeCP2; blue squares indicate DNA methylation compromised the LLPS of MeCP2. c Phase diagrams of MeCP2 with 4×601 DNA or 4×601 5me-DNA. Scale bars, 20 µm. d Comparison of phase diagrams of MeCP2 in c. Value percentage = [Area occupied percentage of Methylation (4×601 5me-DNA with MeCP2)] – [Area occupied percentage of Unmethylation (4×601 DNA with MeCP2)]. e Phase diagrams of MeCP2 with DNA oligos or 5me-DNA oligos. Scale bars, 10 µm. f Comparison of phase diagrams of MeCP2 in e. Value percentage = [Area occupied percentage of Methylation (5me-DNA Oligos with MeCP2)] – [Area occupied percentage of Unmethylation (DNA Oligos with MeCP2)].
RTT nonsense mutations compromise MeCP2-mediated chromatin LLPS
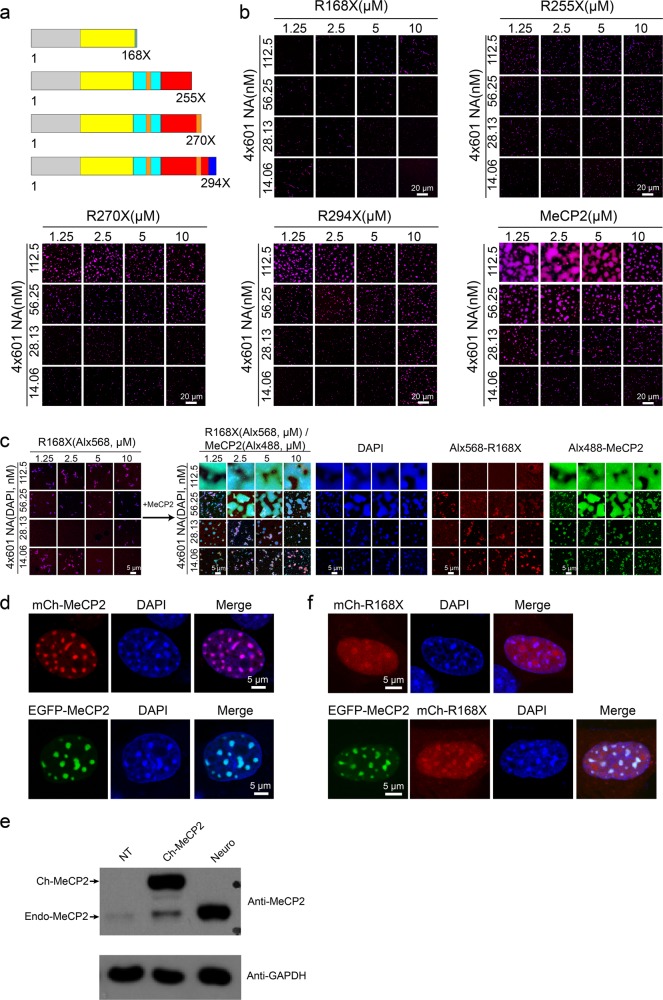
Numerous missense and nonsense mutations of MeCP2 cause Rett syndrome.31 Many of these MeCP2 mutations have been shown to compromise the ability for chromatin compaction and large-scale chromatin organization.24,32 Therefore, we wondered whether the LLPS ability of MeCP2 was also compromised by RTT mutations. The four most prevalent nonsense RTT mutations, R168X, R255X, R270X and R294X (Fig. 3a; Supplementary information, Fig. S4a, b), account for about 92% of all nonsense mutations and approximately 25% of all mutations.33 Of note, clinical studies showed that RTT nonsense mutations closer to the N-terminus cause more severe symptoms than those nearer to the C-terminus.34 Interestingly, the LLPS ability of these RTT truncations with 4×601-NA are largely inversely correlated with the disease severity: R168X failed to undergo LLPS with 4×601-NA at all conditions tested, while R255X, R270X and R294X had progressively increased LLPS ability as the length of the proteins increased (Fig. 3b). The LLPS ability of R294X was lower than FL MeCP2 (486 aa). The result with R168X is consistent with our result for the very similar NTD-MBD fragment (1–180 aa): both were incapable of condensing NA into compacted chromatin structures (Fig. 1b).16 Interestingly, the NTD-MBD-TRD fragment (1–312 aa) shows stronger LLPS ability than R294X (Fig. 3b; Supplementary information, Fig. S2c), which indicates that the region (294–312 aa) located at the C-terminal end of TRD contributes to the LLPS ability of MeCP2 with 4×601-NA. Similar trends in LLPS ability were observed for these four nonsense mutations with DNA substrates (Supplementary information, Fig. S4c, e). However, R294X exhibited a similar LLPS ability (both phase boundary and area occupied in phase diagram) as FL MeCP2 when 4×601 DNA and DNA oligo were used as substrates (Supplementary information, Figs. S1e and S4c–e). These results further demonstrated a potential functional connection between the CTD and LLPS ability of MeCP2 with 4×601-NA and suggest that CTD likely contains chromatin-specific binding sites.17 Since chromatin is the native substrate for MeCP2, the defective LLPS of R294X with 4×601-NA may be relevant to the pathology of RTT.
Fig. 3. RTT-related nonsense mutations affect MeCP2-driven chromatin LLPS.
a Diagrams showing the location of the four main RTT-related MeCP2 nonsense mutations. b Phase diagrams of FL MeCP2 and Rett-related MeCP2 nonsense mutations (shown in Fig. 3a) with 4×601-NA. Scale bars, 20 µm. c Inability of MeCP2 R168X to undergo phase separation with 4×601-NA is rescued by addition of extra FL MeCP2 protein. Left panel, phase diagram of MeCP2 R168X with 4×601-NA. Only merged channels are shown here. Right, phase diagrams after addition of FL MeCP2 into the reaction shown in the left panel. Scale bars, 5 µm. d Location of overexpressed mCherry-MeCP2 or EGFP-MeCP2 to heterochromatic chromocenters marked by DAPI in NIH 3T3 cells. Scale bars, 5 µm. e Western blot analysis to check the protein level of overexpressed MeCP2 and endogenous MeCP2 in 3T3 cells and in hippocampal neurons. Top panel, anti-MeCP2 antibody is used. The upper band is the overexpressed mCherry-MeCP2, and the lower bands are endogenous MeCP2. Bottom panel, anti-GAPDH antibody is used. GAPDH is used as the internal control. NT (no transfected 3T3 cells), mCh-MeCP2 (overexpressed mCherry-MeCP2 in 3T3 cells) in d, neuron (hippocampal neurons) in Supplementary information, Fig. S5b. f Dispersion of overexpressed mCherry-MeCP2-R168X alone or with EGFP-MeCP2 in NIH 3T3 cells. Scale bars, 5 µm.
The MECP2 gene is on the X chromosome. RTT patients, who are chiefly girls, normally have a wild-type (WT) MECP2 allele and a mutated allele. In female cells, one of the two X chromosomes is randomly inactivated. Hence, in RTT patients, the WT allele is inactivated in affected neurons whereas the mutant allele is inactivated in normal neurons. Studies using mouse models have demonstrated that RTT is a reversible (i.e., curable) disorder.35 It has been suggested that a potential way of alleviating RTT is to re-activate the silent WT allele in affected neurons.36 To mimic this scenario, we evaluated whether introduction of WT MeCP2 could rescue the LLPS defects in MeCP2 mutant/NA system. For the nonsense mutations, introduction of WT MeCP2 protein indeed rescued the compromised LLPS of mutant proteins with 4×601-NA in vitro (Fig. 3c; Supplementary information, S5a). We also tested the rescue effect in cells. Here, we used 3T3 cells for protein overexpression. When overexpressed in NIH 3T3 cells, FL MeCP2 mainly localized to chromocenters (Fig. 3d). Western blot was performed to check the protein levels of endogenous and overexpressed MeCP2. Results showed that the protein level of overexpressed MeCP2 was similar to that of endogenous MeCP2 (Fig. 3e). Endogenous MeCP2 also localized to chromocenters in mouse adult neurons (Supplementary information, Fig. S5b). These observations are consistent with previous reports.32,37 We divided the nucleus into three regions according to the DAPI signal: DAPI-deficient region, DAPI-dispersed region, and DAPI-dense region, which largely correspond to nucleolus, euchromatin, and heterochromatin, respectively. We analyzed the average fluorescence signals for labeled proteins and DNA in DAPI-dense regions by using the signals in DAPI-dispersed regions as the background. We found that the DNA and MeCP2 signals in DAPI-dense regions in NIH 3T3 cells or in mouse adult neurons were more than 2-fold higher than the background (Supplementary information, Fig. S5c), which is consistent with the presence of bright DNA-MeCP2-enriched puncta. In contrast, overexpressed R168X protein was dispersed in the nucleus of NIH 3T3 cells, with strong nucleolar localization and weak chromocenter localization (Fig. 3f, upper panels). Interestingly, when 3T3 cells were co-transfected with EGFP-MeCP2 and Cherry-MeCP2 R168X, the FL MeCP2 still showed strong chromocenter localization and R168X partially translocated into chromocenter (Fig. 3f, lower panels). When overexpressed alone in 3T3 cells, R270X colocalizes to chromocenters (Supplementary information, Fig. S5d). When FL MeCP2 and R270X were expressed in 3T3 cells, they both exhibited strong chromocenter localization (Supplementary information, Fig. S5d). These data suggest that the MBD domain alone is insufficient for MeCP2 binding to chromocenters in vivo; other parts of MeCP2 like ID and TRD are also required for its stable binding to chromocenters in 3T3 cells. In addition, our results suggest that intermolecular interactions probably occur between MeCP2 molecules in the presence of DNA or NA since the FL MeCP2 can recruit the truncated proteins to chromatin puncta in vivo and in vitro (Fig. 3c, f; Supplementary information, Fig. S5a).
Missense mutations in MBD compromise MeCP2-mediated chromatin LLPS
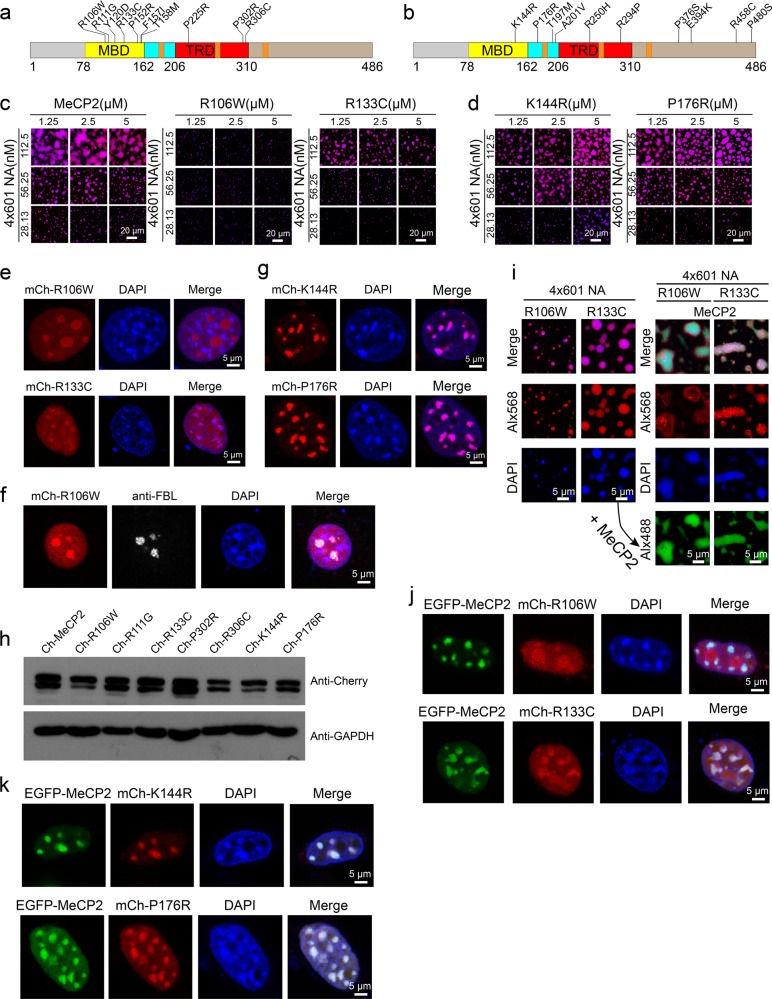
RTT missense mutations are mainly concentrated in two discrete regions: the MBD and the TRD. Clinical roles for mutations in other domains such as the intervening domain (ID) or AT-hook domains are yet to be determined.34 We next explored the effect of missense RTT mutations on the LLPS ability of MeCP2 with 4×601-NA (Fig. 4a; Supplementary information, Fig. S6a). As controls, we also investigated 10 benign polymorphic MeCP2 variants identified from the normal population (Fig. 4b; Supplementary information, Fig. S6b). Our results showed that most RTT missense mutations in the MBD (R106W, R111G, Y120D, R133C, F157I, T158M), with the exception of P152R, reduced the MeCP2-driven LLPS of chromatin in vitro (Fig. 4c; Supplementary information, Fig. S6c), while all the benign variants were indistinguishable from WT MeCP2 (Fig. 4d; Supplementary information, Fig. S6d). Similarly, when expressed in 3T3 cells, most of the MBD RTT missense mutant proteins (R106W, R111G, Y120D, R133C, F157I) did not form condensates, but the P152R mutant showed localization to DAPI-dense chromocenters despite slightly weaker than WT MeCP2 (Fig. 4e; Supplementary information, Figs. S7a and S8f). Interestingly, whereas the RTT MBD missense mutants of MeCP2 did not accumulate in DAPI-dense heterochromatin, they accumulated in the DAPI-deficient region, which is the nucleolus, as judged by fibrillarin labeling (Fig. 4e, f). When the benign polymorphic variants of MeCP2 were expressed in 3T3 cells, they formed condensate patterns indistinguishable from that of WT MeCP2 (Fig. 4g; Supplementary information, Figs. S7b and S8f). We also examined the protein levels of these MeCP2 mutants, and found that they were lower than that of WT MeCP2 (Fig. 4h), consistent with the previous report.38
Fig. 4. RTT-related missense mutations in the MBD compromise MeCP2-driven chromatin LLPS.
a, b Schematic diagrams of the missense mutations analyzed in this study. Ten Rett-related MeCP2 missense mutations in MBD or TRD (a). Ten neutral MeCP2 missense variants, which are not associated with disease symptoms (b). c Phase diagrams of WT MeCP2 and two RTT-related missense mutations in MBD (R106W and R133C) with 4×601-NA. Scale bars, 20 µm. d Phase diagrams of MeCP2 benign variants (K144R and P176R) with 4×601-NA. Scale bars, 20 µm. e Dispersion of overexpressed mCherry-MeCP2-R106W or R133C within the nucleus in NIH 3T3 cells. Scale bars, 5 µm. f In NIH 3T3 cells, overexpressed mCherry-MeCP2-R106W is dispersed in the nucleus and also enriched in the nucleolus (labeled with Fibrillarin (FBL)). Scale bar, 5 µm. g Location of overexpressed mCherry-MeCP2-K144R and P176R to heterochromatic chromocenters marked by DAPI in NIH 3T3 cells. Scale bars, 5 µm. h Western blot analysis to check the protein levels of overexpressed WT MeCP2 and MeCP2 mutants in 3T3 cells. Upper panel, anti-Cherry antibody is used. Lower panel, anti-GAPDH antibody is used. GAPDH is used as the internal control. i Left panels, mCherry-MeCP2-R106W and R133C undergo weak phase separation with 4×601-NA. Right panels, phase separation after addition of WT Alx488-MeCP2 into the reactions shown in the left panels. Scale bars, 5 µm. j Co-expression of EGFP-MeCP2 with mCherry-MeCP2-R106W or R133C in NIH 3T3 cells. Scale bars, 5 µm. k Colocalization of EGFP-MeCP2 with mCherry-MeCP2-K144R or P176R in NIH 3T3 cells. Scale bars, 5 µm.
Next we evaluated whether introduction of WT MeCP2 can rescue the LLPS defects in MeCP2 mutant/NA system. When WT MeCP2 is introduced to pre-formed droplets containing RTT missense mutants (R106W or R133C as shown in Fig. 4i) with 4×601-NA, it can rescue the impairment of LLPS by these RTT missense mutant proteins with NA just as it did to the truncated nonsense mutants (Fig. 4i; Supplementary information, Fig. S9). We further tested whether the in vitro observations of the phase separation behavior of MeCP2 and its variants hold true in cells. When WT MeCP2 and MeCP2 MBD RTT missense mutants were co-expressed in 3T3 cells, WT MeCP2 preferentially located to chromocenters but RTT missense mutants were dispersed in the nucleus and only partially located to chromocenters (Fig. 4j; Supplementary information, Fig. S7c). When WT MeCP2 and benign polymorphic variants were co-expressed in 3T3 cells, both proteins colocalized well to the chromocenters (Fig. 4k; Supplementary information, Fig. S7d). FRAP assays in cells also showed that benign polymorphic variants behaved very similarly to the WT protein, but the MBD RTT missense mutant proteins could not be adequately bleached or the recovery rate was too fast to be detected (Supplementary information, Fig. S8a–c). In addition, when co-expressed with the WT protein, these mutants showed a similar FRAP trajectory as when they were expressed alone (Supplementary information, Fig. S8d, e). These results together demonstrated that the MBD, which has been identified to bind both methylated and unmethylated DNA,5,16 contributes to the LLPS ability of MeCP2 in vitro and its tight association with heterochromatin in vivo. Especially importantly, these results further indicate that re-introduction of WT MeCP2 in mutant cells may restore their normal functions and alleviate disease symptoms.
Binding-induced self-interaction of MeCP2 contributes to its LLPS potential
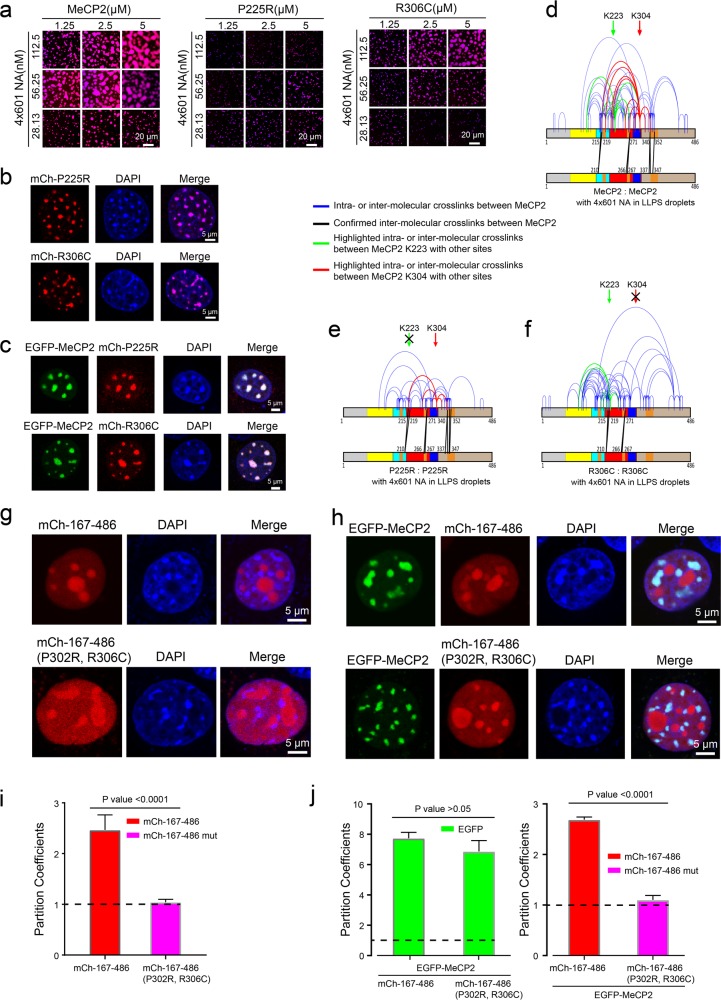
Besides the RTT MBD mutations, two frequent RTT mutations (P302R and R306C) are found in a “basic cluster” region in the NCoR/SMRT interaction domain (NID),39 which is rich in positively charged amino acids and was reported to bind to DNA in vitro.40 Of note, R306C (found in about 5% of RTT patients) was reported to impair the interaction of this region of MeCP2 with DNA.40 NID missense mutations have been shown to prevent MeCP2 from interacting with NCoR/SMRT. However, a recent research showed that blocking the activity of HDAC through mutant NCoR cannot alleviate the toxicity of overexpressed MeCP2,14 indicating that the function of NID missense mutations in RTT might be independent of HDAC recruitment. Hence, we investigated the function of the NID missense mutations on DNA binding.39,40 In this vein, as shown above (Fig. 1i; Supplementary information, Fig. S4c–e), the short region located in the NID (285–309 aa) contributes to the LLPS ability of MeCP2 with 4×601-NA. Consistent with these observations, our results further show that the RTT mutations outside MBD (P225R, P302R and R306C) also cause defective LLPS of MeCP2 with 4×601-NA, although to a lesser degree than the RTT mutations within MBD (Fig. 5a; Supplementary information, Fig. S10a). These observations suggest a critical role of the TRD in the LLPS ability of MeCP2 with NA.
Fig. 5. RTT-related missense mutations in the TRD affect MeCP2-driven chromatin LLPS.
a Phase diagrams of WT MeCP2 and two variants with RTT-related missense mutations, P225R and R306C, in the TRD in Fig. 4a. Scale bars, 20 µm. b Localization of overexpressed mCherry-MeCP2 P225R or R306C in NIH 3T3 cells. Scale bars, 5 µm. c Colocalization of EGFP-MeCP2 with mCherry-MeCP2 P225R or R306C in NIH 3T3 cells. Scale bars, 5 µm. d–f Cross-linking results of MeCP2 within phase-separated droplets formed with 4×601-NA. WT MeCP2 is shown in d and the RTT-related TRD-mutants are shown in e, f. g Dispersion of overexpressed mCherry-MeCP2 167–486 (top panel) or the 167–486 truncation carrying the P302R or R306C mutations (bottom panel) within the nucleus in NIH 3T3 cells. Scale bars, 5 µm. h Co-expression of EGFP-MeCP2 with mCherry-MeCP2 167–486 (top panel) or with the 167–486 truncation carrying the P302R or R306C mutations (bottom panel) in NIH 3T3 cells. Scale bars, 5 µm. i Statistical analysis of the partition coefficients of mCherry-proteins (P value = 0.1924) on the heterochromatin in g. The average fluorescence signals for labeled proteins and DNA in DAPI-dense regions are analyzed by using the signals in DAPI-dispersed regions as the background. j Statistical analysis of the partition coefficients of EGFP-MeCP2 (left panel, P value = 0.0193) and mCherry-proteins (right panel, P value < 0.0001) on the heterochromatin in h.
Interestingly, overexpression experiments show that RTT missense mutations in TRD do not affect the recruitment of MeCP2 mutant proteins to chromocenters in 3T3 cells (Fig. 5b; Supplementary information, Fig. S10c). When co-expressed in 3T3 cells, WT and TRD mutants also colocalized to heterochromatin (Fig. 5c; Supplementary information, Fig. S10d). FRAP assays showed that these TRD mutant proteins exhibit similar recovery dynamics as WT protein in 3T3 cells whether overexpressed alone or co-expressed with the WT protein (Supplementary information, Figs. S8a and S10e–g). Consistent with the minor effect of the R306C mutation on LLPS in vitro and binding to chromocenters in cells, patients with this mutation have delayed onset of symptoms and decreased disease severity compared with patients who have MBD mutations or earlier truncations.34 This observation indicates that the degree of impairment of the LLPS function of MeCP2 is related to the clinical course of RTT pathologies.
The NID was previously reported to be involved in the intermolecular oligomerization of MeCP2 in the presence of DNA and NA.16 To further explore whether the interactions of MeCP2 with itself and its DNA substrates were affected by RTT mutations (P225R, P302R and R306C) during LLPS formation, we performed in vitro chemical cross-linking of proteins coupled with mass spectrometry (CXMS) analysis to investigate the MeCP2-MeCP2 interactions induced by LLPS with DNA or NA (Fig. 5d–f). A number of MeCP2-MeCP2 intermolecular interactions were observed when MeCP2 was bound to 4×601-NA and undergoes LLPS (Fig. 5d). Interestingly, a number of these interactions, especially those involving K223, disappeared when we used the P225R mutant (Fig. 5e), and those involving K304 disappeared when we used the P302R and R306C mutants (Fig. 5f; Supplementary information, Fig. S10b). These results suggest that MeCP2 cooperatively oligomerizes when bound to 4×601-NA, and TRD missense mutations preferentially perturb the oligomerization interaction in their own vicinity. The latter perturbation is likely the cause for the impairment of MeCP2-driven LLPS of NA by these RTT mutations.
To further validate the function of NID-mediated intermolecular interactions, we overexpressed the MeCP2 167–486 truncation in 3T3 cells, and found that it lost its chromocenter localization (Fig. 5g, i). However, when the 167–486 truncation was co-expressed with FL MeCP2 in 3T3 cells, the truncated protein was partially recruited to chromocenters by FL MeCP2 (Fig. 5h, j). We subsequently generated a double mutation in the NID (P302R and R306C) of the 167–486 truncation. When overexpressed alone in 3T3 cells, the mutated 167–486 truncation was dispersed throughout the nucleus (Fig. 5g, i). When this mutated truncation was co-expressed with FL MeCP2, the mutated 167–486 truncation failed to locate to chromocenters (Fig. 5h, j), which suggests that the NID is important for intermolecular interactions between MeCP2 molecules. Together, our results suggest that the TRD domain participates in the MeCP2-MeCP2 interaction, which is critical for MeCP2-mediated chromatin phase separation, and the RTT TRD mutations weaken the intermolecular interaction and compromise MeCP2-mediated LLPS of chromatin.
Competition and incompatibility between MeCP2- and H1-mediated chromatin puncta
Despite their completely different amino acid sequences, and secondary and tertiary structural organization, MeCP2 and linker histone H1 exhibit a striking amount of functional convergence. They both can bind nucleosomes at the linker DNA entry/exit position41 and induce marked compaction of nucleosomal arrays in vitro.37 This suggests that there may be competition between the two proteins for nucleosomal binding sites.8,37 Indeed numerous in vitro experiments showed that MeCP2 can modulate the higher order chromatin structures by competing with H1 and compacting nucleosomal arrays.8,15–17 Furthermore, in vivo studies showed that MeCP2 deficiency results in a doubling of the level of H1 and global changes of chromatin organization in neurons, suggesting that MeCP2 serves as an alternative chromatin architecture protein in neurons.10
Interestingly, it has recently been reported that the highly disordered C-terminal tail of H1 can form condensates with DNA in vitro42 and that H1 can also promote chromatin LLPS in vitro.19 To gain more insights into the molecular mechanism underlying the inverse relationship between these two fundamental chromatin organizers, we extensively investigated the LLPS ability of H1 with its binding partners NA and DNA. While individual solutions of FL H1 or its binding partners are homogeneous (Supplementary information, Figs. S1b and S11a), H1 underwent phase separation with 4×601 DNA or 4×601-NA upon mixing (Fig. 6a; Supplementary information, Fig. S11b). Pairs of puncta fused and relaxed into larger ones upon contact (Supplementary information, Fig. S11c). FRAP experiments showed that H1 is dynamic within puncta and more dynamic than MeCP2 (Fig. 1e; Supplementary information, Fig. S1g and S11d, e).
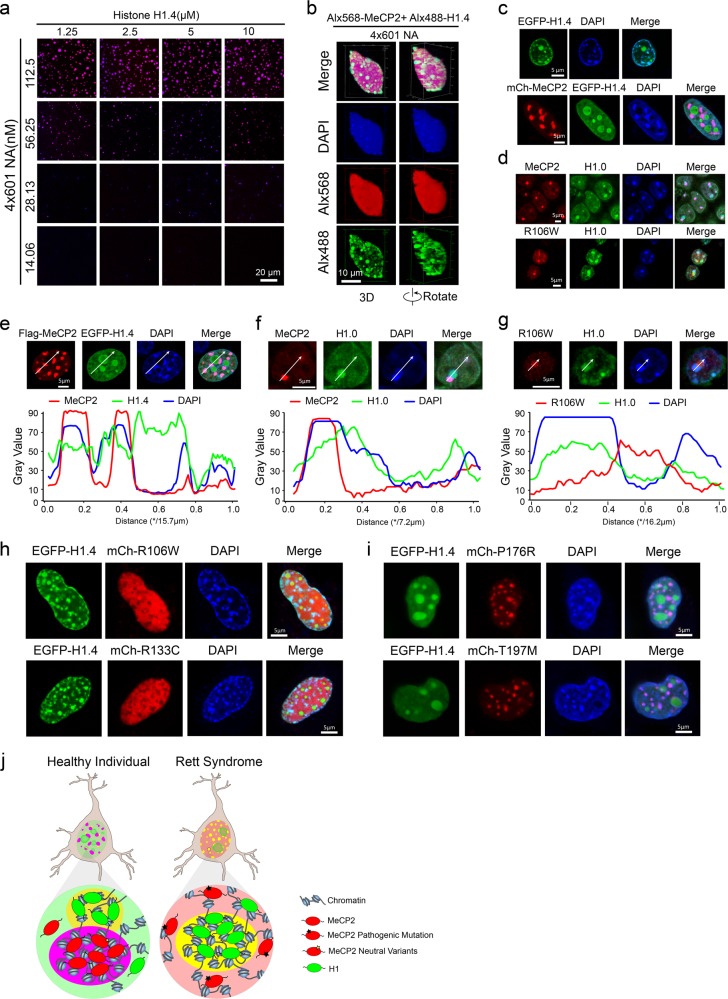
Fig. 6. Competition and incompatibility between MeCP2- and H1-containing chromatin puncta.
a Phase diagram of human linker histone H1.4 with 4×601-NA. Scale bar, 20 µm. b 3D shapes of H1.4-NA droplets within a big MeCP2-NA droplet at a high concentration of 4×601-NA. Scale bar, 10 µm. c Top panel, overexpressed EGFP-H1.4 forms chromatin puncta and colocalizes with heterochromatic chromocenters in NIH 3T3 cells. Bottom panel, co-expression of mCherry-MeCP2 with EGFP-H1.4 in NIH 3T3 cells. mCherry-MeCP2 forms chromatin puncta and colocalizes with heterochromatic chromocenters, whereas H1.4 is dispersed within the nucleus and enriched in the nucleolus. Scale bars, 5 µm. d Top, Immunofluorescence analysis of MeCP2 WT and H1.0 in hippocampal neurons from mice. Bottom, Immunofluorescence analysis of MeCP2 R106W and H1.0 in hippocampal neurons from mice. Scale bars, 5 µm. e Top panel, immunofluorescence images of overexpressed Flag-MeCP2 and EGFP-H1.4 in NIH 3T3 cells, Scale bar, 5 µm. Bottom panel, plots of the red, green and blue pixel intensities along the white arrow in the top panel. f Top panel, immunofluorescence images of MeCP2 and H1.0 in mouse hippocampal neurons. Scale bar, 5 µm. Bottom panel, plots of the red, green and blue pixel intensities along the white arrow in the top panel. g Top panel, immunofluorescence images of MeCP2 R106W mutant and H1.0 in mouse hippocampal neurons. Scale bar, 5 µm. Bottom panel, plots of the red, green and blue pixel intensities along the white arrow in the top panel. h Overexpression of EGFP-H1.4 with the MeCP2 mutants, mCherry-MeCP2 R106W or R133C in NIH 3T3 cells. MeCP2 mutant proteins are dispersed within the nucleus in cells, whereas H1 localizes to heterochromatin. Scale bars, 5 µm. i Overexpression of EGFP-H1.4 with two polymorphic MeCP2 variants, mCherry-MeCP2-P176R or T197M, in NIH 3T3 cells. The MeCP2 benign variants localize to heterochromatin, while H1 mainly localizes within the nucleolus. Scale bars, 5 µm. j Model of the phase distribution of H1 and MeCP2 in neurons. Left, H1 and MeCP2 phases are mutually exclusive but close to each other in healthy neurons. Right, MeCP2 proteins harboring MBD RTT mutations are dispersed in the nucleus, while H1 still forms phase-separated compartments in diseased neurons.
Next, we tested whether MeCP2 and H1 compete with each other in the process of LLPS with chromatin. When we co-mixed H1 and MeCP2 with their binding partners (e.g., 4×601-NA), we found numerous small H1-enriched puncta in one large MeCP2-enriched punctum (Fig. 6b). Interestingly, this engulfment of H1-rich condensates within MeCP2-containing condensates is similar to the co-existence of multiple phases within the nucleolus43 or the co-assembly phenomenon of MEG-3 gel phases and PGL-3 liquid phases of P granules.44 We also found that both MeCP2 and H1 are highly dynamic, because when they were mixed with 4×601-NA, MeCP2 puncta were observed to fuse together, and both fission and fusion of H1 puncta were observed within MeCP2 puncta (Supplementary information, Fig. S11f).
To investigate MeCP2-H1 competition in more details, we titrated H1 protein into pre-formed MeCP2-4×601-NA droplets. When the H1 concentrations were below the critical concentration for phase separation, H1 uniformly partitioned into MeCP2-NA puncta (Supplementary information, Fig. S12a, left panels). However, when the H1 concentration reached the concentration for phase separation, H1 competed with MeCP2 for 4×601-NA and formed distinct small phase-separated droplets within the large MeCP2-NA droplets (Supplementary information, Fig. S12a). Similarly, when MeCP2 protein was titrated into pre-formed H1-4×601-NA droplets, MeCP2 competed with H1 for 4×601-NA and formed condensed droplets that engulfed the H1-4×601-NA droplets (Supplementary information, Fig. S12b). Formation of immiscible puncta also occurred when DNA oligos with or without methylation were used as substrate (Supplementary information, Fig. S12c, d). These data together indicate that the immiscibility of MeCP2- and H1-puncta are due to the intrinsic properties of MeCP2- and H1-chromatin.
To investigate the role of DNA methylation in regulating MeCP2-H1 competition, we established phase diagrams of MeCP2-H1 mixtures, which contained an equal molar ratio of the two proteins, with 4×601-NA (Supplementary information, Fig. S12e) or 4×5me-NA (Supplementary information, Fig. S12f). Comparing images within the two phase diagrams, we found that DNA methylation elevated the relative concentration ratios between MeCP2 and H1 across the whole phase diagrams (Supplementary information, Fig. S12e, f). Similarly, methylated DNA oligo elevated the relative intensities of MeCP2 versus H1 within droplets of their mixture with DNA oligo (Supplementary information, Fig. S12d). These results are consistent with the notion that DNA methylation enhances the binding affinity with MeCP2.
To investigate whether the competition between MeCP2 and H1 also occurs in the nucleus, we transiently expressed these two proteins in NIH 3T3 cells. When expressed alone, H1 formed chromatin condensates that overlapped at least partially with chromocenters (strong DAPI sites) (Fig. 6c; Supplementary information, Fig. S13a). When both H1 and MeCP2 were co-expressed in 3T3 cells, a negative correlation was observed for H1 and MeCP2 localization: MeCP2 formed chromatin condensates with strong DAPI chromocenters, whereas H1 was dispersed throughout the nuclei with a few condensates in the nucleolus (Fig. 6c, e; Supplementary information, Fig. S13b). FRAP assays showed that H1 was slightly more dynamic than MeCP2 when these proteins were overexpressed alone in 3T3 cells (Supplementary information, Fig S13e). The dynamics of H1 and MeCP2 when co-expressed were similar to when they were overexpressed alone in 3T3 cells (Supplementary information, Fig. S13f, g). Endogenous H1 also shows a more dispersed distribution than MeCP2 in nuclei, but endogenous H1 also has some strong DAPI signals very close to the MeCP2 puncta in mouse adult neurons (Fig. 6d, f; Supplementary information, Fig. S13b). Therefore, the overexpression result is similar to the endogenous distribution of MeCP2 and H1, especially H1 also accumulated to the nucleolus regions (Supplementary information, Fig. S13c).
Interestingly, when H1 was co-expressed with the MeCP2 RTT MBD missense mutants R106W and R133C, the mutant MeCP2 proteins dispersed throughout the nuclei, while H1 formed numerous puncta that colocalized with strong DAPI signals (Fig. 6h; Supplementary information, Fig. S13k). This suggests that the RTT MBD mutant proteins loss the ability to compete with H1 in cells. FRAP assays showed that the dynamics of co-overexpressed R106W and R133C with H1 was similar to when each protein was overexpressed alone (Supplementary information, Figs. S8a, c, and S13h). In contrast, benign polymorphic mutants and TRD mutants (P225R, P302R and R306C) behaved similarly to WT MeCP2 when co-expressed with H1 in 3T3 cells (Fig. 6c, i; Supplementary information, Fig. S13i, k). In addition, when the benign mutants or TRD mutants were co-expressed with H1, they exhibited the same recovery dynamics as when they were overexpressed alone (Supplementary information, Figs. S8b, c, and S13g, j). Importantly our immunofluorescence imaging data using adult neurons from a male mouse expressing MeCP2 R106W showed that H1, but not the mutated MeCP2, localized in DAPI-dense heterochromatin region (Fig. 6d, g; Supplementary information, Fig. S13d). Instead, MeCP2 R106W formed separate puncta juxtaposed to heterochromatin ones (Fig. 6d, g; Supplementary information, Fig. S13d).
In summary, we have linked the LLPS-inducing functions of MeCP2 with RTT disease. In healthy neurons, MeCP2 and H1 form distinct heterochromatin domains, which are mutually exclusive but close to each other. RTT missense mutations in the MBD of MeCP2 compromise chromatin phase separation and destroy the MeCP2-heterochromatin domains. Instead of localizing to chromocenters, these mutated MeCP2 proteins mis-localize to the nucleolus and H1 takes over the role of organizing heterochromatin in RTT neurons. RTT-causing point mutations in the TRD of MeCP2 may compromise the chromatin LLPS by disrupting the intermolecular interactions of MeCP2 (Fig. 6i).
Discussion
Heterochromatin is defined epigenetically by DNA methylation and H3K9 methylation, which recruit binding partners such as MeCP2 and HP1, respectively. In addition, linker histone H1, an important chromatin organizer, has been shown to be essential for chromatin architecture, for example heterochromatin formation.45 The diverse biological functions of heterochromatin rely on its ability to compact the underlying genomic DNA and to recruit different types of modulators. Interestingly, recent studies showed that HP1 and H1 drive phase separation of heterochromatin to form liquid droplets, thus providing a novel mechanism for heterochromatin formation.18–22
Compaction and phase separation of chromatin mediated by MeCP2
As an important chromatin organizer, MeCP2 binds to DNA and nucleosomes through different regions and induces efficient chromatin compaction in vitro.15,16,23,37,46,47 However, it is still largely unclear which regions of MeCP2 take part in chromatin compaction. To this end, our AUC assays and EM analysis showed that the MBD-TRD (72–312 aa) region is mainly responsible for chromatin compaction by MeCP2, while the NTD contributes little to this process, and the CTD is necessary for full chromatin compaction. Given the highly disordered sequence of MeCP2 and its ability to bind and compact DNA or nucleosomes, it is very likely that MeCP2 can drive phase separation with DNA or nucleosomes. Although MeCP2 itself does not undergo phase separation, an obvious demixing is observed when MeCP2 is mixed with nucleosomes or DNA under physiological condition. Interestingly, we found that the abilities of MeCP2 and its truncations to induce chromatin phase separation correspond well with their abilities to induce chromatin compaction, which suggests that chromatin compaction may make a major contribution to LLPS during MeCP2-mediated heterochromatin formation.
LLPS is thought to be derived from multivalent interactions between macromolecules and their multivalent substrates or ligands. Therefore, we hypothesized that chromatin phase separation driven by MeCP2 may involve multiple interactions between MeCP2 and chromatin, including MeCP2-MeCP2, MeCP2-DNA, MeCP2-histone, and nucleosome-nucleosome interactions. In this vein, it is noteworthy that MeCP2 is predicted to contain an unusually high number of Molecular Recognition Features (MoRFs),17 which are intrinsically disordered protein regions that likely constitute combinatorial interaction sites for binding a variety of partners.48 A cooperative association between MeCP2 monomers would result in local enrichment of MoRFs, potentially facilitating the local recruitment of binding partners and demixing from solution to form liquid droplets. In line with this hypothesis, and similar to previous observations,17 our in vitro cross-linking experiments showed that although MeCP2 is monomeric in solution, it oligomerizes in conjunction with DNA or NA binding, which is a potential mechanism underlying coordination of chromatin phase separation by MeCP2.
Competition and incompatibility between MeCP2- and H1-mediated chromatin liquid droplets
Previously, both in vitro and in vivo studies revealed an inverse correlation between MeCP2 and linker histone H1 in chromatin binding, chromatin compaction and higher order chromatin organization.7,8,17,41,49,50 Interestingly, our in vitro phase separation experiments showed that histone H1 and MeCP2 can compete with each other in phase separation of chromatin to form mutually exclusive droplets with distinct properties. Again, our in vivo cytological experiments showed that overexpressed H1, like MeCP2, preferentially binds to DAPI-dense heterochromatic chromocenters. However, when co-expressed with MeCP2 in cells, H1 redistributed from DAPI-dense chromocenters to the nucleolus or became dispersed in the nucleus, which is consistent with previous observations that MeCP2 binds chromatin more tightly and is more potent at displacing H1 than vice versa.7,17,49,50
MeCP2 was originally identified as a transcriptional repressor which recognizes and binds preferentially to methylated CpG dinucleotides.5,8,13 Interestingly, our results showed that MeCP2 remains bound to highly methylated chromocenters when the abundance of H1 is increased, which is consistent with previous findings that H1 competes efficiently with nonspecifically bound MeCP2 but not with the tightly bound fraction of MeCP2 on highly methylated heterochromatin.17 Surprisingly, MeCP2, which is abundantly present in the pericentromeric chromocenters, does not associate significantly with the methylated rDNA repeats, whereas H1 does overlap partially with these clusters when H1 and MeCP2 are co-expressed simultaneously in NIH 3T3 cells or in native neurons. This is consistent with previous observations that histone H1 is preferentially associated with the more methylated rDNA genes.51 Our in vitro and in vivo experiments showed that the RTT missense mutations in the MBD, which may abolish the binding of MeCP2 to methylated DNA, result in defective formation of chromatin droplets or condensation by MeCP2. In contrast, the RTT missense mutations in the TRD (P302R and R306C) partially compromise the MeCP2-mediated LLPS of chromatin by disrupting the cooperative MeCP2-MeCP2 interactions, whereas these mutations do not alter the localization of MeCP2 to highly methylated chromocenters. Consistent with this, our in vitro experiments showed that DNA methylation indeed facilitates the LLPS ability of MeCP2 with NA and helps MeCP2 to win the competition with H1 during LLPS with NA. These findings suggest that in neurons, histone H1 and MeCP2 may bind to distinct genomic regions and organize the genome into distinct heterochromatin domains by different mechanisms, including chromatin compaction and phase separation.
RTT-causing mutations compromise MeCP2-driven chromatin phase separation
Mutations in MeCP2 result in Rett syndrome, a severe postnatal neurodevelopmental disorder.4 Clinical genetic studies showed that RTT mutations predominantly locate in two discrete regions of MeCP2, MBD and TRD.32,52 A classic model has been proposed in which MeCP2 represses gene transcription mainly through interacting with methylated DNA via MBD and recruiting HDAC-containing co-repressor complexes by TRD. These interactions are affected by RTT-associated missense mutations.11,12,39 However, it has also been demonstrated that MeCP2 is able to repress transcription by directly compacting chromatin independently of histone deacetylation and DNA methylation.13,14 Recent in vivo studies showed that MeCP2 is also able to reorganize the global heterochromatin architecture into punctate chromocenters.10,16,23–25 These results indicate that MeCP2 may also mediate formation of heterochromatin by LLPS. Indeed, our in vitro and in vivo experiments showed that MeCP2 can drive phase separation of chromatin to form droplets independently of DNA methylation, and almost all RTT mutations, including mutations in MBD, TRD and NID, disturb MeCP2-mediated chromatin LLPS to different extents. In addition, we showed that MeCP2 competes with linker histone H1 to form two incompatible chromatin phase-separated compartments, and MeCP2 variants with RTT mutations in MBD, but not in NID or TRD, fail to form chromatin condensates because they lose the competition with H1. More and more lines of evidence have suggested that LLPS is not only essential for distinct physiological processes, but is also associated with the pathogenesis of many human diseases.53,54 Therefore, our results provide a new perspective on the mechanisms by which LLPS drives formation of distinct heterochromatin foci mediated by MeCP2 and H1, perturbations to the fine balance between these two kinds of chromatin domains may contribute to RTT pathogenesis.
Materials and methods
Nucleosome assembly
Histones and DNA templates of 4 or 12 tandem 177-bp repeats of the 601 sequence were cloned and purified as previously described.55 Histone octamer assembly was performed according to the method of Dyer et al.56 Four histones at equal molar amounts were dialyzed into refolding buffer (2 M NaCl, 10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 5 mM β-mercaptoethanol) and purified by a Superdex 200 column (GE Healthcare, USA). NA were assembled using the salt dialysis method as previously described.57 The reconstitution reaction of octamer and DNA templates was carried out at 4 °C overnight, from the TEN buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 2 M NaCl) diluted by TE (10 mM Tris-HCl, pH 8.0, 1 mM EDTA) to lower concentration of NaCl buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 0.6 M NaCl), followed by a final dialysis step in HE buffer (10 mM HEPES, pH 8.0, 0.1 mM EDTA) for 4 h at 4 °C.
Sedimentation velocity analytical ultracentrifugation
Binding of MeCP2 or its truncations to nucleosome arrays at a ratio of 1:1 was performed overnight at 4 °C in HEN50 buffer (10 mM HEPES, pH 8.0, 0.1 mM EDTA, 50 mM NaCl). Sedimentation experiments were performed on a Beckman Coulter ProteomeLab XL-I using a 4-hole An-60Ti rotor. Chromatin samples with an initial absorbance at 260 nm of approximately 0.3–0.8 were pre-equilibrated for 1.5 h at 20 °C under a vacuum in a centrifuge prior to sedimentation. The absorbance at 260 nm was measured during sedimentation at 20,000× g in 12 mm double-sector cells. Data were analyzed using enhanced van Holde-Weischet analysis and the Ultrascan II software as previously described.58
Electron microscopy
NA were prepared and visualized using metal shadowing electron microscopy as previously described.59 Chromatin samples (30 µg/mL) bound by MeCP2 or its truncations were stained with 2% uranyl acetate according to a previously reported method.59 Samples were examined using a FEI Tecnai G2 Spirit 120 kV transmission electron microscope.
Construction of recombinant plasmids
Human MeCP2 (Gene ID: 4204) and its mutants and truncations were cloned into pET-28a (+) for expression and purification. Human H1.4 (Gene ID: 3008) was cloned into pET-28a (+) plasmid for expression and purification as described previously.57 pEGFP-H1.4 and pmCherry-MeCP2 were also constructed for transient expression in cells. In addition, pEGFP-MeCP2 and pmCherry-MeCP2 mutations were constructed for transient expression in cells.
Protein purification
Plasmids of pET28a-MeCP2 and its truncations and mutations were transferred into BL21(DE3) cells, then cultured at 37 °C overnight. Single clones were picked into LB medium and cultured overnight at 37 °C with shaking at 200 rpm. The culture was inoculated into fresh LB medium at the ratio of 1:50 to expand the culture to OD 0.6, then induced with 0.5 mM IPTG for 4 h at 37 °C. The bacteria were collected by centrifugation at 4000 rpm for 30 min (4 °C), then resuspended with lysis buffer (20 mM Tris-HCl, pH 8.0, 1 mM PMSF, 500 mM NaCl, 5% glycerol, 20 mM Imidazole) followed by sonication on ice. The lysates were centrifuged at 18,000× g for 30 min at 4 °C, then the supernatant was transferred to a new tube and incubated with Ni-NTA resin at 4 °C for 3 h. Ni-NTA beads were washed extensively with wash buffer (20 mM Tris-HCl, pH 8.0, 300 mM NaCl, 5% glycerol, 20 mM Imidazole), and recombinant proteins were eluted with elution buffer (20 mM Tris-HCl, pH 8.0, 300 mM NaCl, 5% glycerol, 100/300/500/1000 mM Imidazole). The eluted fractions were analyzed by SDS-PAGE. Subsequently, the recombinant proteins were further purified on Hitrap Heparin HP columns, and examined by SDS-PAGE. The purified proteins were exchanged via dialysis into storage buffer (20 mM Tris-HCl, pH 8.0, 300 mM NaCl, 10% glycerol and 1 mM DTT) and stored at −80 °C.
pET28a-H1.4 was expressed in the E. coli strain BL21 (DE3) cell (Tiangen), and the protein was purified under denaturing condition (Qiagen, USA) and refolded in buffer (20 mM Tris-HCl, pH 8.0, 0.2 mM EDTA, 10% glycerol, 1 mM DTT, 0.2 mM PMSF and 100 mM KCl).
Protein labeling
All MeCP2 proteins (WT, truncations and mutants) and human linker histone H1.4 were exchanged into reaction buffer (0.1 M sodium bicarbonate buffer, pH 8.3) using a SD200 column. Proteins (10 mg) were labeled by incubating with a 1:1 molar ratio of Alexa Fluor™ 488 or 568 carbox (Thermo Fisher Scientific) for 1 h at room temperature (RT) with continuous stirring. To remove free dye and change the protein stock buffer, the samples were concentrated and loaded onto a SD200 column using storage buffer (20 mM HEPES pH 7.5, 300 mM NaCl, and 1 mM DTT). The labeled proteins were finally stored in storage buffer at −80 °C. For in vitro phase separation experiments, 5% labeled proteins were mixed with unlabeled ones before use.
Synthesis of DNA oligos
A 72-bp DNA oligo (GCCACCGGTGGCTTCTTCTAGCCACCGGTGGC TTCTTCTAGCCACCGGTGGCTTCTTCTAGCCACCGGTGGC) was synthesized with FAM488 labeling. This oligo contains four 12-bp DNA oligo units (GCCACCGGTGGC), and each repeating unit was linked by a random DNA sequence (TTCTTCTA). The 12-bp DNA oligo unit contains a 5-me methylation site. A 72-bp methylated DNA oligo (GCCACC(5me)GGTGGCTTCTTCTAGCCACC(5me)GGTGGCTTCTTCTAGCCACC(5me)GGTGGCTTCTTCTAGCCACC(5me)GGTGGC) was synthesized with FAM488 labeling.
Phase separation assays
Phase separation assays were performed in reaction buffer (20 mM HEPES pH 7.4, 100 mM NaCl), with various protein and nucleosomal array concentrations. In-cell assays were carried out on glass-bottomed 35 mm dishes (In Vitro Scientific), which were coated with 3% bovine serum albumin for 15 min and then washed with Milli Q H2O three times. For in vitro experiments such as FRAP and time-lapse imaging experiments, phase separation was recorded on 384 low-binding multi-well 0.17 mm microscopy plates (In Vitro Scientific) and sealed with optically clear adhesive film. Imaging was performed with a NIKON A1 microscope equipped with a 100× oil immersion objective. NIS-Elements AR Analysis was used to analyze these images.
Fluorescence recovery after photobleaching (FRAP) measurements
In vivo and in vitro FRAP experiments were carried out with a NIKON A1 microscope equipped with a 100× oil immersion objective. Droplets were bleached with a 488- or 561-nm laser pulse (3 repeats, 70% intensity, dwell time 1 s). Recovery from photobleaching was recorded for the indicated time.
Cell transfection and western blot
NIH 3T3 cells were maintained in DMEM (Hyclone) supplemented with 10% FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin. Cells were transfected with DNA Transfection reagent (Biotool), and collected at 36 h post-transfection.
Cells with overexpressed mCherry-MeCP2 and its mutations were collected after 36 h of transfection, Hippocampal neurons were separated from male mice of 8 weeks. Samples were lysed with lysis buffer (10 mM Tris, 1 mM EDTA, 1% SDS), following by water bath sonication. Proteins were separated by SDS-PAGE. Separated proteins were transferred to nitrocellulose (Whatman, 10401196). The membranes were blocked with 5% milk at RT for 1 h, and incubated with primary antibodies overnight at 4 °C. After washing with TBST, the membranes were incubated with secondary antibodies for 1 h at RT followed by washing. Antibodies used are: rabbit anti-MeCP2 (1:5000, Cell Signaling Technology, #3456), mouse anti-GAPDH (1:10,000, ABclonal, AC033), mouse anti-mCherry (1:1000, Abbkine, A02080-1), goat anti-rabbit second antibody (1:10,000, Zhongshan Golden Bridge Bio-technology, ZB-2301), and goat anti-mouse second antibody (1:10,000, Zhongshan Golden Bridge Bio-technology, ZB-2305).
Fluorescence imaging
NIH 3T3 cells were fixed with 4% PFA for 15 min at RT, then washed three times with PBS. DNA was stained with DAPI (Sigma) at RT for 5 min. Slides were then washed with PBS. Slides were mounted with Slowfade Diamond Antifade reagent (Life Technologies), and images were acquired with an OLYMPUS FV1200 microscope. Analysis of fluorescence was performed using the FV10-ASW4.2 Viewer tool.
Generation of MeCP2 R106W mice
Mouse experiments were approved by the Institutional Animal Care and Use Committees at the Institute of Biophysics, Chinese Academy of Sciences. MeCP2 R106W C57BL/6 J mice were generated in our laboratory by pronuclear microinjection with pre-assembled CaS10 protein/sgRNA RNPs and single-stranded donor DNA as described previously.60 The guide RNA sequence used is: 5′-GGACACGAAAGCTTAAACAA-3′. The 97-nt single-stranded oligodeoxynucleotide donor harboring R106W (CGA/TGG) and PAM neighboring deoxynucleotide mutations was synthesized, the sequence is: 5′-GACCGGGGACCTATGTATGATGACCCCACCTTGCCTGAAGGTTGGACGTGGAAACTCAAGCAAAGGAAGTCTGGCCGATCTGCTGGAAAGTATGATGT-3′. Genotyping was performed by PCR products-sequencing method using genome extracted from mouse tail tissue. The brain slices were obtained from an 8-week male MeCP2 R106W mouse.
Immunofluorescence imaging
Brain slides were subjected to antigen retrieval, pretreated with 0.5% Triton X-100 in PBS for 30 min at RT, incubated in blocking solution (CWBIO), then incubated with anti-MeCP2 antibody (Cell Signaling Technology, #3456) and anti-H1.0 antibody (Abcam, ab11079) overnight at 4 °C. The secondary antibodies were Andy Fluor™ 488 Goat Anti-Mouse IgG (H + L) Antibody (Genecopoeia, L109A) for H1 and Andy Fluor™ 594 Goat Anti-Rabbit IgG (H + L) Antibody (Genecopoeia, L120A) for MeCP2. Immunofluorescence images were acquired with an Olympus laser confocal microscope and analyzed with FV10-ASW viewer (Olympus).
DNA methylation assays
The in vitro methylation reaction was performed with CpG methyltransferase M.SssI (New England Biolabs) according to the manufacturer’s recommended protocol. 500 μg DNA was methylated with 400 units of enzyme in 1× NEB buffer 2 containing 160 μM SAM at 37 °C for 4 h.
Dot blot
Methylated DNA (200 ng per sample) was denatured with an equal volume of 0.2 M NaOH for 10 min at 95 °C. The DNA was then neutralized with a double volume of 1 M NH4OAc on ice. 5 ng and 15 ng of each DNA samples were spotted onto an N+ membrane (GE Healthcare). The membrane was blotted at 80 °C for 30 min with dry bath. The N+ membrane was blocked in 5% BSA in TBS-T for 1 h. After three washes in TBS-T, the membrane was incubated with a mouse anti-5-methylcytosine (5-mC) monoclonal antibody (1:1000, Active Motif #39649) in TBS-T at 4 °C overnight. The membrane was then washed for 5 min three times in TBS-T, and then incubated with a secondary sheep anti-mouse antibody (1:5000) in TBS-T for 1 h at RT. The membrane was washed for 5 min three times in TBS-T. The secondary antibody signal was visualized using a chemiluminescence kit (Invitrogen) according to the manufacturer’s instructions.
Chemical cross-linking of proteins coupled with mass spectrometry analysis (CXMS)
About 15 μg of nucleosome arrays containing 16 μg of MeCP2 in a volume of 30 μL (20 mM HEPES, pH 7.4, 100 mM NaCl) was cross-linked with 1 mM DSS (disuccinimidyl suberate, Thermo Fisher Scientific) at RT for 1 h, and then quenched with 20 mM NH4HCO3. For each cross-linked sample, the DNA was digested with 4 units of DNase I (New England Biolabs) at 37 °C for 3 h, and the proteins were precipitated with 4–5 volumes of acetone at −20 °C overnight. After centrifugation, the protein pellet was resuspended in 8 M urea, 100 mM Tris, pH 8.5 and digested with trypsin. LC-MS/MS analysis of the digested samples was performed on an Easy-nLC 1000 UHPLC (Thermo Fisher Scientific) coupled to a Q Exactive HF Orbitrap mass spectrometer (Thermo Fisher Scientific). Peptides were loaded on a pre-column (75 μm inner diameter, 4 cm in length, packed with ODS-AQ 12 nm–10 mm beads from YMC Co., Ltd.) and separated on an analytical column (75 μm inner diameter, 13 cm in length, packed with ReproSil-Pur C18-AQ 1.9 μm 120 A° resin from Dr Maisch GmbH) over a 60-min linear gradient made with buffer A (0.1% formic acid) and buffer B (100% acetonitrile and 0.1% formic acid) as follows: 0–2 min, 0–4% B; 2–52 min, 4%–36% B; 52–55 min 36%–95% B; 55–60 min, 95% B. The flow rate was 250 nL/min. The top 15 most intense precursor ions from each full scan (resolution 60,000) were isolated for HCD MS2 (resolution 15,000; NCE 27) with a dynamic exclusion time of 30 s. Precursors with 1+, 2+, 8+, above 8+ and unassigned charge states were excluded. pLink 261 was used to identify cross-linked peptide pairs with FDR < 5% at the spectrum level, E-value < 1E-7, # of spectrum ≥ 2.
Supplementary information
Acknowledgements
We thank Dr Xiaoqun Wang for helping us to get mouse brain slices, Dr Keping Hu for providing the MeCP2 plasmid as a gift, Master Ting Yao for her help in analytical ultracentrifugation experiments, and Dr Zhaolan Zhou for critical reading and discussion with our manuscript. This work was supported by grants from the Ministry of Science and Technology of China (2017YFA0504202 to G.L.; 2019YFA0508403 to P.L.; 2018YFE0203302 to P.C.), the National Natural Science Foundation of China (31525013, 31630041 and 31521002 to G.L.; 31871443 to P.L.; 31871290 to P.C.). The work was also supported by the Chinese Academy of Sciences (CAS) Strategic Priority Research Program (XDB19040202), the CAS Key Research Program on Frontier Science (QYZDY-SSW-SMC020) and an HHMI International Research Scholar grant (55008737) to G.L. We are particularly grateful to the SLSTU-Nikon Biological Imaging Center (Tsinghua university) for assistance with NIS-Elements AR Analysis, and Center for Bio-imaging & Core Facility for Protein Sciences (Institute of Biophysics, Chinese Academy of Sciences) for fluorescence and electron microscopy imaging data collection.
Author contributions
L.W. carried out phase separation experiments and composed the figures. M.H. was responsible for AUC, EM images and cell fluorescence images. M.-Q.Z. performed CXMS experiments. J.Z. and L.H. generated the mice with the MeCP2 R106W mutation. D.W. helped with protein purification. Y.W. performed the statistical analysis of clinical data. Y.L. took part in the plasmid construction. P.C. helped with CXMS experiments and guided nucleosome assembly experiments. X.B. helped to discuss the project. M.-Q.D. guided CXMS experiments and data interpretation. G.L. and P.L. conceived and supervised the project, analyzed the data and wrote the manuscript with L.W. and M.H.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Liang Wang, Mingli Hu
Contributor Information
Guohong Li, Email: liguohong@sun5.ibp.ac.cn.
Pilong Li, Email: pilongli@mail.tsinghua.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-020-0288-7.
References
- 1.Lyst MJ, Bird A. Rett syndrome: a complex disorder with simple roots. Nat. Rev. Genet. 2015;16:261–275. doi: 10.1038/nrg3897. [DOI] [PubMed] [Google Scholar]
- 2.Hagberg B, Aicardi J, Dias K, Ramos O. A progressive syndrome of autism, dementia, ataxia, and loss of purposeful hand use in girls: Rett’s syndrome: report of 35 cases. Ann. Neurol. 1983;14:471–479. doi: 10.1002/ana.410140412. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong DD. Neuropathology of Rett syndrome. Ment. Retard. Dev. Disabil. Res. Rev. 2002;8:72–76. doi: 10.1002/mrdd.10027. [DOI] [PubMed] [Google Scholar]
- 4.Amir RE, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 5.Lewis JD, et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 6.Meehan RR, Lewis JD, Bird AP. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992;20:5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klose RJ, et al. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol. Cell. 2005;19:667–678. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 8.Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 9.Thambirajah AA, et al. MeCP2 binds to nucleosome free (linker DNA) regions and to H3K9/H3K27 methylated nucleosomes in the brain. Nucleic Acids Res. 2012;40:2884–2897. doi: 10.1093/nar/gkr1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skene PJ, et al. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol. Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones PL, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998;19:187. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 12.Nan X, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 13.Wade PA. Methyl CpG-binding proteins and transcriptional repression. BioEssays. 2001;23:1131–1137. doi: 10.1002/bies.10008. [DOI] [PubMed] [Google Scholar]
- 14.Koerner MV, et al. Toxicity of overexpressed MeCP2 is independent of HDAC3 activity. Genes Dev. 2018;32:1514–1524. doi: 10.1101/gad.320325.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikitina T, et al. Multiple modes of interaction between the methylated DNA binding protein MeCP2 and chromatin. Mol. Cell. Biol. 2007;27:864–877. doi: 10.1128/MCB.01593-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgel PT, et al. Chromatin compaction by human MeCP2. J. Biol. Chem. 2003;278:32181–32188. doi: 10.1074/jbc.M305308200. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh RP, et al. Unique physical properties and interactions of the domains of methylated DNA binding protein 2. Biochemistry. 2010;49:4395–4410. doi: 10.1021/bi9019753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, et al. Histone modifications regulate chromatin compartmentalization by contributing to a phase separation mechanism. Mol. Cell. 2019;76:646–659. doi: 10.1016/j.molcel.2019.08.019. [DOI] [PubMed] [Google Scholar]
- 19.Gibson BA, et al. Organization of chromatin by intrinsic and regulated phase separation. Cell. 2019;179:470–484.e421. doi: 10.1016/j.cell.2019.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larson AG, et al. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature. 2017;547:236–240. doi: 10.1038/nature22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larson AG, Narlikar GJ. The role of phase separation in heterochromatin formation, function, and regulation. Biochemistry. 2018;57:2540–2548. doi: 10.1021/acs.biochem.8b00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strom AR, et al. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241–245. doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brero A, et al. Methyl CpG-binding proteins induce large-scale chromatin reorganization during terminal differentiation. J. Cell Biol. 2005;169:733–743. doi: 10.1083/jcb.200502062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agarwal N, et al. MeCP2 Rett mutations affect large scale chromatin organization. Hum. Mol. Genet. 2011;20:4187–4195. doi: 10.1093/hmg/ddr346. [DOI] [PubMed] [Google Scholar]
- 25.Agarwal N, et al. MeCP2 interacts with HP1 and modulates its heterochromatin association during myogenic differentiation. Nucleic Acids Res. 2007;35:5402–5408. doi: 10.1093/nar/gkm599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guy J, Cheval H, Selfridge J, Bird A. The role of MeCP2 in the brain. Annu. Rev. Cell Dev. Biol. 2011;27:631–652. doi: 10.1146/annurev-cellbio-092910-154121. [DOI] [PubMed] [Google Scholar]
- 27.Baker SA, et al. An AT-hook domain in MeCP2 determines the clinical course of Rett syndrome and related disorders. Cell. 2013;152:984–996. doi: 10.1016/j.cell.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sperlazza MJ, Bilinovich SM, Sinanan LM, Javier FR, Williams DC., Jr. Structural basis of MeCP2 distribution on Non-CpG methylated and hydroxymethylated DNA. J. Mol. Biol. 2017;429:1581–1594. doi: 10.1016/j.jmb.2017.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Damen D, Heumann R. MeCP2 phosphorylation in the brain: from transcription to behavior. Biol. Chem. 2013;394:1595–1605. doi: 10.1515/hsz-2013-0193. [DOI] [PubMed] [Google Scholar]
- 30.Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh RP, Horowitz-Scherer RA, Nikitina T, Gierasch LM, Woodcock CL. Rett syndrome-causing mutations in human MeCP2 result in diverse structural changes that impact folding and DNA interactions. J. Biol. Chem. 2008;283:20523–20534. doi: 10.1074/jbc.M803021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Casas-Delucchi CS, Becker A, Bolius JJ, Cardoso MC. Targeted manipulation of heterochromatin rescues MeCP2 Rett mutants and re-establishes higher order chromatin organization. Nucleic Acids Res. 2012;40:e176. doi: 10.1093/nar/gks784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krishnaraj R, Ho G, Christodoulou J. RettBASE: Rett syndrome database update. Hum. Mutat. 2017;38:922–931. doi: 10.1002/humu.23263. [DOI] [PubMed] [Google Scholar]
- 34.Lombardi LM, Baker SA, Zoghbi HY. MECP2 disorders: from the clinic to mice and back. J. Clin. Investig. 2015;125:2914–2923. doi: 10.1172/JCI78167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shah RR, Bird AP. MeCP2 mutations: progress towards understanding and treating Rett syndrome. Genome Med. 2017;9:17. doi: 10.1186/s13073-017-0411-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu D, Sakurai F, Corey DR. Clonal Rett syndrome cell lines to test compounds for activation of wild-type MeCP2 expression. Bioorg. Med. Chem. Lett. 2011;21:5202–5205. doi: 10.1016/j.bmcl.2011.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosh RP, Horowitz-Scherer RA, Nikitina T, Shlyakhtenko LS, Woodcock CL. MeCP2 binds cooperatively to its substrate and competes with histone H1 for chromatin binding sites. Mol. Cell. Biol. 2010;30:4656–4670. doi: 10.1128/MCB.00379-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tillotson R, Bird A. The molecular basis of MeCP2 function in the brain. J. Mol. Biol. 2019 doi: 10.1016/j.jmb.2019.10.004. [DOI] [PubMed] [Google Scholar]
- 39.Lyst MJ, et al. Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nat. Neurosci. 2013;16:898–902. doi: 10.1038/nn.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heckman, L. D., Chahrour, M. H. & Zoghbi, H. Y. Rett-causing mutations reveal two domains critical for MeCP2 function and for toxicity in MECP2 duplication syndrome mice. Elife3, 10.7554/eLife.02676 (2014). [DOI] [PMC free article] [PubMed]
- 41.Nikitina T, et al. MeCP2-chromatin interactions include the formation of chromatosome-like structures and are altered in mutations causing Rett syndrome. J. Biol. Chem. 2007;282:28237–28245. doi: 10.1074/jbc.M704304200. [DOI] [PubMed] [Google Scholar]
- 42.Turner AL, et al. Highly disordered histone H1-DNA model complexes and their condensates. Proc. Natl. Acad. Sci. USA. 2018;115:11964–11969. doi: 10.1073/pnas.1805943115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feric M, et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165:1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Putnam A, Cassani M, Smith J, Seydoux G. A gel phase promotes condensation of liquid P granules in Caenorhabditis elegans embryos. Nat. Struct. Mol. Biol. 2019;26:220–226. doi: 10.1038/s41594-019-0193-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibbs EB, Kriwacki RW. Linker histones as liquid-like glue for chromatin. Proc. Natl. Acad. Sci. USA. 2018;115:11868–11870. doi: 10.1073/pnas.1816936115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar A, et al. Analysis of protein domains and Rett syndrome mutations indicate that multiple regions influence chromatin-binding dynamics of the chromatin-associated protein MECP2 in vivo. J. Cell Sci. 2008;121:1128–1137. doi: 10.1242/jcs.016865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horike S. [How the methyl-CpG binding protein-related epigenetic disease turns on the genes that produce its symptoms] Tanpakushitsu Kakusan Koso. 2005;50:978–984. [PubMed] [Google Scholar]
- 48.Mohan A, et al. Analysis of molecular recognition features (MoRFs) J. Mol. Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 49.Lever MA, Th’ng JP, Sun X, Hendzel MJ. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature. 2000;408:873–876. doi: 10.1038/35048603. [DOI] [PubMed] [Google Scholar]
- 50.Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 51.Akhmanova A, Verkerk T, Langeveld A, Grosveld F, Galjart N. Characterisation of transcriptionally active and inactive chromatin domains in neurons. J. Cell Sci. 2000;113:4463–4474. doi: 10.1242/jcs.113.24.4463. [DOI] [PubMed] [Google Scholar]
- 52.Della Ragione F, Vacca M, Fioriniello S, Pepe G, D’Esposito M. MECP2, a multi-talented modulator of chromatin architecture. Brief. Funct. Genom. 2016;15:420–431. doi: 10.1093/bfgp/elw023. [DOI] [PubMed] [Google Scholar]
- 53.Pande A, et al. Decrease in protein solubility and cataract formation caused by the Pro23 to Thr mutation in human gamma D-crystallin. Biochemistry. 2005;44:2491–2500. doi: 10.1021/bi0479611. [DOI] [PubMed] [Google Scholar]
- 54.Dorsaz N, Thurston GM, Stradner A, Schurtenberger P, Foffi G. Phase separation in binary eye lens protein mixtures. Soft Matter. 2011;7:1763–1776. [Google Scholar]
- 55.Li G, et al. Highly compacted chromatin formed in vitro reflects the dynamics of transcription activation in vivo. Mol. Cell. 2010;38:41–53. doi: 10.1016/j.molcel.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dyer PN, et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 57.Song F, et al. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344:376–380. doi: 10.1126/science.1251413. [DOI] [PubMed] [Google Scholar]
- 58.Chen P, et al. H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes Dev. 2013;27:2109–2124. doi: 10.1101/gad.222174.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li W, et al. FACT remodels the tetranucleosomal unit of chromatin fibers for gene transcription. Mol. Cell. 2016;64:120–133. doi: 10.1016/j.molcel.2016.08.024. [DOI] [PubMed] [Google Scholar]
- 60.Modzelewski AJ, et al. Efficient mouse genome engineering by CRISPR-EZ technology. Nat. Protoc. 2018;13:1253–1274. doi: 10.1038/nprot.2018.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen ZL, et al. A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides. Nat. Commun. 2019;10:3404. doi: 10.1038/s41467-019-11337-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.