Abstract
Dysregulation of histone deacetylase 6 (HDAC6) can lead to the pathologic states and result in the development of various diseases including cancers and inflammatory diseases. The objective of this study was to elucidate the regulatory role of microRNA-22 (miR-22) in HDAC6-mediated expression of pro-inflammatory cytokines in lipopolysaccharide (LPS)-stimulated macrophages. LPS stimulation induced HDAC6 expression, but suppressed miR-22 expression in macrophages, suggesting possible correlation between HDAC6 and miR-22. Luciferase reporter assays revealed that 3’UTR of HDAC6 was a bona fide target site of miR-22. Transfection of miR-22 mimic significantly inhibited LPS-induced HDAC6 expression, while miR-22 inhibitor further increased LPS-induced HDAC6 expression. LPS-induced activation of NF-κB and AP-1 was inhibited by miR-22 mimic, but further increased by miR-22 inhibitor. LPS-induced expression of pro-inflammatory cytokines such as TNF-α, IL-1β, and IL-6 was inhibited by miR-22 mimic, but further increased by miR-22 inhibitor. Taken together, these data provide evidence that miR-22 can downregulate LPS-induced expression of pro-inflammatory cytokines via suppression of NF-κB and AP-1 axis by targeting HDAC6 in macrophages.
Keywords: Cytokine, HDAC6, LPS, Macrophages, miR-22
INTRODUCTION
Inflammatory immune response is one of first lines of defense against pathogens that involve various immune cells such as neutrophils, monocytes, and macrophages (1). However, dysregulated and chronic activation of inflammatory immune responses can result in various diseases including cancers (2). Macrophages are principal cells that contribute to the inflammation process by producing various pro-inflammatory mediators such as cytokines (3). Elucidation of cellular devices to control expression of inflammatory mediators can provide novel molecular targets against various inflammatory diseases.
Histone deacetylase 6 (HDAC6), a member of the class IIb HDACs, plays an important role in inflammatory immune responses (4, 5). Growing evidences suggest that HDAC6 is a critical player in mediating immune responses upon stimulation with various microbial products, such as lipopolysaccharide (LPS), Clostridium difficile toxin A, and HIV-1 Tat (4,6-8). Deacetylase activity of HDAC6 has also been shown to be involved in LPS-induced activation of macrophages (4) and macrophage infiltration in a mouse model of acute peritonitis (5). TLR4 stimulation with LPS can induce the activation of nuclear factor-kappa B (NF-κB) and activator protein-1 (AP-1) signaling cascades, leading to expression of pro-inflammatory mediators including cytokines in macrophages (9). It has been reported that HDAC6 is a master regulator of the expression of pro-inflammatory mediators by modulating NF-κB and AP-1 axis in macrophages (10).
MicroRNAs (miRNAs) are involved in various biological activity by silencing specific target messenger RNAs. Among them, miR-22 has been reported to be able to regulate innate immune responses by regulating production of inflammatory cytokines in several studies (11-13), suggesting its pro-inflammatory activity. Overexpression of miR-22 can suppress TLR3-mediated expression of pro-inflammatory cytokines via inhibition of interferon regulatory factor-3 and NF-κB by targeting mitochondrial antiviral signaling protein (MAVS) in glial cells (11). Overexpression of miR-22 can exert protective effects against myocardial and cerebral ischemia-reperfusion injury by down-regulating inflammatory cytokines (12, 13). However, miR-22 is involved in Th17 responses by targeting HDAC4 in lung myeloid dendritic cells of smokers (14), suggesting its pro-inflammatory activity. Taken together, these studies suggest that miR-22 can exert anti-inflammatory or pro-inflammatory activities depending on cell type and stimulus.
After screening selected miRNAs targeting HDAC6, several miRNAs including miR-22 were found to be putative regulators of HDAC6 in this study. We focused on miR-22 in the present study due to its immunomodulatory activity (11, 13). Whether HDAC6 might be a candidate target of miR-22 during TLR4-mediated immune responses was then investigated. The role and mechanisms of miR-22 in LPS-induced expression of pro-inflammatory cytokines in macrophages were also explored. Our results revealed that LPS stimulation increased HDAC6 expression with concomitant downregulation of miR-22 in macrophages. Luciferase reporter assays revealed that HDAC6 is a bona fide target site of miR-22. Using miRNA-22 mimic and inhibitor, it was demonstrated that miR-22 could downregulate LPS-induced cytokines TNF-α, IL-1β, and IL-6 via suppression of NF-κB and AP-1 axis by targeting HDAC6 in macrophages.
RESULTS
Suppression of miR-22 expression with concomitant upregulation of HDAC6 in LPS-stimulated macrophages
Previous studies have revealed that HDAC6 is involved in the immune response to bacterial and viral infection (6, 15). In addition, HDAC6 is a master regulator of the expression of pro-inflammatory mediators in macrophages (10). To find out regulatory mechanisms involved in HDAC6-mediated immune responses, microRNAs that could target HDAC6 were screened. Using TargetScan, a miR target prediction database, miR-22 was found to be a putative regulator of HDAC6. Supporting this notion, recent studies have reported that overexpression of miR-22 can reduce the level of HDAC6 protein by targeting 3’UTR of HDAC6 mRNA (16, 17). Thus, there might be a possible correlation between HDAC6 and miR-22 in LPS-stimulated macrophages. To examine the expression pattern of HDAC6 and miR-22 upon LPS stimulation, RAW 264.7 cells were treated with LPS and then levels of miR-22 and HDAC6 expression were analyzed by qRT-PCR and Western blot. As shown in Fig. 1A, the level of miR-22 was decreased, while the level of HDAC6 mRNA was increased in a dose-dependent manner in LPS-stimulated RAW 264.7 cells. Similarly, LPS stimulation increased HDAC6 protein expression with concomitant decrease of acetylated α-tubulin in RAW 264.7 cells (Fig. 1A). These results suggest a reverse correlation between HDAC6 and miR-22 in LPS-stimulated macrophages.
Fig. 1.
Reverse correlation between HDAC6 and miR-22 in LPS-stimulated macrophages and HDAC6 as a direct target gene of miR-22. (A) RAW 264.7 cells were exposed to LPS at various doses for 3 h (for mRNA) or 24 h (for protein). Total RNA was extracted from cells and analyzed for mRNA of HDAC6, miR-22, and β-actin by quantitative RT-PCR using specific primers. Data are shown as mean ± SD of three independent experiments. **P < 0.01, ***P < 0.001, as compared to control cells. Cell lysates were prepared and analyzed for protein expression of HDAC6, acetylated α-tubulin, and β-actin by Western blotting. (B) Schematic representation showing the luciferase reporter carrying wild type or mutant 3’UTR (untranslational region) of HDAC6 and the location of putative miR-22 binding sites in the 3’UTR of HDAC6 mRNA. (C) RAW 264.7 cells were co-transfected with luciferase reporter plasmids and miR-22 mimic or anti-miR-22 at 50 nM concentration. 24 h post-transfection, luciferase activities were evaluated using dual-luciferase assays and expressed as fold induction over the control (vehicle alone). Data are presented as mean ± SD of three independent experiments. ***P < 0.001, †††P < 0.001, as compared to the cells treated with vehicle alone.
HDAC6 is a direct target gene of miR-22 in macrophages
According to computational analysis, HDAC6 has a 7-nt seed region which is complementary to miR-22 within its 3’UTR (Fig. 1B). To determine whether HDAC6 is a direct target of miR-22, we cloned wild type- or mutant 3’UTR of HDAC6 into a luciferase reporter and then performed luciferase reporter assays in RAW 264.7 cells. Cells were transfected with wild type or mutant HDAC6-3’UTR luciferase reporter plasmid and a miR-22 mimic or inhibitor. Overexpression of miR-22 decreased luciferase activity comparing to control group, while inhibition of miR-22 increased luciferase activity (Fig. 1C). However, these effects were not observed when cells were transfected with mutant HDAC6-3’UTR luciferase reporter plasmid. These results suggest that 3’UTR of HDAC6 is a bona fide target site of miR-22 in macrophages.
Effects of miR-22 on LPS-induced HDAC6 expression and subsequent acetylated α-tubulin in macrophages
Based on our finding that miR-22 directly targets HDAC6, we further examined the HDAC6 mRNA level. The decreased and increased HDAC6 mRNA levels were noted in the cells transfected with miR-22 mimic and miR-22 inhibitor, respectively, without LPS stimulation, suggesting that HDAC6 expression can be regulated by both mRNA degradation and translational repression (Fig. 2A and 2B). We next investigated the effect of miR-22 on LPS-induced HDAC6 expression and acetylated α-tubulin levels. Transfection of miR-22 mimic significantly inhibited LPS-induced level of HDAC6 mRNA (Fig. 2A). MiR-22 inhibitor further increased LPS-induced HDAC6 mRNA level (Fig. 2B). Similar to this result, miR-22 mimic significantly inhibited LPS-induced HDAC6 protein expression, with concomitant recovery of reduced level of acetylated α-tubulin (Fig. 2C). However, miR-22 inhibitor further increased LPS-induced HDAC6 protein expression (Fig. 2D).
Fig. 2.

The effect of miR-22 on LPS-induced HDAC6 expression and subsequent acetylated α-tubulin in macrophages. RAW 264.7 cells were transfected with miR-22 mimic (A) or anti-miR-22 (B) at 50 nM concentration. 24 h post-transfection, cells were stimulated with LPS (100 ng/ml) for 3 h. Total RNA was analyzed for mRNA level of HDAC6 and β-actin by qRT-PCR. Data are shown as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001, as compared to the cells treated with vehicle alone. Cells transfected with miR-22 mimic (C) or anti-miR-22 (D) were stimulated with LPS (100 ng/ml) for 24 h. Cell lysates were analyzed for protein levels of HDAC6, acetylated α-tubulin, and β-actin by Western blotting.
Effects of miR-22 on LPS-induced activation of NF-κB and AP-1 in macrophages
Both NF-κB and AP-1 are major transcriptional factors participating in inflammatory immune responses upon LPS stimulation (9). Therefore, we next investigated effects of miR-22 on signaling cascades leading to NF-κB and AP-1 activation. As shown in Fig. 3A, miR-22 overexpression reduced LPS-induced NF-κB p65 phosphorylation and restored LPS-induced IκBα degradation in macrophages. We also observed that miR-22 inhibition further amplified effects of LPS on NF-κB p65 phosphorylation and IκBα degradation (Fig. 3A). Similarly, miR-22 mimic inhibited LPS-induced c-Jun phosphorylation, while miR-22 inhibitor further increased LPS-induced c-Jun phosphorylation (Fig. 3B). In addition, LPS-induced promoter activities of NF-κB (Fig. 3C) and AP-1 (Fig. 3D) were alleviated by miR-22 mimic, but further increased by miR-22 inhibitor. These data suggest that miR-22 can modulate TLR4-mediated immune response by targeting NF-κB and AP-1 signaling cascades.
Fig. 3.
The effect of miR-22 on LPS-induced activation of NF-κB and AP-1 in macrophages. RAW 264.7 cells were transfected with miR-22 mimic or anti-miR-22. 24 h post-transfection, cells were stimulated with LPS (100 ng/ml) for 1 h. Whole cell lysates were prepared and analyzed for phosphor-p65 (p-p65) and IκBα (A) or phosphor-c-Jun (p-c-Jun) (B) by Western blotting using indicated antibodies. Cells were transiently co-transfected with a NF-κB (C) or AP-1 (D) promoter-luciferase plasmid and miR-22 mimic or anti-miR-22. After 24 h, the transfected cells were stimulated with LPS for 24 h, and the luciferase activity was measured. Luciferase activities were normalized to the β-galactosidase activities and expressed as fold induction over the control (vehicle alone). Data are presented as mean ± SD of three independent experiments. †P < 0.05, ***P < 0.001, †††P < 0.001, as compared to the control cells treated with vehicle alone.
Effects of miR-22 on LPS-induced expression of pro-inflammatory cytokines in macrophages
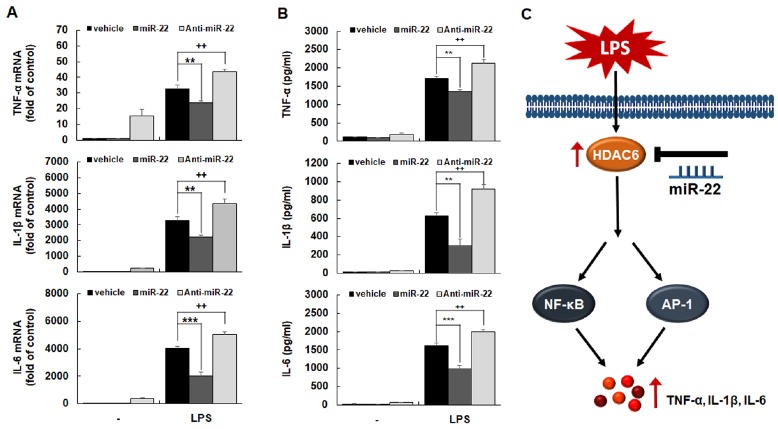
We next investigated the involvement of miR-22 in expression of pro-inflammatory cytokines such TNF-α, IL-1β, and IL-6 in LPS-stimulated macrophages. Cells were transfected with miR-22 mimic or miR-22 inhibitor followed by LPS stimulation. Expression levels of TNF-α, IL-1β, and IL-6 were then evaluated by RT-qPCR and ELISA. As shown in Fig. 4, miR-22 mimic significantly decreased expression levels of TNF-α, IL-1β, and IL-6 at both mRNA (Fig. 4A) and protein (Fig. 4B) levels, while miR-22 inhibitor further increased levels of these cytokines. Therefore, these studies provide evidence that miR-22 could inhibit LPS-induced expression of pro-inflammatory cytokines such as TNF-α, IL-1β, and IL-6 by modulating HDAC6-NF-κB/AP-1 signaling pathways in macrophages (Fig. 4C).
Fig. 4.
The effect of miR-22 on LPS-induced expression of pro-inflammatory cytokines in macrophages. RAW 264.7 cells were transfected with miR-22 mimic or anti-miR-22. 24 h post-transfection, cells were stimulated with LPS (100 ng/ml) for 3 h (for mRNA expression) or 24 h (for protein expression). Total RNA was analyzed by quantitative RT-PCR (A). Culture media of cells were collected and assayed for TNF-α, IL-1β, and IL-6 by ELISA (B). Data are shown as mean ± SD of three independent experiments. **P < 0.01, ††P < 0.01, ***P < 0.001, as compared to the control cells treated with vehicle alone. (C) Schematic diagram showing negative regulation of miR-22 on LPS-induced inflammatory response. Up-regulation of miR-22 decreases LPS-induced HDAC6 expression, and subsequent activation of NF-κB and AP-1, leading to suppression of production of pro-inflammatory chemokines, such as TNF-α, IL-1β, and IL-6, in macrophages.
DISCUSSION
HDAC6 is an important regulator in inflammatory responses upon stimulation with various bacterial and viral products (4,6-8). HDAC6 contributes to the production of inflammatory mediators by modulating NF-κB/AP-1 axis in macrophages (10). Therefore, regulatory strategies targeting HDAC6 provide a rationale to develop novel molecular therapeutics against various inflammatory diseases.
miRNAs play a role as central modulators of immune responses against various pathogens by regulating production of inflammatory mediators (18). In the present study, we screened several miRNAs targeting HDAC6 using TargetScan, and found that some miRNAs were putative regulators of HDAC6. MiR-22 has been reported to possess immunomodulatory activity (11, 13). Therefore, it was chosen for the present study. We observed an inverse correlation between HDAC6 and miR-22 in LPS-stimulated macrophages. Our results showed that 3’UTR of HDAC6 was a bona fide target site of miR-22 in macrophages. Experiments using miRNA-22 mimic and inhibitor revealed that miR-22 could regulate LPS-induced expression of pro-inflammatory cytokines by targeting HDAC6-NF-κB/AP-1 axis in macrophages (Fig. 4C).
Several studies have suggested the presence of an inverse correlation between HDAC6 and miR-22 expression in various biological processes (16, 17, 19, 20). For example, inverse correlation between HDAC6 and miR-22 has been reported in several types of cancer tissues and cells including cervical cancer and cholangiocarcinoma (19, 20). Inverse relationship between HDAC6 and miR-22 expression has also been observed in osteogenic and adipogenic differentiation of human adipose-tissue-derived mesenchymal stem cells (hADMSCs) (16, 17). In the present study, we also observed a significant reverse correlation between HDAC6 and miR-22 expression in LPS-stimulated macrophages. Taken together, these results suggest that the inverse regulation of HDAC6 and miR-22 might play important roles in various biological processes.
Growing evidences suggest that miR-22 can directly regulates expression levels of HDAC6 in various biological contexts. It has been reported that miR-22 can decrease HDAC6 protein level by targeting its 3’UTR, resulting in the regulation of adipogenic/osteogenic differentiation of hADMSCs (16). In a cellular model of osteogenic differentiation, it has been shown that miR‐22 can suppress HDAC6 protein expression by targeting its 3’UTR in human periodontal ligament stem cells, thus affecting the osteogenic differentiation process (17). In addition, miR-22 can down-regulate the level of HDAC6 mRNA by targeting its 3'UTR in cervical cancer cells by acting as a tumor suppressor (19). Taken together, these results suggest that HDAC6 is one of target genes of miR-22.
MiR-22 may regulate innate immune responses via various distinct mechanisms. Previous studies reported that miR-22 plays a critical roles in innate immune responses by directly targeting an inflammatory mediator or regulating signaling pathways involved in the production of inflammatory mediators (21-23). It has been shown that miR-22 can downregulate p38 protein expression by directly binding to the 3’UTR of its mRNA, thus interfering with IL-6 production and Th17 differentiation derived by dendritic cells (21). In addition, miR-22 mediates vascular inflammation by targeting ICAM-1 mRNA in endothelial cells (22). It has been shown that miR-22 is involved in inflammatory response by targeting MCP-1 mRNA in peripheral blood mononuclear cells (23). Along with our data showing that miR-22 downregulates LPS-induced expression of pro-inflammatory cytokines by targeting HDAC6, these results suggest that miR-22 plays a critical role in modulating innate immune responses.
Elucidation of the molecular mechanisms by which miRNAs regulate inflammatory immune responses can provide the insight into strategies for developing therapeutic targets against inflammatory diseases. Our present study reveals the evidence that miR-22 can inhibit LPS-induced inflammatory response by modulating HDAC6-NF-κB/AP-1 axis, leading to suppression of pro-inflammatory cytokines in macrophages.
MATERIALS AND METHODS
Reagents
Antibodies against acetylated α-tubulin, phosphor-p65, IκBα, and phosphor-c-jun were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Antibodies against β-actin and HDAC6 were obtained from Cell Signaling Technology (Beverly, MA, U.S.A.). Oligonucleotide primers for HDAC6, miR-22, TNF-α, IL-1β, IL-6, and β-actin) were obtained from commercially (Bioneer, Seoul, Korea). Lipopolysaccharide (LPS, Escherichia coli 0127:B8) was purchased from Sigma (St. Louis, MO, USA).
Cell culture
RAW 264.7 murine macrophages were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and grown in Dulbecco’s modified Eagle’s medium (Gibco Laboratories, NY, USA) supplemented with 10% fetal bovine serum (Hyclone, Victoria, Australia) and antibiotics (100 U/ml penicillin G, 100 μg/ml streptomycin, Gibco Laboratories, NY, USA) at 37°C in a humidified incubator containing 5% CO2 and 95% air (9).
Western blot analysis
Cell lysates were prepared using a buffer containing 125 mM Tris-HCl pH 6.8, 2% SDS, 10% v/v glycerol. The protein concentrations of the cell lysates were determined using a Bradford assay kit (Bio-Rad, Hercules, CA, USA). Thirty micrograms of proteins were subjected to 10% SDS-PAGE, and transferred to nitrocellulose membranes. The blots were probed with the primary antibody against the target protein at 4°C overnight, followed by incubation with the horseradish peroxidase (HRP)-conjugated secondary antibody (1:10,000 dilution) for 2 h at room temperature. The antibody-reactive bands were visualized by a chemiluminescence kit (Amersham Life Sciences, Parsippany, NJ, USA) (24).
Real-time reverse transcription-polymerase chain reaction (RT-PCR) analysis
Total cellular RNA from cells was isolated using a Trizol reagent kit (Invirogen, Gaithersburg, MD, USA). Two micrograms of total RNA were reverse-transcribed into cDNA using 10,000 U of reverse transcriptase and 0.5 μg/μl oligo-(dT)15 primer (Promega). The resulting cDNA was analyzed by quantitative PCR using iQTM SYBRⓇ Green Supermix (BIO-RAD, Hercules, CA, USA) with the following primer sets. Primer sequences were as follows: TNF-α sense, ACA AGC CTG TAG CCC ACG; TNF-α antisense, TCC AAA GTA GAC CTG CCC; IL-6 sense, CAA GAA AGA CAA AGC CAG AGT CCT T; IL-6 antisense, TGG ATG GTC TTG GTC CTT AGC C; IL-1β sense, TGC AGA GTT CCC CAA CTG GTA CAT C; IL-1β antisense, GTG CTG CCT AAT GTC CCC TTG AAT C; β-actin sense, AGT GTG ACG TTG ACA TCC GTA AAG A; and β-actin antisense, GGA CAG TGA GGC CAG GAT GG. PCR amplification and analysis of PCR products were carried out as previously described (10).
Construction of luciferase reporter plasmids and luciferase reporter assay
In order to analyze the effects of miRNA-22 on the mRNA levels of HDAC6, luciferase reporter plasmids carrying a wild-type or mutant binding sequences in 3’UTR of HDAC6 (accession number NM_001130416; the entire length is 349 bp) were constructed based on the psiCHECKTM-2 vector (Promega, Madison, WI, USA) using the following primers, as previously described (25). Primer sequences were as follows: HDAC6 3’UTR, sense 5’-ATA TGC GAT CGC ACT CAC ACT AAA TCC CAG AC-3’, and antisense 5’-ATT AGC GGC CGC CAG GTT TTG TAG CAA CTC CAA-3’; HDAC6-MUT, sense 5’-TGA GGC CAC AGG CAG GGC CCA ATA GTT CAC-3’, and antisense 5’-GTG AAC TAT TGG GCC CTG CCT GTG GCC TCA-3’. RAW 264.7 cells were seeded at a density of 3 × 105 cells/ml in a 6-well plate. After 24 h of incubation, cells were co-transfected with HDAC6-3’-UTR luciferase reporter plasmid and a miR-22 mimic or inhibitor using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s instructions. The levels of luciferase expression were analyzed using the Dual Luciferase Reporter Assay System (Promega, Madison, WI, USA) following the manufacturer’s instructions. To analyze NF-κB or AP-1 promoter activity, transient transfection into cells were performed with pNF-κB-luc or AP-1-luc vector (Stratagene, La Jolla, CA, USA) and pCMV-β-galactosidase vector (Clontech, Mountain View, CA, USA), as previously described (9).
Enzyme-linked immunosorbent assay (ELISA)
Culture media were harvested and the protein levels of TNF-α, IL-1β, and IL-6 in the culture medium were measured using an ELISA kit (R&D systems, Minneapolis, MN, USA), according to the manufacturer's instructions (26) .
Statistical analysis
Results are presented as the mean ± standard deviation (SD) from three independent experiments. Statistical analysis was performed by one-way analysis of variance, followed by Bonferroni’s test using SigmaPlot 10.0 software (SYSTAT Software Inc, Chicago, IL, USA). Differences were considered significant at *P < 0.05, **P < 0.01 and ***P < 0.001.
ACKNOWLEDGEMENTS
This research was supported by Basic Science Research Program Grants (2018R1D1A3B07050435, 2019R1A6A1A11036849) through the National Research Foundation of Korea, funded by the Ministry of Education.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Vijay K. Toll-like receptors in immunity and inflammatory diseases: Past, present, and future. Int Immunopharmacol. 2018;59:391–412. doi: 10.1016/j.intimp.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat Immunol. 2011;12:715–723. doi: 10.1038/ni.2060. [DOI] [PubMed] [Google Scholar]
- 3.Zhang X, Mosser DM. Macrophage activation by endogenous danger signals. J Pathol. 2008;214:161–178. doi: 10.1002/path.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan B, Xie S, Liu Z, et al. HDAC6 deacetylase activity is critical for lipopolysaccharide-induced activation of macrophages. PLoS One. 2014;9:e110718. doi: 10.1371/journal.pone.0110718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan B, Xie S, Liu Y, et al. Histone deacetylase 6 modulates macrophage infiltration during inflammation. Theranostics. 2018;8:2927–2938. doi: 10.7150/thno.25317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nam HJ, Kang JK, Kim SK, et al. Clostridium difficile toxin A decreases acetylation of tubulin, leading to microtubule depolymerization through activation of histone deacetylase 6, and this mediates acute inflammation. J Biol Chem. 2010;285:32888–32896. doi: 10.1074/jbc.M110.162743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang B, Rao YH, Inoue M, et al. Microtubule acetylation amplifies p38 kinase signalling and anti-inflammatory IL-10 production. Nat Commun. 2014;5:3479. doi: 10.1038/ncomms4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Youn GS, Ju SM, Choi SY, Park J. HDAC6 mediates HIV-1 tat-induced proinflammatory responses by regulating MAPK-NF-kappaB/AP-1 pathways in astrocytes. Glia. 2015;63:1953–1965. doi: 10.1002/glia.22865. [DOI] [PubMed] [Google Scholar]
- 9.Kwon DJ, Bae YS, Ju SM, Youn GS, Choi SY, Park J. Salicortin suppresses lipopolysaccharide-stimulated inflammatory responses via blockade of NF-kappaB and JNK activation in RAW 264.7 macrophages. BMB Rep. 2014;47:318–323. doi: 10.5483/BMBRep.2014.47.6.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Youn GS, Lee KW, Choi SY, Park J. Overexpression of HDAC6 induces pro-inflammatory responses by regulating ROS-MAPK-NF-kappaB/AP-1 signaling pathways in macrophages. Free Radic Biol Med. 2016;97:14–23. doi: 10.1016/j.freeradbiomed.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 11.Wan S, Ashraf U, Ye J, et al. MicroRNA-22 negatively regulates poly(I:C)-triggered type I interferon and inflammatory cytokine production via targeting mitochondrial antiviral signaling protein (MAVS) Oncotarget. 2016;7:76667–76683. doi: 10.18632/oncotarget.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu H, Wu M, Zhao P, Huang Y, Wang W, Yin W. Neuroprotective effects of viral overexpression of microRNA-22 in rat and cell models of cerebral ischemia-reperfusion injury. J Cell Biochem. 2015;116:233–241. doi: 10.1002/jcb.24960. [DOI] [PubMed] [Google Scholar]
- 13.Yang J, Fan Z, Yang J, Ding J, Yang C, Chen L. microRNA-22 attenuates myocardial ischemia-reperfusion injury via an anti-inflammatory mechanism in rats. Exp Ther Med. 2016;12:3249–3255. doi: 10.3892/etm.2016.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu W, You R, Yuan X, et al. The microRNA miR-22 inhibits the histone deacetylase HDAC4 to promote T(H) 17 cell-dependent emphysema. Nat Immunol. 2015;16:1185–1194. doi: 10.1038/ni.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chattopadhyay S, Fensterl V, Zhang Y, Veleeparambil M, Wetzel JL, Sen GC. Inhibition of viral pathogenesis and promotion of the septic shock response to bacterial infection by IRF-3 are regulated by the acetylation and phosphorylation of its coactivators. MBio. 2013;4:e00636–12. doi: 10.1128/mBio.00636-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang S, Wang S, Bian C, et al. Upregulation of miR-22 promotes osteogenic differentiation and inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells by repressing HDAC6 protein expression. Stem Cells Dev. 2012;21:2531–2540. doi: 10.1089/scd.2012.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan GQ, Wang X, Yang F, et al. MicroRNA-22 promoted osteogenic differentiation of human periodontal ligament stem cells by targeting HDAC6. J Cell Biochem. 2017;118:1653–1658. doi: 10.1002/jcb.25931. [DOI] [PubMed] [Google Scholar]
- 18.Hirschberger S, Hinske LC, Kreth S. MiRNAs: dynamic regulators of immune cell functions in inflammation and cancer. Cancer Lett. 2018;431:11–21. doi: 10.1016/j.canlet.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 19.Wongjampa W, Ekalaksananan T, Chopjitt P, et al. Suppression of miR-22, a tumor suppressor in cervical cancer, by human papillomavirus 16 E6 via a p53/miR-22/HDAC6 pathway. PLoS One. 2018;13:e0206644. doi: 10.1371/journal.pone.0206644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mansini AP, Lorenzo Pisarello MJ, Thelen KM, et al. MicroRNA (miR)-433 and miR-22 dysregulations induce histone-deacetylase-6 overexpression and ciliary loss in cholangiocarcinoma. Hepatology. 2018;68:561–573. doi: 10.1002/hep.29832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang X, Liu Y, Mei S, et al. MicroRNA-22 impairs anti-tumor ability of dendritic cells by targeting p38. PLoS One. 2015;10:e0121510. doi: 10.1371/journal.pone.0121510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gidlof O, Sathanoori R, Magistri M, et al. Extracellular uridine triphosphate and adenosine triphosphate attenuate endothelial inflammation through miR-22-mediated ICAM-1 inhibition. J Vasc Res. 2015;52:71–80. doi: 10.1159/000431367. [DOI] [PubMed] [Google Scholar]
- 23.Chen B, Luo L, Zhu W, et al. miR-22 contributes to the pathogenesis of patients with coronary artery disease by targeting MCP-1: An observational study. Medicine (Baltimore) 2016;95:e4418. doi: 10.1097/MD.0000000000004418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.An SY, Youn GS, Kim H, Choi SY, Park J. Celastrol suppresses expression of adhesion molecules and chemokines by inhibiting JNK-STAT1/NF-kappaB activation in poly(I:C)-stimulated astrocytes. BMB Rep. 2017;50:25–30. doi: 10.5483/BMBRep.2017.50.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park JK, Doseff AI, Schmittgen TD. MicroRNAs targeting caspase-3 and -7 in PANC-1 cells. Int J Mol Sci. 2018;19:1206. doi: 10.3390/ijms19041206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jo H, Jang HY, Youn GS, et al. Hindsiipropane B alleviates HIV-1 Tat-induced inflammatory responses by suppressing HDAC6-NADPH oxidase-ROS axis in astrocytes. BMB Rep. 2018;51:394–399. doi: 10.5483/BMBRep.2018.51.8.061. [DOI] [PMC free article] [PubMed] [Google Scholar]