Abstract
l‐Tyrosine (Tyr) is an aromatic amino acid synthesized de novo in plants and microbes downstream of the shikimate pathway. In plants, Tyr and a Tyr pathway intermediate, 4‐hydroxyphenylpyruvate (HPP), are precursors to numerous specialized metabolites, which are crucial for plant and human health. Tyr is synthesized in the plastids by a TyrA family enzyme, arogenate dehydrogenase (ADH/TyrAa), which is feedback inhibited by Tyr. Additionally, many legumes possess prephenate dehydrogenases (PDH/TyrAp), which are insensitive to Tyr and localized to the cytosol. Yet the role of PDH enzymes in legumes is currently unknown. This study isolated and characterized Tnt1‐transposon mutants of MtPDH1 (pdh1) in Medicago truncatula to investigate PDH function. The pdh1 mutants lacked PDH transcript and PDH activity, and displayed little aberrant morphological phenotypes under standard growth conditions, providing genetic evidence that MtPDH1 is responsible for the PDH activity detected in M. truncatula. Though plant PDH enzymes and activity have been specifically found in legumes, nodule number and nitrogenase activity of pdh1 mutants were not significantly reduced compared with wild‐type (Wt) during symbiosis with nitrogen‐fixing bacteria. Although Tyr levels were not significantly different between Wt and mutants under standard conditions, when carbon flux was increased by shikimate precursor feeding, mutants accumulated significantly less Tyr than Wt. These data suggest that MtPDH1 is involved in Tyr biosynthesis when the shikimate pathway is stimulated and possibly linked to unidentified legume‐specific specialized metabolism.
Keywords: legumes, prephenate dehydrogenase, the shikimate pathway, TyrA dehydrogenase, tyrosine
1. INTRODUCTION
l‐Tyrosine (Tyr) is an aromatic amino acid synthesized de novo in plants and microbes, but not animals; thus, humans must acquire Tyr through their diet or by enzymatic conversion of l‐phenylalanine (Phe, Fitzpatrick, 1999). In addition to its involvement in protein synthesis, Tyr and a Tyr‐pathway intermediate 4‐hydroxyphenylpyruvate (HPP) are the precursors to numerous specialized metabolites crucial for plant and animal health (Schenck & Maeda, 2018). Tyr‐derived plant specialized metabolites have roles as photosynthetic electron carriers (plastoquinone, Metz, Nixon, Rogner, Brudvig, & Diner, 1989), pollinator attractants (betalain pigments, Strack, Vogt, & Schliemann, 2003) and defense compounds (dhurrin and rosmarinic acidMøller, 2010; Petersen, 2013). In grasses, Tyr also serves as a precursor to lignin, the main phenolic polymer in plants (Barros et al., 2016; Higuchi, Ito, & Kawamura, 1967; Rosler, Krekel, Amrhein, & Schmid, 1997). Humans have co‐opted some of these natural products to serve nutritional and medicinal roles such as some benzylisoquinoline alkaloids, which have antitussive, analgesic, and antimicrobial activities (Barken, Geller, & Rogosnitzky, 2008; Beaudoin & Facchini, 2014; Kries & O’Connor, 2016), and the antioxidant properties of tocopherols (collectively referred to as vitamin E, Bramley et al., 2000).
The aromatic amino acids (AAAs; Tyr, Phe, and tryptophan [Trp]) are synthesized in the plastids from chorismate, the final product of the shikimate pathway (Maeda & Dudareva, 2012; Tzin & Galili, 2010). Chorismate mutase (CM, EC number 5.4.99.5) catalyzes the committed step of Tyr and Phe synthesis—the pericyclic Claisen rearrangement of chorismate into prephenate (Eberhard et al., 1996; Goers & Jensen, 1984; Kuroki & Conn, 1989; Mobley, Kunkel, & Keith, 1999). Prephenate is converted into Tyr via two reactions, oxidative decarboxylation catalyzed by a TyrA dehydrogenase enzyme and transamination. These reactions can occur in either order, leading to alternative Tyr pathways (Figure 1, Schenck & Maeda, 2018). In most microbes, a prephenate‐specific TyrA dehydrogenase (PDH/TyrAp EC 1.3.1.13, Figure 1) first converts prephenate into HPP followed by transamination to Tyr via Tyr‐aminotransferase (Tyr‐AT or TyrB, EC2.6.1.5). In plants, these reactions occur in the reverse order with transamination of prephenate to form arogenate by a plastidic prephenate aminotransferase (PPA‐AT, EC 2.6.1.78), followed by oxidative decarboxylation catalyzed by a plastidic arogenate‐specific TyrA dehydrogenase (ADH/TyrAa EC 1.3.1.78) producing Tyr (Figure 1). The ADH‐mediated Tyr pathway appears to be essential for normal growth and development in plants as indicated by the severe phenotype of Arabidopsis thaliana adh2/tyra2 knockout mutant, which was further exacerbated by transient suppression of the other ADH1/TyrA1 gene (de Oliveira et al., 2019). PDH and ADH are the key enzymes in their respective pathways, as they compete for substrates that are shared with Phe biosynthesis, and are generally feedback inhibited by Tyr (Connelly & Conn, 1986; Gaines, Byng, Whitaker, & Jensen, 1982; Rippert & Matringe, 2002; Rubin & Jensen, 1979).
Figure 1.
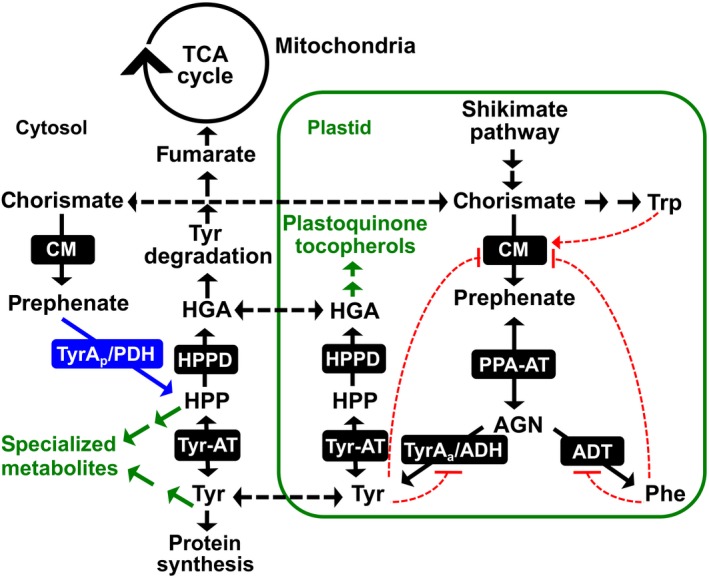
Tyr biosynthesis and associated pathways in legumes. In addition to a highly regulated plastid localized ADH pathway for Tyr biosynthesis, legumes possess a Tyr‐insensitive, cytosolic PDH enzyme (blue). Tyr synthesized in the plastids is exported into the cytosol where it can be incorporated into proteins, enter the Tyr degradation pathway to the TCA cycle or serve as a precursor to specialized metabolism. Black dotted lines denote known or potential transport steps, and red dotted lines represent feedback regulation with arrows meaning induction and hashes inhibition. AGN, arogenate; ADT, arogenate dehydratase; CM, chorismate mutase; HGA, homogentisate; HPP, 4‐hydroxyphenylpyruvate; HPPD, HPP dioxygenase; PPA‐AT, prephenate aminotransferase; TyrAa/ADH, arogenate dehydrogenase; TyrAp/PDH, prephenate dehydrogenase; Tyr‐AT, tyrosine aminotransferase.
PDH activity, which is commonly found in microbes, has been detected in tissue extracts of some plants, all restricted to the legume family (Gamborg & Keeley, 1966; Rubin & Jensen, 1979; Siehl, 1999). In soybean, leaf tissue had the highest PDH activity of all analyzed tissues (Schenck, Chen, Siehl, & Maeda, 2015). Phylogenetic analyses of plant and microbial TyrAs showed that PDH genes are uniquely present in legumes, suggesting that legume PDH enzymes are the result of a recent legume‐specific gene duplication event of a plant ADH rather than horizontal gene transfer from a PDH‐possessing rhizobia (Schenck et al., 2015). However, not all legumes possess PDH, which suggests gene loss in some legumes (Schenck et al., 2015; Schenck, Holland, et al., 2017). PDH recombinant enzymes from Glycine max (soybean; GmPDH1 and GmPDH2) and Medicago truncatula (Medicago; MtPDH1) preferred prephenate over arogenate as their substrates (Schenck et al., 2015; Schenck, Holland, et al., 2017) and, unlike plant ADH enzymes, were insensitive to Tyr inhibition and localized to the cytosol (Schenck et al., 2015; Schenck, Holland, et al., 2017). These recently diverged plant ADH and PDH enzymes were used to identify a single amino acid residue (Asn222 of GmPDH1) of TyrA dehydrogenases that switches TyrA substrate specificity and underlies the evolution of legume PDH enzymes (Schenck, Holland, et al., 2017; Schenck, Men, & Maeda, 2017). While these biochemical and evolutionary studies established that some legumes have an alternative Tyr‐insensitive cytosolic PDH enzyme(s), its in vivo function is unknown.
To address this issue, here we hypothesized and tested four non‐mutually exclusive in planta functions of the PDH enzyme in legumes. Hypothesis I: PDH functions as a part of a redundant Tyr biosynthetic pathway in the cytosol (Figure 1). Although AAA biosynthesis is localized to the plastids (Bickel, Palme, & Schultz, 1978; Jung, Zamir, & Jensen, 1986), some plants including legumes possess cytosolic isoforms of CM and Tyr‐AT, which catalyze immediately up‐ and down‐stream steps of PDH and complete the cytosolic Tyr biosynthetic pathway from chorismate (Figure 1) (D’Amato, Ganson, Gaines, & Jensen, 1984; Ding et al., 2007; Eberhard et al., 1996; Schenck et al., 2015; Wang, Toda, & Maeda, 2016). Also, a cytosolic Phe pathway was recently identified in plants (Qian et al., 2019; Yoo et al., 2013). Hypothesis II: PDH is involved in the production of Tyr or HPP‐derived metabolite(s) (Figure 1). Tyr is the precursor of many plant specialized metabolites (Schenck & Maeda, 2018), and duplicated primary metabolic enzymes can be co‐opted to efficiently provide Tyr and HPP precursors to support their downstream specialized metabolism (Maeda, 2019; Moghe & Last, 2015; Weng, Philippe, & Noel, 2012). Hypothesis III: PDH is involved in the Tyr catabolism pathway to the tricarboxylic acid (TCA) cycle. Tyr catabolism proceeds through HPP, the product of PDH, and feeds intermediates (e.g., fumarate) of the TCA cycle (Figure 1). Thus, HPP produced from PDH may be directly incorporated into the Tyr degradation pathway. Hypothesis IV: PDH is involved in the legume‐rhizobia symbiosis. Many legumes form a symbiotic relationship with rhizobia, and PDH activity is uniquely present in legumes. Furthermore, MtPDH1 expression is upregulated in the nodules after treatment with nitrate (NO3 ‐, Benedito et al., 2008) or phosphinothricin (PPT, Seabra, Pereira, Becker, & Carvalho, 2012), both of which stimulate nodule senescence (Matamoros et al., 1999; Pérez Guerra et al., 2010; Streeter & Wong, 1988). To systematically test the above four hypotheses, this study isolated and characterized pdh mutants using genetics, biochemistry, metabolomics, histochemical staining, microscopy, and gene expression analyses in the model plant M. truncatula, which conveniently has a single PDH gene compared with some other legumes that have multiple copies (Schenck et al., 2015; Schenck, Holland, et al., 2017).
2. MATERIALS AND METHODS
2.1. Plant materials and growth conditions
Medicago truncatula accession R108 wild type and pdh1 mutant seeds were scarified in concentrated sulfuric acid for eight minutes and repeatedly washed with water. Seeds were then surface sterilized with bleach for 1.5 min followed by repeated water washes. Seeds were placed in sterile water for 16 hr at 4°C and then transferred to germination medium (0.5× MS medium, 0.8% agar, 1 μM GA3, pH 7.6), wrapped in aluminum foil and placed at 4°C. After 48 hr, plates with sterilized seeds were moved to 22°C. After 24 hr, the aluminum foil was removed and the germinated seedings were transferred to standard potting soil and placed in a growth chamber (Conviron). Pots were watered with 1x Hoaglands solution when dry, and grown under 12‐hr light:dark cycles with 200 μE light intensity and ~60% humidity.
For extended dark treatments, plants were grown under the above conditions for six weeks. Leaves were then excised and floated in water and returned to the above growth conditions with dark‐treated leaves being covered with a cardboard box. After the treatment, leaves were dried using a kimwipe and used for metabolite extraction.
2.2. Genotyping
A single young leaf from 6‐week‐old plants was placed in 1.7‐ml microcentrifuge tube with 600 μl of DNA extraction buffer (10 mM Tris‐HCl pH 8.0, 25 mM ethylenediaminetetraacetic acid (EDTA), and 0.5% SDS), pulverized with a mini plastic blue pestle, and incubated at 55°C for 15 min. The solution was cooled to room temperature and 200 μl of 5 M ammonium acetate was added, vortexed for 20 s, and centrifuged at 14,000 g for 3 min. The supernatant was transferred to a fresh tube and 600 μL of isopropanol was added, followed by centrifugation at 14,000 g for 1 min. The supernatant was decanted and the pellet washed with 400 μL of 70% ethanol followed by centrifugation at 14,000 g for 1 min. The supernatant was decanted and the pellet dried for 1 hr in a sterile hood. The resulting DNA was dissolved in 50 μl of H2O. Genotyping PCRs (20 μl total) contained 1 μM gene and insertion‐specific primers (Table S2), 1x EconoTaq PLUS master mix (Lucigen), and 1 μl of genomic DNA. DNA was amplified in a thermocycler with the following conditions: an initial denaturation at 95°C for 5 min, 35 cycles of amplification at 95°C for 20 s, 60–65°C for 20 s, 72°C for 60 s, with a final extension at 72°C for 5 min.
2.3. Quantitative reverse transcription PCR (qRT‐PCR)
RNA was extracted from leaves of 6‐week‐old plants. About 50 mg of tissue was pulverized in liquid N2 using a mini pestle. RNA extraction buffer (68 mM sodium citrate, 132 mM citric acid, 1mM EDTA and 2% SDS) was added and vortexed immediately for 10 s, and the tubes were then placed on their sides for 5 min at 22°C. The solution was centrifuged at 12,000 g for 2 min and 400 μl of the supernatant was transferred to a fresh tube. 100 μl of 1 M NaCl was added and mixed by pipetting, followed by addition of 300 μl of chloroform, inverted multiple times and centrifuged at 4°C for 10 min at 12 000 g. The upper phase was transferred to a new tube and an equal volume of isopropanol was added, mixed by inverting and placed at 4°C for 10 min. The solution was then centrifuged at 4°C for 10 min at 12,000 g and the supernatant decanted. The pellet was washed with 70% ethanol, followed by centrifugation for 3 min at 12,000 g and the supernatant decanted and the pellet dried in a sterile hood until all residual ethanol was evaporated. The resulting pellet was redissolved in 25 μl of nuclease‐free H2O (Promega). To remove DNA, the RNA solution was treated with DNase (Turbo DNase, Fisher) following the manufacturer's protocol. The remaining RNA was quantified using a nano‐drop spectrophotometer (Thermo Scientific) and diluted to a 20 ng/μl concentration. RNA was converted into cDNA using reverse transcriptase (Applied biosystems) with an oligo d(T) primer.
For qPCR, cDNA was diluted to 5 ng/μl and additional fivefold dilutions were made to calculate primer efficiency. All primer pair efficiencies were between 90%–100%. cDNA was mixed with GoTaq qPCR master mix (Promega) containing SYBR green and 300 nM of each primer. Reactions were placed in an Stratagene Mx3000P (Agilent) thermocycler using the following PCR cycle an initial denaturation at 95°C for 10 min, 45 cycles of amplification at 95°C for 15 s, 60°C for 30 s, 72°C for 30 s.
For relative quantification, Ct values were extracted for each reaction and used to quantify initial cDNA concentration using 2−∆∆C(t) method normalized to a housekeeping gene (MtPI4K, Kryvoruchko et al., 2016 , Table S2) using the LinRegPCR program (Ruijter et al., 2013). For absolute quantification, pET28a vectors carrying MtPDH1, MtncADH and MtADH were used as the template to amplify a fragment of the corresponding genes. The resulting fragments were purified from 0.8% agarose gels using a QIAquick gel extraction kit (Qiagen) following manufacturer's protocol. The DNA concentration was quantified and repeated fivefold dilutions were made and used to obtain a standard curve in qRT‐PCR with gene‐specific primers (Table S2). Ct values were extracted for each qRT‐PCR reaction and used to quantify initial cDNA concentration by using the linear range created as above for the respective gene.
2.4. Enzyme extraction and ADH and PDH assays
Leaf tissue from 6‐week‐old plants were ground to a fine powder in a prechilled mortar and pestle under liquid N2. Extraction buffer (25 mM HEPES, pH 7, 50 mM KCl, 10% ethylene glycol, 1% polyvinylpyrrolidone (PVP) and 1 mM dithiothreitol (DTT)) was added in a 1:3 ratio of tissue to buffer (w/v). The slurry was centrifuged for 20 min at 4°C, at 20,000 g and the resulting supernatant desalted using a gel filtration column (Sephadex G50‐80 resin, Sigma‐Aldrich) equilibrated with extraction buffer without DTT and PVP. Protein concentrations were determined by a Bradford assay (Bio‐Rad Protein Assay, Bio‐Rad).
The desalted crude enzyme extracts were used in ADH and PDH reactions that contained 25 mM HEPES (pH 7.5), 50 mM KCl, 10% ethylene glycol, 1 mM NADP+, 1 mM substrate (l‐arogenate or prephenate, respectively). Arogenate was prepared by enzymatic conversion from prephenate (Sigma‐Aldrich), as previously reported (Maeda, Yoo, & Dudareva, 2011). Reactions were initiated by addition of enzyme from various sources and incubated at 37°C for 45 min. The resulting assays were injected into HPLC equipped with a ZORBAX SB‐C18 column (Agilent) to directly detect the final product of the assay as described previously (Schenck et al., 2015).
For PDH and ADH activities from various legumes (Figure S9), leaf material was obtained from the University of Wisconsin‐Madison Botany Department greenhouse or from identified trees on campus. Plants were grown under varying light and temperature conditions and leaf material was collected at different developmental stages. Enzyme extractions and ADH and PDH assays were performed as described above.
2.5. Metabolite extraction and detection
Tissue from 6‐week‐old plants was added into 1.7‐ml microcentrifuge tubes with 400 μL of extraction buffer (2:1 (v/v) methanol:chloroform, 0.01% butylated hydroxytoluene (BHT), 100 μM norvaline and 1.25 μg/ml tocol as previously described (Collakova & DellaPenna, 2003), with 3 glass beads (3 mm). Samples were vigorously shaken for 3 min at 1,000 rpm using a genogrinder (MiniG 1600, SPEX SamplePrep). Additional chloroform (125 μl) and water (300 μl) were added and vortexed for 30 s. Samples were centrifuged for 10 min at 20,000 rpm and the polar and non‐polar phases were transferred to new tubes and dried using a vacuum concentrator (Labconco).
For tocopherol analysis, the dried nonpolar phase was resuspended in methanol with 0.01% BHT. Samples were injected into a HPLC (Agilent 1260) equipped with a ZORBAX SB‐C18 column (Agilent) using a 30‐min isocratic elution of 95% methanol, 5% water. Tocopherols in the extractions were detected using fluorescence excitation at 290 nm and emission at 330 nm and compared with authentic standards (Sigma‐Aldrich) and normalized to an internal control (tocol).
For anthocyanin detection, 1 M HCl of methanol was added to the polar phase in a 1:1 ratio. A spectrophotometer was used to measure absorbance at 520 nm. Absolute levels were estimated using an extinction coefficient of anthocyanin absorbance (Lee et al., 2005) of 33,000 L × M−1 × cm−1.
The polar and non‐polar phases were analyzed with GC‐MS. The dried polar phase was redissolved in pyridine with 15.0 mg/ml methoxyamine‐HCl. Samples were vortexed for 30 s followed by sonication for 10 min and incubated for 60 min at 60°C. This was repeated once more, then the samples were derivatized with an equal volume of N‐methyl‐N‐(tert‐butyldimethylsilyl)trifluoroacetamide + 0.1% tert‐butyldimethylchlorosilane (MTBSTFA + 0.01% t‐BDMCS, Sigma‐Aldrich) and incubated for 60 min at 60°C. Samples were then injected into GC‐MS (Trace 1310, ISQ LT, Thermo Scientific). The dried non‐polar phase was redissolved in 800 μl of chloroform with 50 ppm BHT and 500 μl of 1.25 M HCl in methanol. Following incubation at 50°C for 4 hr, samples were completely dried under nitrogen gas. The dried non‐polar phase was resuspended in 70 μl of pyridine and derivatized with 30 μl of MTBSTFA + 0.01% t‐BDMCS (Sigma‐Aldrich). Following transfer to a glass vial, 1 μl of the polar and non‐polar phases were injected onto a 30 m column (TG‐5MS, Thermo Scientific) using a 10:1 split ratio, and an oven ramp method of 5°C per minute for 46 min and held at 300°C for 10 min. Detected compounds were compared with NIST library matches and peak areas based on ion abundance were normalized to an internal standard (norvaline).
2.6. Acetylene reduction assay (ARA)
Plants used for ARA were scarified and germinated as described previously. Seedlings were transferred to 12” square plates containing modified solid Fahräeus medium supplemented with 0.5 mM NH4NO3 (Catoira et al., 2000). Plates were grown under the same conditions as pot grown plants; however, plates were placed at a ~ 60° angle so that the roots would not penetrate the agar. After 5 days of growth on plates, each plant was inoculated with 1 ml of water containing Sinorhizobium meliloti Rm1021 at an OD600 0.02. Plants were then allowed to grow for 14, 21 and 28 dpi at which point nodules were counted and plants were moved to 10‐mL glass jars with 1 ml of sterile water at the bottom. Glass jars were sealed with a rubber stopper, then injected with 1 ml of acetylene gas (10% acetylene final concentration). After 48 hr of incubation at 37°C, 1 ml of the gas from the glass jars was injected into a gas chromatograph (GC‐2010, Shimadzu) to measure the production of ethylene and ARA activity was calculated as described in Hardy, Holsten, Jackson, & Burns, 1968.
2.7. Histochemical staining of lignin composition
For cell wall composition staining, thin cross sections of stem tissue from 6‐week‐old plants were prepared using a razor blade. For Mäule staining, stem sections were placed in a 1% potassium permanganate solution for 5 min, then rinsed with water followed by addition of a 12% HCl (v/v) for 5 min and rinsed with water. A 1.5% solution of sodium bicarbonate was added to facilitate a color change to dark red and visualized using an epifluorescence microscope (Olympus BX60).
For phloroglucinol staining, similar stem cross sections were placed in a well plate with 1 ml of 10% phloroglucinol (w/v) solution in 95% ethanol with 500 μl of 10 N HCl and incubated for 5 min. Stem sections were transferred to a glass slide and washed with water and visualized using a epifluorescence microscope (Olympus BX60).
GUS staining was performed on nodules developed after inoculation with S. meliloti Rm1021 carrying a PnifH::UidA fusion, to localize expression of the bacterial nifH gene, which is required for nitrogen fixation. Nodules were embedded in 4% agarose and 50–100 μM sections were made with a vibratome® 1000 plus (Leica). Sections were immersed in a staining solution (2.5% 5‐Bromo‐4‐chloro‐3‐indoxyl‐beta‐D‐glucuronic (X‐Gluc), 0.2 M sodium phosphate buffer (pH 7), 0.1 M potassium ferricyanide, 0.1 M potassium ferrocyanide, 0.25 M Na2EDTA, and 10% Triton X‐100) and vacuum infiltrated for 10 min, incubated in the dark at 37°C for 30 min, and rinsed with phosphate buffer. Sections were visualized using bright field microscopy.
3. RESULTS
3.1. Isolation of pdh mutants in Medicago truncatula
To investigate the in vivo function of PDH in legumes, two independent homozygous alleles of Tnt1‐transposon mutants of M. truncatula (Tadege et al., 2008) were identified for the MtPDH1 locus (Mt3g071980) (Cheng et al., 2014). The pdh1‐1 and pdh1‐2 mutants carried a transposon insertion in the first and last exons of the MtPDH1 gene, respectively (Figure 2a). MtPDH1 was constitutively expressed across many tissues and under various conditions with the highest and lowest expression being detected in aerial tissues and seeds, respectively (Figure S1). Both alleles did not produce any PDH transcript in the leaves (Figure 2b). The M. truncatula genome contains two ADH genes, MtADH (Mt4g115980) and MtncADH (Mt5g083530) (Schenck, Holland, et al., 2017), but neither were enhanced in the pdh1 mutants (Figure 2b). Consistent with their transcript levels, PDH activity was almost completely abolished in both pdh1 mutants without significant reduction in ADH activity (Figure 2c). After six weeks of growth, pdh1‐1 showed no phenotypic difference from wild‐type (Wt; R108). The pdh1‐2 mutant, on the other hand, had a slight bushy phenotype, which could be due to an unknown secondary transposon insertion(s) (Figure 2d); however, multiple backcrossing attempts were unsuccessful. These data provide genetic evidence that MtPDH1 is responsible for the PDH activity detected in M. truncatula. The minor impacts of eliminating PDH activity on overall plant growth in two independent pdh1 mutants suggest that the PDH enzyme is not essential during standard growth conditions in M. truncatula.
Figure 2.
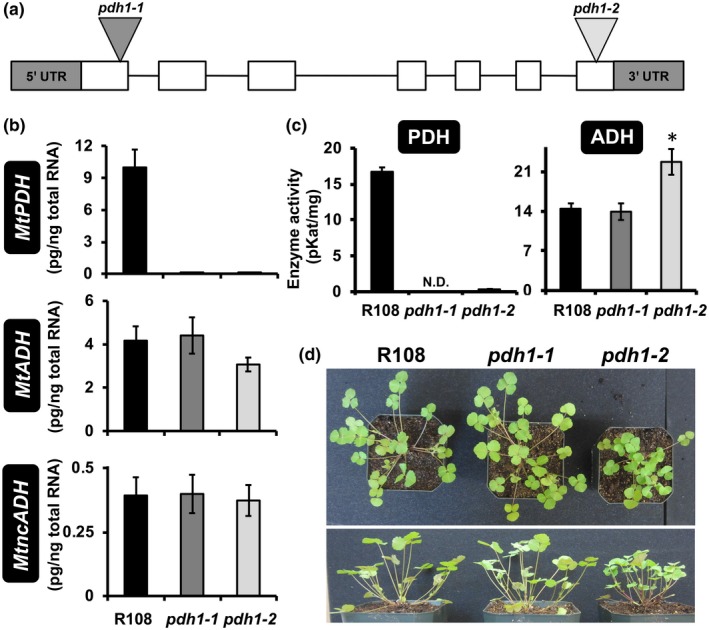
Isolation of Medicago pdh1 mutants. (a) MtPDH1 (Mt3g071980) genomic structure, exons (white boxes) and introns (lines), 5′ and 3′ untranslated regions (UTR, gray boxes). Two Tnt1‐transposon mutants were isolated with insertions in exon one (pdh1‐1) and seven (pdh1‐2). (b) MtPDH1 transcripts were nearly abolished in pdh1‐1 and pdh1‐2, without affecting expression of either ADH homolog. Bars represent average absolute mRNA levels (pg/ng total RNA) ± SEM of three biological replicates. (c) PDH and ADH activity from mutants and wild‐type (Wt, R108). Bars represent average enzymatic activity (pKat/mg) ± SEM of three biological replicates. Significant differences to R108 control are indicated; *p ≤ .05. (d) Phenotype of R108 and mutants after 6‐week growth under standard conditions
3.2. Tyr and Tyr‐derived compounds are unaltered in pdh mutants during standard growth condition
To test the hypothesis that the PDH pathway serves as a redundant Tyr biosynthetic route (hypothesis I), Tyr and Tyr/HPP‐derived compounds were analyzed in Wt and mutants. Metabolites were extracted from root and leaf tissue of 6‐week‐old plants grown under standard conditions. Surprisingly, the levels of Tyr were not significantly different between Wt and mutants in either leaf or root tissue (Figure 3a). Additionally, the levels of Phe and Trp in the leaves and roots were not significantly reduced in mutants as compared to Wt, though a slightly higher Trp level was observed in pdh1‐2 (Figure 3a). Potential effects on tocopherols, HPP‐derived metabolites, were also tested (hypothesis II), but their levels were not significantly different between Wt and mutants in both leaf and root tissue (Figure 3b). We also performed non‐targeted analysis using GC‐MS for both polar and non‐polar metabolites; however, no consistent differences were observed between Wt and mutants (Table S1). Together, these data suggest that under standard growth conditions, the lack of the PDH enzyme had no substantial effects on the overall accumulation of AAAs or Tyr/HPP‐derived metabolites analyzed.
Figure 3.
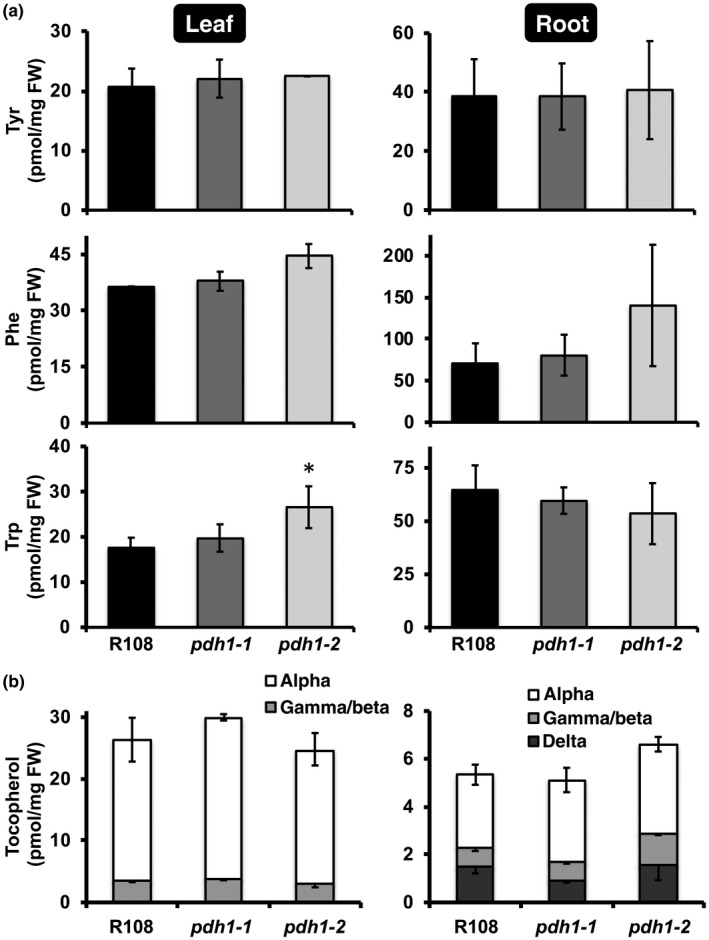
Targeted metabolite analysis of pdh1 mutants and Wt. Leaf and root tissues from 6‐week‐old plants, grown under standard conditions, were used for metabolite extraction. Bars represent average absolute metabolite levels (pmol/mg fresh weight (FW)) ± SEM of three biological replicates. (a) Aromatic amino acid levels in leaf and root tissues. Significant differences to Wt (R108) control are indicated; *p ≤ .05. (b) Tocopherol composition and content from leaf and root tissue
In grasses, upwards of 50% of the total lignin is derived from Tyr, via Tyr ammonia‐lyase (TAL) activity (Barros et al., 2016). Since TAL activity has also been detected in some non‐grass species, including legumes (Beaudoin‐Eagan & Thorpe, 1985; Giebel, 1973; Khan, Prithiviraj, & Smith, 2003), the legume PDH enzyme may synthesize Tyr that is directly incorporated into the phenylpropanoid pathway for downstream products such as lignin (hypothesis II). To test this possibility, stem cross sections from Wt and mutants were stained using two different reagents, Mäule and phloroglucinol, which can detect potential differences in composition or linkages of lignin (Mitra & Loqué, 2014; Pomar, Merino, & Barceló, 2002). Neither staining method showed any obvious differences in stem lignification between Wt and mutants (Figure S2a,b). When phenylpropanoid intermediates, p‐coumarate and ferulate, involved in lignin biosynthesis were analyzed by GC‐MS, they were somewhat reduced in pdh1‐2 but not consistently in pdh1‐1 (Figure S2c). Thus, the lack of PDH does not have substantial impacts on lignin biosynthesis in M. truncatula.
3.3. Less tocopherols are accumulated in pdh1 than Wt under high light treatment
Under various biotic and abiotic stresses, the shikimate pathway is induced, which often leads to accumulation of AAAs (Betz et al., 2009; Dyer, Henstrand, Handa, & Herrmann, 1989; Gilbert, Gadush, Wilson, & Madore, 1998; Zhao, Williams, & Last, 1998). To test the potential role of PDH in Tyr biosynthesis under stress (hypothesis I), Wt and pdh1‐1 were subjected to 48‐hr high light treatment (Gonzali, Mazzucato, & Perata, 2009), which is known to induce production of AAA‐derived antioxidants (Collakova & DellaPenna, 2003). The levels of Tyr were not altered, except at 24 hr when Tyr increased slightly in pdh1‐1 compared with Wt (Figure S3a). As expected, high light treatment enhanced tocopherol accumulation, but to a significantly lesser extent in pdh1‐1 compared with Wt at both 24 and 48 hr (Figure S3b). The levels of anthocyanins, which are also induced under various stresses including high light stress (Collakova & DellaPenna, 2003), were increased after 24 and 48 hr of high light treatment but were not significantly different between Wt and pdh1‐1 (Figure S3c). These results show that the lack of PDH negatively impacts the accumulation of HPP‐derived tocopherols when their production is induced under high light conditions.
3.4. MtPDH1 is co‐expressed with senescence‐related genes but has no major impacts on dark‐induced senescence
To identify potential processes and pathways that are coordinately regulated with PDH, a gene co‐expression analysis with MtPDH1 was performed using the Medicago gene expression atlas (He et al., 2009). MtPDH1 was co‐expressed with genes mainly involved in senescence‐related processes (e.g., nucleases, proteases, and lipases, Figure S1) and the gene encoding HPP dioxygenase (HPPD, Siehl et al., 2014), a senescence‐activated enzyme involved in Tyr catabolism (Figure S5a) (Wang, Toda, Block, & Maeda, 2019). To experimentally test the potential involvement of PDH in senescence, PDH gene expression and enzymatic activity were monitored at different developmental stages during natural leaf senescence (Figure S4a). To define stages of leaf senescence, expression of a senescence marker gene (MtVPE, Pérez Guerra et al., 2010) was monitored, together with loss of chlorophyll in leaves collected at various developmental stages from a single plant (Figure S4b). MtVPE was basally expressed in fully green leaves (defined here as the S1 stage) and gradually induced upon senescence (defined here as S2, S3, and S4; Figure S4b), mirroring the loss of chlorophyll. Fully senescent leaves (S5) were collected but did not yield high‐quality RNA and proteins, and hence could not be further analyzed. The highest MtPDH1 expression was detected in green (S1) leaves, and PDH enzymatic activity was not induced upon senescence (Figure S4c,d). Thus, MtPDH1 is not upregulated during natural senescence in the leaves. Similar to PDH, expression of the two ADH genes in M. truncatula did not follow the developmental pattern of leaf senescence (Figure S4c). Unlike PDH activity, however, ADH enzymatic activity was gradually induced upon senescence (Figure S4d).
Since MtPDH1 was co‐expressed with MtHPPD (Figure S1), and PDH together with HPPD provides a direct route for catabolism of Tyr to homogentisate, eventually leading to fumarate (Figure 1, Figure S5a), we investigated potential impacts of pdh1 deficiency in Tyr catabolism (hypothesis III). The expression of genes encoding all enzymes of the Tyr catabolic pathway was measured in the mutants and Wt under standard growth conditions (Figure S5a, Dixon & Edwards, 2006). The expression of genes encoding the first two steps of the pathway, HPPD and homogentisate dioxygenase (HGO), were not significantly altered in the mutants compared with Wt. The subsequent step, maleylacetoacetate isomerase (MAAI), was induced by twofold and 2.5‐fold in pdh1‐1 and pdh1‐2, respectively (Figure S5b), though the final step in the pathway, fumarylacetoacetate hydrolase (FAH), showed opposite expression patterns in the two mutants (Figure S5b). Thus, no consistent changes in the expression of the Tyr degradation pathway genes, beyond MAAI, were observed in pdh1 mutants.
To further examine Tyr catabolism during leaf senescence under artificial, but controlled, conditions excised leaves from Wt and mutants were exposed to an extended dark treatment (Peng et al., 2015; Wang et al., 2019). Over 7 days, leaves from Wt and mutants responded to dark‐induced senescence similarly with no apparent growth phenotypes (Figure S6a). Furthermore, there were no significant differences in α‐tocopherol levels between Wt and mutants at any time point analyzed (Figure S6b). These data together suggest that the lack of PDH had no substantial effects on Tyr catabolism or metabolism under standard growth conditions and at least under the senescence conditions tested here (Figures S5 & S6).
3.5. Less Tyr is accumulated in pdh mutants following shikimate feeding
Although the steady‐state levels of Tyr were not altered in the mutants (Figure 3a), some of the above data suggest that the PDH enzyme may contribute to Tyr production (hypothesis I) at least under some stress conditions (Figures S1 and S3b). Thus, we hypothesized that the potential role of PDH in Tyr biosynthesis might be manifested when the carbon flux toward the Tyr pathway is elevated. To test this possibility, an intermediate of the upstream shikimate pathway, shikimate, was exogenously fed to Wt and mutants, and the levels of AAAs were analyzed. Excised leaves were floated in a solution containing shikimate or H2O for up to 8 hr, rinsed to remove metabolites in the feeding solution, and leaf metabolites were extracted and analyzed using HPLC and GC‐MS. As more prolonged feeding and higher concentrations of shikimate led to abnormal leaf phenotypes, 8 hr and 25 mM shikimate were selected as an optimal time and concentration for increased carbon flux without pleiotropic effects. After the feeding, shikimate and Phe levels were increased drastically in all genotypes, suggesting that shikimate was taken up and metabolized by the leaves (Figure 4). Tocopherol levels were generally unaffected upon shikimate feeding, likely because the feeding time was not long enough to convert shikimate into tocopherols (Figure 4). Trp levels were reduced in the mutants compared with Wt after feeding with water, and were induced upon shikimate feeding in pdh1‐1 (Figure 4). The unexpected differences observed with Trp levels under standard growth conditions (Figure 3) and the feeding experiments (Figure 4) may reflect unknown stress responses during the feeding experiments. Nevertheless, after 8 hr of feeding, Tyr levels increased by > 39‐fold in Wt, but only by 16‐ and 13‐fold in pdh1‐1 and pdh1‐2, respectively (Figure 4). Repeated 8 hr shikimate feeding experiments with pdh1 mutants and Wt yielded similar Tyr accumulation patterns. The difference in the Tyr accumulation suggests that PDH contributes to Tyr biosynthesis when carbon flow through the shikimate pathway is enhanced.
Figure 4.
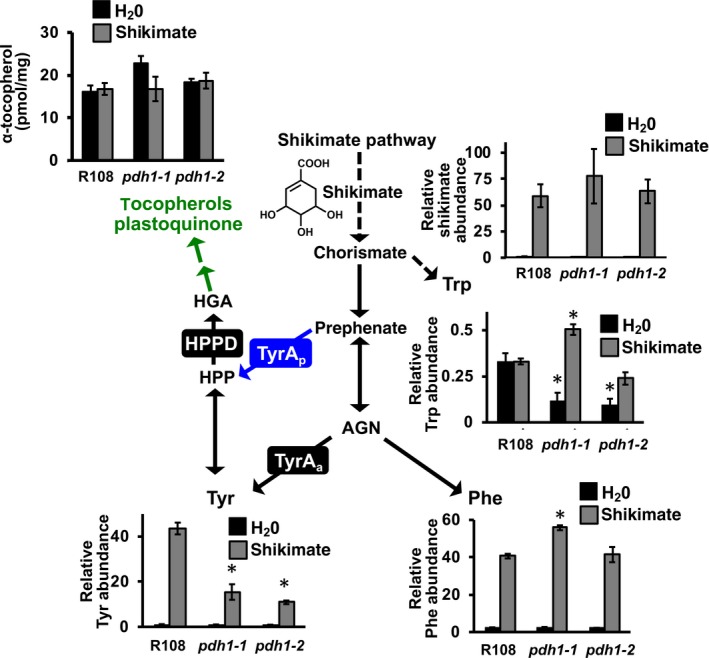
Metabolite analysis after shikimate feeding in Wt and pdh1 mutants. Excised leaves from 6‐week‐old plants were floated on a solution containing H2O (black bars) or 25 mM shikimate (gray bars) for 8 hr under constant light. Leaves were then used for metabolite analysis using GC‐MS (Tyr, Trp, Phe and shikimate) or HPLC (α‐tocopherol). Bars represent average relative metabolite abundance of Tyr, Phe, Trp and shikimate ± SEM of three biological replicates. α‐tocopherol is shown as the average absolute metabolite abundance in pmol/mg FW ± SEM of three biological replicates. Additional metabolites measured following shikimate feeding are shown in Figures S7 and S8. Significant differences to Wt (R108) control are indicated; *p ≤ .05
To determine if global metabolite changes occurred after shikimate feeding, additional amino acids and TCA pathway metabolites were analyzed by GC‐MS. Interestingly, glutamine levels were higher in H2O‐fed Wt, compared with mutants (Figure S7), and upon shikimate feeding, glutamine levels were further significantly elevated in Wt (Figure S7). The levels of TCA cycle intermediates may indicate the functionality of the Tyr catabolic pathway; however, those TCA cycle intermediates analyzed here, including fumarate, were not consistently altered after shikimate feeding in Wt and mutants (Figure S8). Citrate levels were reduced in pdh1‐1 and pdh1‐2 compared with Wt in H2O control treatment; however, after shikimate feeding these differences were no longer apparent (Figure S8). These data did not provide evidence to support the involvement of the PDH enzyme in Tyr catabolism to TCA cycle intermediates.
3.6. PDH has limited role in the legume‐rhizobia symbiosis
To test whether PDH plays a role in legume‐rhizobia symbiosis (hypothesis IV), Wt and pdh1‐1 mutants were grown side‐by‐side on low nitrogen Fahräeus medium and inoculated with a well‐characterized rhizobium of M. truncatula, Sinorhizobium meliloti Rm1021. At various time points (e.g., 14, 21, and 28 days post‐inoculation (dpi)) pdh1‐1 mutants did not display any phenotypic difference from Wt plants, including the number of nodules produced per root (Figure 5a), suggesting that PDH is not essential for nodule development. M. truncatula forms indeterminate nodules that display the four standard zones: I, meristematic; II, infection; III, fixation; and IV, senescence zones in both Wt and pdh1‐1 (Figure 5b, Van de Velde et al., 2006). Expression of the bacterial nifH gene, which is required for nitrogen fixation, was monitored by using a S. meliloti strain expressing a PnifH::UidA fusion to evaluate nodule maturation on pdh1‐1 and Wt plants (Starker, Parra‐Colmenares, Smith, Mitra, & Long, 2006). After 21 and 28 dpi, β‐glucuronidase (GUS) activity and the overall nodule development were not altered between pdh1‐1 and Wt (Figure 5c). To measure nitrogenase activity, acetylene reduction assays (ARA, Wych & Rains, 1978) were also performed at 14, 21, and 28 dpi. Although nitrogenase activity in pdh1‐1 appeared to be slightly reduced compared with Wt at 21 and 28 dpi when nodule senescence might have been initiated, no significant differences were observed (Figure 5d). ARA were repeated at the 21 dpi time point with pdh1‐1 and this time including pdh1‐2. Again, nitrogenase activity was not significantly different between genotypes, even though pdh1‐1 showed a slight reduction, similar to the first experiment (Figure 5e).
Figure 5.
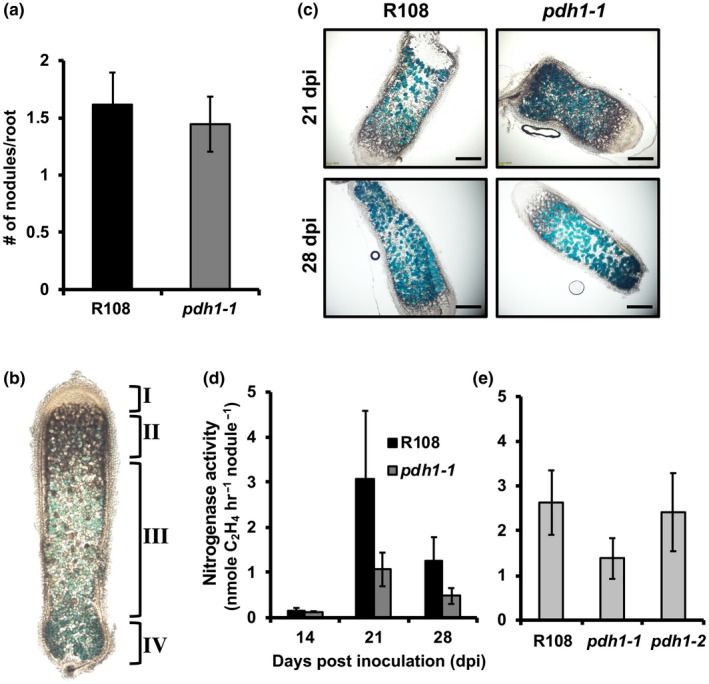
The role of PDH in legume‐rhizobia symbiosis. (a) Wt and pdh1‐1 were grown on plates with low nitrogen Fahräeus medium, inoculated with S. meliloti, and the number of nodules was counted at 14 days post‐inoculation (dpi). Bars represent average number of nodules per root ± SEM., n ≥ 11. (b) Indeterminate nodules from M. truncatula consist of four developmental zones: I, meristem; II, infection; III, fixation; IV, senescence located closest to the root. These four developmental zones are shown on a characteristic nodule (enlarged for visualization) from 28 dpi. GUS staining with PnifH::UidA shows expression of bacterial nifH gene localized to the bacteroids (blue). (c) Thin sections of nodules from 21 and 28 dpi showing expression of PnifH::UidA, highlight bacteroid and nodule development in R108 and pdh1‐1. Approximately equal staining was observed in all nodule developmental zones, suggesting pdh1‐1 is not affected in nodule development or bacteroid number, scale bar = 1 mm. (d) Nitrogen fixation efficiency was measured using an acetylene reduction assay (ARA). Plants were grown under the same conditions as in (a). At 14, 21, and 28 dpi, ethylene production was measured and expressed as the average ± SEM, n > 7. (e) Nitrogen fixation efficiency assay performed in the same way as in (d), but with both mutant alleles and Wt and only at 21 dpi. Activity is expressed as the average ± SEM, n > 4
To test whether the presence and absence of PDH activity positively correlated with those of nodulation, we also measured PDH activity from plant tissue extracts of over twenty species sampled across the legume phylogeny (Azani et al., 2017, Figure S9) and compared with their capacity to form nodules (Afkhami et al., 2018). All legumes sister to previously analyzed G. max (soybean) and M. truncatula (Schenck et al., 2015) showed PDH activity including Arachis ipaensis (peanut) (Figure S9a), which possess a TyrA enzyme with similar ADH and PDH activity (Schenck, Holland, et al., 2017). All of these legumes were previously reported to be able to nodulate (Afkhami et al., 2018). When four legume species in the genistoid crown were analyzed (e.g., Lupinus polyphyllus), PDH activity was not detectable in any of them, despite the successful detection of ADH activity (Figure S9b). Importantly, these legumes were reported to be able to nodulate, suggesting that legume plants having undetectable levels of PDH activity can still maintain nodulation. Out of four legumes from the mimosoid crown that are known to nodulate, three species showed PDH activity (Figure S9c). In contrast, all four early diverging species tested are incapable of nodulating (Afkhami et al., 2018), but showed PDH activity (Figure S9d). The lack of detectable PDH activity (e.g., the genistoid crown) may be simply due to insufficient sensitivity with HPLC‐based assay and/or the use of tissues that do not express PDH. However, detection of PDH activity in species lacking nodulation (e.g., the early diverging lineages, Figure S9d) strongly support the lack of positive correlation between the presence of PDH activity and nodulation across the legume phylogeny. Phylogenetic sampling (Figure S9) together with pdh1 mutant analysis (Figure 5) suggest that PDH is not essential for legume‐rhizobia symbiosis, though more detailed analyses are needed to fully address its contribution to potentially enhance nitrogen fixation within the nodules in some legumes.
4. DISCUSSION
In this study, we sought to understand the function of legume‐specific PDH enzymes through analysis of Tnt1‐transposon mutants of PDH in M. truncatula. Not only is M. truncatula a convenient model system having mutant populations and many genomic/transcriptomic resources, but M. truncatula also has a single PDH gene unlike some other legumes, that is, soybean, having two PDH genes (Schenck et al., 2015). Analysis of two independent pdh1 mutant lines suggests that MtPDH1 is responsible for all the PDH activity in M. truncatula (Figure 1). This observation is consistent with chromatographic separation of PDH from ADH in soybean, which resulted in the detection of a single PDH peak as compared to multiple ADH peaks (Schenck et al., 2015). Our genetic data also support that ncADH, and ADH enzymes do not contribute to PDH activity, consistent with in vitro data of soybean and M. truncatula enzymes (Schenck et al., 2015; Schenck, Holland, et al., 2017).
Surprisingly, the null pdh1 mutants displayed no visible aberrant growth phenotypes (Figure 1). A slight bushy phenotype was observed in the pdh1‐2 allele, but not in pdh1‐1. Thus, here we focused on consistent responses observed in both alleles to avoid potential pleiotropic effects specific to pdh1‐2. The lack of substantial phenotypes in pdh1 is in contrast to the highly compromised growth and leaf development phenotypes of Arabidopsis adh2/tyra2 mutant (de Oliveira et al., 2019). In M. truncatula, we were also unable to recover homozygous mutants of the canonical ADH, which is a single copy gene in M. truncatula as compared to two ADH copies in Arabidopsis (Rippert & Matringe, 2002), further suggesting the essential nature of the ADH‐mediated Tyr pathway in legumes. Thus, the canonical plastid‐localized ADH pathway is the predominant Tyr biosynthetic route and cannot be compensated by the cytosolic PDH pathway in legumes.
Using the pdh1 mutants, this study systematically evaluated potential roles of PDH in Tyr biosynthesis (hypothesis I), biosynthesis of Tyr‐ and HPP‐derived metabolites (II), Tyr catabolism and senescence (III), and legume‐rhizobia symbiosis (IV). The obtained data provide evidence for support of hypothesis I that the PDH enzyme is involved in Tyr biosynthesis when carbon flux is increased through the shikimate pathway. Although Tyr levels were not altered between Wt and mutants under standard conditions (Figure 3a), upon feeding with shikimate, Wt accumulated over twofold more Tyr than pdh1‐1 and pdh1‐2 (Figure 4), suggesting that the PDH enzyme contributes to synthesis of Tyr under enhanced flux through the shikimate pathway. Although AAA biosynthesis is localized to the plastids (Bickel et al., 1978; Jung et al., 1986; Maeda & Dudareva, 2012), some isoforms of the shikimate and AAA pathway enzymes were detected in the cytosol (Ding et al., 2007; Ganson, D’Amato, & Jensen, 1986; Rubin & Jensen, 1985). Cytosolic CM activities have been detected from various plant species including soybean (Goers & Jensen, 1984; Kuroki & Conn, 1989; Mobley et al., 1999; Schenck et al., 2015), which are not regulated by the AAAs, unlike their plastidic counterparts (Eberhard et al., 1996; Mobley et al., 1999; Westfall, Xu, & Jez, 2014). Tyr‐insensitive PDH enzymes provide a cytosolic route for conversion of prephenate into HPP, which can be further transaminated to Tyr by cytosolic Tyr‐AT enzymes (Figure 1) (Wang et al., 2016). Although the PDH‐mediated Tyr pathway is not the predominant route for Tyr biosynthesis, it may provide an alternative route for additional Tyr synthesis under some conditions that increase flux through the shikimate pathway.
The current data also partly support hypothesis II that the PDH enzyme contributes to the production of specialized metabolites derived from Tyr and/or HPP in legumes. Although Tyr/HPP‐derived tocopherols were not altered in the pdh1 mutants under standard growth conditions (Figure 3b), high light‐induced tocopherol accumulation was partially attenuated in the pdh1 mutant (Figure S3). Therefore, the PDH enzyme may also contribute to tocopherol biosynthesis under stress conditions. Furthermore, after shikimate precursor feeding, Wt accumulated more Tyr than mutants (Figure 4), suggesting that the PDH enzyme is involved in synthesizing Tyr and possibly downstream compounds when the shikimate pathway flux is increased. Therefore, the alternative PDH pathway could be used under certain conditions (e.g., wounding or other stresses) to support production of Tyr/HPP‐derived specialized metabolites. Specialized metabolic pathways often emerge through duplication and neofunctionalization of genes from primary metabolic pathways (Moghe & Last, 2015; Schenck & Last, 2019; Weng et al., 2012). The legume PDH enzyme emerged as the result of a recent gene duplication of an ADH gene followed by a shift in substrate specificity from arogenate to prephenate (Schenck, Holland, et al., 2017). Thus, legumes may be able to divert the shikimate pathway flux to provide additional HPP or Tyr in the cytosol for the synthesis of downstream specialized metabolites including tocopherols. To further test this possibility, additional metabolomics experiments were conducted, but did not identify any putative HPP‐derived compounds that were absent or lower in pdh1 mutants than Wt (Table S1). Although there are many Tyr and HPP‐derived specialized metabolites produced in plants (e.g., rosmarinic acid, betalains, dhurrin, and benzylisoquinoline alkaloids), none appear to be specific to the legume lineage or correlate with the distribution of PDH activity (Figure S9) (Schenck & Maeda, 2018). Some species of the legume genus Inga can accumulate Tyr at 20% of leaf dry weight, which deters insect predation (Lokvam, Brenes‐Arguedas, Lee, Coley, & Kursar, 2006). Further analysis identified that Inga species with high PDH expression have hyper‐accumulation of Tyr‐derived specialized metabolites, such as Tyr‐gallates (Coley et al., 2019). Therefore, more comprehensive analyses of Tyr/HPP‐derived metabolites under different conditions, tissue types, and other legume species may identify specialized metabolites downstream of the PDH enzyme in legumes.
The current data did not provide sufficient support for hypothesis III that the PDH pathway potentially contributes to Tyr catabolism and senescence. As Tyr is the most energetic amino acid (Hildebrandt, Nunes Nesi, Araújo, & Braun, 2015), its catabolism is a crucial process in recovering energy. Mutations in Tyr catabolic genes can have dramatic effects on plant development (Han et al., 2013). Tyr is catabolized into fumarate, a TCA pathway intermediate, first by conversion into HPP via a Tyr‐AT enzyme and further through the canonical Tyr catabolism pathway (Figure S5a) (Hildebrandt et al., 2015). Knockout of a soybean HGO did not cause lethality but led to an increase in vitamin E content by twofold, suggesting that a significant amount of carbon flux is diverted into vitamin E production when Tyr catabolism is blocked (Stacey et al., 2016). Furthermore, MtPDH1 was co‐expressed with HPPD, which together provide a direct pathway for homogentisate production. This potential coordination of PDH and HPPD, and cytosolic localization, can bypass three enzymatic steps of the ADH‐mediated Tyr biosynthetic pathway, catalyzed by PPA‐AT, ADH, and Tyr‐AT to enter into the canonical Tyr catabolism pathway (Figures 1 & S5a). Additionally, PDH was co‐expressed with many senescence‐related genes (Figure S1, Van de Velde et al., 2006; Kusaba, Tanaka, & Tanaka, 2013; Xi, Chen, Nakashima, Wang, & Chen, 2013) suggesting that PDH may function when Tyr catabolism is enhanced. Despite these correlative data, however, experiments to test the potential link between PDH and senescence did not provide evidence to support this hypothesis: upon dark‐induced (Figure S6) and natural senescence (Figure S4) conditions that likely stimulate Tyr catabolism, no phenotypic differences were observed between mutants and Wt. Also, genes involved in Tyr catabolism were not consistently altered in mutants as compared to Wt (FigureS5b). Although PDH expression was reduced, PDH enzymatic activity was unaltered (Figure S4c), suggesting that there may be some unknown post‐transcriptional regulation on PDH following the induction of senescence. Thus, further experiments under different conditions (e.g., specific stress that induces Tyr catabolism) are needed to address the potential role of PDH in Tyr catabolism and senescence.
The data obtained in this study failed to directly support the hypothesis IV that the PDH pathway plays a role in legume‐rhizobia symbiosis. Legumes initiate a symbiotic relationship with soil‐dwelling rhizobia when there is insufficient nitrogen (Oldroyd, Murray, Poole, & Downie, 2011). A chemical communication ensues that ultimately results in compatible rhizobia invading legume roots and formation of a new organ, the root nodule (Oldroyd, 2013). In the nodule, rhizobia fix atmospheric dinitrogen into ammonium, which is assimilated by the plant through the glutamine synthetase‐glutamine oxoglutarate aminotransferase (GS‐GOGAT) cycle, in which glutamine and glutamate are key amino acid carriers (Krapp, 2015). Mutations in MtPDH1 did not alter nodule numbers (Figure 5a) or the developmental progression of the nodules (Figure 5c). Furthermore, nitrogenase activity was not significantly affected in mutants at any stage during the symbiotic interaction even when senescence might have been initiated (Figure 5d,e). Alterations in nitrogen assimilation can lead to reduced nodulation in legumes (Matamoros et al., 1999; Streeter & Wong, 1988): for example, plants treated with a GS inhibitor phosphinothricin (PPT) result in loss of nodulation (Seabra et al., 2012) and also stimulate MtPDH1 expression (Figure S1) as well as many other genes. Interestingly, pdh1‐1 andpdh1‐2 had reduced glutamine levels in the H2O‐treated control leaves after 8 hr (Figure S7), which also persisted after shikimate feeding (Figure S7). Reduced glutamine levels in the pdh1 mutants may, in turn, affect the symbiotic efficiency with rhizobia, although no statistically‐significant reduction in nitrogenase activity was observed (Figure 5d,e). Furthermore, PDH activity, which was measured in a phylogenetically diverse group of legumes, does not correlate with ability to nodulate (Afkhami et al., 2018; Azani et al., 2017; Figure S9). Thus, PDH may provide an adaptive advantage to some legumes but is likely not directly involved in the legume‐rhizobia symbiosis.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTION
CAS and HAM conceptualized the project. CAS, JMA, LWS, and HAM designed experiments. CAS, JW, DJ, and KG carried our experiments and interpreted results. KSM and JW isolated Tnt1 mutant lines. CAS and HAM wrote the manuscript. All authors read and edited the manuscript.
Supporting information
Figure S1‐S9
Tables S1‐S2
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the National Science Foundation IOS‐1354971 to H.A.M. and NSF IOS‐1331098 and IOS‐1546742 to JMA. LWS is supported in part by NSF awards 1340058, 1743594, 1139489, and 1639618. The University of Missouri, Office of Research provided initial instrumental and personnel funding for the MU Metabolomics Center. Development of M. truncatula Tnt1 mutant population was, in part, funded by the National Science Foundation, USA (DBI‐0703285 and IOS‐1127155) to KSM.
Schenck CA, Westphal J, Jayaraman D, et al. Role of cytosolic, tyrosine‐insensitive prephenate dehydrogenase in Medicago truncatula . Plant Direct. 2020;4:1–15. 10.1002/pld3.218
The preprint of this article: https://www.biorxiv.org/content/10.1101/768317v1
REFERENCES
- Afkhami, M. E. , Luke Mahler, D. , Burns, J. H. , Weber, M. G. , Wojciechowski, M. F. , Sprent, J. , & Strauss, S. Y. (2018). Symbioses with nitrogen‐fixing bacteria: Nodulation and phylogenetic data across legume genehra. Ecology, 99, 502 10.1002/ecy.2110 [DOI] [PubMed] [Google Scholar]
- Azani, N. , Babineau, M. , Bailey, C. D. , Banks, H. , Barbosa, A. R. , Pinto, R. B. , … Zimmerman, E. (2017). A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny: The Legume Phylogeny Working Group (LPWG). Taxon, 66, 44–77. 10.12705/661.3 [DOI] [Google Scholar]
- Barken, I. , Geller, J. , & Rogosnitzky, M. (2008). Noscapine inhibits human prostate cancer progression and metastasis in a mouse model. Anticancer Research, 28, 3701–3704. [PubMed] [Google Scholar]
- Barros, J. , Serrani‐Yarce, J. C. , Chen, F. , Baxter, D. , Venables, B. J. , & Dixon, R. A. (2016). Role of bifunctional ammonia‐lyase in grass cell wall biosynthesis. Nature Plants, 2, 16050 10.1038/nplants.2016.50 [DOI] [PubMed] [Google Scholar]
- Beaudoin, G. A. W. , & Facchini, P. J. (2014). Benzylisoquinoline alkaloid biosynthesis in opium poppy. Planta, 240, 19–32. 10.1007/s00425-014-2056-8 [DOI] [PubMed] [Google Scholar]
- Beaudoin‐Eagan, L. D. , & Thorpe, T. A. (1985). Tyrosine and phenylalanine ammonia lyase activities during shoot initiation in tobacco callus cultures. Plant Physiology, 78, 438–441. 10.1104/pp.78.3.438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito, V. A. , Torres‐Jerez, I. , Murray, J. D. , Andriankaja, A. , Allen, S. , Kakar, K. , … Udvardi, M. K. (2008). A gene expression atlas of the model legume Medicago truncatula . Plant Journal, 55, 504–513. [DOI] [PubMed] [Google Scholar]
- Betz, G. A. , Gerstner, E. , Stich, S. , Winkler, B. , Welzl, G. , Kremmer, E. , … Ernst, D. (2009). Ozone affects shikimate pathway genes and secondary metabolites in saplings of European beech (Fagus sylvatica L.) grown under greenhouse conditions. Trees, 23, 539–553. 10.1007/s00468-008-0300-1 [DOI] [Google Scholar]
- Bickel, H. , Palme, L. , & Schultz, G. (1978). Incorporation of shikimate and other precursors into aromatic amino acids and prenylquinones of isolated spinach chloroplasts. Phytochemistry, 17, 119–124. 10.1016/S0031-9422(00)89691-0 [DOI] [Google Scholar]
- Bramley, P. M. , Elmadfa, I. , Kafatos, A. , Kelly, F. J. , Manios, Y. , Roxborough, H. E. , … Wagner, K.‐H. (2000). Vitamin E. Journal of the Science of Food and Agriculture, 80, 913–938. [DOI] [Google Scholar]
- Catoira, R. , Galera, C. , de Billy, F. , Penmetsa, R. V. , Journet, E. P. , Maillet, F. , … Dénarié, J. (2000). Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell, 12, 1647–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, X. , Wang, M. , Lee, H.‐K. , Tadege, M. , Ratet, P. , Udvardi, M. , … Wen, J. (2014). An efficient reverse genetics platform in the model legume Medicago truncatula . New Phytologist, 201, 1065–1076. [DOI] [PubMed] [Google Scholar]
- Coley, P. D. , Endara, M.‐J. , Ghabash, G. , Kidner, C. A. , Nicholls, J. A. , Pennington, R. T. , … Kursar, T. A. (2019). Macroevolutionary patterns in overexpression of tyrosine: An anti‐herbivore defence in a speciose tropical tree genus, Inga (Fabaceae). Journal of Ecology, 107, 1620–1632. [Google Scholar]
- Collakova, E. , & DellaPenna, D. (2003). Homogentisate phytyltransferase activity is limiting for tocopherol biosynthesis in Arabidopsis. Plant Physiology, 131, 632–642. 10.1104/pp.015222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly, J. A. , & Conn, E. E. (1986). Tyrosine biosynthesis in Sorghum bicolor: Isolation and regulatory properties of arogenate dehydrogenase. Zeitschrift für Naturforschung C, 41, 69–78. 10.1515/znc-1986-1-212 [DOI] [PubMed] [Google Scholar]
- D’Amato, T. , Ganson, R. J. , Gaines, C. , & Jensen, R. (1984). Subcellular localization of chorismate‐mutase isoenzymes in protoplasts from mesophyll and suspension‐cultured cells of Nicotiana silvestris . Planta, 162, 104–108. 10.1007/BF00410205 [DOI] [PubMed] [Google Scholar]
- de Oliveira, M. V. V. , Jin, X. , Chen, X. , Griffith, D. , Batchu, S. , & Maeda, H. A. (2019). Imbalance of tyrosine by modulating TyrA arogenate dehydrogenases impacts growth and development of Arabidopsis thaliana . Plant Journal, 97, 901–922. [DOI] [PubMed] [Google Scholar]
- Ding, L. , Hofius, D. , Hajirezaei, M.‐R. , Fernie, A. R. , Börnke, F. , & Sonnewald, U. (2007). Functional analysis of the essential bifunctional tobacco enzyme 3‐dehydroquinate dehydratase/shikimate dehydrogenase in transgenic tobacco plants. Journal of Experimental Botany, 58, 2053–2067. 10.1093/jxb/erm059 [DOI] [PubMed] [Google Scholar]
- Dixon, D. P. , & Edwards, R. (2006). Enzymes of tyrosine catabolism in Arabidopsis thaliana . Plant Science, 171, 360–366. 10.1016/j.plantsci.2006.04.008 [DOI] [PubMed] [Google Scholar]
- Dyer, W. E. , Henstrand, J. M. , Handa, A. K. , & Herrmann, K. M. (1989). Wounding induces the first enzyme of the shikimate pathway in Solanaceae. Proceedings of the National Academy of Sciences, USA, 86, 7370–7373. 10.1073/pnas.86.19.7370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard, J. , Ehrler, T. , Epple, P. , Felix, G. , Raesecke, H.‐R. , Amrhein, N. , & Schmid, J. (1996). Cytosolic and plastidic chorismate mutase isozymes from Arabidopsis thaliana: Molecular characterization and enzymatic properties. Plant Journal, 10, 815–821. 10.1046/j.1365-313X.1996.10050815.x [DOI] [PubMed] [Google Scholar]
- Fitzpatrick, P. F. (1999). Tetrahydropterin‐dependent amino acid hydroxylases. Annual Review of Biochemistry, 68, 355–381. 10.1146/annurev.biochem.68.1.355 [DOI] [PubMed] [Google Scholar]
- Gaines, C. G. , Byng, G. S. , Whitaker, R. J. , & Jensen, R. A. (1982). L‐Tyrosine regulation and biosynthesis via arogenate dehydrogenase in suspension‐cultured cells of Nicotiana silvestris Speg. et Comes. Planta, 156, 233–240. 10.1007/BF00393730 [DOI] [PubMed] [Google Scholar]
- Gamborg, O. L. , & Keeley, F. W. (1966). Aromatic metabolism in plants I. A study of the prephenate dehydrogenase from bean plants. Biochimica Et Biophysica Acta, 115, 65–72. 10.1016/0304-4165(66)90049-3 [DOI] [PubMed] [Google Scholar]
- Ganson, R. J. , D’Amato, T. A. , & Jensen, R. A. (1986). The two‐isozyme system of 3‐deoxy‐d‐arabino‐heptulosonate 7‐phosphate synthase in Nicotiana silvestris and other higher plants. Plant Physiology, 82, 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giebel, J. (1973). Phenylalanine and tyrosine ammonia‐lyase activities in potato roots and their significance in potato resistance to Heterodera rostochiensis . Nematologica, 19, 3–6. 10.1163/187529273X00024 [DOI] [Google Scholar]
- Gilbert, G. A. , Gadush, M. V. , Wilson, C. , & Madore, M. A. (1998). Amino acid accumulation in sink and source tissues of Coleus blumei Benth. during salinity stress. Journal of Experimental Botany, 49, 107–114. 10.1093/jxb/49.318.107 [DOI] [Google Scholar]
- Goers, S. K. , & Jensen, R. A. (1984). Separation and characterization of two chorismate‐mutase isoenzymes from Nicotiana silvestris . Planta, 162, 109–116. 10.1007/BF00410206 [DOI] [PubMed] [Google Scholar]
- Gonzali, S. , Mazzucato, A. , & Perata, P. (2009). Purple as a tomato: Towards high anthocyanin tomatoes. Trends in Plant Science, 14, 237–241. 10.1016/j.tplants.2009.02.001 [DOI] [PubMed] [Google Scholar]
- Han, C. , Ren, C. , Zhi, T. , Zhou, Z. , Liu, Y. , Chen, F. , … Xie, D. (2013). Disruption of fumarylacetoacetate hydrolase causes spontaneous cell death under short‐day conditions in Arabidopsis. Plant Physiology, 162, 1956–1964. 10.1104/pp.113.216804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy, R. W. F. , Holsten, R. D. , Jackson, E. K. , & Burns, R. C. (1968). The acetylene‐ethylene assay for N2 fixation: Laboratory and field evaluation. Plant Physiology, 43, 1185–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, J. , Benedito, V. A. , Wang, M. , Murray, J. D. , Zhao, P. X. , Tang, Y. , & Udvardi, M. K. (2009). The Medicago truncatula gene expression atlas web server. BMC Bioinformatics, 10, 441 10.1186/1471-2105-10-441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi, T. , Ito, Y. , & Kawamura, I. (1967). p‐hydroxyphenylpropane component of grass lignin and role of tyrosine‐ammonia lyase in its formation. Phytochemistry, 6, 875–881. 10.1016/S0031-9422(00)86035-5 [DOI] [Google Scholar]
- Hildebrandt, T. M. , Nunes Nesi, A. , Araújo, W. L. , & Braun, H.‐P. (2015). Amino acid catabolism in plants. Mol Plant, 8, 1563–1579. 10.1016/j.molp.2015.09.005 [DOI] [PubMed] [Google Scholar]
- Jung, E. , Zamir, L. O. , & Jensen, R. A. (1986). Chloroplasts of higher plants synthesize L‐phenylalanine via L‐arogenate. Proceedings of the National Academy of Sciences USA, 83, 7231–7235. 10.1073/pnas.83.19.7231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, W. , Prithiviraj, B. , & Smith, D. L. (2003). Chitosan and chitin oligomers increase phenylalanine ammonia‐lyase and tyrosine ammonia‐lyase activities in soybean leaves. Journal of Plant Physiology, 160, 859–863. 10.1078/0176-1617-00905 [DOI] [PubMed] [Google Scholar]
- Krapp, A. (2015). Plant nitrogen assimilation and its regulation: A complex puzzle with missing pieces. Current Opinion in Plant Biology, 25, 115–122. 10.1016/j.pbi.2015.05.010 [DOI] [PubMed] [Google Scholar]
- Kries, H. , & O’Connor, S. E. (2016). Biocatalysts from alkaloid producing plants. Current Opinion in Chemical Biology, 31, 22–30. 10.1016/j.cbpa.2015.12.006 [DOI] [PubMed] [Google Scholar]
- Kryvoruchko, I. S. , Sinharoy, S. , Torres‐Jerez, I. , Sosso, D. , Pislariu, C. I. , Guan, D. , … Udvardi, M. K. (2016). MtSWEET11, a nodule‐specific sucrose transporter of Medicago truncatula . Plant Physiology, 171, 554–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroki, G. W. , & Conn, E. E. (1989). Differential activities of chorismate mutase isozymes in tubers and leaves of Solanum tuberosum L. Plant Physiology, 89, 472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba, M. , Tanaka, A. , & Tanaka, R. (2013). Stay‐green plants: What do they tell us about the molecular mechanism of leaf senescence. Photosynthesis Research, 117, 221–234. 10.1007/s11120-013-9862-x [DOI] [PubMed] [Google Scholar]
- Lee, J. , Durst, R. W. , Wrolstad, R. E. , Eisele, T. , Giusti, M. M. , Hach, J. , … Wightman, J. D. (2005). Determination of total monomeric anthocyanin pigment content of fruit juices, beverages, natural colorants, and wines by the pH differential method: Collaborative study. Journal of AOAC International, 88, 1269–1278. 10.1093/jaoac/88.5.1269 [DOI] [PubMed] [Google Scholar]
- Lokvam, J. , Brenes‐Arguedas, T. , Lee, J. S. , Coley, P. D. , & Kursar, T. A. (2006). Allelochemic function for a primary metabolite: The case of L‐tyrosine hyper‐production in Inga umbellifera (Fabaceae). American Journal of Botany, 93, 1109–1115. [DOI] [PubMed] [Google Scholar]
- Maeda, H. A. (2019). Evolutionary diversification of primary metabolism and Its contribution to plant chemical diversity. Frontiers in Plant Science, 10 10.3389/fpls.2019.00881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, H. , & Dudareva, N. (2012). The shikimate pathway and aromatic amino acid biosynthesis in plants. Annual Review of Plant Biology, 63, 73–105. 10.1146/annurev-arplant-042811-105439 [DOI] [PubMed] [Google Scholar]
- Maeda, H. , Yoo, H. , & Dudareva, N. (2011). Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate. Nature Chemical Biology, 7, 19–22. 10.1038/nchembio.485 [DOI] [PubMed] [Google Scholar]
- Matamoros, M. A. , Baird, L. M. , Escuredo, P. R. , Dalton, D. A. , Minchin, F. R. , Iturbe‐Ormaetxe, I. , … Becana, M. (1999). Stress‐induced legume root nodule senescence. physiological, biochemical, and structural alterations. Plant Physiology, 121, 97–112. 10.1104/pp.121.1.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz, J. G. , Nixon, P. J. , Rogner, M. , Brudvig, G. W. , & Diner, B. A. (1989). Directed alteration of the D1 polypeptide of photosystem II: Evidence that tyrosine‐161 is the redox component, Z, connecting the oxygen‐evolving complex to the primary electron donor, P680. Biochemistry, 28, 6960–6969. 10.1021/bi00443a028 [DOI] [PubMed] [Google Scholar]
- Mitra, P. P. , & Loqué, D. (2014). Histochemical staining of Arabidopsis thaliana secondary cell wall elements. JoVE Journal of Visualized Experiments, e51381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley, E. M. , Kunkel, B. N. , & Keith, B. (1999). Identification, characterization and comparative analysis of a novel chorismate mutase gene in Arabidopsis thaliana . Gene, 240, 115–123. 10.1016/S0378-1119(99)00423-0 [DOI] [PubMed] [Google Scholar]
- Moghe, G. D. , & Last, R. L. (2015). Something old, something new: Conserved enzymes and the evolution of novelty in plant specialized metabolism. Plant Physiology, 169, 1512–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller, B. L. (2010). Functional diversifications of cyanogenic glucosides. Current Opinion in Plant Biology, 13, 338–347. 10.1016/j.pbi.2010.01.009 [DOI] [PubMed] [Google Scholar]
- Oldroyd, G. E. D. (2013). Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nature Reviews Microbiology, 11, 252–263. 10.1038/nrmicro2990 [DOI] [PubMed] [Google Scholar]
- Oldroyd, G. E. D. , Murray, J. D. , Poole, P. S. , & Downie, J. A. (2011). The rules of engagement in the legume‐rhizobial symbiosis. Annual Review of Genetics, 45, 119–144. 10.1146/annurev-genet-110410-132549 [DOI] [PubMed] [Google Scholar]
- Peng, C. , Uygun, S. , Shiu, S.‐H. , & Last, R. L. (2014). The impact of the branched‐chain ketoacid dehydrogenase complex on amino acid homeostasis in Arabidopsis. Plant Physiology, 169, 1807–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez Guerra, J. C. , Coussens, G. , De Keyser, A. , De Rycke, R. , De Bodt, S. , Van De Velde, W. , … Holsters, M. (2010). Comparison of developmental and stress‐induced nodule senescence in Medicago truncatula . Plant Physiology, 152, 1574–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, M. (2013). Rosmarinic acid: new aspects. Phytochemistry Reviews, 12, 207–227. 10.1007/s11101-013-9282-8 [DOI] [Google Scholar]
- Pomar, F. , Merino, F. , & Barceló, A. R. (2002). O‐4‐Linked coniferyl and sinapyl aldehydes in lignifying cell walls are the main targets of the Wiesner (phloroglucinol‐HCl) reaction. Protoplasma, 220, 17–28. 10.1007/s00709-002-0030-y [DOI] [PubMed] [Google Scholar]
- Qian, Y. , Lynch, J. H. , Guo, L. , Rhodes, D. , Morgan, J. A. , & Dudareva, N. (2019). Completion of the cytosolic post‐chorismate phenylalanine biosynthetic pathway in plants. Nature Communications, 10, 15 10.1038/s41467-018-07969-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippert, P. , & Matringe, M. (2002). Purification and kinetic analysis of the two recombinant arogenate dehydrogenase isoforms of Arabidopsis thaliana . European Journal of Biochemistry, 269, 4753–4761. 10.1046/j.1432-1033.2002.03172.x [DOI] [PubMed] [Google Scholar]
- Rosler, J. , Krekel, F. , Amrhein, N. , & Schmid, J. (1997). Maize phenylalanine ammonia‐lyase has tyrosine ammonia‐lyase activity. Plant Physiology, 113, 175–179. 10.1104/pp.113.1.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, J. L. , & Jensen, R. A. (1979). Enzymology of L‐tyrosine biosynthesis in mung bean (Vigna radiata [L.] Wilczek). Plant Physiology, 64, 727–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, J. L. , & Jensen, R. A. (1985). Differentially regulated isozymes of 3‐deoxy‐d‐arabino‐heptulosonate‐7‐phosphate synthase from seedlings of Vigna radiata [L.] Wilczek. Plant Physiology, 79, 711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijter, J. M. , Pfaffl, M. W. , Zhao, S. , Spiess, A. N. , Boggy, G. , Blom, J. , … Vandesompele, J. O. (2013). Evaluation of qPCR curve analysis methods for reliable biomarker discovery: Bias, resolution, precision, and implications. Methods, 59, 32–46. 10.1016/j.ymeth.2012.08.011 [DOI] [PubMed] [Google Scholar]
- Schenck, C. A. , Chen, S. , Siehl, D. L. , & Maeda, H. A. (2015). Non‐plastidic, tyrosine‐insensitive prephenate dehydrogenases from legumes. Nature Chemical Biology, 11, 52–57. 10.1038/nchembio.1693 [DOI] [PubMed] [Google Scholar]
- Schenck, C. A. , Holland, C. K. , Schneider, M. R. , Men, Y. , Lee, S. G. , Jez, J. M. , & Maeda, H. A. (2017a). Molecular basis of the evolution of alternative tyrosine biosynthetic routes in plants. Nature Chemical Biology, 13, 1029–1035. 10.1038/nchembio.2414 [DOI] [PubMed] [Google Scholar]
- Schenck, C. A. , & Last, R. L. (2019). Location, location! cellular relocalization primes specialized metabolic diversification. FEBS Journal, 287(7), 1359–1368. [DOI] [PubMed] [Google Scholar]
- Schenck, C. A. , & Maeda, H. A. (2018). Tyrosine biosynthesis, metabolism, and catabolism in plants. Phytochemistry, 149, 82–102. 10.1016/j.phytochem.2018.02.003 [DOI] [PubMed] [Google Scholar]
- Schenck, C. A. , Men, Y. , & Maeda, H. A. (2017). Conserved molecular mechanism of TyrA dehydrogenase substrate specificity underlying alternative tyrosine biosynthetic pathways in plants and microbes. Frontiers in Molecular Biosciences, 4 10.3389/fmolb.2017.00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabra, A. R. , Pereira, P. A. , Becker, J. D. , & Carvalho, H. G. (2012). Inhibition of glutamine synthetase by phosphinothricin leads to transcriptome reprograming in root nodules of Medicago truncatula . Molecular Plant‐Microbe Interactions, 25, 976–992. [DOI] [PubMed] [Google Scholar]
- Siehl, D. (1999). The biosynthesis of tryptophan, tyrosine, and phenylalanine from chorismate In Singh B. (Ed.), Plant Amino Acids: Biochemistry and Biotechnology (pp. 171–204). New York: CRC Press. [Google Scholar]
- Siehl, D. L. , Tao, Y. , Albert, H. , Dong, Y. , Heckert, M. , Madrigal, A. , … Castle, L. A. (2014). Broad 4‐hydroxyphenylpyruvate dioxygenase inhibitor herbicide tolerance in soybean with an optimized enzyme and expression cassette. Plant Physiology, 166(3), 1162–1176. 10.1104/pp.114.247205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey, M. G. , Cahoon, R. E. , Nguyen, H. T. , Cui, Y. , Sato, S. , Nguyen, C. T. , … Stacey, G. (2016). Identification of homogentisate dioxygenase as a target for vitamin E biofortification in oilseeds. Plant Physiology, 172, 1506–1518. 10.1104/pp.16.00941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starker, C. G. , Parra‐Colmenares, A. L. , Smith, L. , Mitra, R. M. , & Long, S. R. (2006). Nitrogen fixation mutants of Medicago truncatula fail to support plant and bacterial symbiotic gene expression. Plant Physiology, 140, 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack, D. , Vogt, T. , & Schliemann, W. (2003). Recent advances in betalain research. Phytochemistry, 62, 247–269. 10.1016/S0031-9422(02)00564-2 [DOI] [PubMed] [Google Scholar]
- Streeter, J. , & Wong, P. P. (1988). Inhibition of legume nodule formation and N2 fixation by nitrate. Critical Reviews in Plant Sciences, 7, 1–23. [Google Scholar]
- Tadege, M. , Wen, J. , He, J. I. , Tu, H. , Kwak, Y. , Eschstruth, A. , … Mysore, K. S. (2008). Large‐scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula . Plant Journal, 54, 335–347. 10.1111/j.1365-313X.2008.03418.x [DOI] [PubMed] [Google Scholar]
- Tzin, V. , & Galili, G. (2010). New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Molecular Plant, 3, 956–972. 10.1093/mp/ssq048 [DOI] [PubMed] [Google Scholar]
- Van de Velde, W. , Guerra, J. C. P. , De Keyser, A. , De Rycke, R. , Rombauts, S. , Maunoury, N. , … Goormachtig, S. (2006). Aging in legume symbiosis. A molecular view on nodule senescence in Medicago truncatula . Plant Physiology, 141, 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Toda, K. , Block, A. , & Maeda, H. A. (2019). TAT1 and TAT2 tyrosine aminotransferases have both distinct and shared functions in tyrosine metabolism and degradation in Arabidopsis thaliana . Journal of Biological Chemistry, 294, 3563–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Toda, K. , & Maeda, H. A. (2016). Biochemical properties and subcellular localization of tyrosine aminotransferases in Arabidopsis thaliana . Phytochemistry, 132, 16–25. 10.1016/j.phytochem.2016.09.007 [DOI] [PubMed] [Google Scholar]
- Weng, J.‐K. , Philippe, R. N. , & Noel, J. P. (2012). The rise of chemodiversity in plants. Science, 336, 1667–1670. 10.1126/science.1217411 [DOI] [PubMed] [Google Scholar]
- Westfall, C. S. , Xu, A. , & Jez, J. M. (2014). Structural evolution of differential amino acid effector regulation in plant chorismate mutases. Journal of Biological Chemistry, 289, 28619–28628. 10.1074/jbc.M114.591123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wych, R. D. , & Rains, D. W. (1978). Simultaneous measurement of nitrogen fixation estimated by acetylene‐ethylene assay and nitrate absorption by soybeans. Plant Physiology, 62, 443–448. 10.1104/pp.62.3.443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi, J. , Chen, Y. , Nakashima, J. , Wang, S. , & Chen, R. (2013). Medicago truncatula esn1 defines a genetic locus involved in nodule senescence and symbiotic nitrogen fixation. Molecular Plant‐Microbe Interactions, 26, 893–902. [DOI] [PubMed] [Google Scholar]
- Yoo, H. , Widhalm, J. R. , Qian, Y. , Maeda, H. , Cooper, B. R. , Jannasch, A. S. , … Dudareva, N. (2013). An alternative pathway contributes to phenylalanine biosynthesis in plants via a cytosolic tyrosine: Phenylpyruvate aminotransferase. Nature Communications, 4, 1–11. 10.1038/ncomms3833 [DOI] [PubMed] [Google Scholar]
- Zhao, J. , Williams, C. C. , & Last, R. L. (1998). Induction of Arabidopsis tryptophan pathway enzymes and camalexin by amino acid starvation, oxidative stress, and an abiotic elicitor. Plant Cell, 10, 359–370. 10.1105/tpc.10.3.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1‐S9
Tables S1‐S2
Supplementary Material


