Abstract
Protein O-glucosyltransferase 1 (POGLUT1) activity is critical for the Notch signaling pathway, being one of the main enzymes responsible for the glycosylation of the extracellular domain of Notch receptors. A biallelic mutation in the POGLUT1 gene has been reported in one family as cause of an adult-onset limb-girdle muscular dystrophy (LGMD R21; OMIM# 617232). As the result of a collaborative international effort, we have identified the first cohort of 15 patients with LGMD R21, from nine unrelated families coming from different countries, providing a reliable phenotype-genotype and mechanistic insight. Patients carrying novel mutations in POGLUT1 all displayed a clinical picture of limb-girdle muscle weakness. However, the age at onset was broadened from adult to congenital and infantile onset. Moreover, we now report that the unique muscle imaging pattern of “inside-to-outside” fatty degeneration observed in the original cases is indeed a defining feature of POGLUT1 muscular dystrophy. Experiments on muscle biopsies from patients revealed a remarkable and consistent decrease in the level of the NOTCH1 intracellular domain, reduction of the pool of satellite cells (SC), and evidence of α-dystroglycan hypoglycosylation. In vitro biochemical and cell-based assays suggested a pathogenic role of the novel POGLUT1 mutations, leading to reduced enzymatic activity and/or protein stability. The association between the POGLUT1 variants and the muscular phenotype was established by in vivo experiments analyzing the indirect flight muscle development in transgenic Drosophila, showing that the human POGLUT1 mutations reduced its myogenic activity. In line with the well-known role of the Notch pathway in the homeostasis of SC and muscle regeneration, SC-derived myoblasts from patient’s muscle samples showed decreased proliferation and facilitated differentiation. Together, these observations suggest that alterations in SC biology caused by reduced Notch1 signaling result in muscular dystrophy in LGMD R21 patients, likely with additional contribution from α-dystroglycan hypoglycosylation. This study settles the muscular clinical phenotype linked to POGLUT1 mutations and establishes the pathogenic mechanism underlying this muscle disorder. The description of a specific imaging pattern of fatty degeneration and muscle pathology with a decrease of α-dystroglycan glycosylation provides excellent tools which will help diagnose and follow-up the LGMD R21 patients.
Keywords: POGLUT1, satellite cells, Notch, muscle dystrophy, α-dystroglycan
INTRODUCTION
Muscular dystrophies are inherited disorders characterized by progressive weakness due to skeletal muscle degeneration. The long list of disease genes reveals remarkable heterogeneity of underlying molecular mechanisms [29]. We have recently reported a homozygous missense mutation (p.D233E) in the protein O-glucosyltransferase 1 gene, POGLUT1, in four siblings with an autosomal recessive limb-girdle muscular dystrophy (LGMD), OMIM# 617232 [35] named as LGMD2Z or LGMD R21 POGLUT1-related [7, 38]. POGLUT1 specifically adds O-glucose to EGF repeats containing the CxSx(P/A)C consensus sequence, many of which are found in Notch extracellular domains [13, 31]. Glycosylation of the extracellular domain of Notch receptors is essential for the Notch signaling pathway, which controls cell fate during developmental processes in multiple tissues including skeletal [18, 40, 44]. Satellite cells (SC) are muscle-specific adult stem cells, which are primary contributors to skeletal muscle growth and repair [11]. Despite continuous regeneration, the pool of SC is maintained in healthy muscle through asymmetric cell divisions, which give rise both to self-renewing SC and to committed myogenic progenitors [21, 32]. In this process, the Notch pathway is key for maintaining quiescence in SC and for homing of SC-derived myoblasts [6, 9]. No primary defects of SC or Notch have been identified in human muscular dystrophies. The p.D233E POGLUT1 mutation dramatically reduces O-glucosyltransferase activity, leading to impaired Notch signaling, defects in SC proliferation and differentiation, and exhaustion of the SC pool as the primary cause of this novel muscular dystrophy [35].
Our first description of the LGMD R21 was based on the analysis of one family carrying the same homozygous mutation in the POGLUT1 gene [35]. Although the four siblings showed almost identical clinical and radiological features, it was not clear whether the phenotypes observed in those patients would be similar in new patients carrying different mutations, and therefore representative of this newly characterized muscular dystrophy. The description of additional LGMD R21 patients would clarify this question and contribute to the identification of potential biomarkers to facilitate the diagnosis of these patients. Additionally, as the application of next generation sequencing (NGS) techniques is also uncovering an increasing number of variants of unknown significance (VUS), the availability of specific biomarkers would provide insight into their pathogenicity.
Here, we describe 15 patients from nine unrelated families from different countries carrying novel biallelic mutations in POGLUT1, broadening the presentation from congenital to adult onset. Although the clinical picture showed limb-girdle weakness regardless of the age at onset, similar to other LGMDs, muscle imaging showed a very specific and selective pattern of fatty degeneration in all patients. In addition, muscle biopsies and isolated myoblasts from patients with POGLUT1 mutations displayed a remarkable reduction of the NOTCH1 intracellular domain (NICD), PAX7 levels, and α-dystroglycan glycosylation, together with altered proliferation and differentiation, regardless of the severity of the phenotype. These findings not only provide important clues to establish the pathogenic mechanism underlying this muscle disorder, but also describe specific muscle pathology and a characteristic and recognizable radiological phenotype, which will help in diagnosing additional patients and understanding the disease course.
METHODS
Patients and clinical assessment
The responsible neurologist in each participant center performed identification, examination of the patients and their relatives, and muscle biopsy. This study was approved by the Institutional Research Ethic Committee at Hospital Universitario Virgen del Rocío in Sevilla (Spain) as the coordinating center. Prior to inclusion in the study, written informed consent was received from participants in their respective centers for genetic studies, muscle biopsies, and pictures appearing in the manuscript.
Muscle imaging
Axial muscle MRI (1.5 or 3 Tesla scanner) or computerized tomography (CT) scans were performed at the level of the thighs and lower legs. MRI studies included a T1-weighted spin-echo sequence (TR 600–700 ms, TE 30 ms) and a short-time inversion recovery (STIR) sequence (TR 2500–3500 ms, TE 60 ms, TI 150 ms), in 10 mm slices. Fatty degeneration of muscles was identified according to the increased signal in T1 sequences (or hypodensity on CT). STIR sequences were used to evaluate the presence of edema.
Genetic studies
DNA was extracted from blood using standard procedures and patients were studied using different NGS technologies. All variants were confirmed by Sanger sequencing.
Family 1 was reported elsewhere [35]. Patient II.1 from Family 2 was studied with a panel of 4,800 genes associated with inherited diseases (TruSight One Sequencing Panel) using HighSeq Illumina platform. Patient II.2 from Family 3 was studied with a custom made panel of 230 genes associated with muscular dystrophies, using HighSeq and NextSeq Illumina platforms. Samples from patients II.1, II.2 and II.3 from Family 4, II.1 from Family 5, II.4 from Family 6, II.1 from Family 7 (all gathered through the MYO-SEQ project, a research collaboration between academia, patient organizations and industry to apply whole exome sequencing [WES] to patients with unexplained limb-girdle muscle weakness), II.1 and II.2 from Family 8, and II.1 from Family 9 were exome-sequenced at the Broad Institute of Harvard and MIT (Broad Institute, Cambridge, MA, USA), using Illumina exome capture (38 Mb target) and analysed using the Genomics Platform (https://seqr.broadinstitute.org/).
Muscle biopsy
Muscle samples were obtained by open muscle biopsies from the biceps brachii muscle from Family 2 (II.1), Family 3 (II.2), Family 4 (II.1), Family 5 (II.1), Family 6 (II.4) and five aged-matched healthy controls. Control muscles were obtained from healthy individuals undergoing orthopedic surgery. Frozen muscle sections were cross-sectioned on a cryostat at 7μm thickness and mounted on slides for hematoxylin and eosin (H&E) and NADH staining. Stained sections were evaluated with an Olympus BX41 (Tokyo, Japan) equipped with a ColorView IIIu camera (Olympus).
Immunohistochemistry
Cultured primary myoblasts of one patient from Family 2 (II.1) and from three aged-matched healthy controls were fixed with 4% paraformaldehyde in phosphate buffer for 30 min, permeabilized in 0.2% Triton X-100 (Sigma-Aldrich) for 10 min and blocked in 1% BSA/PBS for 45 min. The following primary antibodies were used: mouse monoclonal anti-Pax7 (1:25; DHSB), rabbit polyclonal anti-MyoD (aa211–240) (1:50; LifeSpan Biosciences); rabbit monoclonal anti-Desmin (D93FD5) (1:100; Abcam); mouse monoclonal anti-Myogenin (FD5) (1:100; Abcam) and incubated for 12 h at 4 °C. Frozen muscle samples were fixed with 4% PFA for 15 min, immersed in citrate buffer (pH 6) for 30 min at 80 °C and washed with PBS. Afterward, muscle samples were blocked with 2% non-fat milk + 0.3% Triton X-100 in PBS for 30 min and with 5% BSA + 0.5% Triton X-100 in PBS for 1 h. The following primary antibodies were used: mouse monoclonal anti-Pax7 (1:50; DHSB); mouse monoclonal anti-collagen VI (3C4) (1:1000; Chemicon); and embryonic myosin heavy chain (eMHC) and incubated for 3 days at 4 °C. After washing, samples (myoblasts and muscles) were incubated with the secondary antibodies conjugated with Cy2 and Cy3 fluorophores (1:500; Jackson ImmunoResearch) for 1 h. Finally, the nuclei were stained for 20 min with To-pro-3-Iodide (Topro 3) at 1/1000 in PBS and the slides were coverslipped with fluorescence mounting medium (Dako). Coverslips were imaged on a Zeiss LSM 710 confocal laser scanning microscope using 20x objective with a numerical aperture of 1.3. Images from controls and patients were obtained at the same day and under equal conditions (laser intensities and photomultiplier voltages). Maximal projections of Z-stacked images were analyzed with ImageJ software (RRID:SCR_003070).
Immunoblots
Frozen muscle samples were homogenized in RIPA buffer (20 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% IGEPAL, 0.1% SDS) containing protease inhibitor cocktail (Roche). The lysates were centrifuged at 13,000 rpm at 4°C for 20 min. The supernatant was collected. For enrichment of glycoproteins, 200 μg of protein of the total lysates was mixed with 100 μg of wheat-germ agglutinin (WGA) agarose (Sigma-Aldrich) as described previously (Michele et al, 2002). The protein lysates non-incubated or incubated with WGA agarose beads were separated on 7–10% SDS–PAGE gels and transferred onto PVDF membranes (Millipore). Western blot analysis of equal-protein loading was performed with the following primary antibodies: mouse monoclonal anti-α dystroglycan (IIH6C4) (1:1000; Millipore); mouse monoclonal anti-β dystroglycan (43DAG1/8D5) (1:500; Novocastra); sheep anti-α dystroglycan core (317) (1:200; S. Kroger laboratory); rabbit monoclonal anti-Notch1 (val1744) (1:1000; Cell Signaling); rabbit polyclonal anti-POGLUT1 (1:100; Novus Biologicals) and rabbit polyclonal anti-GAPDH (1:2000; Sigma-Aldrich). Immunoreactivity was detected with secondary antibodies conjugated to horseradish peroxidase (1:5000; Jackson Immuno Research) and developed with SuperSignal West Femto (Thermo Fisher Scientific) using an ImageQuant LAS 4000 MiniGold System (GE Healthcare Life Sciences).
Ligand overlay assays were performed as previously described with minor modifications [26, 43]. Briefly, PVDF membranes were incubated with Engelbreth-Holm-Swarm laminin (Sigma-Aldrich) overnight at 4°C in laminin-binding buffer. Then, the membranes were washed and incubated with monoclonal anti-laminin (LAM-89) (1:100; Sigma-Aldrich) primary antibody and their corresponding secondary antibody. Blots were imaged using the protocol described for western blots.
Primary cell culture
Myoblasts from Family 2 (II.1) and from three age-matched healthy controls were used for the experiments. Fresh muscle samples were minced and cultured in a monolayer. Briefly, the culture medium for the myoblast stage (proliferation medium) contained 75% Dulbecco’s Minimal Essential Medium (DMEM, Invitrogen) and 25% M199 medium (Invitrogen), supplemented with 10% fetal bovine serum (FBS), 10 μg/ml insulin, 2 mM glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, 0.25 μg/ml fungizone, 10 ng/ml epidermal growth factor, and 25 ng/ml fibroblast growth factor. To obtain highly purified myoblasts, each 107 cells were mixed with 20 μl of CD56-coated microbeads (Milteny Biotec, Bergisch Gladbach, Germany) and incubated at 4 °C for 15 min. Unbound microbeads were removed by washing cells in excess PBS buffer followed by centrifugation at 400×g for 10 min. The cell pellet was resuspended in PBS buffer to a concentration of 2 × 108 cells/ml before separation on a midiMACS cell separator (Milteny Biotec, Bergisch Gladbach, Germany).
Biochemical assays
POGLUT1 mutants were generated by standard PCR-based site-directed mutagenesis method with a pcDNA4-based (wild type) WT POGLUT1-MycHis6 expression vector or a pTracer-based POGLUT1-FLAG expression vector as a template and primers specific to each mutation (Supplementary Table 1, online resource). Successful incorporation of the specific mutations was confirmed by DNA Sanger sequencing.
To confirm expression of WT and mutant forms of POGLUT1 proteins, transient transfection of the pcDNA4-based vectors of WT and R183W, Y57C, I129T, R98W, C102F, and W308L mutants of POGLUT1-MycHis was performed in HEK293T cells using PEI as described [39]. An empty vector was used as a negative control. After transfection, the expression and secretion of POGLUT1 proteins in cell lysates and culture media were examined by western blot analysis with an anti-Myc antibody. Revertants were generated by using specific primers (Supplementary Table 1, online resource) and expressed in HEK293T cells to confirm whether the defects of some mutants were due to the intended mutations, and not to undesirable mutations generated during PCR during mutagenesis.
Enzymatic assays were performed with those POGLUT1 proteins that were secreted, the EGF repeat from human factor IX (which contains the CxSx(P/A)C consensus), and UDP-glucose. These assays were performed as previously described with a slight modification [37]. Briefly, 100–200 ng of either POGLUT1-MycHis6 WT, Y57C or R183W were incubated with 10 μM UDP-glucose (Ultra Pure, Promega) in the presence or absence of 10 μM human Factor IX EGF in separate 20 μL reactions containing 50 mM HEPES, pH 6.8, at 37°C for 1 h. Reactions were stopped by the addition of the UDP-Glo™ Detection Reagent. Quantitation of released UDP was performed using the UDP-Glo™ Glycosyl transferase Assay Kit (Promega Corporation) as described by the manufacturer. A standard curve using 0–3.125 μM UDP was performed, and the range of measurements was determined to be in the linear range of detection (in all cases, UDP-glucose consumption was <30% of the starting amount), where the luminescence detected is directly proportional to UDP concentration. After 1 h of incubation, luminescence was measured in triplicate using a Cytation™ 3 imaging reader (BioTek) with the following settings: Shake (Linear 1 sec frequency 567 cpm, 3 mm); Luminescence Endpoint; Integration time, 10 sec; Filter Set 1 (Emission: Full light; Optics: Top, Gain 200), Read Speed: Normal, Delay 100 msec; Extended Dynamic Range; Read Height: 1 mm.
Secretion assay of mouse Notch1 EGF1–18-MycHis6 was performed with co-transfection of WT or mutants of pcDNA4 POGLUT1 in POGLUT1 KO HEK293T cells as described [39]. Briefly, two expression vectors, pSecTag encoding mouse Notch1 EGF1–18-MycHis6 and WT or mutant forms of pcDNA4 POGLUT1-MycHis6, were co-transfected with an IgG expression vector as a secretion control in WT or POGLUT1 KO HEK293T cells (note that the secretion of IgG does not depend on POGLUT1-mediated O-glucosylation). The mouse Notch1 EGF1–18-MycHis proteins and POGLUT1 proteins, or IgG in the culture media and cell lysates were detected by western blot using an anti-Myc antibody (clone 9E10) or anti-human IgG antibody, respectively as described [39].
Transgenic fly generation
To generate the transgenic flies for overexpression of human POGLUT1mutant, a BamHI-XbaI fragment containing the POGLUT1mutant-FLAG ORF was released from pTracer-POGLUT1mutant-FLAG and cloned into the pUASTattB vector [5] double digested with BglII and XbaI sites to generate pUASTattB-POGLUT1mutant-FLAG constructs. After verification by Sanger sequencing, the constructs were integrated into the VK22 docking site by using ΦC31-mediated transgenesis [41], resulting in the generation of UASattB-POGLUT1mutant-FLAG-VK22 transgenes. Embryo injections were performed by Geneti Vision (Houston, USA).
Expression level analysis of human POGLUT1 in Drosophila
Expression levels of WT and mutant POGLUT1 were examined by performing immunostaining in third instar wing imaginal discs overexpressing WT and mutant POGLUT1 driven by dpp-GAL4, which drives expression along the anterior-posterior boundary of the wing disc. Animals were raised at 25°C. UASattB-POGLUT1WT-FLAG-VK22 and UASattB-POGLUT1mutant-FLAG-VK22 animals were crossed with UAS-GFP; dpp-GAL4 animals to obtain UASattB-POGLUT1WT-FLAG-VK22/ UAS-GFP; dpp-GAL4/+ or UASattB-POGLUT1mutant-FLAG-VK22/ UAS-GFP; dpp-GAL4/+ animals. Staining was performed by α-FLAG antibody. The signal intensity of POGLUT1 for each imaginal disc was measured by ImageJ and normalized by the GFP signal intensity and the total area of the disc (also measured by ImageJ).
For Western blotting, proteins were extracted from wing imaginal discs in lysis buffer containing a protease inhibitor cocktail (Promega). The following antibodies were used: mouse α-FLAG M2 1:100 (Sigma), mouse α-tubulin 1:1000 (Santa Cruz Biotechnology Cat# sc-8035, RRID:AB_628408), goat α-mouse-HRP 1:2000 (Jackson Immuno Research Laboratories). Western blots were developed using Pierce ECL Western Blotting Substrates (Thermo Scientific). The bands were detected using an Image Quant LAS 4000 system from GE Healthcare.
Muscle phenotype analysis in Drosophila
The effect of mutations on POGLUT1 activity was assessed in terms of the development of indirect flight muscles in Drosophila [16]. Rescue experiments were performed with Mef2-GAL4 driven overexpression of POGLUT1WT and POGLUT1mutant in rumi79/79 animals, as reported previously [35]. Animals were raised at 25°C until puparium formation. After 25% pupal development, pupal indirect flight muscles were dissected and stained using standard methods. Antibodies used were mouse 22C10 1:50 (DSHB), mouse α‐FLAG M2 1:100 (Sigma), donkey α-rabbit-Cy3 1:500, donkeyα-mouse-Cy3 1:500 (Jackson Immuno Research Laboratories). Confocal imaging was performed on a Leica TCS-SP8 microscope. Myotube length measurements were performed with Amira5.2.2. Mean myotube lengths and standard deviations (SD) were calculated for pupae with the following genotypes: WT control (y w; n=3), rumi79/79 (n=3), and rumi79/79 overexpressing POGLUT1WT (n=4), POGLUT1D233E (n=3), POGLUT1R183W (n=3), POGLUT1Y57C (n=3) and POGLUT1C102F (n=3). One-way ANOVA with Tukey’s multiple comparisons test was used to determine the P-values. Images were processed with Adobe Photoshop CS5; figures were assembled in Adobe Illustrator CS5.
Data availability
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
RESULTS
Novel pathogenic variants in POGLUT1 were identified in multiple patients with recessive LGMD
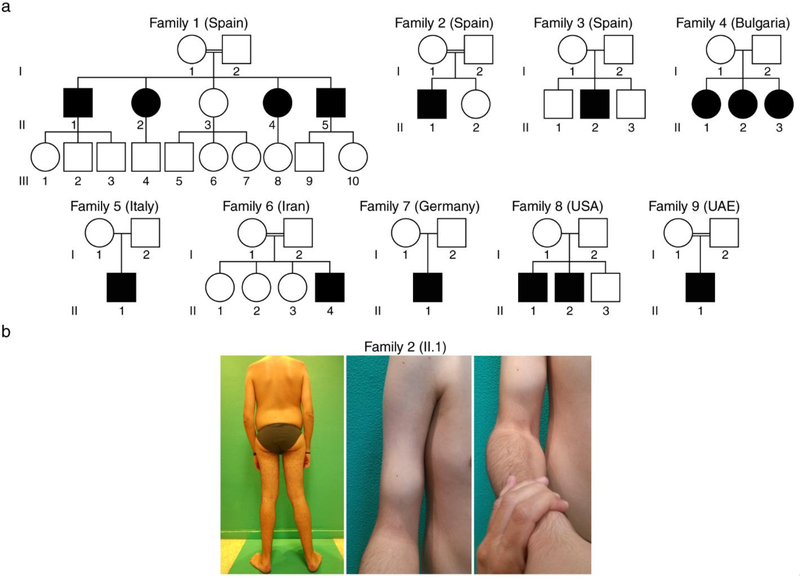
Sequencing data analysis identified nine missense and one nonsense recessive variants (homozygous or compound heterozygous) in the POGLUT1 gene, in 15 patients from nine independent families from seven different countries in Europe (Spain, Bulgaria, Italy, Germany), North America (USA) and Asia (Iran, United Arab Emirates) (Fig. 1a). Consanguinity was recognized in four of the nine families. All mutations were identified and annotated based on the canonical transcript (ENST00000295588). The mutation p.D233E was previously reported as homozygous in patients II.1, II.2, II.4 and II.5 from Family 1 [35]. The variant p.R279Q (previously described in a Dowling-Degos disease [DDD] patient as a heterozygous mutation [30]) was identified as homozygous in patient II.1 from Family 2. The homozygous missense variants p.I129T, p.R98W, p.Y57C, p.W308L, p.C102F (novel variants), and p.R183W (previously described as a DDD mutation) were identified in patients from Families 3, 4, 5, 6, 7, and 9 respectively. In the two affected siblings from Family 8 we identified two novel variants, p.Y156N and the c.552G>A variant leading to a stop codon p.W184*. No asymptomatic carriers were identified in the segregation analysis in any of the families. Study of affected and unaffected relatives confirmed correct segregation of the disease with the homozygous or compound heterozygous state for the POGLUT1 variants.
Figure 1. Patients identified with a LGMD and POGLUT1 missense mutations.
a Pedigrees of the nine independent families with different geographic origins (the family 1 has been reported elsewhere [35] and only the pedigree is included here); b patient II.1 from Family 2 with a congenital form showed scapular winging and elbow contractures (left), weakness and wasting of the biceps brachii (middle) and brachioradialis muscles (right).
Public databases were screened for the novel variants. All variants were either absent (p.I129T, p.W308L and p.W184*) or found in heterozygous state in one (p.R98W and p.C102F) or two individuals (p.Y156N and p.Y57C) out of >60,000 healthy European (Non-Finnish) controls (ExAC, http://exac.broadinstitute.org/).
POGLUT1 mutations are associated with adult, infantile and congenital forms of muscular dystrophy
Here, we report a series of 15 patients with a muscular dystrophy due to different mutations in POGLUT1, the largest series thus far. The patients showed a clinical spectrum of the disease with highly variable age of onset and an overall phenotype of LGMD. The characteristics of the patients are detailed in Table 1. We identified one patient with a congenital presentation, two patients with infantile onset before 3 years of age, and 12 patients with adult onset (58.3% in 2nd decade, 25% in 3rd decade, and 16.6% in 4th-5th decade). The weakness showed a proximal distribution, with a clear predominance in the lower limbs and slow progression. Seven patients (46.6%) with onset in their teens, were confined to wheelchair between the 3th and 5th decade. Mild to moderate scapular winging was present in 11 of the 15 patients. The presenting symptom in the congenital case was hypotonia with lower limb proximal weakness after gait acquisition, and further progression with mild weakness, wasting and contractures of the upper limbs (Fig. 1b). This patient with congenital presentation showed mild facial weakness, ptosis, and nasal voice. Mild to severe respiratory involvement was present in six patients (only two with ventilatory support at night) from the mid-forties onwards, regardless of the age at onset. No significant bulbar, cardiac or other systemic manifestations were detected. The level of serum CK was normal, except for four patients who showed mild elevation ranging from x2 to x6 the normal level.
Table 1.
Clinical data of the patients with recessive POGLUT1 mutations
| Family/Patient | Aminoacid change | Decade at onset | Actual age (y)/ Sex | Phenotype | LL proximal (MRC) | LL distal (MRC) | UL proximal (MRC) | UL distal (MRC) | CK(x nl) | Scapular winging | FVC (%) | Wheelchair /at age | Other signs in theclinical exam |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1/ II.1 | p.D233E/ p.D233E | 2nd | 60/M | LGMD- adult onset | 2 | 5 | 4 | 5 | N | Yes | 29 | Yes/ 50 | |
| 1/ II.2 | p.D233E/ p.D233E | 2nd | 51/F | LGMD- adult onset | 2 | 5 | 4 | 5 | N | Yes | 22 | Yes/ 44 | |
| 1/ II.4 | p.D233E/ p.D233E | 3th | 50/F | LGMD- adult onset | 2 | 5 | 4 | 5 | N | Yes | 91 | No | |
| 1/ II.5 | p.D233E/ p.D233E | 3th | 42/M | LGMD- adult onset | 2 | 5 | 3 | 5 | x3 | Yes | No | ||
| 2/ II.1 | p.D233E/ p.R279Q | 1st | 24/M | Congenital onset | 2 | 5 | 3–4 | 5 | N | Yes | >80 | Yes/22 | Elbow contractureMyopathic faciesArched palate |
| 3/ II.2 | p.I129T/ p.I129T | 1st | 46/M | LGMD- infantile onset | 2 | 5 | 2 | 5 | x2 | Yes | 45 | Yes/ 33 | Ankle contractureCalf hypertrophyScoliosis, hyperlordosis |
| 4/ II.1 | p.R98W/ p.R98W | 5th | 52/F | LGMD- adult onset | 4- | 5 | 4 | 5 | N | Yes | 72 | No | |
| 4/ II.2 | p.R98W/ p.R98W | 3rd | 47/F | LGMD- adult onset | 2 | 5 | 4 | 5 | N | Yes | 53 | No | |
| 4/ II.3 | p.R98W/ p.R98W | 3rd | 50/F | LGMD- adult onset | 2 | 5 | 4 | 5 | N | Yes | 76 | No | |
| 5/ II.1 | p.Y57C/ p.Y57C | 3rd | 38/F | LGMD- adult onset | 2–3 | 5 | 4 | 5 | N | Yes | >80 | No | Hyperlordosis |
| 6/ II.4 | p.W308L/ p.W308L | 2nd | 22/M | LGMD- adult onset | 2–3 | 5 | 5 | 5 | x6 | No | 86 | No | |
| 7/ II.1 | p.C102F/ p.C102F | 4th | 59/M | LGMD- adult onset | 2–3 | 5 | 3–4 | 5 | x3 | Yes | 85 | No | |
| 8/ II.1 | p.Y156N/ p.W184* | 2nd | 41/M | LGMD- adult onset | 2–3 | 4 | 4 | 5 | x4 | Yes | 86 | Yes/38 | |
| 8/ II.2 | p.Y156N/ p.W184* | 2nd | 35/M | LGMD- adult onset | 2–3 | 5 | 2–3 | 4–5 | x10 | Yes | 27 | Yes/32 | |
| 9/ II.1 | p.R183W/ p.R183W | 1st | 19/M | LGMD- infantile onset | 2–3 | 4–5 | 2 | 5 | x2 | Yes | 41 | Yes/13 | NystagmusRetinitis pigmentosa |
y= Years; M= Male; F= Female; LGMD= Limb Girdle Muscular Dystrophy; LL= Lower limb; UL= Upper limit; MRC= Medical Research Council scale for manual muscle testing; CK= Creatine kinase; nl= Normal limit; FVC= Forced vital Capacity
A characteristic pattern in muscle imaging
Muscle imaging studies of the lower limbs were available from 11 of 15 patients. The muscle MRI or CT depicted a so called “from inside-to-outside” pattern of fatty degeneration in all the patients (Fig. 2). This special pattern of partial degeneration of the muscles, starting in the inner segments, specifically involved the vastus lateralis, medialis and intermedius, the posterior thigh compartment and the gastrocnemius muscles. This radiological pattern was more evident during the first stages of the disease, but was still observed even at the end stages in the vastus lateralis muscle. The lower leg muscles were better preserved than thigh muscles, even when the thigh showed an advanced degeneration (see Family 3/II.2 in Fig. 2). Remarkably, this muscle imaging pattern was found in all the patients.
Figure 2. Pattern of muscle MRI associated to POGLUT1 mutations.
The muscle MRI images display a pattern of muscle fatty infiltration showing the internal parts of the muscles more severely degenerated compared to the external areas, especially in the thigh muscles (for example, asterisks in the vastus lateralis, semimembranosus, semitendinosus and biceps femoris muscles of patient II.1, Family 7), making up what we called an “inside-to outside” pattern. This pattern progressively disappears during the disease progression, but it was still recognizable in the vastus lateralis muscle when the rest of the thigh muscles had reached the end stage (arrows). The lower leg muscles were better preserved even when the thigh muscles showed an advanced degeneration (arrowheads). For each patient, the upper row is thigh and the lower row is lower leg.
Reduction of α-dystroglycan glycosylation and level of PAX7 and NICD in muscle biopsy
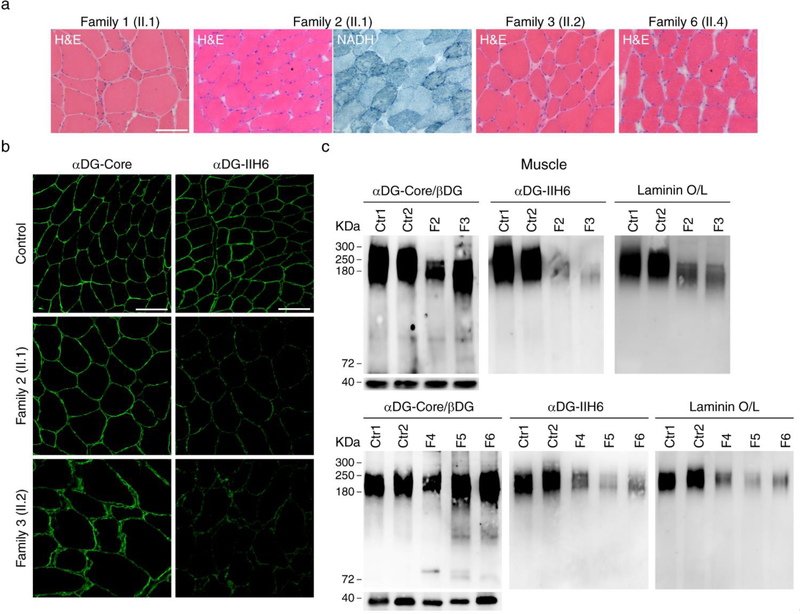
We studied the muscle biopsy from nine of the 15 patients (belonging to six of the nine families). A dystrophic pattern was observed in biopsies from the vastus lateralis muscle obtained from patient II.1 from Family 1 (Fig. 3a) and patient II.2 from Family 3. The remaining biopsies were taken from the biceps brachii muscle (much less affected than others to avoid side alterations due to the dystrophic process in order to isolate myoblasts), showing mild myopathic features and lobulated fibers in some cases (Fig. 3a).
Figure 3. Decreased α-dystroglycan glycosylation and laminin binding in muscle.
a A biopsy from vastus lateralis muscle in patient II.1 from family 1 and II.2 from family 3 showed moderate dystrophic features with atrophic fibers and increased endomysial connective tissue; the muscle biopsies from biceps brachii in patient II.1 from family 2 and patient II.4 from Family 6 showed the usual myopathic features as internal nuclei and variable fiber size, together with lobulated fibers defined by small subsarcolemmal aggregates extending into the interior of the muscle fiber (asterisks); b immunostaning studies showed reduced signal using an antibody against glycosylated α-dystroglycan (IIH6) while the signal was the same as control samples using an antibody against the α-dystroglycan core, supporting α-dystroglycan hypoglycosylation; c Western blots corroborated the decreased α-dystroglycan glycosylation in all the muscle samples available, which showed a reduced level of glycosylated α-dystroglycan and low α-dystroglycan binding to laminin. Scale bar, 100 μm.
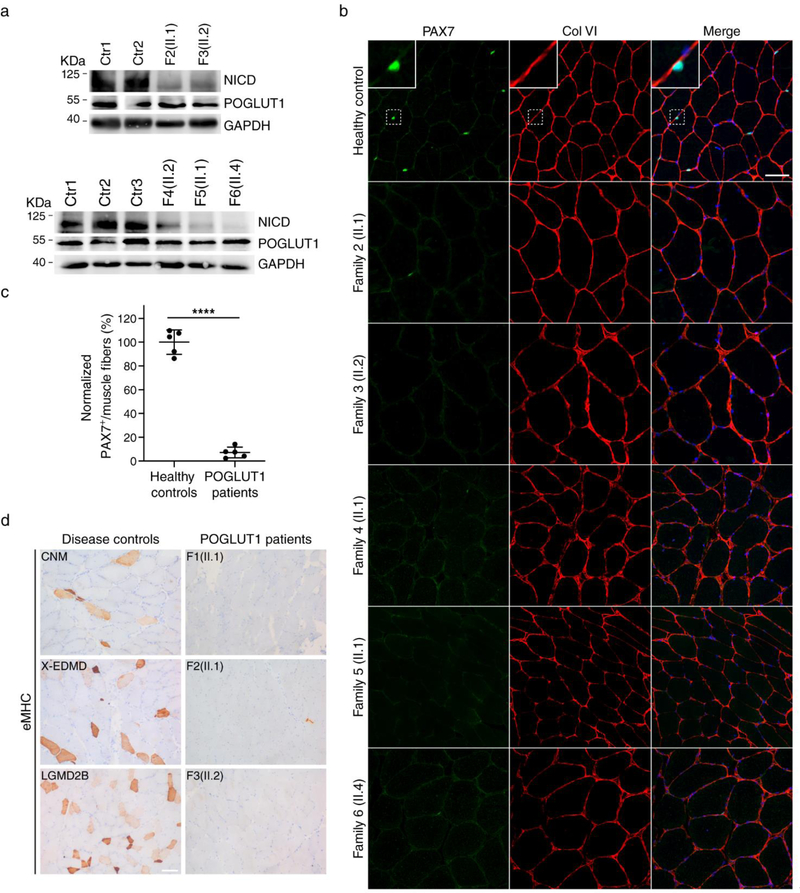
Immunofluorescence staining with an antibody against glycosylated α-dystroglycan revealed a reduction of the glycosylated form of α-dystroglycan at the sarcolemma in all muscle biopsies, in contrast to the normal staining by antibodies against the α-dystroglycan core (Fig. 3a). Consistently, Western blots showed a reduction in α-dystroglycan glycosylation and small decrease in the molecular weight of glycosylated α-dystroglycan in skeletal muscle, together with a diminished laminin-binding activity in the ligand overlay assay (Fig. 3b). Western blots also demonstrated that the NOTCH1-intracellular-domain (NICD) level in skeletal muscle was decreased in patients compared to controls (Fig. 4a), which supported the hypothesis that the novel mutations affect Notch activity, as it was demonstrated for p.D233E. In accordance with the critical role of Notch signaling in regulating SC maintenance, we found a reduced number of PAX7+ cells in the muscle of patients (Fig. 4b, c). To evaluate the regenerative activity, muscle sections were stained with an antibody to eMHC, which is expressed in regenerating myofibers. We found no staining of eMHC or a few scattered, eMHC+ myofibers in patients with POGLUT1 mutations, in contrast to disease controls (including muscle dystrophies and congenital myopathies), which showed many eMHC+ myofibers (Fig. 4d).
Figure 4. Reduction of activated Notch1 and pool of satellite cells in muscle.
a Western blot of patient’s muscle showed a reduced level of NICD in LGMD R21 compared to controls; b immunostaining using antibodies against PAX7 (green) and COLVI (red), with Topro 3 staining (blue), identified SC in the correct position below the basement membrane, both in patients and controls; c quantification of PAX7+ cells revealed that the number of SC in LGMD R21 patients (n = 5) showed a significant reduction compared to healthy controls (n = 5). Average SCs of 4–7 independent fields per muscle biopsy from each patient and control was scored. ****P<0.0001. Student’s t-test. Error bars indicate SD. Scale bar, 50 μm (Supplementary Source Data 1, online resource); d eMHC immunohistochemical staining of skeletal muscle revealed a large number of positive regenerating fibers in the disease controls (including LGMD2B and centronuclear myopathies [CNM]), while POGLUT1 patients showed no positive cells (patients F1/II.1, F3/II.2) or, at most, a few scattered positive cells (patient F2/II.1). Scale bar, 100 μm.
In vitro pathogenic role of novel POGLUT1 mutations
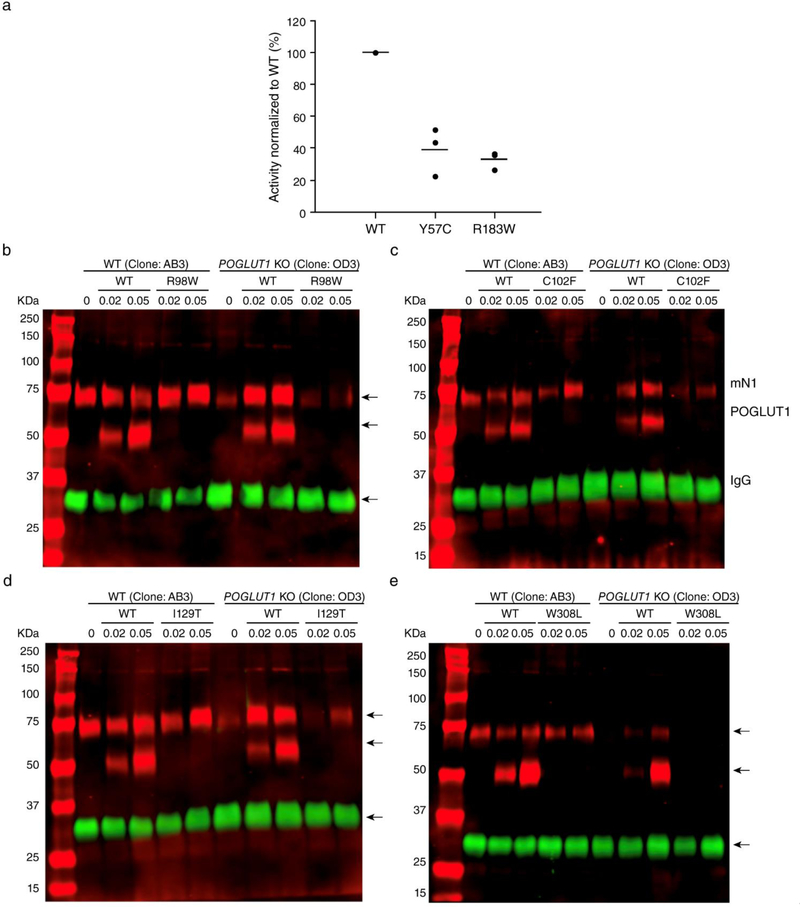
The enzymatic activity of POGLUT1 is required for proper Notch signaling [1]. Although the molecular mechanism(s) by which O-glucose glycans added by POGLUT1 regulate Notch-receptor activation is not fully understood, we recently reported that addition of O-glucose glycans to NOTCH1 is necessary for its proper trafficking to the cell surface, where NOTCH1 binds to Notch ligands [39]. It is possible that the mutations found in this study could affect the enzymatic activity of or destabilize the POGLUT1 protein. In order to examine whether POGLUT1 proteins with each mutation are properly folded, stable proteins, we tested the expression and secretion of each mutant by transient transfection in HEK293T cells. Among the mutants tested, the R183W and Y57C mutants were secreted to culture media similarly to WT POGLUT1, indicating that these mutations did not destabilize POGLUT1 proteins (Supplementary Fig. 1a, online resource). Notably, in vitro POGLUT1 activity assay with the normalized amounts of the WT, and the R183W and Y57C mutants of POGLUT1 proteins purified from the culture media showed much lower POGLUT1 activities compared to WT POGLUT1 (Fig. 5a). The levels of secretion of the p.I129T, p.R98W, p.C102F, and p.W308L mutants were much lower than that of WT POGLUT1 (Supplementary Fig. 1a, online resource). However, the revertants of p.I129T, p.R98W, p.C102F, and p.W308L showed a secretion level comparable to WT POGLUT1, confirming that the secretion defects of these four mutants were due to their corresponding mutation in POGLUT1, rather than any undesirable mutations generated during PCR for mutagenesis (Supplementary Fig. 1b, online resource). Since the quality control system in the endoplasmic reticulum prevents secretion of unfolded proteins, reduced secretion compared to WT suggests a defect in folding. Thus, these results suggested that the p.I129T, p.R98W, p.C102F, and p.W308L mutations destabilized POGLUT1 proteins. The p.I129T mutant was not detected in cell lysates either, suggesting it causes a more severe destabilization of the protein, at least in HEK293T cells. Alternatively, the mutations could cause defective trafficking.
Figure 5. Patient mutations reduce POGLUT1’s enzymatic activity or its ability to rescue Notch1 EGF1–18 secretion from POGLUT1 KO cells.
a In vitro POGLUT1 enzymatic assays showed that the activities of the R183W and Y57C mutant versions of POGLUT1 are lower than the activity of WT POGLUT1 (from 3 biological replicates; each assay was performed in triplicate; Supplementary Source Data 2, online resource); b-e Notch1 EGF1–18-MycHis secretion from WT HEK293T cells (clone AB3) or POGLUT1 KO HEK293T cells (clone OD3). Secretion was rescued fully by co-expression of plasmids encoding WT, but not by the indicated mutant versions of POGLUT1. Numbers below refer to micrograms of POGLUT1 plasmid used. A plasmid encoding IgG was co-transfected as secretion control, since IgG is not affected by loss of POGLUT1. Red channel, anti-Myc antibody. Green channel, anti-IgG antibody. Migration positions of Notch1 EGF1–18-MycHis (mN1), POGLUT1-Myc-His, and IgG are indicated.
Since the p.I129T, p.R98W, p.C102F, and p.W308L mutants of POGLUT1 could not be produced in sufficient quantities for in vitro assays, we examined whether these mutants could rescue loss of Notch1 EGF1–18-MycHis secretion from POGLUT1 KO HEK293T cells [39]. As expected, secretion of Notch1 EGF1–18-MycHis from POGLUT1 KO cells (clone OD3) was significantly lower than that from WT HEK293T cells (clone AB3), but secretion could be rescued by co-expression of WT POGLUT1. Co-expression of the p.I129T, p.R98W, p.C102F, and p.W308L mutants of POGLUT1 showed little or no rescue of Notch1 EGF1–18-MycHis secretion when the equivalent amounts of DNA were transfected into the POGLUT1 KO cells (Fig. 5b-e). These results suggest that the p.R98W, p.C102F, p.I129T, and p.W308L mutants of POGLUT1 have little or no ability to rescue secretion of Notch1 EGF1–18-MycHis in these assays.
Muscular dystrophy mutations in human POGLUT1 reduce its myogenic activity in transgenic flies.
We next examined the effects of a selected group of the new mutations on the expression level and activity of human POGLUT1 in transgenic flies. Immunostaining with α-FLAG antibody followed by normalization based on GFP expression and wing disc size indicated that p.D233E, p.R183W, p.I129T and p.Y57C mutations did not significantly change the POGLUT1 expression level compared to WT hPOGLUT1 (Supplementary Fig. 2a, b, c, e, f, and i, online resource). In contrast, p.R98W, p.W308L and p.C102F mutations showed significant decrease in expression level compared to WT POGLUT1 (Supplementary Fig. 2a, d, g, h, and i, online resource). These results were confirmed by Western blotting on wing imaginal disc extracts from third instar larvae probed by an α-POGLUT1 antibody (Supplementary Fig. 2j, online resource).
To examine the in vivo effects of the POGLUT1 mutations identified in muscular dystrophy patients, we used a Drosophila myogenesis rescue assay which we have previously set up [35]. During early pupal development, six rather long dorsal longitudinal indirect flight muscles form on each side of the Drosophila thorax in a Notch pathway-dependent manner [16]. Animals homozygous for the catalytically-inactive allele of the Drosophila rumi (rumi79/79 [1]) show severe defects in myogenesis, resulting in shortening of indirect flight muscle fibers (Fig. 6a, b, i). These defects were rescued by overexpression of WT hPOGLUT1 in muscle progenitors, as the average myotube length of rumi79/79; Mef2>POGLUT1WT-FLAG was not statistically different from that of control pupae (Fig. 6c, i). The POGLUT1-D233E mutant protein showed a partial rescue (Fig. 6d, i), as reported earlier [35]. Interestingly, the other three mutations whose expression levels were comparable with p.D233E and WT POGLUT1 (p.R183W, p.I129T and p.Y57C; Supplementary Fig. 2, online resource), also resulted in a partial rescue, with average myotubes lengths very similar to the D233E-expressing pupae (Fig. 6e, f, g, i). We also performed rescue experiments with one of the mutations causing significantly decreased POGLUT1 expression and found that expression of POGLUT1C102F with Mef2-GAL4 did not recue the muscle phenotype in rumi79/79 animals (Fig. 6h, i). We conclude that all of the mutations examined in our assay decrease the ability of human POGLUT1 to rescue the loss of fly Rumi/Poglut1 during myogenesis, even those mutations that do not affect the expression level of POGLUT1.
Figure 6. Mutations in human POGLUT1 reduce its myogenic activity in transgenic flies.
a-b Staining of Drosophila indirect flight muscles at 25% pupal development in y w (control) and rumi79/79 animals using α−22C10 antibody (red); c-h staining of Drosophila indirect flight muscles at 25% pupal development in rumi79/79 animals upon Mef2-GAL4 driven overexpression of FLAG-tagged WT and indicated mutant human POGLUT1 with α-FLAG antibody (red). All animals were raised at 25°C. Scale bar, 200 μm. i Graph shows the quantification to myotube length in the indicated genotypes. Horizontal bars represent Mean ± SD. Average length of 3–6 myotubes per animal from 3–4 animals per genotype was scored. Significance is ascribed as *P<0.05 and ****P<0.0001 compared to rumi79/79, and $ $ $P<0.001 and $ $ $ $P<0.0001 compared to y w. One-way ANOVA with Dunnett’s multiple comparisons test was used to determine the P-values (Supplementary Source Data 3, online resource).
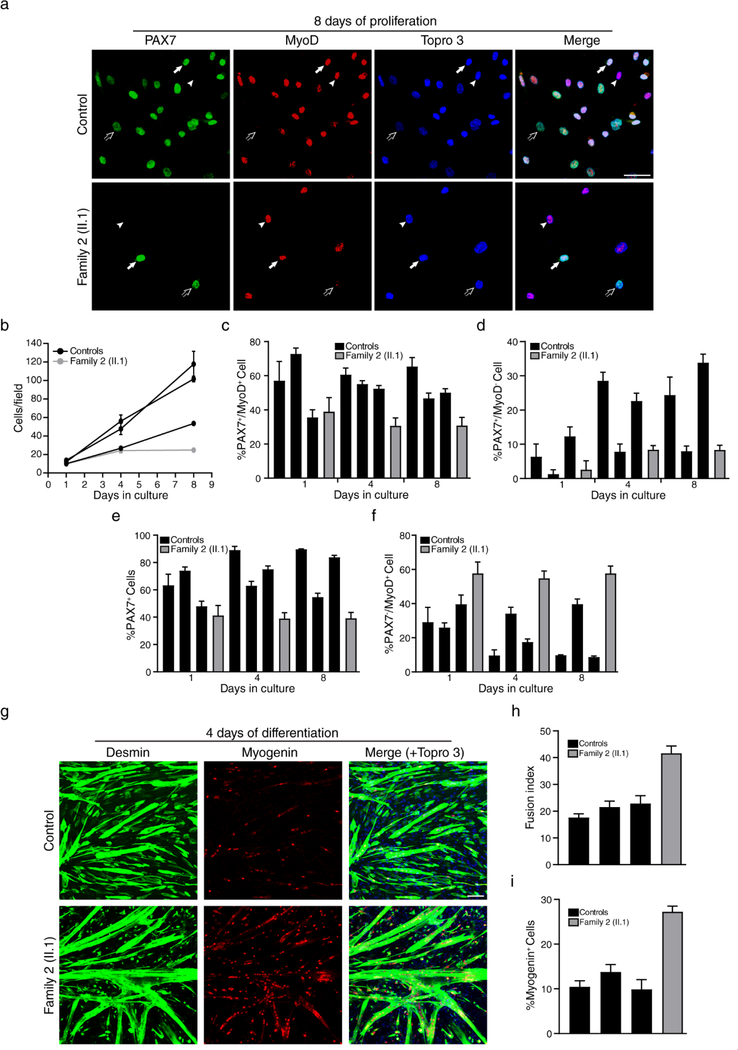
Myoblast culture from patient muscles showed decreased proliferation and premature differentiation
We next studied if the novel recessive POGLUT1 mutations alter myoblast proliferation and differentiation. To address this point, primary myoblasts from the congenital case carrying the compound heterozygous mutations p.D233E/ p.R279Q were cultured and compared to healthy controls (Fig. 7). The cell proliferation rate was lower in the patient’s myoblasts compared to controls (Fig. 7a, b). The percentage of proliferating (PAX7+MyoD+) and self-renewing (PAX7+MyoD−) cells were lower in the patient than in controls, while the percentage of differentiating cells (PAX7−MyoD+) was increased in the patient (Fig. 7c-f). Next, confluent myoblasts were cultured in differentiation medium, in which myoblasts start to elongate and fuse to form myotubes (Fig. 7g). Myogenin expression and fusion index were increased in the patient’s myoblasts (Fig. 7h, i). Our data support the conclusion that slow proliferation and facilitated myogenic differentiation underlies POGLUT1-related muscular dystrophy.
Figure 7. D233E/R279Q primary myoblasts exhibit decreased proliferation and increased differentiation.
a-f Proliferation assays examined proliferating (PAX7+ MyoD+, arrows), self-renewing (PAX7+ MyoD−, open arrows), and differentiating (PAX7− MyoD+, arrowheads) myoblasts from p.D233E/ p.R279Q patient (n = 1) and healthy controls (n = 3) at 1, 4, and 8 days growing in proliferation medium. a Immunofluorescence showed staining for MyoD (red), PAX7 (green) and for nuclear staining TOPRO-3 (blue); b the proliferation rate was reduced in p.D233E/ p.R279Q myoblasts compared with controls; c-e the percentage of proliferating cells, self-renewing cells and PAX7+ cells in the patient were also reduced; f the percentage of differentiating cells was higher in the patient. Average cell type of 14–24 independent fields per culture from 3 different controls and the p.D233E/p.R279Q patient was scored and represented in bars. Error bars indicate SEM. Scale bar, 50 μm. g-i When primary myoblasts from D233E/R279Q patient (n = 1) and healthy controls (n = 3) reached confluence, proliferation medium was replaced with differentiation medium and the myoblasts started to fuse into myotubes, which were analyzed after 4 days of differentiation; g immunofluorescence showed double staining of primary myoblasts, desmin (red) and myogenin (green), and nuclei were counterstained with Topro 3 (blue); h the fusion index and i the expression of myogenin were increased in p.D233E/ p.R279Q myoblasts compared with controls. The data support that differentiation process was facilitated in patient’s muscle. Average fusion index and myogenin positive nuclei of 5–9 independent fields per culture from 3 different controls and the p.D233E/p.R279Q patient was scored and represented in bars. Error bars indicate SEM. Scale bar, 100 μm (Supplementary Source Data 4, online resource).
DISCUSSION
In this study, we report the largest cohort to date of patients with muscular dystrophy due to recessive mutations in the POGLUT1 gene (LGMD R21). The study of this large group of patients with POGLUT1 mutations has allowed us to better define the phenotype and to identify a highly specific radiological biomarker. In addition, our current data support an underlying pathogenic mechanism with primary involvement of the Notch signaling pathway and alteration of SC homeostasis and muscle regeneration, together with a reduction of α-dystroglycan glycosylation.
The overall phenotype of the POGLUT1-related muscular dystrophy fulfilled criteria of a LGMD. Although there is phenotypic overlap with other LGMDs, some specific features in LGMD R21 patients can help establish the diagnosis. The first symptom in both adult and childhood cases was weakness of the proximal lower limb muscles. The thigh muscles were the most severely affected muscles throughout the disease course in all the patients. Scapular winging was present in almost all the patients. Respiratory involvement was not the rule, although it may appear at later stages of the disease. The congenital case presented with a similar distribution of muscle weakness with faster and more severe progression. They showed some additional features that were not observed in the adult forms, such as facial involvement, ptosis and mild contractures. There was no relevant intra- or inter-familial variability of clinical features within the adult onset group. The most relevant variability among the whole cohort was observed in the age at onset, ranging from the 1st to the 5th decade, identifying congenital, infantile and adult onset patients. Another diagnostic hint was that most cases showed normal CK levels. This could serve as a differential diagnosis to other autosomal recessive LGMDs, most of which usually show very high CK level, as e.g. dysferlinopathy (LGMD R2) [23, 28], sarcoglycanopathies (LGMD R3–6) [36] or dystroglycanopathies [27].
Certainly, the most striking characteristic associated with POGLUT1 mutations was the muscle imaging, especially at the level of the thigh. The muscle imaging pattern resembled an “inside-to-outside” fatty degeneration because the most affected segment was the inner region of each muscle in the anterior and posterior compartment of the thigh. In recent years, muscle MRI has been adopted as a key step in the diagnosis and follow-up of several muscle disorders, as it can identify specific patterns of muscle involvement that are helpful in the differential diagnostic approach [2, 12, 14, 15, 28]. In general, finding uncommon radiological signs in cases with a certain myopathy increases the sensitivity and specificity of the muscle MRI, as in the collagen-VI (COLVI)-related myopathies [8, 24]. Our data establish LGMD R21 as another form of muscular dystrophy which can greatly benefit from muscle MRI as a diagnostic tool. A thorough observation of the imaging changes is recommended to avoid confusion between the pattern displayed by POGLUT1 and COLVI mutations. In COLVI mutations, the vastus lateralis muscle shows that the initial degenerative changes are most evident on the outer areas of the muscles while in POGLUT1 mutations is the opposite, the fatty degeneration firstly appears in the inside, and additionally this pattern is found not only in the vastus but also in the muscles of the posterior thigh compartment. Review articles of radiological patterns in LGMDs [25, 42] did not show the pattern observed in LGMD R21 patients, although reports of MRI studies in dystroglycanopathies are scarce [3, 10, 15]. Thus, the muscle MRI pattern caused by POGLUT1 mutations seems to be highly sensitive and specific because it was found in all the patients, and it did not match the patterns described in other forms of muscular dystrophies. Therefore, we propose that muscle MRI should be considered as a very useful tool to screen patients with a LGMD R21.
Among the 10 mutations described in POGLUT1, nine were missense mutations (seven novel and two previously described) distributed throughout the gene. Biochemistry, cell culture and fly experiments indicated that the mutations, even those whose function could not be predicted based on the structure, have a major impact on POGLUT1 activity. The effects of some of these POGLUT1 mutations were also predicted on the basis of the structure description (Supplementary Fig. 3a, b, online resource). The structures of both human and Drosophila POGLUT1 in complex with EGF repeats and donor substrate have been described [22, 45]. Based on these structural studies, R279Q is predicted to interfere with donor binding in human POGLUT1; Y57C mutation likely disrupts the proper disulfide bonding pattern in POGLUT1 since it is close to one of the disulfide bonds; and C102F mutation disrupts a disulfide bond which stabilizes a loop that contains the aromatic residue F104, likely critical for EGF recognition through a hydrophobic interaction [22, 45]. Therefore, it is reasonable to predict that these three mutations reduce the enzymatic activity of POGLUT1. It was difficult to predict the effects of the remaining mutations on the structure and activity of POGLUT1, as it was for the previously reported D233E mutation [35]. Additional functional assays were necessary to confirm the pathogenicity of the novel mutations. In vitro O-glucosyltransferase assays, combined with rescue experiments in mammalian cells and Drosophila demonstrated that eight of the nine missense variants significantly affect the function of human POGLUT1. Consistently, patients carrying any of those recessive pathogenic variants also showed α-dystroglycan hypoglycosylation, reduced level of NICD and PAX7 in the muscle biopsy and a supportive muscle MRI pattern (Supplementary Table 2, online resource). The remaining mutations were the truncated form W184* and the missense Y156N, found in compound heterozygosity in the same patient. The W184* mutation resulted in the loss of the substrate binding site, so it should be inactive (Supplementary Fig. 3b, online resource). Moreover, the muscle MRI pattern of the carrier patient was the same as in the rest of the patients, suggesting that these mutations were pathogenic as well.
Altogether, the in vitro and in vivo functional experiments were helpful in exploring the impact of the POGLUT1 mutations on skeletal muscle function. Although they offered accurate results confirming the pathogenicity of novel POGLUT1 variants, they were complex and time-consuming experiments that may nevertheless need to be considered in a diagnostic context when identifying VUS. Muscle biopsy analysis might be a more straightforward approach to uncover findings strongly suggestive of POGLUT1 mutations. We analyzed muscle biopsies carrying seven out of nine novel mutations described in our cohort of patients, and all of them showed α-dystroglycan hypoglycosylation associated with a reduction of NICD and PAX7 expression. To our knowledge, NICD levels in skeletal muscle have not been systematically tested in other muscle disorders. In the mdx mouse model of Duchenne muscular dystrophy (DMD), a reduction in the number of SCs due to Notch signaling deficiency and decline in SC regenerative capacity has been described [19, 33], however the muscle biopsies from DMD patients have shown an increase in the SC number compared to controls [4, 20]. Severe SC loss has also been demonstrated in the TRIM32-related myopathies, however, no associated alteration in α-dystroglycan glycosylation was observed in this form of myopathy [34]. Therefore, we suggest that when a patient with a recessive form of LGMD shows a combination of α-dystroglycan hypoglycosylation, reduction of PAX7+ cells and NICD levels in the muscle biopsy, and the above-mentioned muscle MRI pattern, a diagnosis of LGMD R21 should be considered and the POGLUT1 gene should be sequenced.
α-Dystroglycan in muscles from LGMD R21 patients showed an immature LARGE-mediated glycosylation, likely as a consequence of the abnormal facilitation of the differentiation process as it has been previously demonstrated [17, 35]. The hypoglycosylation in the first LGMD R21 family described was not seen in fibroblasts, supporting that rather than a primary defect in α-dystroglycan glycosylation (which would presumably be observed in all cell types), it is a secondary defect. This α-dystroglycan hypoglycosylation in the muscle biopsy could lead the pathologist or clinician guiding the diagnostic process to limit their search for mutations in the 18 genes involved in dystroglycanopathies [7, 26, 27]. Our findings suggest that presence of α-dystroglycan hypoglycosylation in an LGMD patient does not necessarily mean that the responsible mutation is in dystroglycan-specific glycosyltransferases, as mutations in regulators of the Notch signaling pathway like POGLUT1 might result in the same phenotype.
In summary, we have described the defining features of the muscular dystrophy due to recessive POGLUT1 mutations, which may facilitate future diagnoses of LGMD R21, namely a highly specific “inside-to-outside” muscle MRI pattern and α-dystroglycan hypoglycosylation observed in muscle biopsy. We have also described a set of functional assays that could provide insight into the pathogenicity of novel POGLUT1 variants.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Stephan Kröger (Münich University) for kind donation of the antibody against the α-dystroglycan core (clone no. 317); Developmental Studies Hybridoma Bank for the 22C10 antibody; Bloomington Drosophila stock center (NIH P40OD018537) for fly strains; and Confocal Microscopy Core of the BCM IDDRC (1U54HD083092; the Eunice Kennedy NICHD) and Sandra Donkervoort (NINDS/NIH) for help with genetic studies.
FUNDING
This work was supported in part by the Instituto de Salud Carlos III and FEDER (FIS PI16–01843 to C. Paradas and JR15/00042 to M. Cabrera-Serrano), the Consejería de Salud, Junta de Andalucía (PI-0085–2016 and PE-S1275 to C. Paradas, and B-0005–2017 to M. Cabrera-Serrano), NIH/NIGMS (R01GM084135 and R35GM130317 to H. Jafar-Nejad, and R01GM061126 to R.S. Haltiwanger), JSPS KAKENHI Grants-in-Aid for Research Activity Start-up and Scientific Research (B) (JP17H06743 and JP19H03176 to H. Takeuchi), and Takeda Science Foundation and Daiichi Sankyo Foundation of Life Science (to H. Takeuchi). MYO-SEQ has been supported by Sanofi Genzyme, Ultragenyx, the LGMD2I Research Fund, Samantha J Brazzo Foundation, LGMD2D Foundation, Kurt+Peter Foundation, Muscular Dystrophy UK and Coalition to Cure Calpain 3. Work in CGB’s group is supported by NINDS/NIH intramural funds.
Footnotes
COMPETING INTERESTS
Authors declare no competing financial and/or non-financial interests in relation to the work described
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Institutional Research Ethic Committee at Hospital Universitario Virgen del Rocío in Sevilla (Spain). Written informed consent was received from participants, prior to inclusion in the study, for genetic studies, for muscle biopsies, and for pictures appearing in the manuscript.
CONSENT FOR PUBLICATION
All authors consented to publication of this paper.
The authors have declared that no conflict of interest exists
REFERENCES
- 1.Acar M, Jafar-Nejad H, Takeuchi H, Rajan A, Ibrani D, Rana NA, Pan H, Haltiwanger RS, Bellen HJ (2008) Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell 132: 247–258 Doi 10.1016/j.cell.2007.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso-Jimenez A, Kroon R, Alejaldre-Monforte A, Nunez-Peralta C, Horlings CGC, van Engelen BGM, Olive M, Gonzalez L, Verges-Gil E, Paradas C et al. (2019) Muscle MRI in a large cohort of patients with oculopharyngeal muscular dystrophy. Journal of neurology, neurosurgery, and psychiatry: Doi 10.1136/jnnp-2018-319578 [DOI] [PubMed] [Google Scholar]
- 3.Balcin H, Palmio J, Penttila S, Nennesmo I, Lindfors M, Solders G, Udd B (2017) Late-onset limb-girdle muscular dystrophy caused by GMPPB mutations. Neuromuscular disorders : NMD 27: 627–630 Doi 10.1016/j.nmd.2017.04.006 [DOI] [PubMed] [Google Scholar]
- 4.Bankole LC, Feasson L, Ponsot E, Kadi F (2013) Fibre type-specific satellite cell content in two models of muscle disease. Histopathology 63: 826–832 Doi 10.1111/his.12231 [DOI] [PubMed] [Google Scholar]
- 5.Bischof J, Maeda RK, Hediger M, Karch F, Basler K (2007) An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proceedings of the National Academy of Sciences of the United States of America 104: 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjornson CR, Cheung TH, Liu L, Tripathi PV, Steeper KM, Rando TA (2012) Notch signaling is necessary to maintain quiescence in adult muscle stem cells. Stem Cells 30: 232–242 Doi 10.1002/stem.773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonne G, Rivier F, Hamroun D (2018) The 2019 version of the gene table of neuromuscular disorders (nuclear genome). Neuromuscular disorders : NMD 28: 1031–1063 Doi 10.1016/j.nmd.2018.09.006 [DOI] [PubMed] [Google Scholar]
- 8.Bonnemann CG (2011) The collagen VI-related myopathies: muscle meets its matrix. Nature reviews Neurology 7: 379–390 Doi 10.1038/nrneurol.2011.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brohl D, Vasyutina E, Czajkowski MT, Griger J, Rassek C, Rahn HP, Purfurst B, Wende H, Birchmeier C (2012) Colonization of the satellite cell niche by skeletal muscle progenitor cells depends on Notch signals. Developmental cell 23: 469–481 Doi 10.1016/j.devcel.2012.07.014 [DOI] [PubMed] [Google Scholar]
- 10.Cirak S, Foley AR, Herrmann R, Willer T, Yau S, Stevens E, Torelli S, Brodd L, Kamynina A, Vondracek P et al. (2013) ISPD gene mutations are a common cause of congenital and limb-girdle muscular dystrophies. Brain : a journal of neurology 136: 269–281 Doi 10.1093/brain/aws312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins CA, Olsen I, Zammit PS, Heslop L, Petrie A, Partridge TA, Morgan JE (2005) Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell 122: 289–301 Doi 10.1016/j.cell.2005.05.010 [DOI] [PubMed] [Google Scholar]
- 12.Diaz-Manera J, Fernandez-Torron R, J LL, James MK, Mayhew A, Smith FE, Moore UR, Blamire AM, Carlier PG, Rufibach L et al. (2018) Muscle MRI in patients with dysferlinopathy: pattern recognition and implications for clinical trials. Journal of neurology, neurosurgery, and psychiatry 89: 1071–1081 Doi 10.1136/jnnp-2017-317488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez-Valdivia R, Takeuchi H, Samarghandi A, Lopez M, Leonardi J, Haltiwanger RS, Jafar-Nejad H (2011) Regulation of mammalian Notch signaling and embryonic development by the protein O-glucosyltransferase Rumi. Development 138: 1925–1934 Doi 10.1242/dev.060020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Figueroa-Bonaparte S, Llauger J, Segovia S, Belmonte I, Pedrosa I, Montiel E, Montesinos P, Sanchez-Gonzalez J, Alonso-Jimenez A, Gallardo E et al. (2018) Quantitative muscle MRI to follow up late onset Pompe patients: a prospective study. Scientific reports 8: 10898 Doi 10.1038/s41598-018-29170-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer D, Walter MC, Kesper K, Petersen JA, Aurino S, Nigro V, Kubisch C, Meindl T, Lochmuller H, Wilhelm K et al. (2005) Diagnostic value of muscle MRI in differentiating LGMD2I from other LGMDs. Journal of neurology 252: 538–547 Doi 10.1007/s00415-005-0684-4 [DOI] [PubMed] [Google Scholar]
- 16.Gildor B, Schejter ED, Shilo BZ (2012) Bidirectional Notch activation represses fusion competence in swarming adult Drosophila myoblasts. Development 139: 4040–4050 Doi 10.1242/dev.077495 [DOI] [PubMed] [Google Scholar]
- 17.Goddeeris MM, Wu B, Venzke D, Yoshida-Moriguchi T, Saito F, Matsumura K, Moore SA, Campbell KP (2013) LARGE glycans on dystroglycan function as a tunable matrix scaffold to prevent dystrophy. Nature 503: 136–140 Doi 10.1038/nature12605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harvey BM, Haltiwanger RS (2018) Regulation of Notch Function by O-Glycosylation. Advances in experimental medicine and biology 1066: 59–78 Doi 10.1007/978-3-319-89512-3_4 [DOI] [PubMed] [Google Scholar]
- 19.Jiang C, Wen Y, Kuroda K, Hannon K, Rudnicki MA, Kuang S (2014) Notch signaling deficiency underlies age-dependent depletion of satellite cells in muscular dystrophy. Disease models & mechanisms 7: 997–1004 Doi 10.1242/dmm.015917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kottlors M, Kirschner J (2010) Elevated satellite cell number in Duchenne muscular dystrophy. Cell and tissue research 340: 541–548 Doi 10.1007/s00441-010-0976-6 [DOI] [PubMed] [Google Scholar]
- 21.Kuang S, Kuroda K, Le Grand F, Rudnicki MA (2007) Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell 129: 999–1010 Doi 10.1016/j.cell.2007.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Fischer M, Satkunarajah M, Zhou D, Withers SG, Rini JM (2017) Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1). Nature communications 8: 185 Doi 10.1038/s41467-017-00255-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Aoki M, Illa I, Wu C, Fardeau M, Angelini C, Serrano C, Urtizberea JA, Hentati F, Hamida MB et al. (1998) Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nature genetics 20: 31–36 Doi 10.1038/1682 [DOI] [PubMed] [Google Scholar]
- 24.Mercuri E, Lampe A, Allsop J, Knight R, Pane M, Kinali M, Bonnemann C, Flanigan K, Lapini I, Bushby K et al. (2005) Muscle MRI in Ullrich congenital muscular dystrophy and Bethlem myopathy. Neuromuscular disorders : NMD 15: 303–310 Doi 10.1016/j.nmd.2005.01.004 [DOI] [PubMed] [Google Scholar]
- 25.Mercuri E, Pichiecchio A, Allsop J, Messina S, Pane M, Muntoni F (2007) Muscle MRI in inherited neuromuscular disorders: past, present, and future. Journal of magnetic resonance imaging : JMRI 25: 433–440 Doi 10.1002/jmri.20804 [DOI] [PubMed] [Google Scholar]
- 26.Michele DE, Barresi R, Kanagawa M, Saito F, Cohn RD, Satz JS, Dollar J, Nishino I, Kelley RI, Somer H et al. (2002) Post-translational disruption of dystroglycan-ligand interactions in congenital muscular dystrophies. Nature 418: 417–422 Doi 10.1038/nature00837 [DOI] [PubMed] [Google Scholar]
- 27.Muntoni F, Torelli S, Wells DJ, Brown SC (2011) Muscular dystrophies due to glycosylation defects: diagnosis and therapeutic strategies. Curr Opin Neurol 24: 437–442 Doi 10.1097/WCO.0b013e32834a95e3 [DOI] [PubMed] [Google Scholar]
- 28.Paradas C, Gonzalez-Quereda L, De Luna N, Gallardo E, Garcia-Consuegra I, Gomez H, Cabello A, Illa I, Gallano P (2009) A new phenotype of dysferlinopathy with congenital onset. Neuromuscular disorders : NMD 19: 21–25 Doi 10.1016/j.nmd.2008.09.015 [DOI] [PubMed] [Google Scholar]
- 29.Rahimov F, Kunkel LM (2013) The cell biology of disease: cellular and molecular mechanisms underlying muscular dystrophy. J Cell Biol 201: 499–510 Doi 10.1083/jcb.201212142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ralser DJ, Takeuchi H, Fritz G, Basmanav FB, Effern M, Sivalingam S, El-Shabrawi-Caelen L, Degirmentepe EN, Kocaturk E, Singh M et al. (2019) Altered Notch Signaling in Dowling-Degos Disease: Additional Mutations in POGLUT1 and Further Insights into Disease Pathogenesis. The Journal of investigative dermatology 139: 960–964 Doi 10.1016/j.jid.2018.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rana NA, Nita-Lazar A, Takeuchi H, Kakuda S, Luther KB, Haltiwanger RS (2011) O-glucose trisaccharide is present at high but variable stoichiometry at multiple sites on mouse Notch1. The Journal of biological chemistry 286: 31623–31637 Doi 10.1074/jbc.M111.268243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM (2008) Self-renewal and expansion of single transplanted muscle stem cells. Nature 456: 502–506 Doi 10.1038/nature07384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sacco A, Mourkioti F, Tran R, Choi J, Llewellyn M, Kraft P, Shkreli M, Delp S, Pomerantz JH, Artandi SE et al. (2010) Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell 143: 1059–1071 Doi 10.1016/j.cell.2010.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Servian-Morilla E, Cabrera-Serrano M, Rivas-Infante E, Carvajal A, Lamont PJ, Pelayo-Negro AL, Ravenscroft G, Junckerstorff R, Dyke JM, Fletcher S et al. (2019) Altered myogenesis and premature senescence underlie human TRIM32-related myopathy. Acta neuropathologica communications 7: 30 Doi 10.1186/s40478-019-0683-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Servian-Morilla E, Takeuchi H, Lee TV, Clarimon J, Mavillard F, Area-Gomez E, Rivas E, Nieto-Gonzalez JL, Rivero MC, Cabrera-Serrano M et al. (2016) A POGLUT1 mutation causes a muscular dystrophy with reduced Notch signaling and satellite cell loss. EMBO molecular medicine 8: 1289–1309 Doi 10.15252/emmm.201505815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sewry CA, Taylor J, Anderson LV, Ozawa E, Pogue R, Piccolo F, Bushby K, Dubowitz V, Muntoni F (1996) Abnormalities in alpha-, beta- and gamma-sarcoglycan in patients with limb-girdle muscular dystrophy. Neuromuscular disorders : NMD 6: 467–474 [DOI] [PubMed] [Google Scholar]
- 37.Sheikh MO, Halmo SM, Patel S, Middleton D, Takeuchi H, Schafer CM, West CM, Haltiwanger RS, Avci FY, Moremen KW et al. (2017) Rapid screening of sugar-nucleotide donor specificities of putative glycosyltransferases. Glycobiology 27: 206–212 Doi 10.1093/glycob/cww114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Straub V, Murphy A, Udd B, LGMD workshop study group (2018) 229th ENMC international workshop: Limb girdle muscular dystrophies - Nomenclature and reformed classification Naarden, the Netherlands, 17–19 March 2017. Neuromuscular disorders : NMD 28: 702–710 Doi 10.1016/j.nmd.2018.05.007 [DOI] [PubMed] [Google Scholar]
- 39.Takeuchi H, Yu H, Hao H, Takeuchi M, Ito A, Li H, Haltiwanger RS (2017) O-Glycosylation modulates the stability of epidermal growth factor-like repeats and thereby regulates Notch trafficking. The Journal of biological chemistry 292: 15964–15973 Doi 10.1074/jbc.M117.800102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varshney S, Stanley P (2018) Multiple roles for O-glycans in Notch signalling. FEBS letters 592: 3819–3834 Doi 10.1002/1873-3468.13251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Venken KJ, He Y, Hoskins RA, Bellen HJ (2006) P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314: 1747–1751 [DOI] [PubMed] [Google Scholar]
- 42.Wattjes MP, Kley RA, Fischer D (2010) Neuromuscular imaging in inherited muscle diseases. European radiology 20: 2447–2460 Doi 10.1007/s00330-010-1799-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willer T, Lee H, Lommel M, Yoshida-Moriguchi T, de Bernabe DB, Venzke D, Cirak S, Schachter H, Vajsar J, Voit T et al. (2012) ISPD loss-of-function mutations disrupt dystroglycan O-mannosylation and cause Walker-Warburg syndrome. Nature genetics 44: 575–580 Doi 10.1038/ng.2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu H, Takeuchi H (2019) Protein O-glucosylation: another essential role of glucose in biology. Current opinion in structural biology 56: 64–71 Doi 10.1016/j.sbi.2018.12.001 [DOI] [PubMed] [Google Scholar]
- 45.Yu H, Takeuchi H, Takeuchi M, Liu Q, Kantharia J, Haltiwanger RS, Li H (2016) Structural analysis of Notch-regulating Rumi reveals basis for pathogenic mutations. Nature chemical biology 12: 735–740 Doi 10.1038/nchembio.2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.