Abstract
Background
Mutations involving the closely linked GJB2 and GJB6 at the DFNB1 locus are a common genetic cause of profound congenital hearing loss in many populations. In some deaf GJB2 heterozygotes, a 309 kb deletion involving the GJB6 has been found to be the cause for hearing loss when inherited in trans to a GJB2 mutation.
Methods
We screened 2,376 probands from a National DNA Repository of deaf individuals.
Results
Fifty‐two of 318 heterozygous probands with pathogenic GJB2 sequence variants had a GJB6 deletion. Additionally, eight probands had an isolated heterozygous GJB6 deletion that did not explain their hearing loss. In two deaf subjects, including one proband, a homozygous GJB6 deletion was the cause for their hearing loss, a rare occurrence not reported to date.
Conclusion
This study represents the largest US cohort of deaf individuals harboring GJB2 and GJB6 variants, including unique subsets of families with deaf parents. Testing additional members to clarify the phase of GJB2/GJB6 variants in multiplex families was crucial in interpreting clinical significance of the variants in the proband. It highlights the importance of determining the phase of GJB2/GJB6 variants when interpreting molecular test results especially in multiplex families with assortative mating.
Keywords: GJB2/GJB6 variant, hearing loss, interpretation of results, unique family structure
In some deaf GJB2 heterozygotes, a 309 kb deletion involving the GJB6 gene has been found to be the cause for hearing loss when inherited in trans to a GJB2 mutation. We screened 2,376 probands from a National DNA Repository of deaf individuals. Fifty‐two of 318 heterozygous probands with pathogenic GJB2 sequence variants had a GJB6 deletion, eight probands had an isolated heterozygous GJB6 deletion that did not explain hearing loss, and two deaf subjects had a homozygous GJB6 deletion that caused their hearing loss.

1. INTRODUCTION
Deafness is an etiologically heterogeneous trait with many recognized genetic and environmental causes (Kremer, 2019; Nance, 2003; Tekin, Arnos, & Pandya, 2001). The DFNB1 locus on 13q12 harbors two homologous genes, GJB2 (OMIM: 121011) and GJB6 (OMIM: 604418) that code for the connexin 26 and 30 subunits of gap junction proteins, respectively. Although initially these proteins were thought to participate in the recycling of potassium ions from the hair cells back into the cochlear endolymph (Steel & Bussoli, 1999), recent literature suggests the digenic mutations reduce endocochlear potential and/or result in cochlear developmental disorder (Mei et al., 2017; Wingard & Zhao, 2015; Zhao, 2017). DFNB1 mutations are the most common genetic cause of severe to profound congenital hearing loss (HL) in many populations (Chan & Chang, 2014; Kenneson, Van Naarden Braun, & Boyle, 2002). The carrier frequency for GJB2 mutation in North Americans has been reported to be as high as 3.5% (Green et al., 1999). The GJB6 was first implicated as a cause for deafness in a single family demonstrating dominant inheritance segregating a one base pair substitution, designated DFNA3 (Grifa et al., 1999). In 2002, del Castillo et al. along with other groups (Lerer et al., 2001; Pallares‐Ruiz, Blanchet, Mondain, Claustres, & Roux, 2002) reported a large deletion involving the 5′ noncoding region of GJB6 that extended into coding region of the gene, which when present in trans with a GJB2 mutation, explained the HL in 67% of their deaf GJB2 heterozygotes. This deletion known as del(GJB6‐D13S1830) was initially described as a 342 kb deletion, however, it is currently estimated at 309 kb (del Castillo et al., 2005). The reported frequency of this deletion among deaf individuals has ranged from as high as 15% in Southern France to between 9% and 5% in Spain, Israel, and United Kingdom, to as low as 1.4% in Italy and Belgium (Marlin et al., 2005). Its incidence in Eastern Europe and other nations is reported to be lower or not present (Adhikary et al., 2015; Frei et al., 2004; Wonkam, Ngo, & Ngogang, 2015). A second smaller deletion in the 5′ untranslated region of GJB6 [del(GJB6‐D13S1854)] was subsequently identified in other deaf GJB2 heterozygotes, mainly in Spain and the UK (del Castillo et al., 2005).
Both proteins are expressed in the inner ear where they form homomeric or heteromeric connexons in largely overlapping regions of the supporting cells of the cochlea (Chen, Chen, Zhu, Liang, & Zhao, 2014; Forge, Marziano, Casalotti, Becker, & Jagger, 2003; Wang et al., 2009; Wingard & Zhao, 2015; Zhao & Santos‐Sacchi, 2000; Zhao & Yu, 2006). Connexin 26 (Cx26) plays a role in the normal development of the auditory sensory epithelium and Connexin 30 (Cx30) is essential for normal repair after sensory hair cell loss (Jagger & Forge, 2015). The deletions in the coding portion of GJB6, and less so the deletion in the 5′ untranslated region of GJB6 result in HL in a substantial proportion of the deaf GJB2 heterozygotes. Studies suggest this phenotype to be secondary to the disruption of a cis‐acting regulatory element for GJB2, in contrast to the prior hypothesis suggesting the contribution of GJB6 haploinsufficiency (Boulay et al., 2013; Rodriguez‐Paris & Schrijver, 2009).
We report on results from screening 2,376 deaf probands from a North American National Repository, the largest sample cohort studied to date (Pandya et al., 2003), and present the frequency of pathogenic variants involving the two genes, and the reported audiologic phenotype in individuals with sequence changes in both the GJB2 and GJB6. We also report some unique pedigrees with the GJB6 deletion that highlight pitfalls and caution needed in interpreting results on deaf probands tested in isolation, especially when they have both parents with hearing loss.
2. METHODS AND MATERIALS
2.1. Patient ascertainment
Deaf probands and their family members were ascertained for this study through several sources, including the Annual Survey of Deaf and Hard of Hearing Children and Youth, conducted at the Gallaudet Research Institute of Gallaudet University (GU), an institution of higher education for the deaf and hard of hearing. The Annual Survey collected educational, etiologic and audiologic data, as well as demographic information such as race, parental mating type, and hearing status of siblings on a nationwide sample of nearly 50,000 deaf and hard of hearing students who receive special education services because of their HL (Pandya et al., 2003). Participants were also recruited through the Gallaudet University Alumni Association (Arnos et al., 2008) and the genetics clinics held at GU and the Virginia Commonwealth University (VCU). All participants completed the informed consent process approved by the VCU and GU Institutional Review Board.
2.2. Molecular testing
DNA was extracted from peripheral blood samples using Pure Gene (Gentra Systems) protocols. Samples from all probands were screened for mutations in exon 1 and 2 of GJB2 by cycle sequencing as described in Pandya et al., 2003. The product was subjected to cycle sequencing with the ABI PRISM Big Dye Terminator cycle sequencing kit. Forward and reverse sequences were analyzed for mutations using Phred, Phrap, and Consed software suite (Gordon, Abajian, & Green, 1998).
The del(GJB6‐D13S1830) deletion was tested using primers described by del Castillo et al. (2003). Briefly, the breakpoint junction and an internal control sequence outside the deletion were amplified simultaneously to test for the deletion in all deaf probands. Individuals heterozygous for the deletion yielded a 460 bp product (breakpoint junction) along with a 360 bp fragment (internal control) from the nondeleted chromosome. Individuals who did not carry the deletion showed only the 360 bp band.
To confirm the presence of a homozygous deletion, all samples showing the breakpoint junction product were tested further using primers for exon 1 of GJB6 along with an internal control. The primers used for GJB6 exon 1 were; Forward E1: 5′‐ATG GAT TGG GGG ACG CTG CA ‐3′. Reverse E1 5′ CCA CCA CTA GGA TCA TGA CTC GGA‐ 3′. Individuals homozygous for the deletion showed no amplification product for exon 1 of GJB6. Amplification of the internal control verified that the PCR reaction was working.
The del(GJB6‐D13S1853) deletion was also tested using primers described by del Castillo et al. (2005). The probands were tested and were negative for changes in the MT‐RNR1 for the m.1555A>G variant, and MT‐TS1 for the m.7445A>C/T/G variant.
2.3. Audiologic studies
Audiometric data were extracted from copies of available audiograms. Only the results of audiometric testing performed in soundproof booths according to ANSI 1969 standards were used for the analysis. Coded data included pure tone averages for all available frequencies, speech reception thresholds, and word recognition scores. A determination was also made about the bilateral symmetry of the audiograms based on the criteria of Liu and Xu (1994).
3. RESULTS
DNA samples from 2,376 deaf probands were screened for the GJB2 and GJB6 mutations as described above. Overall, 22.85% probands carried two pathogenic GJB2 variants that are most likely the cause for their HL. An additional 13.38% of the probands were heterozygous for a single pathogenic GJB2 variant, and 16.35% (52/318) of these apparent heterozygotes carried the del(GJB6‐D13S1830) deletion (Table 1). Seven other probands who were not heterozygous for a GJB2 variant carried the del(GJB6‐D13S1830) deletion, one proband had the GJB6‐D13S1830 deletion in addition to two pathogenic variants in the GJB2 and one proband was homozygous only for the GJB6 deletion. Among individuals with a GJB2 mutation and the del(GJB6‐D13S1830) deletion, 38 (71.7%) were from multiplex families and more than half had deaf parents. Although the overwhelming majority of them were Caucasians (79%), we also observed the GJB6 deletion in probands who were Hispanic (4%), African American (2%), Filipino (2%), and East Indian (2%) (Table 2). In 69.81% (37/53) of the digenic compound heterozygotes the GJB2 mutation was the common c.35delG allele, but the GJB6 deletion was also observed in combination with p.V37I, p.V84M, p.S85Y, p.R143W, c.167delT, c.312del14, and the p.R184P mutant alleles.
Table 1.
Frequency of GJB2 and GJB6 mutations in deaf probands
| GJB2 (Cx26) | GJB6 (Cx30) | Totals | ||
|---|---|---|---|---|
| wt/wt | wt/del | del/del | ||
| wt/wt | 1507 | 7 | 1 | 1515 |
| mut/wt | 266 | 52 | 0 | 318 |
| mut/mut | 542 | 1 | 0 | 543 |
| Totals | 2,315 | 60 | 1 | 2,376 |
Genotypic distribution of del(GJB6‐D13S1830) and pathogenic GJB2 variants in our cohort of deaf probands.
Table 2.
Proband characteristics
| Ethnicity | Cx30 del/wt | Gender | Family mating type | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Cx26 | mut/wt | mut/mut | wt/wt | M | F | DxD | DxH | HxH | |
| Caucasian | 43 | 1 | 6 | 21 | 29 | 25 | 2 | 23 | |
| Ashkenazi Jew | 4 | — | — | 2 | 2 | 1 | — | 3 | |
| South Asian | 2 | — | — | 1 | 1 | — | 1 | 1 | |
| African | 1 | — | — | 1 | — | — | — | 1 | |
| Eastern European | 1 | — | — | 1 | — | 1 | — | — | |
| Hispanic | 1 | — | 1 | — | 2 | 1 | — | 1 | |
| Totals | 52 | 1 | 7 | 26 | 34 | 28 | 3 | 29 | |
Characteristics of the 60 probands with the Cx30 deletion del(GJB6‐D13S1830).
Mut: Pathogenic sequence variant.
DxD: Both parents of the proband have hearing loss.
DxH: Only one parent of the proband has hearing loss.
HxH: Neither parent of the proband has hearing loss.
Figure 1a–c illustrate a few families from the study cohort with several unusual features. In addition to a deaf Caucasian proband homozygous for the del(GJB6‐D13S1830) deletion, we also identified a deaf Caucasian parent in one of our families with the homozygous GJB6 deletion (Figure 1a‐ NSDF 27‐202). Of note, both parents were deaf in this family, and all their children were deaf (noncomplementary mating). This raised the suspicion that a second deafness gene was segregating in the family. Figure 1a ‐ NSDF 2024 illustrates a second family with a few deaf individuals that were initially identified as heterozygous carriers for the c.167delT variant in GJB2 until we could test them for the GJB6 deletion to determine the etiology for the HL.
Figure 1.
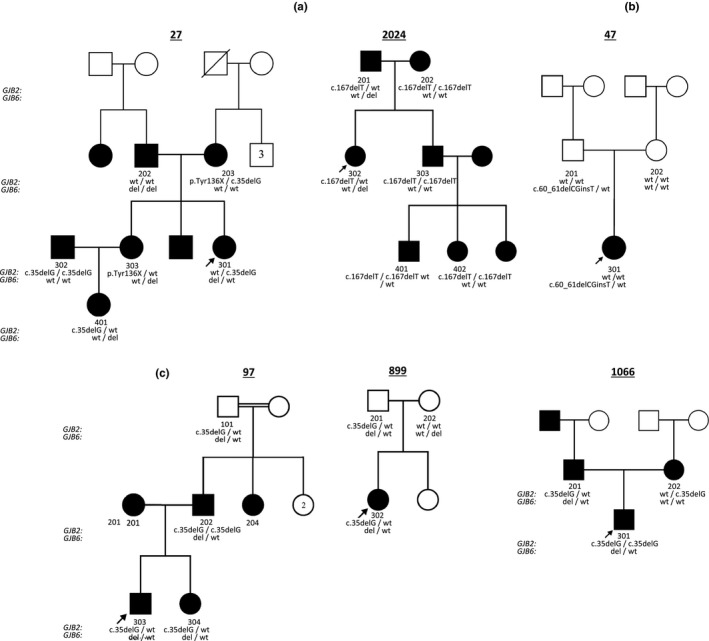
Representative Families from cohort with the digenic variants in both GJB2 and GJB6. (a) Compound heterozygous state in simplex and multiplex families with HL. (b) Simplex family with a 2bp frameshift deletion without GJB2 mutations that may not be the cause of HL. (c) Families requiring testing of additional members for result interpretation in proband. GJB6 del = del(GJB6‐D13S1830)
In three probands (97‐303, 899‐302, 1066‐301) who were heterozygous or homozygous for variants in both GJB2 and GJB6, studying additional members in the family provided valuable information on the phase of these two variants (Figure 1c). Additional members tested including 97‐101 and 899‐201 had variants in GJB2 and GJB6 with normal hearing. In contrast, 1066‐201 has HL but it appears that the GJB2 and GJB6 variants are in cis and his hearing loss may be from a different etiology. We hypothesize that 97‐303 and 97‐304 could either have inherited the 35delG variant from their father and the GJB6 deletion from their mother (97‐201 not available for testing), with DFNB1 as an etiology for HL or they inherited the 35delG andGJB6 deletion in cis from their father and the deafness is due to an unrelated etiology yet to be determined. Individual 899‐302 most likely inherited the DFNB1 changes in cis from his father that would not explain his HL.
A 5‐year‐old child with mild‐to‐moderate progressive hearing loss (Figure 1b) had negative deletion testing for GJB6 but had a frameshift deletion involving a single guanine residue at position 61 resulting in a truncated protein with the introduction of a stop at codon 34. Her father carried the same deletion but had normal hearing by testing. The initial interpretation was to ascribe the moderate HL in the child to the frameshift mutation and postulate reduced penetrance in her hearing father. However further testing by NGS for additional HL genes has identified a more plausible explanation for her phenotype that is being investigated as the etiology for her HL.
No proband was found to carry the shorter del(GJB6‐D13S1854) deletion reported by del Castillo et al. (2005).
The average hearing thresholds in 38 probands with digenic pathogenic variants in the GJB2 and GJB6, and six probands heterozygous for the GJB6 deletion alone, are shown across various frequencies in Figure 2. The average degree of HL in all the probands is in the severe range with no statistically significant difference among those with a c.35delG variant as compared to those with a different GJB2 pathogenic variant. The average thresholds in probands with digenic change are consistently in severe to profound range across most frequencies.
Figure 2.
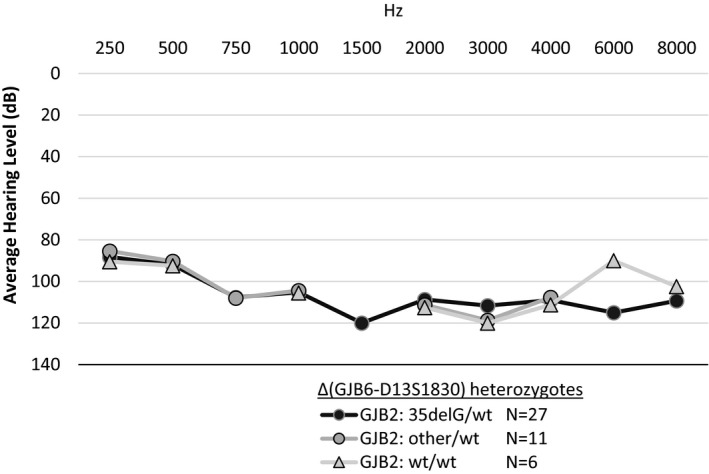
Average hearing threshold in deaf probands with del(GJB6‐D13S1830) deletion in trans with GJB2 variant, or GJB6 deletion by itself
4. DISCUSSION
In 2002, del Castillo et al. reported compelling evidence that deafness can result from the presence of a 342 kb (revised as 309 kb) deletion that included the coding region of GJB6, when present in trans with a pathogenic GJB2 allele. The coinheritance of this deletion was shown to be the apparent cause of the HL in 67% of deaf GJB2 heterozygotes in Spain (del Castillo et al., 2002), and was reported to be the second most frequent mutation apart from the c.35delG allele in patients with DFNB1 deafness (Marlin et al., 2005).
Our study of GJB6 deletion represents one of the largest series of deaf probands studied to date in North America. Digenic compound heterozygotes for the del(GJB6‐D13S1830) deletion explained HL in 16.35% of deaf GJB2 heterozygotes, which is lower compared to reports from other Western European countries (Cama et al., 2009; Gualandi et al., 2004; Feldmann et al., 2004; Santos et al., 2005). This difference is likely due to the smaller cohort size of deaf individuals studied in comparison to the present study. The del(GJB6‐D13S1830) deletion occurred most commonly with the c.35delG mutation in the GJB2, although it is reported in association with other GJB2 pathogenic alleles.
The probands with digenic changes in the present study were often from multiplex families, many times with both parents having HL, which is in contrast to the other large studies (Feldmann et al., 2004; Marlin et al., 2005). In our sample, 71.6% of the digenic probands were from a multiplex family and a unique finding was that 65.7% of these probands had both deaf parents.
In the United States, 80%–90% of individuals with profound prelingual deafness who communicate by sign language select similarly affected marriage partners (Nance, 2003). This linguistic homogamy has created a substantially separate mating pool in which, the frequency of genes for deafness may be amplified. Nance and Kearsey (2004) showed that when combined with relaxed selection among the deaf, this mechanism will preferentially amplify mutations at the commonest locus for recessive deafness and could have doubled the frequency of connexin deafness in this country during the past two centuries.
Families in which both parents are deaf also pose a challenge with interpretation of molecular test results in deaf probands. When the results for the proband are interpreted out of context to the parental genotype (Figure 1c) it was misleading to assume the digenic variant as the etiology for HL. These families have clearly highlighted that the two variants when present in cis tend to be of little clinical significance. The corollary is that the identification of GJB2/GJB6 variants in a deaf individual should not be assumed to be in trans or the cause of their HL.
Overall, the del(GJB6‐D13S1830) deletion allele was present in 2.1% of all deaf probands, which is similar to the frequency reported by several other studies (Table 3). There also appears to be notable differences in the frequency of the del(GJB6‐D13S1830) deletion, as depicted in studies in different populations. Although the frequency of the del(GJB6‐D13S1830) deletion in Brazil, France, Czechia, Russia, and Argentina is high (Bliznets et al., 2012; Dalamón et al., 2005; Marlin et al., 2005; Piatto, Bertollo, Sartorato, & Maniglia, 2004; Seeman et al., 2005), it is rare or infrequently observed in Austria and Morocco and Syria (Abidi et al., 2007; Frei et al., 2004; Gazzaz et al., 2005; Zaidieh, Habbal, & Monem, 2015). In contrast, the GJB6 deletion was not identified in many regions including India, Taiwan, Cameroon, Sicily, Mexico, Iran, Macedonia, and Turkey (Adhikary et al., 2015; Amorini et al., 2015; Bhalla, Sharma, Khandelwal, Panda, & Khular, 2009, 2011; Bosch et al., 2014; Esmaeili, Bonyandi, & Nejadkazem, 2007; Hernandez‐Juarez et al., 2014; Naddafnia, Noormohammadi, Irani, & Salahshoorifar, 2019; Sirmaci, Akcayoz‐Duman, & Tekin, 2006; Sukarova Stefanovska, Cakar, Filipce, & Plaseska Karanfilska, 2012; Wonkam et al., 2015; Yang et al., 2007).
Table 3.
Review of del(GJB6‐D13S1830) frequency reported in literature
| Country | Subjects (No.) | Total no. of subjects with a GJB6 deletion | Total no. of GJB2 heterozygotes tested for a GJB6 deletion | Freq. of GJB6 deletion | Study | |
|---|---|---|---|---|---|---|
| Freq. of GJB6 deletions among GJB2 heterozygotes | In all deaf probands | |||||
| Czechia | 13 | 1 | 13 | 8% | 8% | Seeman et al. (2005) |
| Belgium | 15 | 7 | 9 | 78% | 47% | Stinckens et al. (2004)a |
| Germany | 25 | 2 | 4 | 50% | 8% | Bolz et al. (2004) |
| Russia | 30 | 2 | 2 | 7% | 0% | Bliznets et al. (2012) |
| Switzerland | 32 | 1 | 8 | 13% | 3% | Gürtler et al. (2008) |
| Brazil | 33 | 3 | 6 | 50% | 9% | Piatto et al. (2004) |
| Russia | 35 | 1 | 1 | 3% | 3% | Pshennikova et al. (2017) |
| Spain | 38 | 1 | 7 | 14% | 3% | Gallo‐Teran et al. (2005) |
| Venezuela | 40 | 1 | 9 | 11% | 3% | Utrera et al. (2007) |
| Syria | 41 | 1 | 2 | 0% | 2% | Zaidieh et al. (2015) |
| Argentina | 46 | 4 | 17 | 18% | 9% | Dalamón et al. (2005) |
| Hungary | 47 | 2 | 2 | 4% | 4% | Tóth et al. (2007) |
| Italy | 59 | 2 | 9 | 22% | 3% | Gualandi et al. (2004) |
| U.S | 68 | 2 | 27 | 7% | 3% | Erbe et al. (2004) |
| North America | 95 | 2 | 9 | 22% | 2% | Schimmenti et al. (2008) |
| U.S | 108 | 2 | 30 | 7% | 2% | Wu et al. (2003) |
| Denmark | 165 | 2 | 9 | 22% | 1% | Grønskov et al. (2004) |
| Belarus | 213 | 3 | 3 | 1% | 1% | Danilenko et al. (2012) |
| Netherlands | 222 | 6 | 14 | 29% | 3% | Santos et al. (2005) |
| France | 255 | 16 | 29 | 52% | 6% | Feldmann et al. (2004) |
| France | 256 | 25 | NR | NR | ‐ | Marlin et al. (2005)b |
| Brazil | 300 | 3 | 31 | 10% | 1% | Batissoco et al. (2009) |
| Italy | 376 | 6 | 30 | 20% | 2% | Cama et al. (2009) |
| Spain | 422 | 45 | 33 | 67% | 5% | del Castillo et al. (2002) |
| Argentina | 476 | 5 | 9 | 1% | 2% | Dalamón et al. (2013) |
| Brazil | 600 | 2 | 46 | 4% | 0% | da Silva‐Costa et al. (2011) |
| North America | 888 | 9 | NA | NA | 1% | Putcha et al. (2007) |
| North America | 2,376 | 61 | 318 | 22% | 3% | Current study |
Literature review spanning 2003—present, reporting subjects with a del(GJB6‐D13S1830) presented by ascending cohort size
Abbreviation: NR, not reported.
Single family.
Reported on the basis of the presence of compound heterozygous mutations in GJB2, which includes the association of one GJB2 mutation with a GJB6 deletion.
Most studies from the United States, including our report, have identified deaf individuals with the GJB6 deletion in trans in heterozygous carriers for the GJB2 variant. The frequency of the GJB6 deletion in the general population is not well characterized. Fitzgerald et al. (2004) did not identify the GJB6 deletion in nearly 2,000 blood spots from a diverse ethnic group of newborns presumably with normal hearing, screened in New York State. These data correlate with the reported allele frequency of 0.0001863 in the gnomAD database. It also explains the rare occurrence of individuals carrying the deletion in a homozygous state, as the expected frequency would be ~1 in 28,500,000. In some individuals, the GJB6 deletion would be in cis with a GJB2 variant.
The mechanism by which the del(GJB6‐D13S1830) deletion causes HL, when present in trans with a GJB2 mutation, remains uncertain and could result from an alteration of the expression of the normal GJB2 located in cis to the deletion (del Castillo et al., 2005, 2003), or from haplo‐insufficiency of the GJB6 product or both. The expression of GJB2 was shown to be affected by the del(GJB6‐D13S1830) deletion in a cell specific manner in sweat glands by immuno‐histochemical staining, suggesting the disruption of GJB2 cis‐regulatory elements located within the deletion that control expression of Cx26 in cells (Common et al., 2005). Wilch et al. (2006) have also reported an interesting family with HL segregating a novel DFNB1 allele characterized by significant reduction in expression of both GJB2 and GJB6, suggesting the presence of as‐yet unidentified cis‐regulatory element. Other studies demonstrating abolished GJB2 expression due to an in cis GJB6 deletion (Rodriguez‐Paris & Schrijver, 2009) and the finding of normal hearing prior to postnatal day 30 in a Cx30 knock‐out mouse model (Cx30 Δ/Δ) (Boulay et al., 2013) favors the mechanistic hypothesis that the del(GJB6‐D13S1830) eliminates a putative cis‐regulatory element in the deleted region. At present, based on our results of the frameshift variant in GJB6 not being the etiology of HL, caution needs to be exercised with results obtained by targeted sequencing of GJB6 or whole exome sequencing in individuals with HL.
The more severe audiologic findings in digenic probands is difficult to reconcile with the sole theory of a putative cis‐regulatory element in the deleted region, and it could suggest that both the GJB6 and GJB2 product may make a contribution to the HL. The degree of HL in individuals with a pathogenic GJB2 sequence variant and a GJB6 deletion is more profound than those with bi‐allelic change in GJB2 as previously observed (Pandya et al., 2003), and as reported in other studies (Marlin et al., 2005; Santos et al., 2005; Snoeckx et al., 2005). Homozygous deletion of GJB6, although rare, was found in one proband and another relative of a proband in our study which results in severe to profound HL, similar to the mouse model (Chen et al., 2014; Marziano, Casalotti, Portelli, Becker, & Forge, 2003; Mei et al., 2017). Unlike in humans, the GJB2/GJB6 double heterozygous mice also show decreased endocochlear potential but have moderate HL (Mei et al., 2017; Teubner et al., 2003).
Whether individuals with the GJB2 and GJB6 changes in cis could have a milder HL remains to be determined, as we have identified family members (Figure 1c) with this molecular complement with both normal hearing and HL.
In conclusion, we report the del(GJB6‐D13S1830) deletion in 16.35% of all GJB2 heterozygotes as a cause for their HL. The overall frequency of the digenic mutations was 2.1% among the 2,376 deaf probands studied. There was one proband and a relative of another proband with the homozygous del(GJB6‐D13S1830) deletion not reported previously. The audiologic profile of digenic probands revealed a profound HL. This body of evidence therefore suggests that the HL in digenic heterozygotes may result at least in part from a deficiency of the GJB6 product in addition to any effect the deletion has on the regulation of the adjacent normal GJB2 locus.
CONFLICT OF INTEREST
Arti Pandya, Alexander O'Brien, Michael Kovasala, Guney Bademci, Mustafa Tekin, and Kathleen S. Arnos have nothing to declare.
ACKNOWLEDGMENTS
We are grateful to the patients and their families for their participation.
Pandya A, O'Brien A, Kovasala M, Bademci G, Tekin M, Arnos KS. Analyses of del(GJB6‐D13S1830) and del(GJB6‐D13S1834) deletions in a large cohort with hearing loss: Caveats to interpretation of molecular test results in multiplex families. Mol Genet Genomic Med. 2020;8:e1171 10.1002/mgg3.1171
REFERENCES
- Abidi, O. , Boulouiz, R. , Nahili, H. , Ridal, M. , Alami, M. N. , Tlili, A. , … Hassar, M. (2007). GJB2 (connexin 26) gene mutations in moroccan patients with autosomal recessive non-syndromic hearing loss and carrier frequency of the common GJB2–35delG mutation. International Journal of Pediatric Otorhinolaryngology, 71(8), 1239–1245. [DOI] [PubMed] [Google Scholar]
- Adhikary, B. , Ghosh, S. , Paul, S. , Bankura, B. , Pattanayak, A. K. , Biswas, S. , … Das, M. (2015). Spectrum and frequency of GJB2, GJB6 and SLC26A4 gene mutations among nonsyndromic hearing loss patients in eastern part of India. Gene, 573(2), 239–245. 10.1016/j.gene.2015.07.050 [DOI] [PubMed] [Google Scholar]
- Amorini, M. , Romeo, P. , Bruno, R. , Galletti, F. , Di Bella, C. , Longo, P. , … Rigoli, L. (2015). Prevalence of deafness‐associated connexin‐26 (GJB2) and connexin‐30 (GJB6) pathogenic alleles in a large patient cohort from Eastern Sicily. Annals of Human Genetics, 79(5), 341–349. 10.1111/ahg.12120 [DOI] [PubMed] [Google Scholar]
- Arnos, K. S. , Welch, K. O. , Tekin, M. , Norris, V. W. , Blanton, S. H. , Pandya, A. , & Nance, W. E. (2008). A comparative analysis of the genetic epidemiology of deafness in the United States in two sets of pedigrees collected more than a century apart. The American Journal of Human Genetics, 83(2), 200–207. 10.1016/j.ajhg.2008.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batissoco, A. C. , Abreu‐Silva, R. S. , Braga, M. C. C. , Lezirovitz, K. , Della‐Rosa, V. , Alfredo, T. , … Mingroni‐Netto, R. C. (2009). Prevalence of GJB2 (connexin‐26) and GJB6 (connexin‐30) mutations in a cohort of 300 Brazilian hearing‐impaired individuals: Implications for diagnosis and genetic counseling. Ear and Hearing, 30(1), 1–7. 10.1097/AUD.0b013e31819144ad [DOI] [PubMed] [Google Scholar]
- Bhalla, S. , Sharma, R. , Khandelwal, G. , Panda, N. K. , & Khullar, M. (2009). Low incidence of GJB2, GJB6 and mitochondrial DNA mutations in north indian patients with non‐syndromic hearing impairment. Biochemical and Biophysical Research Communications, 385(3), 445–448. 10.1016/j.bbrc.2009.05.083 [DOI] [PubMed] [Google Scholar]
- Bhalla, S. , Sharma, R. , Khandelwal, G. , Panda, N. K. , & Khullar, M. (2011). Absence of GJB6 mutations in Indian patients with non‐syndromic hearing loss. International Journal of Pediatric Otorhinolaryngology, 75(3), 356–359. 10.1016/j.ijporl.2010.12.003 [DOI] [PubMed] [Google Scholar]
- Bliznets, E. A. , Galkina, V. A. , Matiushchenko, G. N. , Kisina, A. G. , Markova, T. G. , & Poliakov, A. V. (2012). Changes in the connexin 26 (GJB2) gene in Russian patients with hearing disorders: Results of long‐term molecular diagnostics of hereditary nonsyndromic deafness. Genetika, 48(1), 112–124. [PubMed] [Google Scholar]
- Bolz, H. , Schade, G. , Ehmer, S. , Kothe, C. , Hess, M. , & Gal, A. (2004). Phenotypic variability of non‐syndromic hearing loss in patients heterozygous for both c.35delG of GJB2 and the 342‐kb deletion involving GJB6. Hearing Research, 188(1–2), 42–46. 10.1016/S0378-5955(03)00346-0 [DOI] [PubMed] [Google Scholar]
- Bosch, J. , Lebeko, K. , Nziale, J. J. N. , Dandara, C. , Makubalo, N. , & Wonkam, A. (2014). In search of genetic markers for nonsyndromic deafness in Africa: A study in Cameroonians and Black South Africans with the GJB6 and GJA1 candidate genes. OMICS: A Journal of Integrative Biology, 18(7), 481–485. 10.1089/omi.2013.0166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulay, A.‐C. , del Castillo, F. J. , Giraudet, F. , Hamard, G. , Giaume, C. , Petit, C. , … Cohen‐Salmon, M. (2013). Hearing is normal without connexin30. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 33(2), 430–434. 10.1523/JNEUROSCI.4240-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cama, E. , Melchionda, S. , Palladino, T. , Carella, M. , Santarelli, R. , Genovese, E. , … Arslan, E. (2009). Hearing loss features in GJB2 biallelic mutations and GJB2/GJB6 digenic inheritance in a large Italian cohort. International Journal of Audiology, 48(1), 12–17. 10.1080/14992020802400654 [DOI] [PubMed] [Google Scholar]
- Chan, D. K. , & Chang, K. W. (2014). GJB2‐associated hearing loss: Systematic review of worldwide prevalence, genotype, and auditory phenotype. The Laryngoscope, 124(2), E3–E53. 10.1002/lary.24332 [DOI] [PubMed] [Google Scholar]
- Chen, J. , Chen, J. , Zhu, Y. , Liang, C. , & Zhao, H. (2014). Deafness induced by connexin 26 (GJB2) deficiency is not determined by endocochlear potential (EP) reduction but is associated with cochlear developmental disorders. Biochemical and Biophysical Research Communications, 448(1), 28–32. 10.1016/j.bbrc.2014.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Common, J. E. A. , Bitner‐Glindzicz, M. , O'Toole, E. A. , Barnes, M. R. , Jenkins, L. , Forge, A. , & Kelsell, D. P. (2005). Specific loss of connexin 26 expression in ductal sweat gland epithelium associated with the deletion mutation del(GJB6‐D13S1830). Clinical & Experimental Dermatology, 30(6), 688–693. 10.1111/j.1365-2230.2005.01878.x [DOI] [PubMed] [Google Scholar]
- da Silva‐Costa, S. M. , Martins, F. T. A. , Pereira, T. , Pomilio, M. C. , Marques‐de‐Faria, A. P. , & Sartorato, E. L. (2011). Searching for digenic inheritance in deaf Brazilian individuals using the multiplex ligation‐dependent probe amplification technique. Genetic Testing and Molecular Biomarkers, 15(12), 849–853. 10.1089/gtmb.2011.0034 [DOI] [PubMed] [Google Scholar]
- Dalamón, V. , Béhèran, A. , Diamante, V. , Diamante, F. , Pallares, N. , & Elgoyhen, A. B. (2005). Prevalence of GJB2 mutations and the del(GJB6‐D13S1830) in Argentinean non‐syndromic deaf patients. Hearing Research, 207(1), 43–49. 10.1016/j.heares.2005.04.012 [DOI] [PubMed] [Google Scholar]
- Dalamón, V. , Florencia Wernert, M. , Lotersztein, V. , Craig, P. O. , Diamante, R. R. , Barteik, M. E. , … Elgoyhen, A. B. (2013). Identification of four novel connexin 26 mutations in non‐syndromic deaf patients: Genotype–phenotype analysis in moderate cases. Molecular Biology Reports, 40(12), 6945–6955. 10.1007/s11033-013-2814-x [DOI] [PubMed] [Google Scholar]
- Danilenko, N. , Merkulava, E. , Siniauskaya, M. , Olejnik, O. , Levaya‐Smaliak, A. , Kushniarevich, A. , … Davydenko, O. (2012). Spectrum of genetic changes in patients with non‐syndromic hearing impairment and extremely high carrier frequency of 35delG GJB2 mutation in Belarus. PLoS ONE, 7(5), e36354 10.1371/journal.pone.0036354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Castillo, F. J. , Rodriguez‐Ballesteros, M. , Alvarez, A. , Hutchin, T. , Leonardi, E. , de Oliveira, C. A. , … del Castillo, I. (2005). A novel deletion involving the connexin‐30 gene, del(GJB6‐d13s1854), found in trans with mutations in the GJB2 gene (connexin‐26) in subjects with DFNB1 non‐syndromic hearing impairment. Journal of Medical Genetics, 42(7), 588–594. 10.1136/jmg.2004.028324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Castillo, I. , Moreno‐Pelayo, M. A. , del Castillo, F. J. , Brownstein, Z. , Marlin, S. , Adina, Q. , … Moreno, F. (2003). Prevalence and evolutionary origins of the del(GJB6‐D13S1830) mutation in the DFNB1 locus in hearing‐impaired subjects: A multicenter study. The American Journal of Human Genetics, 73(6), 1452–1458. 10.1086/380205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Castillo, I. , Villamar, M. , Moreno‐Pelayo, M. A. , del Castillo, F. J. , Álvarez, A. , Tellería, D. , … Moreno, F. (2002). A deletion involving the connexin 30 gene in nonsyndromic hearing impairment. New England Journal of Medicine, 346(4), 243–249. 10.1056/NEJMoa012052 [DOI] [PubMed] [Google Scholar]
- Erbe, C. B. , Harris, K. C. , Runge‐Samuelson, C. L. , Flanary, V. A. , & Wackym, P. A. (2004). Connexin 26 and connexin 30 mutations in children with nonsyndromic hearing loss. The Laryngoscope, 114(4), 607–611. 10.1097/00005537-200404000-00003 [DOI] [PubMed] [Google Scholar]
- Esmaeili, M. , Bonyadi, M. , & Nejadkazem, M. (2007). Common mutation analysis of GJB2 and GJB6 genes in affected families with autosomal recessive non‐syndromic hearing loss from Iran: Simultaneous detection of two common mutations (35delG/del(GJB6‐D13S1830)) in the DFNB1‐related deafness. International Journal of Pediatric Otorhinolaryngology, 71(6), 869–873. 10.1016/j.ijporl.2007.02.007 [DOI] [PubMed] [Google Scholar]
- Feldmann, D. , Denoyelle, F. , Chauvin, P. , Garabédian, E. , Couderc, R. , Odent, S. , … Marlin, S. (2004). Large deletion of the GJB6 gene in deaf patients heterozygous for the GJB2 gene mutation: Genotypic and phenotypic analysis. American Journal of Medical Genetics Part A, 127A(3), 263–267. 10.1002/ajmg.a.20588 [DOI] [PubMed] [Google Scholar]
- Fitzgerald, T. , Duva, S. , Ostrer, H. , Pass, K. , Oddoux, C. , Ruben, R. , & Caggana, M. (2004). The frequency of GJB2 and GJB6 mutations in the New York state newborn population: Feasibility of genetic screening for hearing defects. Clinical Genetics, 65(4), 338–342. 10.1111/j.1399-0004.2004.00233.x [DOI] [PubMed] [Google Scholar]
- Forge, A. , Marziano, N. K. , Casalotti, S. O. , Becker, D. L. , & Jagger, D. (2003). The inner ear contains heteromeric channels composed of Cx26 and Cx30 and deafness‐related mutations in Cx26 have a dominant negative effect on Cx30. Cell Communication and Adhesion, 10(4), 341–346. 10.1080/714040450 [DOI] [PubMed] [Google Scholar]
- Frei, K. , Ramsebner, R. , Lucas, T. , Baumgartner, W. , Schoefer, C. , Wachtler, F. J. , & Kirschhofer, K. (2004). Screening for monogenetic del(GJB6‐D13S1830) and digenic del(GJB6‐D13S1830)/GJB2 patterns of inheritance in deaf individuals from Eastern Austria. Hearing Research, 196(1), 115–118. 10.1016/j.heares.2004.07.001 [DOI] [PubMed] [Google Scholar]
- Gallo‐Teran, J. , Morales‐Angulo, C. , Rodriguez‐Ballesteros, M. , Moreno‐Pelayo, M. A. , & Moreno, F. (2005). Prevalence of the 35delG mutation in the GJB2 gene, del(GJB6‐D13S1830) in the GJB6 gene, Q829X in the OTOF gene and A1555G in the mitochondrial 12S rRNA gene in subjects with non‐syndromic sensorineural hearing impairment of congenital/childhood onset. Acta Otorrinolaringologica Espanola, 56(10), 463–468. 10.1016/s0001-6519(05)78649-0 [DOI] [PubMed] [Google Scholar]
- Gazzaz, B. , Weil, D. , Raïs, L. , Akhyat, O. , Azeddoug, H. , & Nadifi, S. (2005). Autosomal recessive and sporadic deafness in morocco: High frequency of the 35delG GJB2 mutation and absence of the 342‐kb GJB6 variant. Hearing Research, 210(1), 80–84. 10.1016/j.heares.2005.08.001 [DOI] [PubMed] [Google Scholar]
- Gordon, D. , Abajian, C. , & Green, P. (1998). Consed: A graphical tool for sequence finishing. Genome Research, 8(3), 195–202. 10.1101/gr.8.3.195 [DOI] [PubMed] [Google Scholar]
- Green, G. E. , Scott, D. A. , McDonald, J. M. , Woodworth, G. G. , Sheffield, V. C. , & Smith, R. J. H. (1999). Carrier rates in the midwestern United States for GJB2 mutations causing inherited deafness. JAMA, 281(23), 2211–2216. 10.1001/jama.281.23.2211 [DOI] [PubMed] [Google Scholar]
- Grifa, A. , Wagner, C. A. , D'Ambrosio, L. , Melchionda, S. , Bernardi, F. , Lopez‐Bigas, N. , … Gasparini, P. (1999). Mutations in GJB6 cause nonsyndromic autosomal dominant deafness at DFNA3 locus. Nature Genetics, 23(1), 16–18. 10.1038/12612 [DOI] [PubMed] [Google Scholar]
- Grønskov, K. , Larsen, L. A. , Rendtorff, N. D. , Parving, A. , Nørgaard‐Pedersen, B. , & Brøndum‐Nielsen, K. (2004). GJB2 and GJB6 mutations in 165 Danish patients showing non‐syndromic hearing impairment. Genetic Testing, 8(2), 181–184. 10.1089/gte.2004.8.181 [DOI] [PubMed] [Google Scholar]
- Gualandi, F. , Ravani, A. , Berto, A. , Burdo, S. , Trevisi, P. , Ferlini, A. , … Calzolari, E. (2004). Occurrence of del(GIB6 ‐D13S1830) mutation in italian non‐syndromic hearing loss patients carrying a single GJB2 mutated allele. Acta Oto‐Laryngologica, 124(Supplement 552), 29–34. 10.1080/03655230410017166 [DOI] [PubMed] [Google Scholar]
- Gürtler, N. , Egenter, C. , Bösch, N. , & Plasilova, M. (2008). Mutation analysis of the Cx26, Cx30, and Cx31 genes in autosomal recessive nonsyndromic hearing impairment. Acta Oto‐Laryngologica, 128(10), 1056–1062. 10.1080/00016480701854727 [DOI] [PubMed] [Google Scholar]
- Hernández‐Juárez, A. A. , Lugo‐Trampe, J. D. J. , Campos‐Acevedo, L. D. , Lugo‐Trampe, A. , Treviño‐González, J. L. , de‐la‐Cruz‐Ávila, I. , & Martínez‐de‐Villarreal, L. E. (2014). GJB2 and GJB6 mutations are an infrequent cause of autosomal‐recessive nonsyndromic hearing loss in residents of Mexico. International Journal of Pediatric Otorhinolaryngology, 78(12), 2107–2112. 10.1016/j.ijporl.2014.09.016 [DOI] [PubMed] [Google Scholar]
- Jagger, D. , & Forge, A. (2015). Connexins and gap junctions in the inner ear – It's not just about K+ recycling. Cell and Tissue Research, 360(3), 633–644. 10.1007/s00441-014-2029-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenneson, A. , Van Naarden Braun, K. , & Boyle, C. (2002). GJB2 (connexin 26) variants and nonsyndromic sensorineural hearing loss: A HuGE review. Genetics in Medicine: Official Journal of the American College of Medical Genetics, 4(4), 258–274. 10.1097/00125817-200207000-00004 [DOI] [PubMed] [Google Scholar]
- Kremer, H. (2019). Hereditary hearing loss; about the known and the unknown. Hearing Research, 376, 58–68. 10.1016/j.heares.2019.01.003 [DOI] [PubMed] [Google Scholar]
- Lerer, I. , Sagi, M. , Ben‐Neriah, Z. , Wang, T. , Levi, H. , & Abeliovich, D. (2001). A deletion mutation in GJB6 cooperating with a GJB2 mutation in trans in non‐syndromic deafness: A novel founder mutation in Ashkenazi Jews. Human Mutation, 18(5), 460 10.1002/humu.1222 [DOI] [PubMed] [Google Scholar]
- Liu, X. , & Xu, L. (1994). Nonsyndromic hearing loss: An analysis of audiograms. Annals of Otology, Rhinology & Laryngology, 103(6), 428–433. 10.1177/000348949410300602 [DOI] [PubMed] [Google Scholar]
- Marlin, S. , Feldmann, D. , Blons, H. , Loundon, N. , Rouillon, I. , Albert, S. , … Denoyelle, F. (2005). GJB2 and GJB6 mutations: Genotypic and phenotypic correlations in a large cohort of hearing‐impaired patients. Archives of Otolaryngology‐Head & Neck Surgery, 131(6), 481–487. 10.1001/archotol.131.6.481 [DOI] [PubMed] [Google Scholar]
- Marziano, N. K. , Casalotti, S. O. , Portelli, A. E. , Becker, D. L. , & Forge, A. (2003). Mutations in the gene for connexin 26 (GJB2) that cause hearing loss have a dominant negative effect on connexin 30. Human Molecular Genetics, 12(8), 805–812. 10.1093/hmg/ddg076 [DOI] [PubMed] [Google Scholar]
- Mei, L. , Chen, J. , Zong, L. , Zhu, Y. , Liang, C. , Jones, R. O. , & Zhao, H. (2017). A deafness mechanism of digenic Cx26 (GJB2) and Cx30 (GJB6) mutations: Reduction of endocochlear potential by impairment of heterogeneous gap junctional function in the cochlear lateral wall. Neurobiology of Disease, 108, 195–203. 10.1016/j.nbd.2017.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naddafnia, H. , Noormohammadi, Z. , Irani, S. , & Salahshoorifar, I. (2019). Frequency of GJB2 mutations, GJB6‐D13S1830 and GJB6‐D13S1854 deletions among patients with non‐syndromic hearing loss from the central region of Iran. Molecular Genetics & Genomic Medicine, 7, e00780 10.1002/mgg3.780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance, W. E. (2003). The genetics of deafness. Mental Retardation and Developmental Disabilities Research Reviews, 9(2), 109–119. 10.1002/mrdd.10067 [DOI] [PubMed] [Google Scholar]
- Nance, W. E. , & Kearsey, M. J. (2004). Relevance of connexin deafness (DFNB1) to human evolution. American Journal of Human Genetics, 74(6), 1081–1087. 10.1086/420979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallares‐Ruiz, N. , Blanchet, P. , Mondain, M. , Claustres, M. , & Roux, A. (2002). A large deletion including most of GJB6 in recessive non syndromic deafness: A digenic effect. European Journal of Human Genetics: EJHG, 10(1), 72–76. 10.1038/sj.ejhg.5200762 [DOI] [PubMed] [Google Scholar]
- Pandya, A. , Arnos, K. S. , Xia, X. J. , Welch, K. O. , Blanton, S. H. , Friedman, T. B. , … Nance, W. E. (2003). Frequency and distribution of GJB2 (connexin 26) and GJB6 (connexin 30) mutations in a large North American repository of deaf probands. Genetics in Medicine, 5(4), 295–303. 10.1097/01.GIM.0000078026.01140.68 [DOI] [PubMed] [Google Scholar]
- Piatto, V. B. , Bertollo, E. M. G. , Sartorato, E. L. , & Maniglia, J. V. (2004). Prevalence of the GJB2 mutations and the del(GJB6‐D13S1830) mutation in Brazilian patients with deafness. Hearing Research, 196(1–2), 87–93. 10.1016/j.heares.2004.05.007 [DOI] [PubMed] [Google Scholar]
- Pshennikova, V. G. , Barashkov, N. A. , Solovyev, A. V. , Romanov, G. P. , Diakonov, E. E. , Sazonov, N. N. , … Dzhemileva, L. U. (2017). Analysis of GJB6 (Cx30) and GJB3 (Cx31) genes in deaf patients with monoallelic mutations in GJB2 (Cx26) gene in the Sakha Republic (Yakutia). Russian Journal of Genetics, 53(6), 688–697. 10.1134/s1022795417030103 [DOI] [Google Scholar]
- Putcha, G. V. , Bejjani, B. A. , Bleoo, S. , Booker, J. K. , Carey, J. C. , Carson, N. , … Schrijver, I. (2007). A multicenter study of the frequency and distribution of GJB2 and GJB6 mutations in a large North American cohort. Genetics in Medicine, 9(7), 413–426. 10.1097/GIM.0b013e3180a03276 [DOI] [PubMed] [Google Scholar]
- Rodriguez‐Paris, J. , & Schrijver, I. (2009). The digenic hypothesis unraveled: The GJB6 del(GJB6‐D13S1830) mutation causes allele‐specific loss of GJB2 expression in cis. Biochemical and Biophysical Research Communications, 389(2), 354–359. 10.1016/j.bbrc.2009.08.152 [DOI] [PubMed] [Google Scholar]
- Santos, R. L. P. , Aulchenko, Y. S. , Huygen, P. L. M. , Donk, K. P. V. D. , Wijs, I. J. D. , Kemperman, M. H. , … Cremers, C. W. R. J. (2005). Hearing impairment in Dutch patients with connexin 26 (GJB2) and connexin 30 (GJB6) mutations. International Journal of Pediatric Otorhinolaryngology, 69(2), 165–174. 10.1016/j.ijporl.2004.08.015 [DOI] [PubMed] [Google Scholar]
- Schimmenti, L. A. , Martinez, A. , Telatar, M. , Lai, C.‐H. , Shapiro, N. , Fox, M. , … Palmer, C. G. S. (2008). Infant hearing loss and connexin testing in a diverse population. Genetics in Medicine, 10(7), 517–524. 10.1097/GIM.0b013e31817708fa [DOI] [PubMed] [Google Scholar]
- Seeman, P. , Bendova, O. , Raskova, D. , Malikova, M. , Groh, D. , & Kabelka, Z. (2005). Double heterozygosity with mutations involving both the GJB2 and GJB6 genes is a possible, but very rare, cause of congenital deafness in the Czech population. Annals of Human Genetics, 69(1), 9–14. 10.1046/j.1529-8817.2003.00120.x [DOI] [PubMed] [Google Scholar]
- Sirmaci, A. , Akcayoz‐Duman, D. , & Tekin, M. (2006). The c.IVS1+1G>A mutation in the GJB2 gene is prevalent and large deletions involving the GJB6 gene are not present in the Turkish population. Journal of Genetics, 85(3), 213–216. 10.1007/bf02935334 [DOI] [PubMed] [Google Scholar]
- Snoeckx, R. L. , Huygen, P. L. M. , Feldmann, D. , Marlin, S. , Denoyelle, F. , Waligora, J. , … Van Camp, G. (2005). GJB2 mutations and degree of hearing loss: A multicenter study. The American Journal of Human Genetics, 77(6), 945–957. 10.1086/497996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel, K. P. , & Bussoli, T. J. (1999). Deafness genes: Expressions of surprise. Trends in Genetics, 15(6), 207–211. 10.1016/S0168-9525(99)01753-9 [DOI] [PubMed] [Google Scholar]
- Stinckens, C. , Kremer, H. , van Wijk, E. , Standaert, L. , Hoefsloot, L. H. , Fryns, J. P. , … Cremers, C. W. (2004). Longitudinal phenotypic analysis in patients with connexin 26 (GJB2)(DFNB1) and connexin 30 (GJB6) mutations. Annals of Otology, Rhinology & Laryngology, 113(7), 587–593. 10.1177/000348940411300714 [DOI] [PubMed] [Google Scholar]
- Sukarova Stefanovska, E. , Plaseska Karanfilska, D. , Cakar, M. , & Filipce, I. (2012). Genetics of non syndromic hearing loss in the Republic of Macedonia. Balkan Journal of Medical Genetics, 15(Supplement), 57–59. 10.2478/v10034-012-0020-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekin, M. , Arnos, K. S. , & Pandya, A. (2001). Advances in hereditary deafness. The Lancet, 358(9287), 1082–1090. 10.1016/S0140-6736(01)06186-4 [DOI] [PubMed] [Google Scholar]
- Teubner, B. , Michel, V. , Pesch, J. , Lautermann, J. , Cohen‐Salmon, M. , Söhl, G. , … Willecke, K. (2003). Connexin30 (Gjb6)‐deficiency causes severe hearing impairment and lack of endocochlear potential. Human Molecular Genetics, 12(1), 13–21. 10.1093/hmg/ddg001 [DOI] [PubMed] [Google Scholar]
- Tóth, T. , Kupka, S. , Haack, B. , Fazakas, F. , Muszbek, L. , Blin, N. , … Sziklai, I. (2007). Coincidence of mutations in different connexin genes in Hungarian patients. International Journal of Molecular Medicine, 20(3), 315–321. 10.3892/ijmm.20.3.315 [DOI] [PubMed] [Google Scholar]
- Utrera, R. , Ridaura, V. , Rodriguez, Y. , Rojas, M. J. , Mago, L. , Angeli, S. , & Henriquez, O. (2007). Detection of the 35delG/GJB2 and del(GJB6‐D13S1830) mutations in Venezuelan patients with autosomal recessive nonsyndromic hearing loss. Genetic Testing, 11(4), 347–352. 1089/gte.2006.0526 [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Chang, Q. , Tang, W. , Sun, Y. , Zhou, B. , Li, H. , & Lin, X. (2009). Targeted connexin26 ablation arrests postnatal development of the organ of Corti. Biochemical and Biophysical Research Communications, 385(1), 33–37. 10.1016/j.bbrc.2009.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilch, E. , Zhu, M. , Burkhart, K. B. , Regier, M. , Elfenbein, J. L. , Fisher, R. A. , & Friderici, K. H. (2006). Expression of GJB2 and GJB6 is reduced in a novel DFNB1 allele. The American Journal of Human Genetics, 79(1), 174–179. 10.1086/505333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingard, J. C. , & Zhao, H. (2015). Cellular and deafness mechanisms underlying connexin mutation‐induced hearing loss ‐ A common hereditary deafness. Frontiers in Cellular Neuroscience, 9, 202 10.3389/fncel.2015.00202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonkam, A. , Ngo Bitoungui, V. J. , & Ngogang, J. (2015). Perspectives in genetics and sickle cell disease prevention in Africa: Beyond the preliminary data from Cameroon. Public Health Genomics, 18(4), 237–241. 10.1159/000431020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, B. , Kenna, M. , Lip, V. , Irons, M. , & Platt, O. (2003). Use of a multiplex PCR/sequencing strategy to detect both connexin 30 (GJB6) 342 kb deletion and connexin 26 (GJB2) mutations in cases of childhood deafness. American Journal of Medical Genetics Part A, 121(2), 102–108. 10.1002/ajmg.a.20210 [DOI] [PubMed] [Google Scholar]
- Yang, J. , Huang, S. , Chou, K. , Liao, P. , Su, C. , & Li, S. (2007). Identification of mutations in members of the connexin gene family as a cause of nonsyndromic deafness in Taiwan. Audiology and Neurotology, 12(3), 198–208. 10.1159/000099024 [DOI] [PubMed] [Google Scholar]
- Zaidieh, T. , Habbal, W. , & Monem, F. (2015). Screening of GJB6 gene large deletions among Syrians with congenital hearing impairment. Genetic Testing and Molecular Biomarkers, 19(7), 405–407. 10.1089/gtmb.2015.0019 [DOI] [PubMed] [Google Scholar]
- Zhao, H. (2017). Hypothesis of K ‐recycling defect is not a primary deafness mechanism for Cx26 (GJB2) deficiency. Frontiers in Molecular Neuroscience, 10, 162 10.3389/fnmol.2017.00162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, H. B. , & Santos‐Sacchi, J. (2000). Voltage gating of gap junctions in cochlear supporting cells: Evidence for nonhomotypic channels. The Journal of Membrane Biology, 175(1), 17–24. 10.1007/s002320001051 [DOI] [PubMed] [Google Scholar]
- Zhao, H. , & Yu, N. (2006). Distinct and gradient distributions of connexin26 and connexin30 in the cochlear sensory epithelium of guinea pigs. Journal of Comparative Neurology, 499(3), 506–518. 10.1002/cne.21113 [DOI] [PMC free article] [PubMed] [Google Scholar]


