Abstract
Background
Studies have suggested that micro‐RNAs (miRNAs) can function as an oncogene or a tumor suppressor in cancers. However, the role of MIR‐138‐5P (613394) in prostate cancer (PCa) remains unclear.
Methods
Expression level of MIR‐138‐5P in PCa cell lines and normal cell line was analyzed with the quantitative real‐time PCR method. Cell counting kit‐8 assay, colony formation assay, wound‐healing assay, and transwell invasion assay were performed to analyze the biological functions of MIR‐138‐5P.
Results
We showed MIR‐138‐5P expression level was significantly decreased in PCa cell lines compared with the normal cell line. Overexpression of MIR‐138‐5P inhibits PCa cell proliferation, colony formation, cell migration, and cell invasion in vitro. Mechanistically, we showed Forkhead box C1 (FOXC1, 601090) was a direct target for MIR‐138‐5P in PCa. We confirmed that overexpression of FOXC1 partially reversed the effects of MIR‐138‐5P on PCa cell behaviors.
Conclusions
Collectively, we showed that MIR‐138‐5P functions as a tumor suppressor gene in PCa via targeting FOXC1.
Keywords: FOXC1, MIR‐138‐5P, prostate cancer, tumor suppressor gene
FOXC1 overexpression could increase PCa cell proliferation, colony formation, migration, and invasion. Overexpression of FOXC1 could attenuated the effects of miR‐138‐5p on PCa cell behaviors.
1. INTRODUCTION
Prostate cancer (PCa) is a big health threat for men worldwide with increasing incidence (Castillejos‐Molina & Gabilondo‐Navarro, 2016). What is worse, the pathogenesis of PCa remains largely unknown, which results in the lack of treatment options, leading to worse overall survival of PCa patients (Murillo‐Garzón & Kypta, 2017). Hence, further investigations are needed to understand the mechanisms behind PCa progression.
Noncoding RNAs (ncRNAs) including microRNA (miRNA), long noncoding RNA (lncRNA), and circular RNA (circRNA) have been revealed to function as crucial roles in PCa progression (Greene et al., 2019; Kanwal, Plaga, Liu, Shukla, & Gupta, 2017; Wu, Xiao, Zhou, Zhou, & Yan, 2019). Increasing evidence suggested that ncRNAs can be developed as biomarkers for prognosis prediction and treatment (Greene et al., 2019; Kanwal et al., 2017; Wu et al., 2019). miRNAs are short RNAs (18–24 nucleotides in length) without protein‐coding capability (Ha & Kim, 2014). Dual functions of miRNAs in cancers have been reported, as some miRNA can promote cancer development, while some of them can inhibit tumorigenesis (Suzuki, Maruyama, Yamamoto, & Kai, 2013). By 3′‐untranlated region (3′‐UTR) binding, miRNAs can regulate multiple gene expressions, associated signaling pathways, and eventually affect the hallmarks of cancer (Acunzo, Romano, Wernicke, & Croce, 2015).
MIR‐138‐5P (613394, NC_000003.12) is an miRNA that reported to function as a tumor suppressive role in several human cancers. For instance, MIR‐138‐5P was revealed to be a decreased expression in colorectal cancer, and its low expression was significantly correlated with advanced tumor stage and poor overall survival (Zhao et al., 2016). Additionally, programmed cell death ligand 1 was identified as a direct target for MIR‐138‐5P, indicating MIR‐138‐5P has the potential to regulate immune response (Zhao et al., 2016). Besides that, MIR‐138‐5P was also identified to be a reduced expression in pancreatic cancer and suppressed autophagy and tumor growth through regulating the SIRTUIN 1(604479)/forkhead box O1 (136533)/RAS‐ASSOCIATED PROTEIN RAB7 (602298) axis (Tian, Guo, Yu, Sun, & Jiang, 2017). In non‐small‐cell lung, MIR‐138‐5P was found could affect the response of cancer cell to cisplatin through regulating ATG7 (608760; Pan, Chen, Shen, & Tantai, 2019). Moreover, lncRNA RP11‐476D10.1 (600138) was revealed and could function as a sponge for MIR‐138‐5P to regulate leucine‐rich repeat kinase 2 (609007) expression in papillary thyroid carcinoma (Zhao, Zhao, Li, & Zhong, 2019). Overexpression of MIR‐138‐5P was shown to promote apoptosis and autophagy of papillary thyroid carcinoma cells (Zhao, Zhao, et al., 2019). There is a previous work to indicate MIR‐138 could modulate PCa migration and invasion but did not indicate whether it was the 5p or the 3p strand (Yu, Wang, Li, Yang, & Tang, 2018). However, the biological roles of MIR‐138‐5P in PCa remain to be explored.
Forkhead box C1 (FOXC1, 601090, NG_009368.1), located at chromosome 6p25, is a transcription factor that belongs to the FOX gene family (Nishimura et al., 1998). FOXC1 has found to not only have crucial roles in normal physiological and pathological conditions but also shown to be important mediators for cancer progression (Han et al., 2017). For example, the knockdown of FOXC1 was found to inhibit glioma cell epithelial‑to‑mesenchymal transition via regulating the β‐catenin signaling (Cao et al., 2019).
In this work, we explored the expression level of MIR‐138‐5P in PCa cell lines. Furthermore, gain‐of‐function experiments were performed to investigate the biological roles of MIR‐138‐5P in PCa. Moreover, we predicated the targets of MIR‐138‐5P at TargetScan and FOXC was selected for the following analyses.
2. MATERIALS AND METHODS
2.1. Cell culture
PCa cell lines (PC3 and DU145) were seeded into F‐12K medium in supplement with 10% fetal bovine serum (FBS) purchased from Thermo Fisher Scientific, Inc. Normal prostate epithelial cells (RWPE‐1) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FBS obtained from Thermo Fisher Scientific, Inc. The incubation atmosphere was maintained at 37°C containing 5% of CO2. All these cell lines used were purchased from Cell Bank of Chinese Academy of Sciences.
2.2. Cell treatment
MIR‐138‐5Pmimic (5′‐AGCUGGUGUUGUGAAUCAGGCCG‐3′) and the corresponding negative control (NC‐miR, 5′‐GCGGUCGUGCAGUGCGUGAUAUA‐3′) were synthesized by RiboBio Inc. MIR‐138‐5P overexpression was accompanied by transfecting MIR‐138‐5P mimic into PCa cell lines using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific, Inc.). pcDNA3.1 contains the coding sequence of FOXC1 (pcFOXC1) and an empty vector were purchased from GenScript. Transfection was also conducted using Lipofectamine 2000 using the manufacturer's instruction.
2.3. Quantitative real‐time PCR (RT‐qPCR)
RNA from the cells was isolated with TRIZOL reagent (Beyotime). After concentration determination, RNA was reverse transcribed into complementary DNA with PrimeScript RT Reagent (Takara). RT‐qPCR was conducted at ABI 7500 system (Applied Biosystems) using SYBR Green Mix (Takara). The method of 2 − ΔΔCt was used to measure the relative expression level of MIR‐138‐5P or FOXC1 using U6 small nuclear RNA (U6 snRNA, 180692) or glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH, 138400) as endogenous control. Primers used were as follows: MIR‐138‐5P: Forward: 5′‐GCGAGCTGGTGTTGTGAATC‐3′, Reverse: 5′‐AGTGCAGGGTCCGAGGTATT‐3′; U6 snRNA: Forward: 5′‐CTCGCTTCGGCAGCACA‐3′, Reverse: 5′‐AACGCTTCACGAATTTGCGT‐3′; FOXC1: Forward: 5′‐CGGTATCCAGCCAGTCTCTGTACG‐3′, Reverse: 5′‐GTTCGGCTTTGAGGGTGTGTC‐3′; GAPDH: Forward: 5′‐GGAGGGAGATCCCTCCAAAAT‐3′, Reverse: 5′‐GGCTGTTGTCATACTTCTCATGG‐3′. Experiments were repeated in triplicate.
2.4. Cell counting kit‐8 (CCK‐8) assay
Cells in the density of 1 × 104 cells/well were seeded into 96‐well plates. After 0, 24, 48, or 72 hr incubation, CCK‐8 reagent purchased from Beyotime was filled into the plate and further incubated for 4 hr. Optical density in each well was analyzed at a wavelength of 450 nm. Experiments were repeated in triplicate.
2.5. Colony formation assay
Cells were incubated into 6‐well plate at the density of 800 cells/well. Colonies generated from the cultured cells were fixed with methanol, stained with crystal violet, and then counted under the microscope. Experiments were repeated in triplicate.
2.6. Cell migration assay
Cells were seeded in 6‐well plates and incubated until about 80% confluence. The pipette tip was used to create a wound at the cell surface. Thereafter, the cells were washed with PBS hree times to remove cell debris. After incubation for 0 or 48 hr, cell images were captured under the microscope to evaluate the effects of MIR‐138‐5P or FOXC1 expression on cell migration. Experiments were repeated in triplicate.
2.7. Cell invasion assay
In this experiment, 1 × 105 cells in serum‐free medium were filled into the upper chamber of the Matrigel‐coated insert. The lower chamber was filled with medium contains FBS. After incubation for 48 hr, noninvasive cells were removed with cotton. Then, invasive cells were fixed with methanol, stained using crystal violet, and counted under microscope. Experiments were repeated in triplicate.
2.8. Bioinformatic analysis
TargetScan was used to detect the putative targets for MIR‐138‐5P. Among all these targets, FOXC1 was selected for the following analyses.
2.9. Luciferase activity reporter assay
To validate the direct connection of MIR‐138‐5P and FOXC1, luciferase activity reporter assay was performed. The wild‐type (wt) or mutant (mt) 3′‐UTR sequence of FOXC1 was inserted into psiCHECK‐2 to obtain wt‐FOXC1 or mt‐FOXC1 luciferase vectors. For the luciferase activity reporter assay, cells were co‐transfected with luciferase vectors and miRNAs using Lipofectamine 2000. After transfection for 48 hr, relative luciferase activity was measured with a dual‐luciferase reporter kit (Promega) with Renilla luciferase activity as the internal control. Experiments were repeated in triplicate.
2.10. Statistical analyses
Data were obtained from three independent experiments and then expressed as mean ± SD after analyses using GraphPad Prism 6.0 (GraphPad Software). Differences in groups were assessed with Student's t test or one‐way ANOVA and Tukey post hoc test. p value less than .05 was considered as a statistically significant difference.
3. RESULTS
3.1. Decreased expression of MIR‐138‐5P in PCa cells
To explore the function of MIR‐138‐5P, we first analyzed its expression level in PCa cell lines and in the normal cell line. We found the MIR‐138‐5P expression level was significantly decreased in PCa cells compared with the normal cell line (Figure 1).
Figure 1.
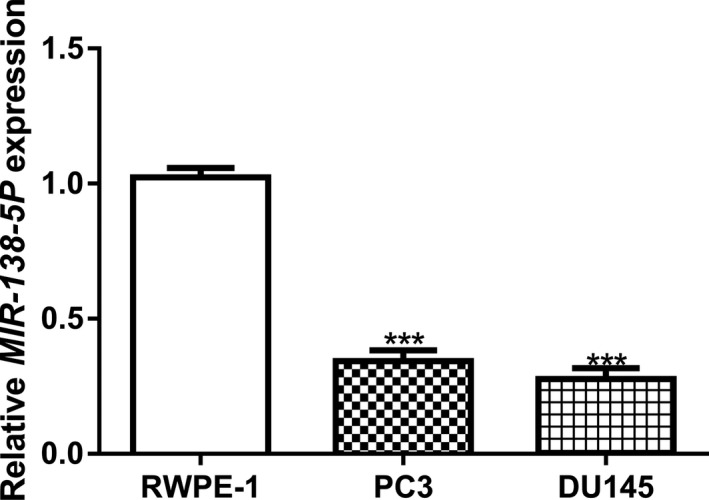
MIR‐138‐5P expression was decreased in PCa cells compared with normal cell line. MIR‐138‐5P: MICRORNA‐138‐5P (***p < .001). PCa, prostate cancer
3.2. MIR‐138‐5P overexpression inhibits PCa cell growth, migration, and invasion
We then detected the roles of MIR‐138‐5P overexpression on PCa cells. Introduction of MIR‐138‐5P mimic significantly increased MIR‐138‐5P levels in PCa cells (Figure 2a). CCK‐8 assay and wound‐healing assay revealed that the overexpression of MIR‐138‐5P inhibits PCa cell proliferation and colony formation (Figure 2b,c). Moreover, we found in PCa cells transfected with MIR‐138‐5P mimic cell migration and invasion abilities were significantly inhibited (Figure 2d,e).
Figure 2.
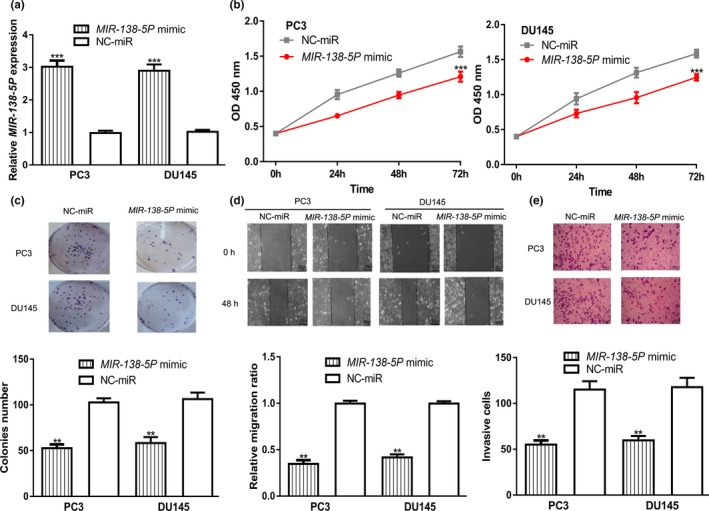
Transfection of MIR‐138‐5P mimic inhibited PCa cell proliferation, colony formation, cell migration, and cell invasion. (a) MIR‐138‐5P expression, (b) Cell proliferation, (c) Colony formation, (d) Cell migration, and (e) Cell invasion in PCa cells transfected with MIR‐138‐5P mimic or NC‐miR (**p < .01, ***p < .001). MIR‐138‐5P, MICRORNA‐138‐5P; NC‐miR, negative control microRNA; PCa, prostate cancer
3.3. MIR‐138‐5P can interact with FOXC1
Subsequently, we are interested to explore the targets of MIR‐138‐5P using TargetScan. We found FOXC1 was a putative target for MIR‐138‐5P (Figure 3a). Luciferase activity reporter assay validated the direct connection of MIR‐138‐5P and the 3′‐UTR of FOXC1 (Figure 3b). The RT‐qPCR analysis results indicated that overexpression of MIR‐138‐5P could inhibit the expression level of FOXC1 in PCa cells (Figure 3c).
Figure 3.
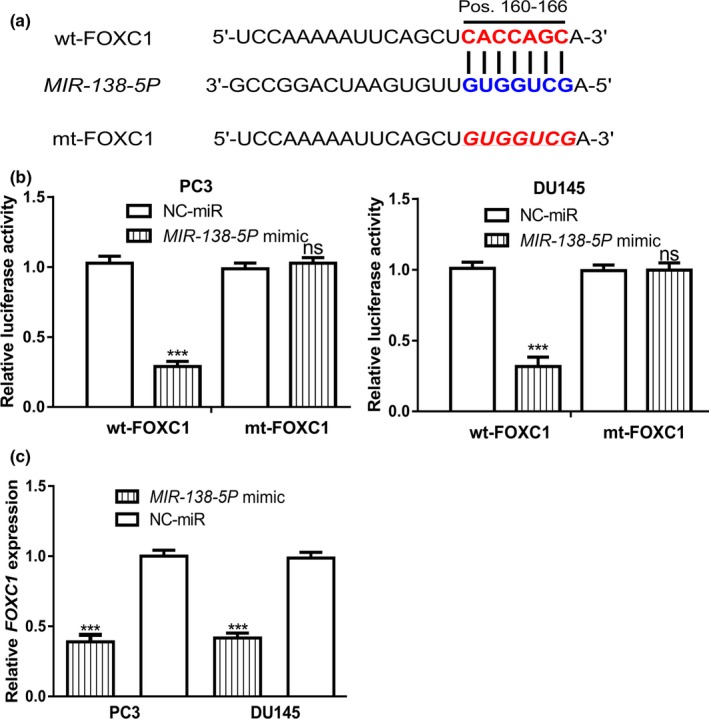
Direct interaction of MIR‐138‐5P and FOXC1. (a) Binding scheme of MIR‐138‐5P and the 3’‐UTR of FOXC1. (b) Relative luciferase activity in PCa cells transfected with wt/mt‐FOXC1 and MIR‐138‐5P mimic or NC‐miR. (c) Relative FOXC1 expression level in PCa cells transfected with MIR‐138‐5P mimic or NC‐miR (ns, not significant; ***p < .001). FOXC1, Forkhead box C1; MIR‐138‐5P, MICRORNA‐138‐5P; mt, mutant; NC‐miR, negative control microRNA; PCa, prostate cancer; UTR, untranslated region; wt, wild‐type
3.4. MIR‐138‐5P regulates PCa cell malignancy behaviors via targeting FOXC1
To clarify the relationship between MIR‐138‐5P and FOXC1, rescue experiments were conducted. We showed pcFOXC1 transfection increased the levels of FOXC1 in PCa cell (Figure 4a). In addition, we found the effects of MIR‐138‐5P mimic on FOXC1 expression can be reversed by pcFOXC1 (Figure 4a). In vitro functional experiments showed that overexpression of FOXC1 promoted PCa cell proliferation, colony formation, cell migration, and cell invasion (Figure 4b–e). Moreover, we showed that overexpression of FOXC1 partially abolished the effects of MIR‐138‐5P mimic on PCa cell behaviors (Figure 4b–e).
Figure 4.
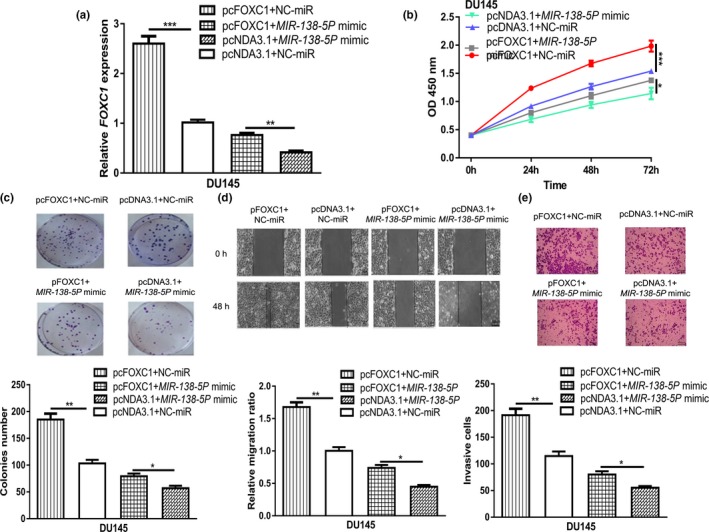
MIR‐138‐5P inhibits PCa progression by directly interacting with FOXC1. (a) MIR‐138‐5P expression, (b) Cell proliferation, (c) Colony formation, (d) Cell migration, and (e) Cell invasion in PCa cells transfected with pcFOXC1+NC‐miR, pcFOXC1+MIR‐138‐5P mimic, pcNDA3.1+NC‐miR, or pcNDA3.1+MIR‐138‐5P mimic (*p < .05, **p < .01, ***p < .001). FOXC1, Forkhead box C1; MIR‐138‐5P, MICRORNA‐138‐5P; NC‐miR: negative control microRNA; PCa, prostate cancer
4. DISCUSSION
miRNAs were reported to play crucial roles in the prevention or promotion of cancer progression (Acunzo et al., 2015; Suzuki et al., 2013). The miRNAs that promote carcinogenesis are the oncomiRs, while these can inhibit tumorigenesis were termed as tumor suppressive miRNAs. To date, numerous miRNAs have been identified to be aberrantly expressed in the development of PCa. For example, MIR‐301A‐3P (615675) was revealed to be an elevated expression in PCa tissues along with several cell lines (Fan, Wang, Huo, & Wang, 2019). In addition, they found PCa cell proliferation and invasion can be stimulated by MIR‐301A‐3P through regulating the expression of runt‑related transcription factor 3 (600210; Fan et al., 2019). Besides that, MIR‐198 (605547) was revealed to be a decreased expression in both PCa tissues and cell lines (Ray et al., 2019). In addition, the tumor growth ability can be inhibited in vitro and in vivo by the MIR‐198/Mindbomb E3 ubiquitin protein ligase 1 (608677) regulatory axis (Ray et al., 2019).
In this study, we showed that MIR‐138‐5P expression was decreased in PCa cell lines compared with the normal cell line. Previous studies demonstrated that MIR‐138‐5P regulates cancer migration, invasion, and epithelial–mesenchymal transition (Zhao, Ling, Li, Hou, & Zhao, 2019). Hence, we also explored the role of MIR‐138‐5P on cell behavior. Here we showed the restored expression of MIR‐138‐5P inhibits PCa cell proliferation, colony formation, cell migration, and cell invasion, indicating that MIR‐138‐5P functions as a tumor suppressive in PCa. Our work presented here is similar to the role of MIR‐138‐5P presented in other cancer types (Pan et al., 2019; Tian et al., 2017; Zhao, Zhao, et al., 2019). Several targets for MIR‐138‐5P have been identified in previous studies, which has helped us to understand the role of MIR‐138‐5P in human cancers. Hence, we also explored the potential target forMIR‐138‐5P in this work. Combining the results of bioinformatic analysis, luciferase activity reporter assay, and RT‐qPCR analysis, we found FOXC1 was a putative target for MIR‐138‐5P. Functionally we showed that the overexpression of FOXC1 could promote PCa cell malignancy behavior. Importantly, rescue experiments found overexpression of FOXC1 could partially reverse the effects of MIR‐138‐5P on PCa cells. This work provides novel evidence regarding the mechanisms behind the progression of PCa, which could provide novel targets for cancer treatment. However, the limitation in this work was that we did not explore the function of MIR‐139‐5P/FOXC1 axis in animal model, which we believe should be performed in the future.
Collectively, our work established the tumor suppressive role of MIR‐138‐5P in PCa. FOXC1 was identified as the novel target for MIR‐138‐5P, through which MIR‐138‐5P exerts the inhibitory effects on PCa cells. The validated MIR‐138‐5P and FOXC1 axis could help us to develop novel targets for PCa treatment.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Huang H, Xiong Y, Wu Z, et al. MIR‐138‐5P inhibits the progression of prostate cancer by targeting FOXC1 . Mol Genet Genomic Med. 2020;8:e1193 10.1002/mgg3.1193
REFERENCE
- Acunzo, M. , Romano, G. , Wernicke, D. , & Croce, C. M. (2015). MicroRNA and cancer‐a brief overview. Advances in Biological Regulation, 57, 1–9. 10.1016/j.jbior.2014.09.013 [DOI] [PubMed] [Google Scholar]
- Cao, Q. , Wang, X. , Shi, Y. , Zhang, M. , Yang, J. , Dong, M. , … Wang, P. (2019). FOXC1 silencing inhibits the epithelial‐to‐mesenchymal transition of glioma cells: Involvement of β‐catenin signaling. Molecular Medicine Reports, 19(1), 251–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejos‐Molina, R. A. , & Gabilondo‐Navarro, F. B. (2016). Prostate cancer. Salud Publica de Mexico, 58(2), 279–284. 10.21149/spm.v58i2.7797 [DOI] [PubMed] [Google Scholar]
- Fan, L. , Wang, Y. , Huo, W. , & Wang, W. H. (2019). MicroRNA‐301a‐3p overexpression promotes cell invasion and proliferation by targeting runt‐related transcription factor 3 in prostate cancer. Molecular Medicine Reports, 20(4), 3755–3763. 10.3892/mmr.2019.10650 [DOI] [PubMed] [Google Scholar]
- Greene, J. , Baird, A.‐M. , Casey, O. , Brady, L. , Blackshields, G. , Lim, M. , … Finn, S. P. (2019). Circular RNAs are differentially expressed in prostate cancer and are potentially associated with resistance to enzalutamide. Scientific Reports, 9(1), 10739 10.1038/s41598-019-47189-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha, M. , & Kim, V. N. (2014). Regulation of microRNA biogenesis. Nature Reviews Molecular Cell Biology, 15(8), 509–524. 10.1038/nrm3838 [DOI] [PubMed] [Google Scholar]
- Han, B. , Bhowmick, N. , Qu, Y. , Chung, S. , Giuliano, A. E. , & Cui, X. (2017). FOXC1: An emerging marker and therapeutic target for cancer. Oncogene, 36(28), 3957–3963. 10.1038/onc.2017.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwal, R. , Plaga, A. R. , Liu, X. , Shukla, G. C. , & Gupta, S. (2017). MicroRNAs in prostate cancer: Functional role as biomarkers. Cancer Letters, 407, 9–20. 10.1016/j.canlet.2017.08.011 [DOI] [PubMed] [Google Scholar]
- Murillo‐Garzón, V. , & Kypta, R. (2017). WNT signalling in prostate cancer. Nature Reviews Urology, 14(11), 683–696. 10.1038/nrurol.2017.144 [DOI] [PubMed] [Google Scholar]
- Nishimura, D. Y. , Swiderski, R. E. , Alward, W. L. M. , Searby, C. C. , Patil, S. R. , Bennet, S. R. , … Sheffield, V. C. (1998). The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nature Genetics, 19, 140–147. 10.1038/493 [DOI] [PubMed] [Google Scholar]
- Pan, X. , Chen, Y. , Shen, Y. , & Tantai, J. (2019). Knockdown of TRIM65 inhibits autophagy and cisplatin resistance in A549/DDP cells by regulating miR‐138‐5p/ATG7. Cell Death & Disease, 10(6), 429 10.1038/s41419-019-1660-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, J. , Hoey, C. , Huang, X. , Jeon, J. , Taeb, S. , Downes, M. , … Liu, S. (2019). MicroRNA‐198 suppresses prostate tumorigenesis by targeting MIB1. Oncology Reports, 42(3), 1047–1056. 10.3892/or.2019.7234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, H. , Maruyama, R. , Yamamoto, E. , & Kai, M. (2013). Epigenetic alteration and microRNA dysregulation in cancer. Frontiers in Genetics, 4, 258 10.3389/fgene.2013.00258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, S. , Guo, X. , Yu, C. , Sun, C. , & Jiang, J. (2017). miR‐138‐5p suppresses autophagy in pancreatic cancer by targeting SIRT1. Oncotarget, 8(7), 11071–11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X. , Xiao, Y. , Zhou, Y. , Zhou, Z. , & Yan, W. (2019). lncRNA SNHG20 promotes prostate cancer migration and invasion via targeting the miR‐6516‐5p/SCGB2A1 axis. American Journal of Translational Research, 11(8), 5162–5169. [PMC free article] [PubMed] [Google Scholar]
- Yu, Z. , Wang, Z. , Li, F. , Yang, J. , & Tang, L. (2018). miR‐138 modulates prostate cancer cell invasion and migration via Wnt/β‐catenin pathway. Molecular Medicine Reports, 17(2), 3140–3145. [DOI] [PubMed] [Google Scholar]
- Zhao, C. , Ling, X. , Li, X. , Hou, X. , & Zhao, D. (2019). MicroRNA‐138‐5p inhibits cell migration, invasion and EMT in breast cancer by directly targeting RHBDD1. Breast Cancer, 26(6), 817–825. 10.1007/s12282-019-00989-w [DOI] [PubMed] [Google Scholar]
- Zhao, L. , Yu, H. , Yi, S. , Peng, X. , Su, P. , Xiao, Z. , … Shen, S. (2016). The tumor suppressor miR‐138‐5p targets PD‐L1 in colorectal cancer. Oncotarget, 7(29), 45370–45384. 10.18632/oncotarget.9659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Zhao, L. , Li, J. , & Zhong, L. (2019). Silencing of long noncoding RNA RP11‐476D10.1 enhances apoptosis and autophagy while inhibiting proliferation of papillary thyroid carcinoma cells via microRNA‐138‐5p‐dependent inhibition of LRRK2. Journal of Cellular Physiology, 234(11), 20980–20991. [DOI] [PubMed] [Google Scholar]


