Abstract
Background
Lipid traits have been inconsistently linked to risk of non-Hodgkin lymphoma (NHL). We examined the association of genetically predicted lipid traits with risk of diffuse large B-cell lymphoma (DLBCL), chronic lymphocytic leukemia (CLL), follicular (FL) and marginal zone (MZL) lymphoma using Mendelian randomization (MR) analysis.
Methods
GWAS data from the InterLymph Consortium were available for 2661 DLBCLs, 2179 CLLs, 2142 FLs, 824 MZLs, and 6221 controls. SNPs associated (P<5×10−8) with high-density lipoprotein (HDL, N=164), low-density lipoprotein (LDL, N=137), total cholesterol (TC, N=161), and triglycerides (TG, N=123) were used as instrumental variables (IV), explaining 14.6%, 27.7%, 16.8% and 12.8% of phenotypic variation, respectively. Associations between each lipid trait and NHL subtype were calculated using the MR inverse variance-weighted method, estimating odds ratios (OR) per standard deviation, and 95% confidence intervals (CI).
Results
HDL was positively associated with DLBCL (OR=1.14, CI:1.00–1.30) and MZL (OR=1.09, CI:1.01–1.18), while TG was inversely associated with MZL risk (OR=0.90, CI:0.83–0.99) all at nominal significance (P<0.05). A positive trend was observed for HDL with FL risk (OR=1.08, CI:0.99–1.19; P=0.087). No associations were noteworthy after adjusting for multiple testing.
Conclusions
We did not find evidence of a clear or strong association of these lipid traits with the most common NHL subtypes. While these IVs have been previously linked to other cancers, our findings do not support any causal associations with these NHL subtypes.
Impact
Our results suggest that prior reported inverse associations of lipid traits are not likely to be causal and could represent reverse causality or confounding.
Introduction
Lipid traits such as high-density lipoprotein (HDL), low-density lipoprotein (LDL), total cholesterol (TC), and triglyceride (TG) have been suggested as non-Hodgkin lymphoma (NHL) risk factors; however, results are inconclusive. Of the strongest studies addressing this hypothesis(1–3), a nested case-control study from the Multi-Ethnic Cohort (275 NHL cases and 549 controls) found inverse associations of TC and HDL, but not LDL or TG, with NHL risk(1). In the Alpha-Tocopherol Beta-Carotene Cancer Prevention Study cohort study, HDL was inversely associated with NHL risk during the first 10 years of follow-up, but not with diagnoses after 10 years of follow-up(2). Recently, a large case-control study from the Cancer Research Network examined the relationship of cholesterol with lymphomagenesis in the 10 years prior to lymphoma diagnosis and found that lymphoma cases had lower estimated TC, HDL, and LDL levels than controls, but this was mainly observed in the 3–4 years prior to diagnosis/index date(3). The authors concluded that low cholesterol could indicate an altered systemic metabolic profile associated with the natural history of lymphoma pre-diagnosis and a potential biomarker of subclinical disease. However, it is not established if the observed inverse association between TC and HDL and risk of NHL is a result of protective actions of these lipids and lipoproteins, confounding or reverse causation.
Currently, single nucleotide polymorphisms (SNPs) associated with lipid traits explain 12–28% of the total variation in these traits in populations of European ancestry(4). Here we apply a Mendelian randomization (MR) analysis to examine the possibility of a causal relationship between four genetically predicted lipid traits and the risk of four common NHL subtypes: diffuse large B-cell lymphoma (DLBCL), chronic lymphocytic leukemia (CLL), follicular lymphoma (FL), and marginal zone lymphoma (MZL).
Materials and Methods
GWAS data from the InterLymph Consortium were available for 2661 DLBCLs, 2179 CLLs, 2142 FLs, 824 MZLs, and 6221 controls of European descent(5–8). SNPs associated (P<5×10−8) with HDL (N=164), LDL (N=137), TC (N=161), and TG (N=123) that were identified through the Global Lipids Genetics Consortium, were used as instrumental variables (IV)(4). SNPs were not in strong linkage disequilibrium (r2<0.05). MR estimates for the association between each lipid trait and NHL subtype were calculated using the inverse variance-weighted (IVW), simple median, and weighted median methods, after testing for evidence of pleiotropy using MR-Egger regression to test for violation of the standard IV assumptions(9). Associations were reported as odds ratios (OR) per standard deviation increase in the MR genetic risk score along with 95% confidence intervals (CI). We defined statistical significance by a Bonferroni-corrected threshold of P<0.003 (0.05/16=0.003, 16 comparisons of 4 lipid traits across 4 NHL subtypes).
Results
In our study sample, there was no evidence of violation of the assumptions for the associations tested using MR-Egger regression. We found at nominal significance (P<0.05) that genetically predicted HDL was positively associated with DLBCL (ORIVW=1.14, CI:1.00–1.30, P=0.049 , Figure 1.A), and MZL (ORIVW=1.09, CI:1.01–1.18, P=0.027, Figure 1.B); while TG was inversely associated with risk of MZL (ORIVW=0.90, CI:0.83–0.99, P=0.025, Figure 1.C) (Table 1). In addition, we observed a suggestive positive trend for genetically predicted HDL and FL risk (ORIVW=1.08, CI:0.99–1.19; P=0.087, Figure 1.D)(Table 1). Using the simple median and weighted median methods did not change the conclusions (Figure 1.A–D). No associations were noteworthy after adjusting for multiple testing.
Figure 1: HDL and TC SNP-specific effects for risk of lymphoma subtypes.
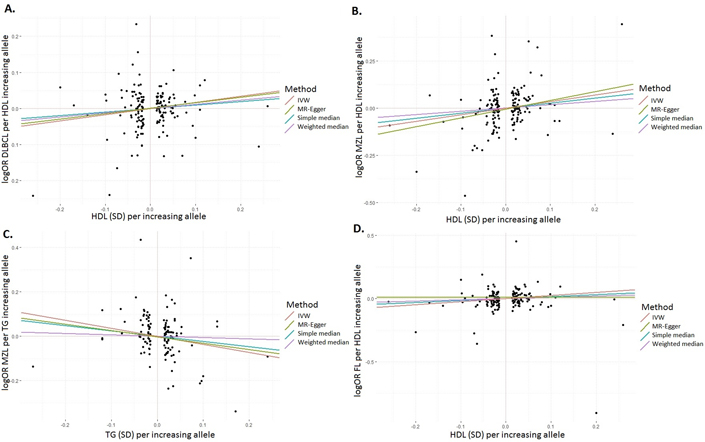
Scatter plots for lipid traits and lymphoma associations for SNPs and four MR estimates (IVW, MR-Egger, simple median, and weighted median): (A) HDL and DLBCL association; (B) HDL and MZL association; (C) TG and MZL association; (D) HDL and FL association; DLBCL, diffuse large B-cell lymphoma; MZL, marginal zone lymphoma; FL, follicular lymphoma; HDL, high-density lipoprotein; TG, triglyceride; SD, standard error; SNP, single nucleotide polymorphism; MR, mendelian randomization; IVW, inverse-variance weighted
Table 1:
Associations between B-cell non-Hodgkin lymphoma subtypes per standard deviation increase in the MR genetic risk score for each lipid trait
| MR-Egger | Inverse-weighted variance | ||||||
|---|---|---|---|---|---|---|---|
| OR per SD increase | 95% CI | P | OR per SD increase | 95% CI | P | ||
| DLBCL | HDL | 1.14 | 0.94–1.38 | 0.176 | 1.14 | 1.00–1.30 | 0.049 |
| LDL | 0.78 | 0.53–1.15 | 0.211 | 0.96 | 0.74–1.24 | 0.738 | |
| TC | 0.91 | 0.74–1.12 | 0.381 | 1.05 | 0.91–1.22 | 0.488 | |
| TG | 1.02 | 0.81–1.30 | 0.862 | 1.08 | 0.92–1.25 | 0.337 | |
| CLL | HDL | 0.97 | 0.81–1.17 | 0.761 | 1.09 | 0.96–1.23 | 0.192 |
| LDL | 0.95 | 0.71–1.26 | 0.708 | 1.00 | 0.82–1.23 | 0.983 | |
| TC | 0.93 | 0.76–1.14 | 0.511 | 0.96 | 0.84–1.11 | 0.597 | |
| TG | 0.95 | 0.82–1.09 | 0.438 | 0.94 | 0.84–1.05 | 0.292 | |
| FL | HDL | 1.04 | 0.93–1.17 | 0.506 | 1.08 | 0.99–1.19 | 0.087 |
| LDL | 1.03 | 0.83–1.28 | 0.779 | 1.04 | 0.89–1.22 | 0.585 | |
| TC | 1.05 | 0.91–1.22 | 0.516 | 1.07 | 0.96–1.18 | 0.219 | |
| TG | 0.98 | 0.84–1.14 | 0.777 | 1.01 | 0.90–1.13 | 0.864 | |
| MZL | HDL | 1.09 | 0.98–1.21 | 0.123 | 1.09 | 1.01–1.18 | 0.027 |
| LDL | 0.94 | 0.77–1.15 | 0.556 | 0.90 | 0.78–1.03 | 0.137 | |
| TC | 0.98 | 0.88–1.10 | 0.771 | 0.98 | 0.90–1.06 | 0.560 | |
| TG | 0.84 | 0.73–0.96 | 0.008 | 0.90 | 0.83–0.99 | 0.025 | |
OR, odds ratio; SD, standard deviation; CI, confidence interval; MR, Mendelian randomization; DLBCL, diffuse large B-cell lymphoma; CLL, chronic lymphocytic leukemia; FL, follicular lymphoma; MZL, marginal zone lymphoma; HDL, high-density lipoprotein; LDL, low-density lipoprotein; TC, total cholesterol; TG, triglyceride;
Discussion
Our large study of NHL found no evidence of a causal association for these lipid traits with the most common B-cell NHL subtypes. The amount of variance accounted for by these SNPs for the lipid traits is larger than for many MR studies, and the IVs have been previously associated with other cancers such as colorectal and prostate cancers. MR is an important tool for appraising causality in epidemiology and may be even more important for establishing null results(9). We found no robust association between the genetic variants associated with the lipid traits and risk of any of the NHL subtypes, suggesting that there might be very little or no effect of lipid traits on these NHL subtypes. We realize that our null findings may be due to a lack of power, although at an exposure prevalence of 50% we had >99% power to detect a RR (as small as 0.70 for DLBCL, CLL and FL (each with over 2000 cases) and MZL (with over 800 cases) with a type I error rate of 0.003. In addition, MR results can be biased if the assumptions are violated, although these biases would be unlikely to move the effect estimate to zero when there is a true (non-zero) effect; in order for this to happen, the biases would have to align perfectly(9). Our results are in agreement with most studies that have assessed history of hyperlipidemia or statin use with risk of NHL and suggest that published inverse associations could be due to reverse causality or confounding.
Acknowledgments
Dr. Kleinstern was supported by the National Institutes of Health grant, R25 CA92049 (Mayo Cancer Genetic Epidemiology Training Program). Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award number R01 CA200703 (J.R. Cerhan).
Support for individual studies:
ATBC – Intramural Research Program of the National Institutes of Health, NCI, Division of Cancer Epidemiology and Genetics.
BC – Canadian Institutes for Health Research (CIHR).
CPSII – The American Cancer Society funds the creation, maintenance, and updating of the CPSII cohort. The authors thank the CPS-II participants and Study Management Group for their invaluable contributions to this research. The authors would also like to acknowledge the contribution to this study from central cancer registries supported through the Centers for Disease Control and Prevention National Program of Cancer Registries, and cancer registries supported by the National Cancer Institute Surveillance Epidemiology and End Results program.
ELCCS – Leukaemia & Lymphoma Research.
ENGELA – Fondation ARC pour la Recherche sur le Cancer. Fondation de France. French Agency for Food, Environmental and Occupational Health & Safety (ANSES), the French National Cancer Institute (INCa).
EPIC – Coordinated Action (Contract #006438, SP23-CT-2005–006438). HuGeF (Human Genetics Foundation), Torino, Italy.
EPILYMPH – European Commission (grant references QLK4-CT-2000–00422 and FOOD-CT-2006–023103); the Spanish Ministry of Health (grant references CIBERESP, PI11/01810, RCESP C03/09, RTICESP C03/10 and RTIC RD06/0020/0095), the Marató de TV3 Foundation (grant reference 051210), the Agència de Gestiód’AjutsUniversitarisi de Recerca – Generalitat de Catalunya (grant reference 2009SGR1465) who had no role in the data collection, analysis or interpretation of the results; the NIH (contract NO1-CO-12400); the Compagnia di San Paolo—Programma Oncologia; the Federal Office for Radiation Protection grants StSch4261 and StSch4420, the José Carreras Leukemia Foundation grant DJCLS-R12/23, the German Federal Ministry for Education and Research (BMBF-01-EO-1303); the Health Research Board, Ireland and Cancer Research Ireland; Czech Republic supported by MH CZ – DRO (MMCI, 00209805) and RECAMO, CZ.1.05/2.1.00/03.0101; Fondation de France and Association de Recherche Contre le Cancer.
Spanish Ministry of Economy and Competitiveness - Carlos III Institute of Health cofunded by FEDER funds/European Regional Develpment Fund (ERDF) - a way to build Europe. Grant numbers: PI17/01280 and PI14/01219;
Disclaimer: Where authors are identified as personnel of the International Agency for Research on Cancer / World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer / World Health Organization.
Centro de Investigación Biomédica en Red: Epidemiología y Salud Pública (CIBERESP, Spain)
Grant sponsor: Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR), CERCA Programme / Generalitat de Catalunya for institutional support. Grant number: 2017SGR1085
GEC – National Institutes of Health CA 118444.
GELA – The French National Cancer Institute (INCa).
HPFS – The HPFS was supported in part by National Institutes of Health grants UM1 CA167552, R01 CA149445 and R01 CA098122. We would like to thank the participants and staff of the Health Professionals Follow-up Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data. The study protocol was approved by the institutional review boards of the Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health, and those of participating registries as required.
Iowa-Mayo SPORE – National Institutes of Health (CA97274). NCI Specialized Programs of Research Excellence (SPORE) in Human Cancer (P50 CA97274). Molecular Epidemiology of Non-Hodgkin Lymphoma Survival (R01 CA129539). Henry J. Predolin Foundation.
Italian GxE – Italian Ministry for Education, University and Research Research (PRIN 2007 prot. 2007WEJLZB, PRIN 2009 prot. 20092ZELR2); the Italian Association for Cancer Research (AIRC, Investigator Grant 11855).
Mayo Clinic Case-Control – National Institutes of Health (R01 CA92153). National Center for Advancing Translational Science (UL1 TR000135)
MCCS – The Melbourne Collaborative Cohort Study recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further supported by Australian NHMRC grants 209057, 251553 and 504711 and by infrastructure provided by Cancer Council Victoria.
MD Anderson – Institutional support to the Center for Translational and Public Health Genomics.
MSKCC – Geoffrey Beene Cancer Research Grant, Lymphoma Foundation (LF5541). Barbara K. Lipman Lymphoma Research Fund (74419). Robert and Kate Niehaus Clinical Cancer Genetics Research Initiative (57470), U01 HG007033. ENCODE, U01 HG007033.
NCI-SEER – Intramural Research Program of the National Cancer Institute, National Institutes of Health, and Public Health Service (N01-PC-65064,N01-PC-67008, N01-PC-67009, N01-PC-67010, N02-PC-71105).
NHS – The NHS was supported in part by National Institutes of Health grants UM1 CA186107, PO1 CA87969, R01 CA49449, R01 CA149445, R01 CA098122 and R01 CA134958. We would like to thank the participants and staff of the Nurses’ Health Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data. The study protocol was approved by the institutional review boards of the Brigham and Women’s Hospital and Harvard T.H. Chan School of Public Health, and those of participating registries as required.
NSW – was supported by grants from the Australian National Health and Medical Research Council (ID990920), the Cancer Council NSW, and the University of Sydney Faculty of Medicine.
NYUWHS - National Cancer Institute (UM1CA182934, P30 CA016087). National Institute of Environmental Health Sciences (ES000260).
PLCO - This research was supported by the Intramural Research Program of the National Cancer Institute and by contracts from the Division of Cancer Prevention, National Cancer Institute, NIH, DHHS.
SCALE – Swedish Cancer Society (2009/659). Stockholm County Council (20110209) and the Strategic Research Program in Epidemiology at Karolinska Institute. Swedish Cancer Society grant (02 6661). Danish Cancer Research Foundation Grant. Lundbeck Foundation Grant (R19-A2364). Danish Cancer Society Grant (DP 08–155). National Institutes of Health (5R01 CA69669–02). Plan Denmark.
UCSF – The UCSF studies were supported by the NCI, National Institutes of Health, CA1046282 and CA154643. The collection of cancer incidence data used in this study was supported by the California Department of Health Services as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885; the National Cancer Institute’s Surveillance, Epidemiology, and End Results Program under contract HHSN261201000140C awarded to the Cancer Prevention Institute of California, contract HHSN261201000035C awarded to the University of Southern California, and contract HHSN261201000034C awarded to the Public Health Institute; and the Centers for Disease Control and Prevention’s National Program of Cancer Registries, under agreement #1U58 DP000807–01 awarded to the Public Health Institute. The ideas and opinions expressed herein are those of the authors, and endorsement by the State of California, the California Department of Health Services, the National Cancer Institute, or the Centers for Disease Control and Prevention or their contractors and subcontractors is not intended nor should be inferred.
UTAH - National Institutes of Health CA134674. Partial support for data collection at the Utah site was made possible by the Utah Population Database (UPDB) and the Utah Cancer Registry (UCR). Partial support for all datasets within the UPDB is provided by the Huntsman Cancer Institute (HCI) and the HCI Comprehensive Cancer Center Support grant, P30 CA42014. The UCR is supported in part by NIH contract HHSN261201000026C from the National Cancer Institute SEER Program with additional support from the Utah State Department of Health and the University of Utah.
WHI - The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201600018C, HHSN268201600001C, HHSN268201600002C, HHSN268201600003C, and HHSN268201600004C. The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A full listing of WHI investigators can be found at: http://www.whi.org/researchers/Documents%20%20Write%20a%20Paper/WHI%20Investigator%20Long%20List.pdf
YALE – National Cancer Institute (CA62006).
References
- 1.Morimoto Y, Conroy SM, Ollberding NJ, Henning SM, Franke AA, Wilkens LR, et al. Erythrocyte membrane fatty acid composition, serum lipids, and non-Hodgkin’s lymphoma risk in a nested case–control study: the multiethnic cohort. Cancer Causes Control. 2012;23:1693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim U, Gayles T, Katki HA, Stolzenberg-Solomon R, Weinstein SJ, Pietinen P, et al. Serum High-Density Lipoprotein Cholesterol and Risk of Non-Hodgkin Lymphoma. Cancer Res. 2007;67. [DOI] [PubMed] [Google Scholar]
- 3.Alford SH, Divine G, Chao C, Habel LA, Janakiraman N, Wang Y, et al. Serum cholesterol trajectories in the 10 years prior to lymphoma diagnosis. Cancer Causes Control. 2018;29:143–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu DJ, Peloso GM, Yu H, Butterworth AS, Wang X, Mahajan A, et al. Exome-wide association study of plasma lipids in >300,000 individuals. Nat Genet. 2017;49:1758–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerhan JR, Berndt SI, Vijai J, Ghesquières H, McKay J, Wang SS, et al. Genome-wide association study identifies multiple susceptibility loci for diffuse large B cell lymphoma. Nat Genet. 2014;46:1233–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berndt SI, Camp NJ, Skibola CF, Vijai J, Wang Z, Gu J, et al. Meta-analysis of genome-wide association studies discovers multiple loci for chronic lymphocytic leukemia Nat Commun. Nature Publishing Group; 2016;7:10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skibola CF, Berndt SI, Vijai J, Conde L, Wang Z, Yeager M, et al. Genome-wide association study identifies five susceptibility loci for follicular lymphoma outside the HLA region. Am J Hum Genet. Elsevier; 2014;95:462–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vijai J, Wang Z, Berndt SI, Skibola CF, Slager SL, de Sanjose S, et al. A genome-wide association study of marginal zone lymphoma shows association to the HLA region. Nat Commun. 2015;6:5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.VanderWeele TJ, Tchetgen Tchetgen EJ, Cornelis M, Kraft P. Methodological Challenges in Mendelian Randomization. Epidemiology. 2014;25:427–35. [DOI] [PMC free article] [PubMed] [Google Scholar]


