Abstract
Background
The success of multi-site collaborative research relies on effective data collection, harmonization and aggregation strategies. Data Coordination Centers (DCCs) serve to facilitate the implementation of these strategies. The utility of a DCC can be particularly relevant for research on rare diseases where collaboration from multiple sites to amass large aggregate data sets is essential. However, approaches to building a DCC have been scarcely documented.
Methods and Materials
The Li-Fraumeni Exploration (LiFE) Consortium’s DCC was created using multiple open source packages, including LAM/G Application (Linux, Apache, MySQL, Grails), Extraction-Transformation-Loading (ETL) Pentaho Data Integration Tool, and the Saiku-Mondrian client. This document serves as a resource for building a rare disease DCC for multi-institutional collaborative research.
Results
The primary scientific and technological objective to create an online central repository into which data from all participating sites could be deposited, harmonized, aggregated, disseminated, and analyzed was completed. The cohort now include 2,193 participants from 6 contributing sites, including 1,354 individuals from families with a pathogenic or likely variant in TP53. Data on cancer diagnoses are also available. Challenges and lessons learned are summarized.
Conclusion
The methods leveraged mitigate challenges associated with successfully developing a DCC’s technical infrastructure, data harmonization efforts, communications, and software development and applications.
Impact
These methods can serve as a framework in establishing other collaborative research efforts. Data from the consortium will serve as a great resource for collaborative research to improve knowledge on, and the ability to care for, individuals and families with Li-Fraumeni Syndrome.
Keywords: Collaborative Research, Data Coordinating Center (DCC), Data Aggregation, Data Harmonization, Web-Based Collaborative Research Resource, Li-Fraumeni Syndrome, Li-Fraumeni Exploration Consortium, TP53
INTRODUCTION
Epidemiological research frequently requires large data collection efforts to establish the natural history of a disease. Collaborative research, especially for rare diseases, is often needed to achieve sufficiently large sample sizes to allow for meaningful study design, and interpretations of findings. However, collaborative research can present technical challenges to researchers with limited access to information technology and bioinformatics resources. Data Coordinating Centers (DCCs) offer a technical and strategic solution that helps mitigate the administrative, computational, and infrastructure burden from individual researchers seeking to conduct multi-site collaborative research and enable the implementation of projects based on aggregate data (1).
A DCC is designed, among other functions, to manage the administrative aspects of building a shared central data repository for multiple research sites by implementing effective infrastructures for communication, data collection and data harmonization, and establishment of a central data repository (2). Experience from large epidemiology consortia proves that data harmonization is vital to the development of an aggregated database since submitting institutions often collect and record data differently (3). DCCs must also have the technical expertise to build a highly curated centralized data repository, with embedded statistical analysis tools, and capability for interactive data query and visualization tools.
There is a dearth of published guidance available for the building of a successful DCC for research consortia, especially those involving rare diseases (1). To address this issue, the National Cancer Institute (NCI), Division of Cancer Control and Population Sciences (DCCPS), Epidemiology and Genomics Research Program (EGRP) provided funding for a pilot project to establish a DCC to support the Li-Fraumeni Exploration (LiFE) Consortium. The EGRP supports national and international research consortia focusing on interdisciplinary and translational research of rare cancers. The objective of this project was to expedite collaborative research across Cancer Epidemiology Consortia (CEC) supported by the NCI and other agencies. Specifically, this was to be accomplished through a demonstration project aimed at establishing an efficient and cost-effective DCC for CEC focusing on rare cancers, and establishing guidelines and Standard Operating Procedures for the creation of such DCC to be used by established, emerging and future rare cancers CECs. The DCC to be created was tasked with providing support for data collection, harmonization, management, distribution and access to the CEC investigators and, through an approved process, to collaborators external to the CEC.
The Li-Fraumeni Exploration (LiFE) Consortium was created with the mission to foster communication among investigators, and to promote collaborative research projects to advance our understanding of Li-Fraumeni Syndrome (LFS) and its impact on affected families. The LiFE Consortium also provides a platform for joint activities between professionals and patients and families to promote support, education and awareness (4).
Li-Fraumeni syndrome (LFS, OMIM#151623) is a rare autosomal dominant cancer predisposition syndrome (5)associated with germline pathogenic variants in the TP53 tumor suppressor gene (6, 7) and is characterized by a high lifetime risk of developing a wide spectrum of childhood and adult onset cancers with osteosarcoma, soft-tissue sarcomas (STS), early-onset breast cancer, brain tumors, leukemia, and adrenal cortical carcinoma (ACC) being the core cancers (8–11). As more families with TP53 mutations are identified, the LFS cancer spectrum has expanded to include melanoma, lung, gastrointestinal tract, thyroid, ovarian, and other cancers (8, 11–13). Cumulative cancer risk associated with LFS has been estimated to be ~50% by age 40 and up to 90% by age 60 (9, 14) with females having higher risk than males (9, 15–17). Clinical diagnostic criteria for classic LFS kindred include a person with sarcoma diagnosed before age 45, with a first-degree relative with any cancer before age 45 and another first- or second-degree relative with a sarcoma at any age or another cancer before age 45 (18). The less stringent Li-Fraumeni-like (LFL) criteria expand the proband’s cancer type to include childhood cancers, brain cancers, and ACC, and extend the relatives’ age at diagnosis to <60 years (19, 20). Germline TP53 pathogenic variants are identified in ~70% of families meeting the classic LFS diagnostic criteria (21, 22) and ~40% of families meeting the LFL diagnostic criteria (19). The frequency of de novo pathogenic variants in TP53 is estimated to be between 7% and 20% (23). Guidelines for TP53 genetic testing have also been developed (8, 24–28). Although progress has been made since LFS was first described in 1969, many questions related to the natural history of this condition and effective clinical management remain unanswered. Given the rarity of LFS, the assembling of information from multiple research institutions to build a larger dataset is essential in the effort to improve understanding of the condition and patient care.
MATERIALS & METHODS
A contract was established with Enterprise Science and Computing (ESAC), Inc., a Biomedical Research Data Management and Health Information Technology company, to build and manage the LiFE DCC. Seven LiFE Consortium participating institutions in the United States, Canada, Brazil, and the United Kingdom were involved in the initial effort to develop a data repository.
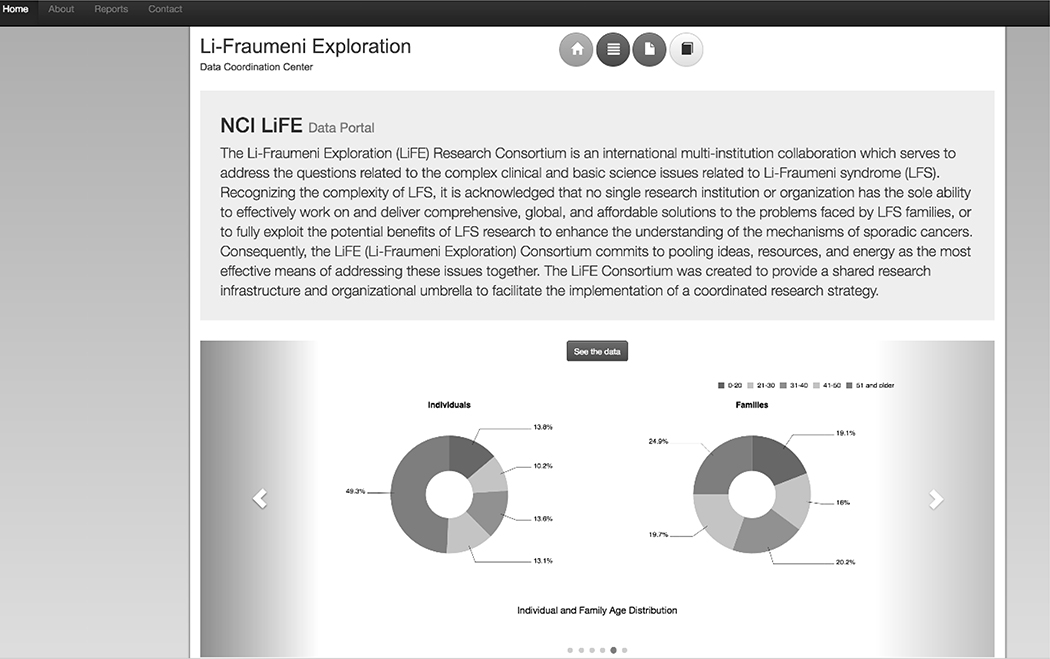
Creating the LiFE DCC involved establishing a technical working group and training on the functioning and processes of the repository. A web-based query tool was also built into the repository for summary data. The LiFE DCC web portal was designed to house and manage consortium-related public and semi-public information (Figure 1).
Figure 1:
The LiFE DCC Web Portal homepage as viewed by the public. The top left tabs are the “Home” page, the “About” the LiFE DCC page, a “Reports” page with summary overview data of what is housed within the DCC, and the “Contact” page providing contact information for the DCC. The colored buttons in the top center of home page link again to the “Home”, “About”, and “Reports” pages (yellow, red, and dark blue respectively). The grey button with a text book symbol at the end links to the “LiFE DCC Data Dictionary” with definitions and validation rules for all data elements implemented in the DCC central repository. The top right corner has a “Login” button which allows DCC contributors with the appropriate credentials to access the query interface for the entirety of the database. The carousel at the bottom provides a visual representation of the summary data made available to the public on the “Reports” page. Once logged in with the appropriate credentials, a fourth button with stack symbol appears in the top center buttons panel as well as in the summary paragraph panel called “Analyze the Data”. Clicking either link takes the user to the Saiku query tool.
Collaboration and Communication Infrastructure Development
A Steering Committee, consisting of Principal Investigators (PIs) from each participating site, Project Leads from ESAC, Inc., and a representative from EGRP, was established at the outset. A Technical Working Group, with research database administrators from each contributing site, was also formed at the beginning of this project. The Steering Committee made decisions regarding all aspects of the development and implementation of the DCC, including data elements, data formatting, inclusion criteria, and prioritization of consortium-related activities. Steering Committee members worked with each other and within their institutions to implement the appropriate agreements, policies and procedures for data transfer, housing, and use. The Steering Committee also determined the approval process permitting access to data in the LiFE DCC central repository.
The Technical Working Group and ESAC Project Leads carried out the implementation data harmonization, including the data elements collected, data formats, values of each data field, and validation rules for data submitted to the LiFE DCC. The Technical Working Group was also responsible for determining submission methods, creating data dictionaries used to standardize submissions across sites, implementing quality control procedures, and keeping validation logs. Once the technical details of creating the harmonized LiFE DCC central repository were determined, data from each site were prepared in accordance with the established rules, and submitted to ESAC, Inc.
One of the barriers to the development of a DCC is establishing coordinated communications among consortium members and with the DCC (29). The LiFE consortium initially included participants located in Brazil, Canada, the United Kingdom (UK), and the United State (US), as well as the LiFE DCC Project Leads at ESAC, Inc., and the EGRP representative. The communication barrier has eased with the availability and accessibility of various web-based communication applications. For the LiFE DCC, communications were facilitated using email correspondence, online meetings, weekly to semi-weekly teleconferences and web conferences, and an online customized Wiki space.
In anticipation of the multiple documents and resources to be shared, the LiFE DCC developed a password-protected online Wiki space to organize and provide members with easy access to all consortium-related information and documents. The functionalities of the online Wiki space included sharing and downloading documents, storing files, listing due dates for specific tasks, and posting of meeting minutes and other relevant information. The Wiki also served as a space for LiFE Consortium members to communicate between teleconferences. The Wiki space was vital in organizing all pertinent information and documentation in one convenient and easily accessible online location.
Data Submission, Harmonization, Management, Quality Control and Assurance
A Data Transfer Agreement (DTA) was drafted by the NCI’s Technology Transfer Team to be executed between each participating LiFE Consortium site and ESAC, Inc. The DTA outlined the handling of data by the DCC and the duration of the agreement. Data could only be submitted to the DCC after the DTA between the contributing site and ESAC, Inc. had been executed.
A Data Use Agreement (DUA) was also created by the NCI’s Technology Transfer Team to define the responsibilities of the contributing LiFE Consortium sites with respect to data use, sharing, protection, and accreditation. The DUA was executed amongst the participating LiFE sites, allowing each site access to the aggregate harmonized data housed in the central repository.
Data harmonization is a vital undertaking for a successful DCC. The establishment of a common data dictionary is essential to organizing all data within a DCC (30). Each consortium site may have its own format and values for each data element. In order to facilitate the harmonization of data among the sites, Technical Working Group members submitted a list of all the data elements collected, and the permissible values for each element, contained in their respective databases in a Microsoft Excel (or equivalent) spreadsheet. The DCC then created a table to map and tally all common data elements (CDE) across the submitting sites. The CDE table was compared against existing data standards, namely, the Cancer Data Standards Registry (caDSR), and the United States Health Information Knowledgebase (USHIK). Existing standardized definitions/coding rules for data elements on the CDE list were adopted where possible. Even though similar data were collected, how they were recorded and coded varied significantly from site to site. Especially challenging are the data on cancer diagnosis, as the information was collected in various formats, including text and ICD codes. There was no standard for how the text was recorded. All data submitted for the cancer diagnosis fields were reviewed by a member of the Steering Committee and mapped to a specific diagnosis.
Based on the CDE analysis, a proposed Minimum Data Set (MDS) was assembled, and approved by the Steering Committee, consisting of twenty data elements which were common to at least three of the seven participating LiFE sites. An additional 27 optional data elements were considered, with 13 added to the final set of elements to be included in the central repository. Coding and validation rules for each data element were determined by the Technical Working Group (TWG) and recorded in the Data Dictionary. Rules for transforming the values of each data element to conform with the format of the central repository were also established. Test cases from all sites were sent securely via an open-source secure File Transfer Protocol (sFTP) client (FileZilla https://filezilla-project.org/client_features.php) to test the implementation of the LiFE DCC Data Dictionary. Once the fidelity of the transferred test data was assured, all sites with established DTAs submitted all data in accordance with the finalized data dictionary. The sites submitted their data over the sFTP client, or via encrypted email.
Manual Quality Assurance (QA) and automated Quality Control (QC) measures were implemented by the DCC to ensure the fidelity of the submitted and transformed data. Manual QA checks included mapping submitted data elements and data formats to the DCC data dictionary equivalents, comparing counts before and after data harmonization and transformation, and confirming the integrity of the data transformed into the database from twenty randomly selected cases from each submitted dataset. All datasets were double checked to ensure that no Personal or Protected Health Information (PHI) was inadvertently submitted to, and stored within, the DCC.
The automated QC solution uses a rule-based validation system based on the LiFE DCC Data Dictionary and is implemented via a Pentaho Data Integration tool within the data Extraction, Transformation, and Loading (ETL) process. This facilitates error detection and correction for data submitted. Validation rules comprise both standard checks on allowable values, and a crosscheck of related database elements for logical and scientific consistency (e.g. Vital Status vs. Year of Death). Validation errors were collected and logged within the database when the submitted data could not be harmonized.
The flexible architecture of the LiFE database allows for exceptions to the validation rules as determined by the Steering Committee. Certain data were allowed into the LiFE database despite validation errors. For example, the format for the “Cancer Diagnosis” field was initially built to accept an ICD-9/10 value; however, this information was not readily available for all subjects in the sites’ datasets. Thus, the validation rules for this field were revised to accept cancer diagnoses text values as well. This permissible violation allowed for the data to be captured, but raised validation error flags in the QC process
For any errors or inconsistencies noted, the DCC contacted the respective site and requested a resubmission of the data. All errors and consistencies identified were satisfactorily addressed. Each site also received their transformed data after each data load for comparison against their own submitted files for any discrepancies, independent of the LiFE DCC QC and QA procedures.
Data Access
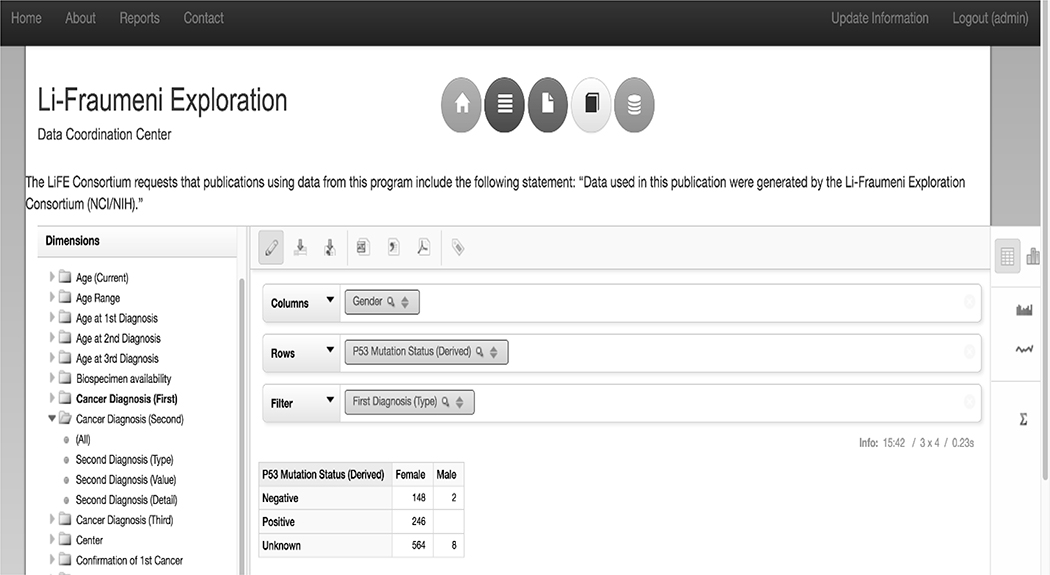
In order to facilitate access to the central repository housed at the LiFE DCC, the DCC built a web-portal consisting of a publicly viewable section, and a user-restricted access component. The publicly viewable content on the web portal is comprised of information about LFS, the consortium, and real-time summary level descriptive data providing an overview of the data housed at the DCC. The user-restricted portion of the web portal, requiring login with username and password provided by the LiFE DCC upon approval by the Steering Committee, contains a web-interface called Saiku which allows for real-time querying of the LiFE database (Figure 2). This tool includes features allowing for drag-and-drop queries of elements in the database with immediate data population and chart generation, and graphic visualization of the data in a style selected by the individual researcher (Figure 2). There are several filtering options available when making queries to generate highly specific sets of data. All data sets and graphics can be downloaded for further analysis. The publicly viewable summary level descriptive data tables, graphs, and charts are generated and displayed live using the Saiku query and visualization interface (Figure 3). Investigators external to the LiFE DCC participating sites have the ability to request tailored datasets from the central repository on the web portal. Tailored data request requirements and dataset delivery protocols were developed by the Steering Committee.
Figure 2:
The Saiku query tool allowing for authorized users to drag and drop data elements from the left Dimensions menu into columns (Gender) and rows (P53 Mutation Status (Derived)), and generate data tables. Filter capabilities (First Diagnosis (Type) = Breast Cancers) allow queries to be further narrowed down to only datasets of interest. Tables can be exported as CSV, Excel, or PDF files using the buttons above the “Column” field. Data can also be visualized in several ways utilizing the visualization tools on the right side bar.
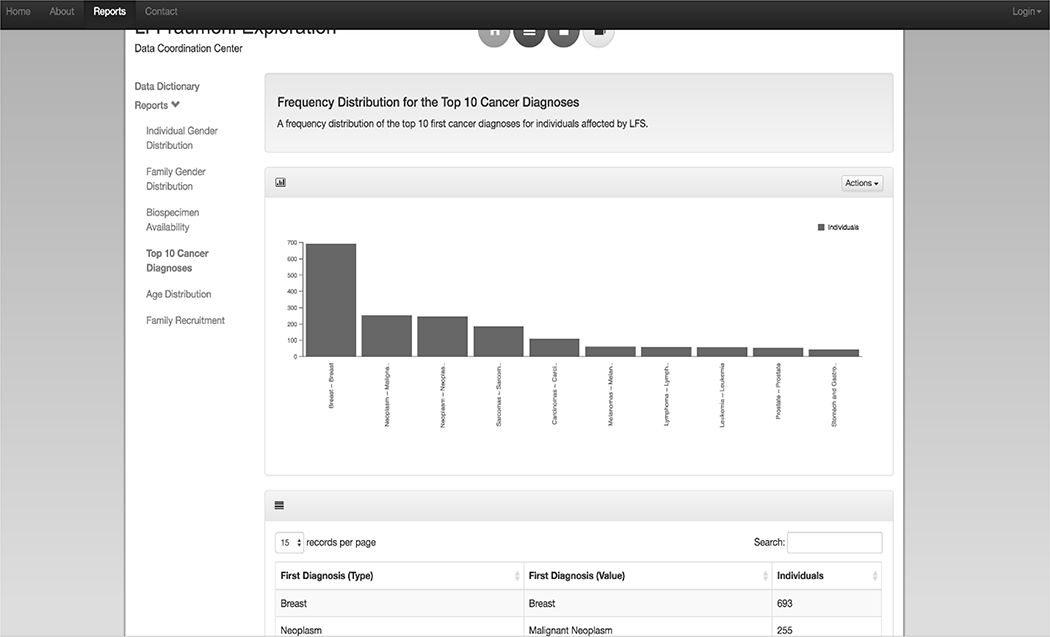
Figure 3:
The “Reports” page of the public LiFE DCC Web Portal provides summary level data tables and interactive visualization tools. A link to the LiFE DCC Data Dictionary is included as well. The descriptive data tables on display include “Individual Gender Distribution”, “Family Gender Distribution”, and “Biospecimen Availability” by p53 mutation status, a frequency distribution of the “Top 10 Cancer Diagnoses” for individuals affected by LFS, “Age Distribution” by individual and family within the repository, and “Family Recruitment” by country and vital status.
Technical and Software Specifications
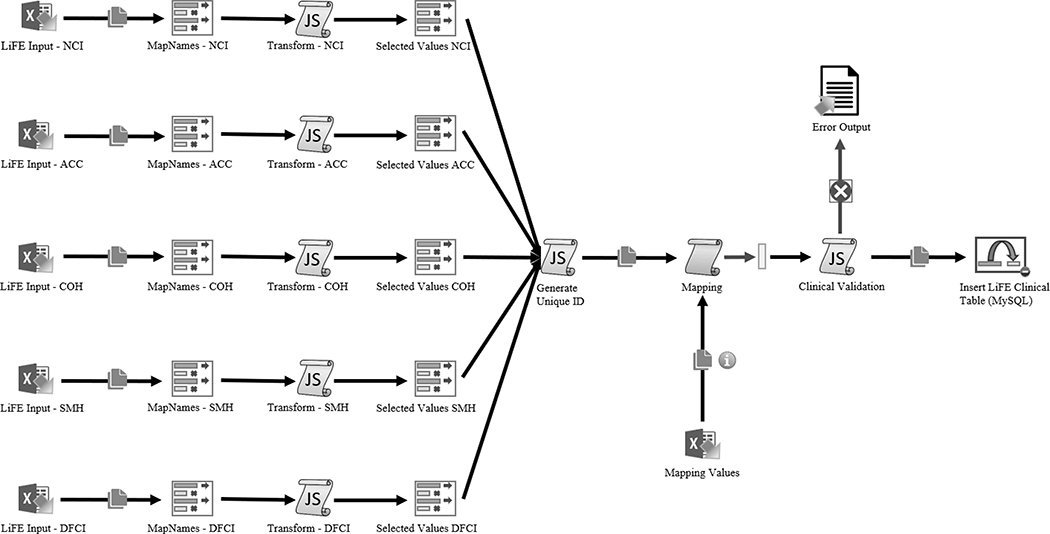
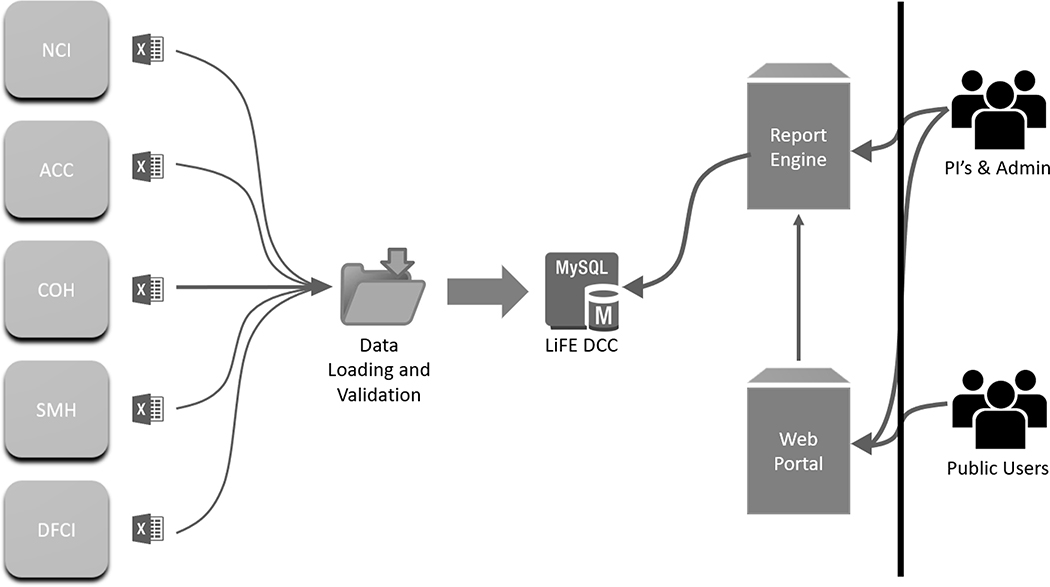
The LiFE DCC was created using open source packages: LAM/G Application (Linux, Apache, MySQL https://www.mysql.com/, Grails http://grails.org), Extraction-Transformation-Loading (ETL) Pentaho Data Integration Tool (https://www.hitachivantara.com/en-us/products/data-management-analytics/pentaho-platform.html), and the Saiku-Mondrian client (http://community.meteorite.bi, http://mondrian.pentaho.com/documentation/schema.php) for queries made of the central repository. The submitted data went through the ETL process and were loaded into the MySQL Database. The Pentaho Data Integration Tool’s ETL Process (Figure 4) facilitates harmonization and aggregation of data submitted by the LiFE sites to the LiFE DCC. The extraction step maps all columns from the submitted excel sheets to the LiFE DCC Data Dictionary elements. The data were then loaded into memory for transformation. The transformation step assigned each submitted case with a unique LiFE DCC ID, mapped all submitted values to the LiFE DCC Data Dictionary, and confirmed there were no validation errors or failed records for each case submitted. Data loading to the central repository is the final step in the automated QC and relational database population process. Manual QA steps are then performed by the DCC before moving the data to production.
Figure 4:
Visual overview of the Extraction, Transformation, and Data Loading (ETL) Process. Files are submitted as excel sheets. The extraction step maps all columns from the submitted excel sheets to the LiFE DCC Data Dictionary elements. The data are then loaded into memory for transformation. The transformation step assigns each submitted case with a unique LiFE DCC ID, maps all submitted values to the LiFE DCC Data Dictionary, and confirms there are no clinical validation errors or failed records for each case submitted. Data loading to the central repository is the final step in the automated QC and relational MySQL database population process.
The Saiku server is an open-source server capable of rendering reports and datasets based on Mondrian schema definition. The Saiku server collects queries from the user, pulls this data from the MySQL database, and reports the results in any format requested by the user including tables, charts, graphs, and other data visualization options (Figure 5). All reports created in Saiku are rendered in the web portal. Data are accessed through a RESTful interface running on an Amazon Web Server M3 - large instance.
Figure 5:
A visual overview of the data submission to display process. Sites submit their data in an XLS format to the DCC which is run through the Pentaho Data Integration Extraction-Transformation-Loading (ETL) tool, and the harmonized data is stored in a LiFE DCC MySQL relational database. The Saiku Report Engine collects queries from researchers with access to the tool, and reports data from the MySQL database. Access to the Saiku Report Engine is conferred via a password protected login through the public facing LiFE DCC web portal.
RESULTS
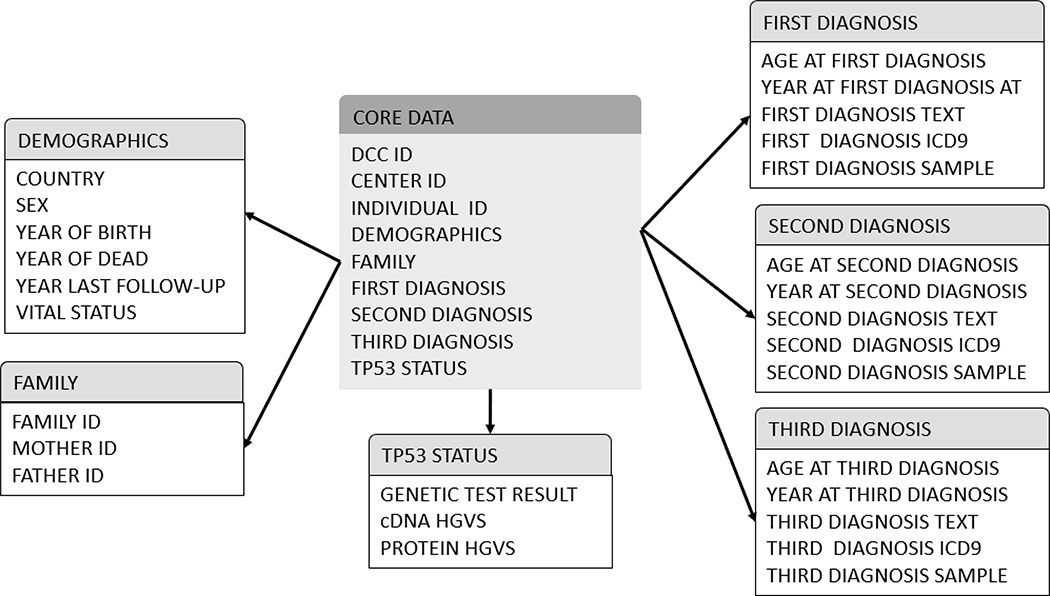
The Life DCC pilot project was completed in July 2016. Table 1 and Figure 3 show the totals for all cases submitted by each contributing LiFE site and illustrate some examples of the graphics that are publicly available. There are 644 families with a TP53 pathogenic or likely pathogenic (P/LP) variant. Among these families, there are 1,354 individuals who tested positive, ages ranging from 0 to 94 years, 499 who tested negative for the familial P/LP variant, with ages ranging from 6 to 89, and 205 untested individuals, ages ranging from 1 to 90. Approximately 71% (965/1354) of individuals who tested positive for the familial TP53 P/LP variant had been diagnosed with at least one cancer. Approximately 37% of the individuals are White, 29% are Hispanic/Latino, and 2.5% are Black. There are also 287 families which meet the classic LFS or LFL diagnostic criteria, but in which no TP53 P/LP variant has been identified. In 25 families, a TP53 variant of uncertain significance (VUS) was identified (Table 1). The clinical data collected included the required and optional data elements. Currently the database contains 48 data elements, including information on contributing centers, demographics, TP53 status, and cancer diagnosis (Figure 6).
Table 1:
Number of individuals and families included in the LiFE DCC database
| Families | TP53 P/LP Variants (N=644) | TP53 VUS (N=25) | TP53 Negative (N=287) | ||
|---|---|---|---|---|---|
| Carriers | Non-carriers | Untested | |||
| Number of individuals | 1354 | 499 | 205 | 38 | 302 |
|
Contributing sites Hospital A.C. Camargo – Fundação Prudente (ACCC) City of Hope (COH) Dana-Farber Cancer Institute (DFCI) Hospital for Sick Children (HSC) MD Anderson Cancer Center (MDA) National Cancer Institute (NCI) Saint Mary’s Hospital (SMH) St. Jude |
208 91 155 Pending 343 317 203 37 |
237 3 47 Pending 146 0 46 20 |
0 0 0 Pending 57 0 135 13 |
3 4 6 Pending 0 25 0 0 |
74 119 107 Pending 0 2 0 0 |
|
Age, mean
(range) |
35.5 (0–94) |
47.3 (6–89) |
39.3 (1–90) |
44.2 (17–73) |
36.4 (4–68) |
|
Race/Ethnicity Non-Hispanic White Non-Hispanic Black Hispanic/Latino Other/Not Specified |
617 33 312 392 |
104 12 269 114 |
44 6 8 147 |
25 0 6 7 |
90 6 113 93 |
|
History of cancer Yes No |
965 389 |
54 445 |
107 98 |
23 14 |
261 41 |
DCC: Data Coordination Center
P/LP: pathogenic or likely pathogenic; VUS: variant of uncertain significance
Figure 6:
Database Star Schema of core data points collected, by category, including: demographic, family, TP53 mutation details, and diagnoses.
As mandated by the EGRP pilot project, the DCC was established to support the creation of a database and communication infrastructure to facilitate collaborative research. Once these goals were reached, the database was transferred from ESAC, Inc. to City of Hope Comprehensive Cancer Center (COH; a contributing LiFE site) who assumed full responsibility for the management and further development of the resource. ESAC, Inc.’s LiFE DCC Project Leads and Technical Team worked closely with COH’s team members to configure all technical specifications required to host the database and web-portal (http://life-dcc.org/), and provided training on all processes required to collect, transform, and upload the data to the database.
Each collaborating site is provided with the data dictionary and associated data entry form, which includes a pedigree-level relational data format, where each person is assigned an identification number (ID) and is connected to family members via their associated Mother ID and Father ID, as per standard practice for genetic research collaborations. Once the Data Transfer Agreement has been completed between the DCC at COH and the collaborating site, the site can submit their data. Data are verified by DCC staff once received and imported into the database. The collaborating site is provided with access to their own data folder so that they can verify that their data were uploaded as expected, and make updates as needed. Alternatively to sending data to the DCC, the collaborating site can enter the data directly into their own folder.
The DCC is now fully functional and is open to any research group that can contribute genotypic and minimally required phenotypic and epidemiological data from at least five individuals with LFS with or without a cancer diagnosis. Groups with smaller number of participants are encouraged to participate through partnership with a larger group.
Several collaborative studies have resulted from the LiFE consortium, with two publications on the phenotype of TP53-associated breast cancers (31) and findings at baseline for surveillance utilizing rapid sequence whole-body Magnetic Resonant Imaging (wb-MRI) among individuals with LFS (32). The LiFE DCC has provided data supporting multiple on-going projects, including a successful R01 grant proposal (R01 CA242218) attempting to address the NCI Provocative Questions around understanding penetrance and modifiers of inherited TP53 (PIs: Weitzel, Garber and Amos) for the Li-Fraumeni & TP53: Understanding & Progress (LiFT UP) study (liftup@coh.org). The DCC has also provided potential sample size data for another recently approved concept on a metformin prevention trial under development at the NCI. More sites have joined since the creation of the database. As intended, data from LiFE centralized repository database have been utilized by researchers both internal and external to the consortium. One example of the research being carried out by external investigators using data from the LiFE DCC is a project examining the phenotype for LFS-associated breast cancers. The data is being used to validate previous observations that Her2/neu amplified breast cancer was seen more frequently among TP53 carriers, and then incorporate phenotype into a multifactorial variant classification algorithm. The LiFE consortium was able to contribute data on 246 TP53-associated breast cancers, and a manuscript is under review. A better understanding of the phenotype of the syndrome has the potential to improve the clinical indications for testing and risk management.
DISCUSSION
Establishing a high-quality database is an essential undertaking for any successful collaborative research. This is especially true in rare diseases where data are collected by multiple centers over many years, and the types and methods of data collection might change over time. A DCC can help facilitate data aggregation and harmonization, and ensure the quality of the database created. Here we described the administrative and technical practices of the LiFE DCC, which worked to establish a centralized database. This project met the goal of the EGRP to establish a DCC for a CEC focusing on rare cancers. The operating procedures outlined in this paper could be utilized by rare cancer CEC for the establishment of a similar DCC.
In addition to the challenges with the coordination of communication and the creation of the data dictionary discussed above, other challenges encountered during the building of the database and, subsequently, its maintenance, include international data transfer agreements and the ability to add data field afterwards. International institutions might have different requirements which would need to be considered. Furthermore, the NCI funded the creation of the database and DCC, but once that was accomplished, a member institution (in this case, COH) needed to assume responsibility for maintenance, and small grants from the patient advocacy group LFS Association helped support a partial effort coordinator. Further, it was determined by the steering committee that a pedigree-driven relational database would be most helpful, thus Progeny (Delray Beach, Florida) was incorporated into the database.
During the course of creating the DCC and the subsequent planning for utilization of the data, several lessons were learned. First, various contributing sites bring different perspectives in terms of data collection. It is essential to define all fields in the data dictionary, and the functionality of the database must be flexible to adapt to evolving permissible values for each field, as well as to new data fields. It is also critical to put in place early in the process plans for continual funding and protocols for data requests and data use.
A goal of a centralized database is ease of access. The LiFE database that was created allowed individual PIs to conduct analyses on available data, including the ability to query records and create charts and figures. No previous coding experience or programming knowledge is required to use the Saiku query interface. Similarly, many clinical cancer genetics programs use the Progeny interface, which helps facilitate data query.
This query-capable multi-site collaborative central data repository serves as a valuable tool for LFS-related research. The web-based application of data analysis and visualization tools allows for the ability to access aggregate data from anywhere. The LiFE database will help to advance our collaborative research efforts in LFS, and ultimately help advance the care of individuals and families burdened by this condition.
Key Messages.
Technological solution for developing a collaborative web-based centralized database for an international cancer epidemiology research consortium: the Li-Fraumeni Exploration (LiFE) Consortium.
Approach and benefit to developing Data Coordinating Centers (DCCs) for multi-site collaborative research projects.
Methodologies and strategies for data harmonization for a multi-site research consortium for collaborative projects.
Establishment of the LiFE database provisioned with data analysis and visualization tools available online allowing LFS researchers to conduct LFS collaborative research.
ACKNOWLEDGEMENTS
This work was supported by the National Cancer Institute at the National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN261201500214P. The Li-Fraumeni Syndrome Association provided partial support to City of Hope for maintaining the LiFE DCC.
Research reported in this publication was also supported by the National Cancer Institute of the National Institutes of Health under Award Number R01CA242218. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The authors declare no potential conflicts of interest
REFERENCES
- 1.Rolland B, Smith BR, Potter JD. Coordinating centers in cancer epidemiology research: the Asia Cohort Consortium coordinating center. Cancer Epidemiol Biomarkers Prev. 2011;20(10):2115–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blumenstein BA, James KE, Lind BK, Mitchell HE. Functions and organization of coordinating centers for multicenter studies. Control Clin Trials. 1995;16(2 Suppl):4S–29S. [DOI] [PubMed] [Google Scholar]
- 3.Fortier I, Burton PR, Robson PJ, Ferretti V, Little J, L’Heureux F, et al. Quality, quantity and harmony: the DataSHaPER approach to integrating data across bioclinical studies. Int J Epidemiol. 2010;39(5):1383–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mai PL, Malkin D, Garber JE, Schiffman JD, Weitzel JN, Strong LC, et al. Li-Fraumeni syndrome: report of a clinical research workshop and creation of a research consortium. Cancer Genet. 2012;205(10):479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li FP, Fraumeni JFJ. Soft-tissue sarcomas, breast cancer, and other neoplasms: A familial syndrome? Ann Intern Med. 1969;71(4):747–52. [DOI] [PubMed] [Google Scholar]
- 6.Malkin D, Li FP, Strong LC, Fraumeni JF Jr., Nelson CE, Kim DH, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250(4985):1233–8. [DOI] [PubMed] [Google Scholar]
- 7.Srivastava S, Zou ZQ, Pirollo K, Blattner W, Chang EH. Germ-line transmission of a mutated p53 gene in a cancer-prone family with Li-Fraumeni syndrome. Nature. 1990;348(6303):747–9. [DOI] [PubMed] [Google Scholar]
- 8.Bougeard G, Renaux-Petel M, Flaman J-M, Charbonnier C, Fermey P, Belotti M, et al. Revisiting Li-Fraumeni Syndrome from TP53 mutation carriers. J Clin Oncol. 2015;33(21):2345–52. [DOI] [PubMed] [Google Scholar]
- 9.Mai PL, Best AF, Peters JA, DeCastro RM, Khincha PP, Loud JT, et al. Risks of first and subsequent cancers among TP53 mutation carriers in the National Cancer Institute Li-Fraumeni syndrome cohort. Cancer. 2016;122(23):3673–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamihara J, Rana HQ, Garber JE. Germline TP53 mutations and the changing landscape of Li–Fraumeni Syndrome. Hum Mutat. 2014;35(6):654–62. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez KD, Noltner KA, Buzin CH, Gu D, Wen-Fong CY, Nguyen VQ, et al. Beyond Li Fraumeni Syndrome: clinical characteristics of families with p53 germline mutations. J Clin Oncol. 2009;27(8):1250–6. [DOI] [PubMed] [Google Scholar]
- 12.Nichols KE, Malkin D, Garber JE, Fraumeni JF, Li FP. Germ-line p53 mutations predispose to a wide spectrum of early-onset cancers. Cancer Epidemiol Biomarkers Prev. 2001;10(2):83–7. [PubMed] [Google Scholar]
- 13.Ruijs MW, Verhoef S, Rookus MA, Pruntel R, van der Hout AH, Hogervorst FB, et al. TP53 germline mutation testing in 180 families suspected of Li-Fraumeni syndrome: mutation detection rate and relative frequency of cancers in different familial phenotypes. J Med Genet. 2010;47(6):421–8. [DOI] [PubMed] [Google Scholar]
- 14.Lustbader ED, Williams WR, Bondy ML, Strom S, Strong LC. Segregation analysis of cancer in families of childhood soft-tissue-sarcoma patients. Am J Hum Genet. 1992;51(2):344–56. [PMC free article] [PubMed] [Google Scholar]
- 15.Wu C-C, Shete S, Amos CI, Strong LC. Joint effects of germ-line p53 mutation and sex on cancer risk in Li-Fraumeni Syndrome. Cancer Res. 2006;66(16):8287–92. [DOI] [PubMed] [Google Scholar]
- 16.Fang S, Krahe R, Bachinski LL, Zhang B, Amos CI, Strong LC. Sex-specific effect of the TP53 PIN3 polymorphism on cancer risk in a cohort study of TP53 germline mutation carriers. Hum Genet. 2011;130(6):789–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang SJ, Lozano G, Amos CI, Strong LC. Germline p53 mutations in a cohort with childhood sarcoma: sex differences in cancer risk. Am J Hum Genet. 2003;72(4):975–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li FP, Fraumeni JF Jr., Mulvihill JJ, Blattner WA, Dreyfus MG, Tucker MA, et al. A cancer family syndrome in twenty-four kindreds. Cancer Res. 1988;48(18):5358–62. [PubMed] [Google Scholar]
- 19.Birch JM, Hartley AL, Tricker KJ, Prosser J, Condie A, Kelsey AM, et al. Prevalence and diversity of constitutional mutations in the p53 gene among 21 Li-Fraumeni families. Cancer Res. 1994;54(5):1298–304. [PubMed] [Google Scholar]
- 20.Eeles RA. Germline mutations in the TP53 gene. Cancer Surv. 1995;25:101–24. [PubMed] [Google Scholar]
- 21.Varley JM. Germline TP53 mutations and Li-Fraumeni syndrome. Hum Mutat. 2003;21(3):313–20. [DOI] [PubMed] [Google Scholar]
- 22.Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat. 2002;19(6):607–14. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez KD, Buzin CH, Noltner KA, Gu D, Li W, Malkin D, et al. High frequency of de novo mutations in Li–Fraumeni syndrome. J Med Genet. 2009;46(10):689–93. [DOI] [PubMed] [Google Scholar]
- 24.Chompret A, Abel A, Stoppa-Lyonnet D, Brugieres L, Pages S, Feunteun J, et al. Sensitivity and predictive value of criteria for p53 germline mutation screening. J Med Genet. 2001;38(1):43–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tinat J, Bougeard G, Baert-Desurmont S, Vasseur S, Martin C, Bouvignies E, et al. 2009 version of the Chompret criteria for Li Fraumeni syndrome. J Clin Oncol. 2009;27(26):e108–9; author reply e10. [DOI] [PubMed] [Google Scholar]
- 26.Bougeard G, Sesboue R, Baert-Desurmont S, Vasseur S, Martin C, Tinat J, et al. Molecular basis of the Li-Fraumeni syndrome: an update from the French LFS families. J Med Genet. 2008;45(8):535–8. [DOI] [PubMed] [Google Scholar]
- 27.McCuaig JM, Armel SR, Novokmet A, Ginsburg OM, Demsky R, Narod SA, et al. Routine TP53 testing for breast cancer under age 30: ready for prime time? Fam Cancer. 2012;11(4):607–13. [DOI] [PubMed] [Google Scholar]
- 28.National Comprehensive Cancer Network (NCCN). Genetic/familial high-risk assessment: breast and ovarian. Version 3.2019. [Available from: https://www.nccn.org/professionals/physician_gls/pdf/genetics_screening.pdf].
- 29.Lawrence K Walking the Tightrope: The Balancing Acts of a Large e-Research Project. Computer Supported Cooperative Work. 152006 p. 385–411. [Google Scholar]
- 30.McGarvey PB, Ladwa S, Oberti M, Dragomir AD, Hedlund EK, Tanenbaum DM, et al. Informatics and data quality at collaborative multicenter Breast and Colon Cancer Family Registries. J Am Med Inform Assoc. 2012;19(e1):e125–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masciari S, Dillon DA, Rath M, Robson M, Weitzel JN, Balmana J, et al. Breast cancer phenotype in women with TP53 germline mutations: a Li-Fraumeni syndrome consortium effort. Breast Cancer Res Treat. 2012;133(3):1125–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ballinger ML, Best A, Mai PL, Khincha PP, Loud JT, Peters JA, et al. Baseline surveillance in Li-Fraumeni Syndrome using Whole-Body Magnetic Resonance Imaging: A meta-analysis. JAMA oncology. 2017;3(12):1634–9. [DOI] [PMC free article] [PubMed] [Google Scholar]