Highlights
-
•
This study applied a logistic growth model with parameters estimated by a non-linear least squares (NLS) method to daily new COVID-19 cases.
-
•
The model fitted time-series data exceedingly well for the whole of China, eleven selected Chinese provinces and municipalities, South Korea and Iran.
-
•
This study provided key estimates and a 95% confidence interval for parameters K, r, and P 0.
-
•
This study found that the growth rates of outbreaks differed between provinces in China and between South Korea and Iran.
-
•
As of March 13, 2020, this study's model suggested that countries such as the U.S.A., France, Italy, and Spain were still in the early stages of outbreaks.
Keywords: COVID-19, Logistic growth model, Non-linear least squares, China
Abstract
Objective
As the coronavirus disease 2019 (COVID-19) pandemic continues to proliferate globally, this paper shares the findings of modelling the outbreak in China at both provincial and national levels. This paper examines the applicability of the logistic growth model, with implications for the study of the COVID-19 pandemic and other infectious diseases.
Methods
An NLS (Non-Linear Least Squares) method was employed to estimate the parameters of a differentiated logistic growth function using new daily COVID-19 cases in multiple regions in China and in other selected countries. The estimation was based upon training data from January 20, 2020 to March 13, 2020. A restriction test was subsequently implemented to examine whether a designated parameter was identical among regions or countries, and the diagnosis of residuals was also conducted. The model's goodness of fit was checked using testing data from March 14, 2020 to April 18, 2020.
Results
The model presented in this paper fitted time-series data exceedingly well for the whole of China, its eleven selected provinces and municipalities, and two other countries - South Korea and Iran - and provided estimates of key parameters. This study rejected the null hypothesis that the growth rates of outbreaks were the same among ten selected non-Hubei provinces in China, as well as between South Korea and Iran. The study found that the model did not provide reliable estimates for countries that were in the early stages of outbreaks. Furthermore, this study concured that the R2 values might vary and mislead when compared between different portions of the same non-linear curve. In addition, the study identified the existence of heteroskedasticity and positive serial correlation within residuals in some provinces and countries.
Conclusions
The findings suggest that there is potential for this model to contribute to better public health policy in combatting COVID-19. The model does so by providing a simple logistic framework for retrospectively analyzing outbreaks in regions that have already experienced a maximal proliferation in cases. Based upon statistical findings, this study also outlines certain challenges in modelling and their implications for the results.
1. Introduction
An outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a zoonotic coronavirus similar to severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome-related coronavirus (MERS-CoV), has rapidly spread across China and various regions of the world. As of April 17, 2020, the cumulative numbers of confirmed cases had reached 82 719 in China (NHCPRC, 2020) and 2 074 529 in 210 countries and territories worldwide (World Health Organization, 2020).
In light of these recent developments, the scientific community has sought understanding of coronavirus disease 2019 (COVID-19), the disease caused by SARS-CoV-2, and many have undertaken statistical and modelling approaches. The R 0 value for virus transmissibility has been evaluated through stochastic Markov chain Monte Carlo (MCMC) methods (Wu et al., 2020a), a mathematical incidence decay and exponential adjustment (IDEA) model (Majumder and Mandl, 2020), and a statistical exponential growth model adopting the serial interval from severe acute respiratory syndrome (SARS) (Zhao et al., 2020). Researchers have also utilized several models to generate short-term forecasts for cumulative case counts (Roosa et al., 2020), and have developed a ‘susceptible, un-quarantined infected, quarantined infected, confirmed infected’ (SUQC) model to characterize the dynamics of outbreaks (Zhao and Chen, 2020).
This study applied a logistic growth function with parameters estimated by a non-linear least squares (NLS) method to model and analyze time-series data from eleven provinces and municipalities in China (Anhui, Beijing, Chongqing, Guangdong, Henan, Hubei, Hunan, Jiangsu, Jiangxi, Shanghai, and Zhejiang) and nine other countries (Iran, South Korea, France, Germany, the U.S.A., Italy, Spain, Singapore and Japan). The implications of the results for the study of infectious diseases are discussed.
2. Methods
Devised by Belgian mathematician Pierre-François Verhulst (1804–1849) and corroborated by others in later years, the logistic function has become one of the essential tools for bio-assays and has been increasingly applied in a variety of fields, including statistics, economics, and epidemiology (Cramer, 2004). Specifically, it has been used to model population growth in a region and bacterial growth in a broth, and has been implemented in binary decision-making processes in economics and finance.
The equation of the logistic function, following a common sigmoid curve, takes the mathematical form
| (1) |
where P(t), or the number of cumulative cases of COVID-19, is expressed as a function of time, t, with parameters . To be exact, K represents carrying capacity, P 0 represents the initial value of the function at t = 0, and r represents the growth rate, or the speed of proliferation.
This study did not, however, use a logistic function to directly estimate a model for cumulative cases. Previous studies have suggested that fitting deterministic models to cumulative cases, due to serial correlation in the error terms (measurement errors), creates biased parameters and overfitting of the model to data, and underestimates the uncertainty associated with parameters (King et al., 2015). Therefore, the derivative of the logistic growth function was adopted to model the daily new cases. The general differential logistic equation takes the following form:
| (2) |
Substituting Equation (1) into Equation (2) gives the logistic differential equation
| (3) |
Specifically, the number of observed daily new cases, I(t), is equal to f(t,β) plus an error term ε, as shown in the statistical model below, where t = 1, 2, …T, and T is the number of observations.
| (4) |
Equation (4) is the key equation for modelling time-series data. The study assumed that the error terms were independent and identically distributed (i.i.d.), and used the NLS method to estimate β by minimizing the residual sum of squares, .
After estimation, this study set , or, as the predicted value at a given t using estimated parameters . The residual is defined as . The study defined , and , representing the total sum of squares, regression sum of squares, and the residual (error) sum of squares, respectively. According to the statistical identity TSS = RSS + ESS (Pindyck and Rubinfeld, 1991), the coefficient of determination is expressed as
| (5) |
The study calculated the F-statistic value through , given that n is the number of parameters, or three, T is the number of observations, and that F asymptotically follows an F-distribution with n and (T–n) degrees of freedom.
This study established matrix X as equivalent to the partial derivative of f(t·β) or such that . The estimate of the asymptotic covariance matrix of β is (Greene, 1997), wherein MSE is the estimate of the residual variance, equal to . The confidence interval of is determined by equating
| (6) |
wherein α is the significance level, substituted with 5% when calculating the 95% confidence interval (95% CI).1
In order to test certain restrictions upon parameters β, this study compared the ESS of a free model (ESS f) to the ESS of a restricted model (ESS r) and calculated a test static F R with q constraints on β, where
| (7) |
The test asymptotically follows an F-distribution with q and (T–n) degrees of freedom (Schabenberger and Pierce, 2002).
Estimation was completed using the SAS software package. The method of optimization utilized a Gauss–Newton algorithm, which is advantageous as it does not require a second derivative and converged quickly with this study's estimations. The Gauss–Newton algorithm iteratively finds the values of parameters β that minimize the sum of squares of the residuals. It starts from an initial estimate β (0) and proceeds by iterations expressed in terms of and β, which are column matrices, and in terms of s, which is the iteration step during the optimization process.
The daily times-series data of cumulative COVID-19 cases from January 20, 2020 to April 18, 2020 were retrieved from the National Health Commission of the People's Republic of China and its respective health commissions in the selected eleven provinces and municipalities. Time-series data for the nine other countries up until April 18, 2020 were obtained through WIND DATA, a leading financial data services provider in China.
This study took measures to adjust the time-series data for cumulative cases in China. It should be noted that on February 12, 2020, clinical evidence and radiographic confirmation were introduced into the diagnosis guidelines for new cases, causing a jump of nearly 15 000 new cases in Hubei, China.2 On April 16, 2020, health officials announced a one-time re-adjustment of the number of cumulative cases in Wuhan, Hubei — an increase by 325 cases — which were originally omitted from the public amidst the epidemic. For data consistency, this study removed all cases that were added on those two days. To further ensure consistency, this study removed all confirmed cases that were imported from abroad (1575 cases nationally, including 741 cases in many of the ten non-Hubei provinces of China). In addition, 305 cases in the prison system reported on February 20, 2020 (271 in Hubei and 34 in Zhejiang) where the spread was relatively independent and not within the coverage of the provisional health authorities were also removed (Wu et al., 2020b).
The time-series data for daily new cases in China, derived from cumulative cases, were split into two time periods for training and testing. The study fit a logistic growth model for time-series data from January 20, 2020 up until March 13, 2020 (defined as training data), when the manuscript was originally written. It estimated all parameters and related statistics of the model based upon the training data only. The estimated model was then fitted to the time-series data from March 14, 2020 to April 18, 2020 (defined as testing data).
The regression model took the assumption of i.i.d. error terms. This study performed several residual diagnosis tests to check whether such an assumption was appropriate for the fitted errors or not. Previous papers have found that using the raw residuals, w(t), of non-linear models for diagnosis may be misleading, as they may have non-zero means and different variances. These papers have, consequently, suggested that the use of alternative residuals, referred to as projected residuals, may overcome many of the shortcomings of the raw residuals (Cook and Tsai, 1985). To test whether the error terms in the regression model were independent as per the suggestion by Cook and Tsai, a Durbin–Watson (DW) statistic (Durbin and Watson, 1950) was employed for both the raw residuals and projected residuals.3 Similarly, to test for homoskedasticity, this study carried out the White test (White, 1980) for both types of residuals.4
3. Results and discussion
Equation (4) was first fitted for the training data for the whole of China, as well as for Hubei Province and non-Hubei provinces; the results are displayed in Table 1 and Fig. 1 . The same model was then fitted for training data from ten selected non-Hubei provinces and municipalities, with the results shown in Table 2 and Fig. 2 . The model was subsequently fitted for the training data from the nine other countries as well, for which the results are shown in Table 3 and Fig. 3 . After the parameters of the model had been estimated, they were fitted in the corresponding testing data, and the results are illustrated in Tables 1, 2 and 3 and Figs. 1, 2 and 3.5
Table 1.
Modelling results for national, Hubei, and non-Hubei time-series data
| Regional classification | T | 95% CI of |
95% CI of |
F statistic | Approx. Pr > F | Estimated date of maximal increase |
R2 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lower | Upper | Lower | Upper | Train | Test | Total | ||||||||
| National | 53 | 71 954.6 | 64 640.2 | 79 268.9 | 0.1927 | 0.1683 | 0.2170 | 0.2170 | 213.77 | <0.0001 | 2020/2/7 | 0.929 | 0.851 | 0.929 |
| Hubei | 52 | 58 221.3 | 51 319.0 | 65 123.5 | 0.1984 | 0.1691 | 0.2277 | 1156 | 151.5 | <0.0001 | 2020/2/9 | 0.903 | 13.271 | 0.903 |
| Non-Hubei | 53 | 13 426.1 | 12 810.6 | 14 041.6 | 0.2386 | 0.2248 | 0.2524 | 530.7 | 1116.33 | <0.0001 | 2020/2/2 | 0.985 | 0.000 | 0.986 |
CI, confidence interval.
Table 1 shows the modelling results of Equation (4) estimated for time-series data of new COVID-19 cases in China, Hubei, and non-Hubei provinces. While the study includes the calculations for the 95% confidence interval for P0, it is not provided within Tables 1, 2 and 3 due to space limitations. The training period lasted from January 20, 2020 to March 13, 2020, whereas the testing period lasted from March 14, 2020 to April 18, 2020. The ‘Train’ column includes the training data, the ‘Test’ column includes the testing data, and the ‘Total’ column includes the training and testing data combined.
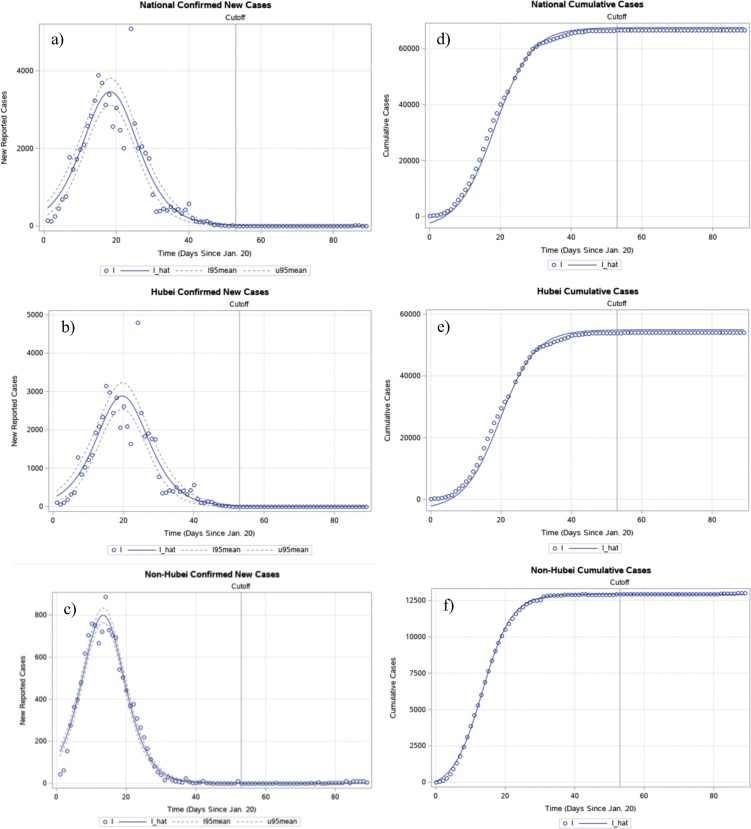
Fig. 1.
Graphical representation of the modelling results in Table 1. The estimated logistic growth function and actual values for new reported cases nationally (a), in Hubei Province (b), and in non-Hubei provinces (c), and the estimated and actual values for cumulative cases nationally (d), in Hubei Province (e), and in non-Hubei provinces (f) are represented. I is the observed value and I_hat is . Graphing Equation (1) using estimated parameters from Equation (3) involved more than plugging parameters in. When integration occurs, one needs to add some constant C, and therefore this study added a constant that was the difference between the mean of all observed data points and that of all predicted data points. The cutoff day, namely March 13, 2020, distinguished the training period from the testing period. The dotted lines of l95mean and u95mean show the 95% confidence interval of the mean predicted value at a given time (t).
Table 2.
Modelling results for ten selected non-Hubei provinces and municipalities in China
| Province/municipality | T | Date of first observation | 95% CI of |
95% CI of |
F statistic | Approx. Pr > F | Estimated date of maximal increase |
R2 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lower | Upper | Lower | Upper | Train | Test | Total | |||||||||
| Anhui | 51 | 22-Jan | 1052.8 | 973.7 | 1131.9 | 0.2518 | 0.2279 | 0.2757 | 41.49 | 416.86 | <0.0001 | 2020/2/3 | 0.963 | 0.000 | 0.963 |
| Beijing | 50 | 23-Jan | 415.5 | 369.3 | 461.8 | 0.2304 | 0.1974 | 0.2634 | 38.869 | 232.06 | <0.0001 | 2020/2/2 | 0.937 | 0.000 | 0.935 |
| Chongqing | 51 | 23-Jan | 695.4 | 601.1 | 789.6 | 0.1764 | 0.1463 | 0.2064 | 109.90 | 212.24 | <0.0001 | 2020/1/31 | 0.930 | 0.000 | 0.930 |
| Guangdong | 54 | 19-Jan | 1347.2 | 1243.2 | 1451.3 | 0.2846 | 0.2572 | 0.312 | 24.89 | 372.66 | <0.0001 | 2020/2/2 | 0.956 | 0.000 | 0.955 |
| Hunan | 51 | 22-Jan | 1116.7 | 1027.2 | 1206.2 | 0.2438 | 0.2188 | 0.2689 | 76.54 | 404.48 | <0.0001 | 2020/2/1 | 0.962 | 0.000 | 0.962 |
| Henan | 50 | 23-Jan | 1366.1 | 1269.9 | 1462.3 | 0.2507 | 0.2281 | 0.2733 | 89.91 | 519.54 | <0.0001 | 2020/2/2 | 0.971 | 0.000 | 0.971 |
| Jiangsu | 51 | 22-Jan | 707 | 657.9 | 756.1 | 0.2124 | 0.1935 | 0.2313 | 47.69 | 534.04 | <0.0001 | 2020/2/2 | 0.971 | 0.000 | 0.971 |
| Jiangxi | 52 | 21-Jan | 983.7 | 890.7 | 1076.7 | 0.2725 | 0.2402 | 0.3048 | 26.59 | 254.12 | <0.0001 | 2020/2/3 | 0.940 | 0.000 | 0.940 |
| Shanghai | 53 | 20-Jan | 345.7 | 313.9 | 377.6 | 0.2625 | 0.2319 | 0.2932 | 14.33 | 278.07 | <0.0001 | 2020/2/1 | 0.943 | 0.000 | 0.943 |
| Zhejiang | 50 | 23-Jan | 1241.7 | 1054.7 | 1428.7 | 0.3053 | 0.2462 | 0.3644 | 98.836 | 120.92 | <0.0001 | 2020/1/31 | 0.885 | 0.000 | 0.885 |
CI, confidence interval.
Table 2 shows the modelling results of Equation (4) estimated for time-series data of new COVID-19 cases in ten non-Hubei provinces. The ten provinces chosen were those that had the highest numbers of cumulative cases or those that were significant to China's economy. The ‘Train’ column includes the training data, the ‘Test’ column includes the testing data, and the ‘Total’ column includes the training and testing data combined.
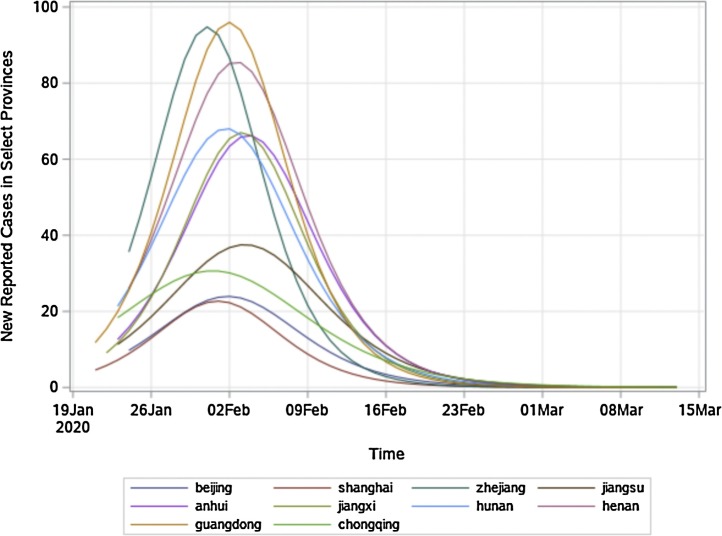
Fig. 2.
Graphical representation of the modelling results in Table 2 for time-series data of ten provinces in China. Due to space limitations, this figure does not present the scatter plot data of observed values for each province/municipality. The scatter plot and fitted curves for each province/municipality have been drawn, and they are available upon request.
Table 3.
Modelling results for nine selected countries
| Country | T | Date of first observation | 95% CI of |
95% CI of |
F statistic | Estimated date of maximal increase |
R2 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lower | Upper | Lower | Upper | Train | Test | Total | ||||||||
| Iran | 23 | 19-Feb | 19 604.5 | 10378.3 | 28830.8 | 0.2151 | 0.1237 | 0.3066 | 197.20 | 79.87 | 11-Mar | 0.923 | 0.030 | 0.085 |
| South Korea | 53 | 20-Jan | 8080 | 7126.2 | 9033.8 | 0.3610 | 0.3082 | 0.4138 | 0.0026 | 157.34 | 1-Mar | 0.904 | 0.007 | 0.858 |
| France | 49 | 24-Jan | 57 021.5 | −1.26E7 | 12718836 | 5.3E–4 | −1.133 | 1.1343 | 51 631 | 0.35 | Convergence criterion unmet | |||
| Germany | 45 | 28-Jan | 70 247.7 | −5.84E6 | 5.98E6 | 9.4E–4 | −0.979 | 0.9814 | 64 905 | 0.36 | Convergence criterion unmet | |||
| U.S.A | 51 | 22-Jan | 5530.3 | −4.728E9 | 4.7282E9 | 5.6E–3 | −132.6 | 132.6 | 5444.5 | 0.04 | Convergence criterion unmet | |||
| Italy | 41 | 1-Feb | 42 896.4 | −1.449E8 | 1.4498E8 | 5.4E–3 | −14.10 | 14.11 | 12 572 | 0.85 | Convergence criterion unmet | |||
| Spain | 41 | 1-Feb | 49 407.6 | −2.498E9 | 2.4981E9 | 6.2E–3 | −22.53 | 22.55 | 47 464 | 0.31 | Convergence criterion unmet | |||
| Singapore | 50 | 23-Jan | 15 679.8 | −5.834E7 | 58373740 | 0.0104 | −0.779 | 0.8007 | 118.3 | 10.29 | Convergence criterion unmet | |||
| Japan | 50 | 20-Jan | 29 78.6 | −5022426 | 5028383 | 0.0070 | −1.602 | 1.616 | 157.0 | 7.13 | Convergence criterion unmet | |||
CI, confidence interval.
Table 3 shows the modeling results of Equation (4) estimated for time-series data of new COVID-19 cases in nine nations. Apart from Iran and South Korea, the model failed to reach optimal results for the other seven nations. Six of the listed nations (France, Germany, Iran, Italy, Spain, and the USA) represented those with the most cumulative cases as of March 13, 2020. The three others (Japan, South Korea, and Singapore) are nations that are situated close to China and underwent early outbreaks. The ‘Train’ column includes the training data, the ‘Test’ column includes the testing data, and the ‘Total’ column includes the training and testing data combined.
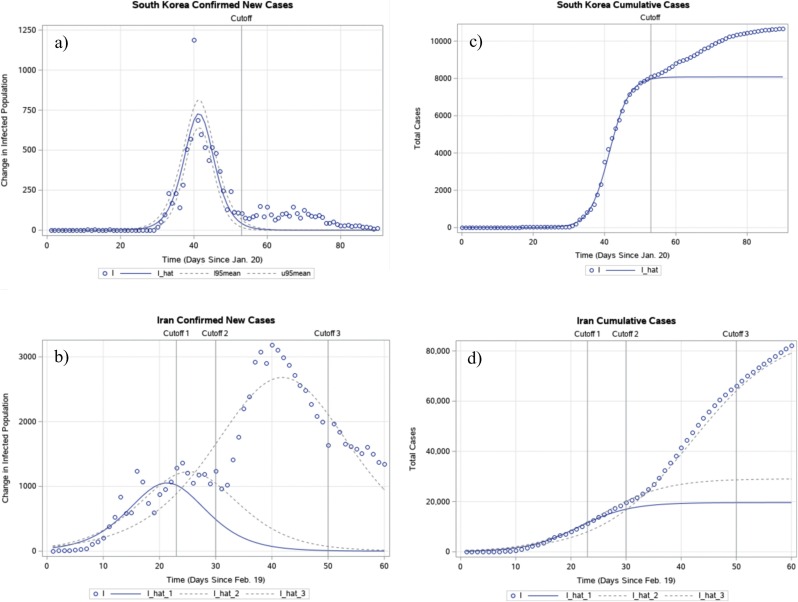
Fig. 3.
Graphical representation of the modelling results in Table 3 for time-series data of South Korea and Iran. The estimated logistic growth function and actual values for new cases in South Korea (a) and Iran (b), and the estimated growth function and actual values for cumulative cases in South Korea (c) and Iran (d) are represented. I is the observed value and I_hat is . As in Fig. 1, this study added a constant that was the difference between the mean of all observed data points and that of all predicted data points in the integration process. The cutoff day in (a) and (c), namely March 13, 2020, distinguished the training period from the testing period. This is also the case for cutoff 1 in graphs (c) and (d). However, for Iran, this study developed two other models, shown with dotted lines, based on different cutoff dates at t = 30 (cutoff 2, March 21, 2020) and t = 50 (cutoff 3, April 9, 2020).
Based on Figures 1, 2 and 3, the regression model fits the testing data well. However, its R 2 values in the testing period, as indicated in Table 1, Table 2, do not show consistent results. This is due to the fact that the testing and training time-series data inherently occupy two distinct portions of the non-linear curve, wherein the training period covers the peak of the outbreak with most of the cases, and the testing period covers only the flat, right-end tail with few cases. This study identified that when the predicted value reached close to 0 in the testing period and new cases became sporadic, the R 2 value ended up unstable - sometimes almost 0 and other times surpassing 1 - a finding also consistent with Greene (Greene, 1997). As a result, the R 2 may be different between distinct sections of the non-linear curve, and may be misleading when it is used to compare goodness-of-fit between these portions.6
The model may not carry the strikingly high R 2 values that appear when fitted to cumulative data. However, the results avoid the problem of bias and the underestimation of uncertainty, yielding more realistic estimates. Notably, as shown in Fig. 1, national COVID-19 cases will be around 71 954.6 (95% CI 64 640.2–79 268.9), the Hubei, China cases will be around 58 221.3 (95% CI 51 319–65 123.5), and the non-Hubei, China cases will be around 13 426.1 (95% CI 12 810.6–14 041.6). It is important to note that the confidence intervals exclude the cases removed during the data adjustment process, as mentioned in the Methods section.
While parameters K and P 0 reflect innate regional differences in population and size, this study demonstrated that the growth rate r might differ between provinces in China resulting from variations in local control measures and policies. Through the restriction test, the study showed that the calculated test statistic was 3.84863 with a corresponding p-value of 0.00010, and rejected the null hypothesis that r was the same among provinces and municipalities (Table 4 ). These findings corroborate previous studies showing that the degree of success in the control of proliferation has been influenced by a host of factors, ranging from individual patient response (Lau et al., 2016) to control and precautionary measures taken. More specifically, these may include differences in quarantine protocols, city-wide lockdowns, and travel restrictions, as well as distinctions in local cultures and behavior. Therefore, there is potential for others to study COVID-19 proliferation in various regions in China with the aim of strengthening viral prevention measures as cases surge internationally.
Table 4.
Parameter restriction test results
| Restriction test | Restricted parameter | ESSr | ESSf | q | T–n | Test statistic | p-Value | Test results |
|---|---|---|---|---|---|---|---|---|
| 10 Provinces/Municipalities | r | 26 777.3 | 24 985.5 | 9 | 483 | 3.84863 | 0.00010 | Reject null hypothesis |
| South Korea – Iran | r | 1 370 869 | 1 185 307 | 1 | 70 | 10.9586 | 0.00083 | Reject null hypothesis |
Table 4 shows the restriction test results on growth rate r. The null hypothesis was that the growth rates were the same among the ten provinces or the same between South Korea and Iran. ESSr is the residual sum of squares with restrictions, ESSf is the residual sum of squares without restrictions, and qrepresents the number of restrictions imposed upon the growth rates. The Test statistic is calculated based upon Equation (7).
In regards to the modelling process for the nine countries listed in Table 3, the model failed to generate reliable estimates for seven out of the nine countries when no clear, discernable date of maximal increase in COVID-19 cases existed according to the data as of March 13, 2020. Consequently, when a model based on Equation (4) failed to reach optimal results, the corresponding region was likely in the early stages of an outbreak as of March 13, 2020.7 In the two nations that did yield reliable estimates, South Korea and Iran, this study predicted COVID-19 cases to reach 8080 (95% CI 7126.2–9033.8) and 19 604.5 (95% CI 10 378.3–28 830.8), respectively. A restriction test was also conducted for the growth rate r between these two countries, the results of which are displayed in Table 4. The test statistic was 10.9586 and the p-value was 0.00083, and this study, again, rejected the null hypothesis that r was the same between South Korea and Iran. In the case of South Korea, the regression model provided a good fit in the training data, but the fitness in the testing period was less satisfactory because the model failed to capture the additional new cases in the testing period. Modelling the data for Iran was a significantly more complicated process. Fig. 3 indicates a multitude of peaks in the time-series data of confirmed new cases of COVID-19 in Iran. As a result, the initial logistic model built using data up to March 13, 2020 failed to capture new peaks after that date. When estimating the regression model using more data and a cutoff date set at t = 30 and t = 50 (March 21, 2020 and April 9, 2020, respectively), this study demonstrated how the logistic regression model had evolved and fitted the data better as new information became available.
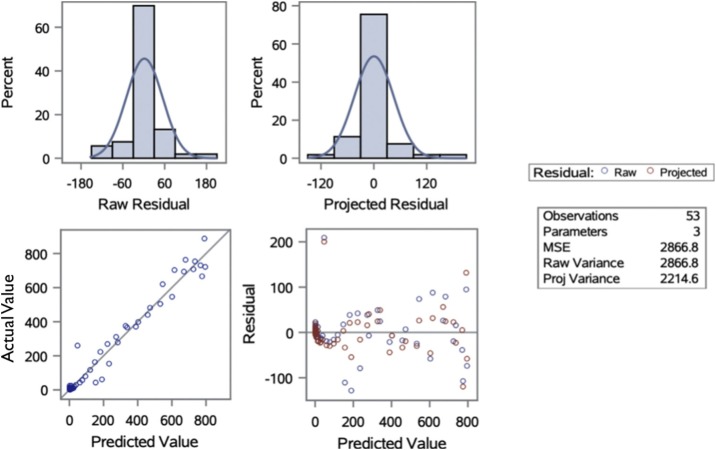
The results of the residual diagnosis tests from Table 5 and Fig. 4 show both the raw and projected residuals have their distributions centered around 0, and their predicted values closely follow a 45-degree line with their corresponding actual values. This study identifies the existence of positive serial correlation in two of the ten selected non-Hubei provinces (Zhejiang and Jiangsu) and in Iran based on either the raw or projected residuals. In the rest of the non-Hubei provinces of China as well as in South Korea, the DW test failed to reject the null hypothesis of no serial correlation in residuals. Furthermore, with the exception of Beijing, Guangdong and Shanghai, China, this study identified the existence of heteroscedasticity in all other selected non-Hubei provinces and cities, suggesting the variance in error terms largely existed and that it may vary depending on different stages of the proliferation. Accordingly, whereas this study's estimates are still consistent, they are not efficient (Pindyck and Rubinfeld, 1991). The findings suggest that further research may be needed to develop more efficient estimators of the model.
Table 5.
Durbin–Watson and White test results
| Country/province | Type of residual | Durbin–Watson test |
White test |
||
|---|---|---|---|---|---|
| DW statistic | Result | Chi-square value | Result | ||
| National | Raw | 1.796 | Fail to reject | 3.198 | Fail to reject |
| Projected | 2.123 | Fail to reject | 2.558 | Fail to reject | |
| Hubei | Raw | 1.911 | Fail to reject | 3.198 | Fail to reject |
| Projected | 2.196 | Fail to reject | 2.558 | Fail to reject | |
| Non-Hubei | Raw | 1.043 | Reject | 19.244 | Reject |
| Projected | 1.629 | Fail to reject | 5.120 | Fail to reject | |
| Anhui | Raw | 2.280 | Fail to reject | 14.810 | Reject |
| Projected | 2.461 | Fail to reject | 16.203 | Reject | |
| Beijing | Raw | 2.041 | Fail to reject | 3.855 | Fail to reject |
| Projected | 2.232 | Fail to reject | 3.295 | Fail to reject | |
| Chongqing | Raw | 1.941 | Fail to reject | 21.915 | Reject |
| Projected | 2.090 | Fail to reject | 21.986 | Reject | |
| Guangdong | Raw | 2.120 | Fail to reject | 4.0863 | Fail to reject |
| Projected | 2.482 | Fail to reject | 5.406 | Fail to reject | |
| Hunan | Raw | 1.993 | Fail to reject | 6.115 | Reject |
| Projected | 2.190 | Fail to reject | 7.609 | Reject | |
| Henan | Raw | 1.937 | Fail to reject | 13.720 | Reject |
| Projected | 2.259 | Fail to reject | 20.010 | Reject | |
| Jiangsu | Raw | 0.767 | Reject | 6.980 | Reject |
| Projected | 1.531 | Indeterminate | 9.280 | Reject | |
| Jiangxi | Raw | 1.549 | Indeterminate | 8.200 | Reject |
| Projected | 1.826 | Fail to reject | 8.112 | Reject | |
| Shanghai | Raw | 1.850 | Fail to reject | 1.850 | Fail to reject |
| Projected | 1.901 | Fail to reject | 1.901 | Fail to reject | |
| Zhejiang | Raw | 1.301 | Reject | 20.455 | Reject |
| Projected | 1.650 | Fail to reject | 23.465 | Reject | |
| Iran | Raw | 1.193 | Reject | 3.712 | Reject |
| Projected | 2.088 | Fail to reject | 5.329 | Reject | |
| South Korea | Raw | 1.776 | Fail to reject | 2.253 | Fail to reject |
| Projected | 2.100 | Fail to reject | 2.798 | Fail to reject | |
Table 5 shows the results of the Durbin–Watson test and White test conducted on both raw and projected residuals. The critical value for the DW test with the number of explanatory variables being 1 was 1.51 to 1.59, and ‘Reject’ indicated serial correlation issues. The critical value for the White test was 5.99, and ‘Reject’ indicated heteroskedasticity. Both the DW test and White test were conducted at the 5% significance level. The residual diagnosis was not performed for the seven other nations not listed in this table due to the lack of optimal model estimates.
Fig. 4.
Residual diagnosis for the non-Hubei regression model. The results in this figure are based on the non-Hubei time-series data; the results for other regions are available upon request.
In conclusion, this simple, three-parameter logistic growth function for reported COVID-19 cases estimated by an NLS method presents certain insights for current and future studies of outbreaks. This study's findings demonstrate that the model fitted the data in China exceedingly well, and the study was able to provide estimates for COVID-19 cases and compare the speed of proliferation among regions. However, the model failed to provide estimates for outbreaks in their early stages, and only yielded results after there was a definitive day of maximal increase in cases. Conducting a restriction test on the r parameter, the study found that between provinces in China and between other countries alike, the growth rates of COVID-19 differed and the study conjectured that this was due to disparities in local public health policies, societal behavior, patient response, etc. Accordingly, there is potential for this model to contribute to formulating better policies towards combatting COVID-19 by retrospectively analyzing the outbreaks in regions such as provinces in China. This study also observed that in non-linear regressions, the R 2 value varied between different sections of the non-linear curve, and that the existence of heteroscedasticity and serial correlation in some provinces and countries warrant further research.
In summary, the study's findings show that in a relatively isolated environment such as China, where control measures are consistently strict and regulatory, the logistic regression model fits very well. However, when other factors become prevalent — such as diverging public health control practices and imported cases from abroad — the proliferation of infectious diseases may complicate research methods and a single logistic growth model may not suffice.
Declarations
Funding: Not applicable.
Ethical approval and consent to participate: The need for ethical approval or individual consent was not applicable.
Availability of data and materials: All data and materials used in this work are publicly available.
Consent for publication: Not applicable.
Conflict of interest: No conflict of interest to declare.
Acknowledgements
The author would like to thank Professor Wenbin Chen of Fudan University for mathematical and modeling advice and Dr. Chaohui Dong of the National Healthcare Security Administration of China for epidemic disease advice. The author is also indebted to the anonymous reviewers for their indispensable feedback.
Footnotes
For a more detailed discussion on the estimation of non-linear regression models and properties of estimated parameters, please refer to Chapter 10 (Greene, 1997) and Chapter 5 (Davidson and MacKinnon, 1993).
These cases were unaccounted for prior to the new change in diagnosing and reporting COVID-19 cases. As a result, there were 14 840 new cases in Hubei and only 312 new cases in other regions in China reported on February 12, 2020.
For more information on the projected residuals, please refer to the aforementioned paper by Cook and Tsai. This study employed SAS to calculate the projected residuals.
Specifically, the White test in this study developed a regression of the square of the residuals based on the independent variables t and t2, in which the R2 of the regression multiplied by the number of observations follows a Chi-square distribution with two degrees of freedom.
A similar approach can be undertaken to estimate new COVID-19 deaths, and when this study did so it found that the model also fitted the data exceedingly well. The study observed that the maximal increase in deaths lagged behind that in cases by 6–12 days in China. Knowing that there is a strong correlation between deaths and cases, the study chose not to fully develop models for new COVID-19 deaths.
The high R2 mainly constitutes the peak period of an outbreak when many new cases are present. When calculating the overall R2 for both the testing and training period combined, as shown in Tables 1, 2 and 3, it was found that the total R2 was similar to that of the training period.
Singapore may be an exception. While its outbreak began as early as late January of 2020, the model did not fit the data well. This indicates a further need to study the outbreak in Singapore.
References
- Cook R.D., Tsai C.L. Residuals in Nonlinear Regression. Biometrika. 1985;72:23–29. [Google Scholar]
- Cramer J.S. The early origins of the logit model. Stud Hist Philos Sci Part C Stud Hist Philos Biol Biomed Sci. 2004;35(4):613–626. [Google Scholar]
- Davidson J., MacKinnon J. Oxford University Press; New York: 1993. Estimation and Inference in Econometrics. [Google Scholar]
- Durbin J., Watson G.S. Testing for Serial Correlation in Least Squares Regression: I. Biometrika. 1950;37(3/4):409–428. [PubMed] [Google Scholar]
- Greene W.H. Third Edit. Prentice-Hall; 1997. Econometric Analysis. [Google Scholar]
- King A.A., De Cellés M.D., Magpantay F.M.G., Rohani P. Avoidable errors in the modelling of outbreaks of emerging pathogens, with special reference to Ebola. Proc R Soc B Biol Sci. 2015;282(1806):0–6. doi: 10.1098/rspb.2015.0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau G., Benhamou Y., Chen G., Li J., Shao Q., Ji D. Efficacy and safety of 3-week response-guided triple direct-acting antiviral therapy for chronic hepatitis C infection: a phase 2, open-label, proof-of-concept study. Lancet Gastroenterol Hepatol [Internet] 2016;1(2):97–104. doi: 10.1016/S2468-1253(16)30015-2. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder M., Mandl K.D. Early Transmissibility Assessment of a Novel Coronavirus in Wuhan, China. SSRN Electron J. 2020 [Google Scholar]
- National Health Commission of the People's Republic of China. Newest Updates Regarding COVID-19 Nationally [Internet]. 2020 [cited 2020 Mar 21]. Available from: http://www.nhc.gov.cn/xcs/yqtb/202003/be74d71b2f784cae917cc830f244caa9.shtml.
- Pindyck R.S., Rubinfeld D.L. In: Econometric Models & Economic Forecasts. Third Edit. Stratford S.D., Richmond L., editors. McGraw-Hill; 1991. [Google Scholar]
- Roosa K., Lee Y., Luo R., Kirpich A., Rothenberg R., Hyman J.M. Real-time forecasts of the COVID-19 epidemic in China from February 5th to February 24th, 2020. Infect Dis Model [Internet] 2020;5:256–263. doi: 10.1016/j.idm.2020.02.002. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schabenberger O., Pierce F.J. CRC Press; Boca Raton, FL: 2002. Contemporary Statistical Models for the Plant and Soil Sciences. [Google Scholar]
- White H. A Heteroskedasticity-Consistent Covariance Matrix Estimator and a Direct Test for Heteroskedasticity. Econometrica. 1980;48:817–838. [Google Scholar]
- World Health Organization . 2020. COVID-19 Situation Report 61 [Internet] Available from: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200321-sitrep-61-covid-19.pdf?.sfvrsn=f201f85c_2. [Google Scholar]
- Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan. China: a modelling study. Lancet [Internet]. 2020;395(10225):689–697. doi: 10.1016/S0140-6736(20)30260-9. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Darcet D., Wang Q., Sornette D. 2020. Generalized logistic growth modeling of the COVID-19 outbreak in 29 provinces in China and in the rest of the world. Available from: http://arxiv.org/abs/2003.05681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Chen H. Modeling the epidemic dynamics and control of COVID-19 outbreak in China. Quant Biol. 2020:1–9. doi: 10.1007/s40484-020-0199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Lin Q., Ran J., Musa S.S., Yang G., Wang W. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: A data-driven analysis in the early phase of the outbreak. Int J Infect Dis [Internet]. 2020;92:214–217. doi: 10.1016/j.ijid.2020.01.050. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]