Abstract
Coronavirus disease 2019 (COVID-19) is an emerging infectious disease that was first reported in Wuhan, China, and has subsequently spread worldwide. In the absence of any antiviral or immunomodulatory therapies, the disease is spreading at an alarming rate. A possibility of a resurgence of COVID-19 in places where lockdowns have already worked is also developing. Thus, for controlling COVID-19, vaccines may be a better option than drugs. An mRNA-based anti-COVID-19 candidate vaccine has entered a phase 1 clinical trial. However, its efficacy and potency have to be evaluated and validated. Since vaccines have high failure rates, as an alternative, we are presenting a new, designed multi-peptide subunit-based epitope vaccine against COVID-19. The recombinant vaccine construct comprises an adjuvant, cytotoxic T-lymphocyte (CTL), helper T-lymphocyte (HTL), and B-cell epitopes joined by linkers. The computational data suggest that the vaccine is non-toxic, non-allergenic, thermostable, with the capability to elicit a humoral and cell-mediated immune response. The stabilization of the vaccine construct is validated with molecular dynamics simulation studies. This unique vaccine is made up of 33 highly antigenic epitopes from three proteins that have a prominent role in host-receptor recognition, viral entry, and pathogenicity. We advocate this vaccine must be synthesized and tested urgently as a public health priority.
Keywords: Adjuvant, COVID-19, Immunogenic epitopes, Peptide vaccine, SARS-CoV-2, Subunit vaccine
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) belongs to the genus Betacoronavirus of the Coronaviridae family and is identified as the pathogen of Coronavirus disease 2019 (COVID-19) [1]. The epicenter of the COVID-19 coronavirus outbreak was the central Chinese city of Wuhan, from where it spread globally. On January 30, 2020, the World Health Organization officially declared the COVID-19 epidemic as a public health emergency of international concern. Human to human transmission occurs through droplets, contact, and fomites. People with COVID-19 show symptoms of fever, cough, muscle aches, headache, and diarrhea. The principal feature of the severe disease is acute onset of hypoxemic respiratory failure with bilateral infiltrates.
The virus genome has been sequenced that allowed the development of diagnostic tests and research into vaccines and therapeutics [1,2]. A specific RT-PCR-based test has been developed that is in use for clinical diagnoses [3]. The abundance of publications in the first three months of 2020 indicates the intensive scientific effort to address both molecular mechanisms and therapeutic routes for treating COVID-19 [4]. More than 200 clinical trials are currently underway to test novel and repurposed compounds against SARS-CoV-2 [5,6]. Certain drugs, including hydroxychloroquine, chloroquine, and remdesivir, are being tested in clinical trials [[7], [8], [9]]. One small study reported that combination therapy of hydroxychloroquine with azithromycin reduced the detection of viral RNA compared to control [10,11]. A recent open-label trial with two protease inhibitors, lopinavir, and ritonavir, failed [12]. Several inactivated vaccines, viral vectored vaccines (adenovirus vector, ankara vector), nanoparticle-based vaccines, fusion-protein based vaccines, adjuvanted vaccines, recombinant protein, and DNA vaccines, as well as live-attenuated vaccines, are also being developed and tested, but these vaccines are many months away from the market [[13], [14], [15], [16]]. A phase 1 clinical trial of Moderna's mRNA-based SARS- CoV-2 candidate vaccine, mRNA-1273, has started on March 16, 2020 [[17], [18], [19]]. However, this is the first of several steps in the clinical trial process for evaluating the potential benefits of the vaccine.
The SARS-CoV-2 consists of single, positive-stranded RNA and four structural proteins: a spike glycoprotein (S), a membrane glycoprotein (M), an envelope protein (E), and a nucleocapsid protein (N) [20]. To enter the host cells, the virus uses a densely glycosylated spike protein that binds to the angiotensin-converting enzyme 2 (ACE2) receptor with high affinity [21,22]. Structural and biochemical studies suggest that the RBD has an ultra-high binding affinity to the human ACE2 receptor [23]. Few groups have designed subunit vaccines against SARS-CoV-2; however, their workflow involved either use of single protein for vaccine design [24,25] or used only CTL epitopes without considering the importance of B-cell or HTL epitopes [26]. Some subunit-vaccines are also in preclinical trials [27,28]. Here, we focused on designing a multi-epitope-based subunit vaccine against SARS-CoV-2 using 33 highly antigenic epitopes. We believe that experimental evaluation may result in a novel and immunogenic vaccine that may confer protection against SARS-CoV-2 infection.
2. Methods
2.1. Screening of antigenic proteins
The protein sequences of SARS-CoV-2 were retrieved from the NCBI database (https://www.ncbi.nlm.nih.gov/nuccore/MN996531.1/) for subunit vaccine development (Table 1 ) [29]. Each of these proteins was screened for their average antigenic propensity using the antigenic peptides prediction tool (http://imed.med.ucm.es/Tools/antigenic.pl). Proteins with an antigenic probability score of greater than 0.8 were considered for vaccine construction.
Table 1.
Antigenicity score of the selected proteins.
| Proteins | Accession no. | Predicted Order of Antigenicity |
|---|---|---|
| Nucleocapsid protein | QHR63298.1 | 0.9871 |
| Membrane glycoprotein | QHR63293.1 | 1.0532 |
| Surface spike glycoprotein | QHR63290.1 | 1.0412 |
2.2. Prediction of helper T-lymphocyte (HTL), cytotoxic T-lymphocyte (CTL) and B-cell epitopes
The helper T-lymphocyte (HTL) epitopes for the selected SARS-CoV-2 proteins were predicted using the MHC-II epitope prediction tool from the Immune Epitope Database (IEDB, http://tools.iedb.org/mhcii/). Selected epitopes had the lowest percentile rank and IC50 values. Additionally, these epitopes were checked by the IFN epitope server (http://crdd.osdd.net/raghava/ifnepitope/) for the capability to induce Th1 type immune response accompanied by IFN-ϒ production. Cytotoxic T-lymphocyte (CTL) epitopes for the screened proteins were predicted using the NetCTL1.2 server (http://www.cbs.dtu.dk/services/NetCTL/). B-cell epitopes for the screened SARS-CoV-2 proteins were predicted using the ABCPred server (http://crdd.osdd.net/raghava/abcpred/). The prediction of the toxic/non-toxic nature of all the selected HTL, CTL, and B-cell epitopes was checked using the ToxinPred module (http://crdd.osdd.net/raghava/toxinpred/multi_submit.php).
2.3. Construction of the multi-epitope subunit vaccine
The vaccine subunit was designed by adding an adjuvant, HTL, CTL, and B-cell epitopes connected by specific linkers to provide adequate separation of epitopes in vivo. EAAAK linker was used to join the adjuvant and HTL. Intra HTL, Intra CTL, and B-cell epitopes were joined using GPGPG, AAY, and KK, respectively. To enhance the immunogenicity of the vaccine construct, the TLR3 agonist, human β-defensin 1 (Uniprot ID: P60022), was used as the adjuvant.
2.4. Immunogenicity and allergenicity prediction
The immunogenicity of the vaccine was determined using the VaxiJen server (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) and ANTIGENpro module of SCRATCH protein predictor (http://scratch.proteomics.ics.uci.edu/). The allergenicity of the vaccine was checked using AllerTOP v2.0 (http://www.ddg-pharmfac.net/AllerTOP/) and AlgPred Server (http://crdd.osdd.net/raghava/algpred/).
2.5. Determination of physicochemical properties
The physiochemical characteristics of the vaccine were determined using the ProtParam tool of the ExPASy database server (http://web.expasy.org/protparam/).
2.6. Structure prediction, validation, and docking with the receptor
The secondary structure of the subunit vaccine construct was predicted using PSIPred 4.0 Protein Sequence Analysis Workbench (http://bioinf.cs.ucl.ac.uk/psipred/), while the tertiary structure was predicted by de novo structure prediction-based trRosetta modeling suite. trRosetta uses a deep residual neural network to predict the inter-residue distance and orientation distributions of the input sequence [39]. Then it converts predicted distance and orientation distributions into smooth restraints to build 3D structure models-based on direct energy minimization. The model of the vaccine construct with the best TM-score was validated by PROCHEK v.3.5 (https://servicesn.mbi.ucla.edu/PROCHECK/) and ProSA (https://prosa.services.came.sbg.ac.at/prosa.php) web servers. Vaccine-receptor docking was performed by the ClusPro web server to determine the binding affinity of the vaccine with the TLR3 receptor (PDB ID: 3ULV).
2.7. Molecular dynamics simulations
Molecular dynamics (MD) simulation is an effective method to study the molecular interactions and dynamics of the vaccine-TLR3 complex. The complex structure of the vaccine-TLR3 was initially optimized using Schrödinger Maestro (Schrödinger Release 2016–4: Maestro, Schrödinger, New York) and subsequently used as the starting structure for MD simulations. First, hydrogen atoms were added to the complex, which was then solvated in an octahedral box of a simple point charge (SPC) water in the center at least 1.0 nm from the box edge. The system was subsequently electrostatically neutralized by the addition of appropriate counter ions. MD simulation was carried out with GROMACS 5.1.2 software package using the gromos96 54A7 force-field. A standard MD simulation protocol started with 50,000 steps of energy minimization until no notable change of energy was observed, followed by a heating step from 0 to 300 K in 200 ps (canonical ensemble) and 1000 ps at 300 K (isobaric-isothermal ensemble) by constant temperature equilibration. During this, Parrinello-Rahman barostat pressure coupling was used to avoid the impact of velocity. As a final step of the simulation, 40 ns production run was carried out at 300 K with periodic boundary conditions in the NPT ensemble with modified Berendsen temperature coupling and at a constant pressure of 1 atm. Further, the LINCS algorithm, along with the Particle‐mesh Ewald method, was used for the calculation of long‐range electrostatic forces. Fourier grid spacing and Coulomb radius were set at 0.16 and 1.4 nm, respectively, during the simulations. The van der Waals (VDW) interactions were limited to 1.4 nm, and structures were saved at every 10 ps for structural and dynamic analysis.
2.8. MD simulation-based analyses
The analysis from MD simulation was performed as described earlier [30]. Briefly, the backbone RMSD and root mean square fluctuation (RMSF) of the vaccine-TLR3 complex from the MD simulation trajectory was analyzed using gmx rms and gmx rmsf utilities of GROMACS, respectively. The radius of gyration, which represents the compactness of the vaccine-TLR3 complex, was analyzed using the gmx gyrate tool of GROMACS utilities. Hydrogen bonds were determined using the hydrogen bonds module of visual molecular dynamics (VMD) throughout the 40 ns duration. The intermolecular and intramolecular interactions formed between TLR3 and the vaccine subunit were computed and visualized in Molecular Operating Environment software. Several snapshots of the vaccine-TLR3 complexes were extracted from the MD simulations, and various interactions formed among them were computed.
Essential dynamics (ED), which represent the principal motion directions by a set of eigenvectors, was performed from the MD simulation trajectory of vaccine-TLR3 complex. In this analysis, a variance/covariance matrix was constructed by calculating the eigenvectors and eigenvalues, and their projection along the first two principal components was monitored by principal component analysis (PCA). The eigenvalues associated with each of the eigenvectors of the vaccine-TLR3 complex were used to calculate the percentage of variability.
2.9. Codon adaptation and in-silico cloning
Java Codon Adaptation Tool (JCAT) (http://www.jcat.de/) was used for codon optimization of the vaccine sequence to test high-level expression of the vaccine in E. coli strain K12. NEBcutter (http://nc2.neb.com/NEBcutter2/) was used for the selection of restriction enzyme cleavage sites, and the expression vector pET28a(+) was selected. In silico clone of the vaccine was designed using the SnapGene 1.1.3 restriction cloning tool.
3. Results
3.1. Screening of antigenic proteins
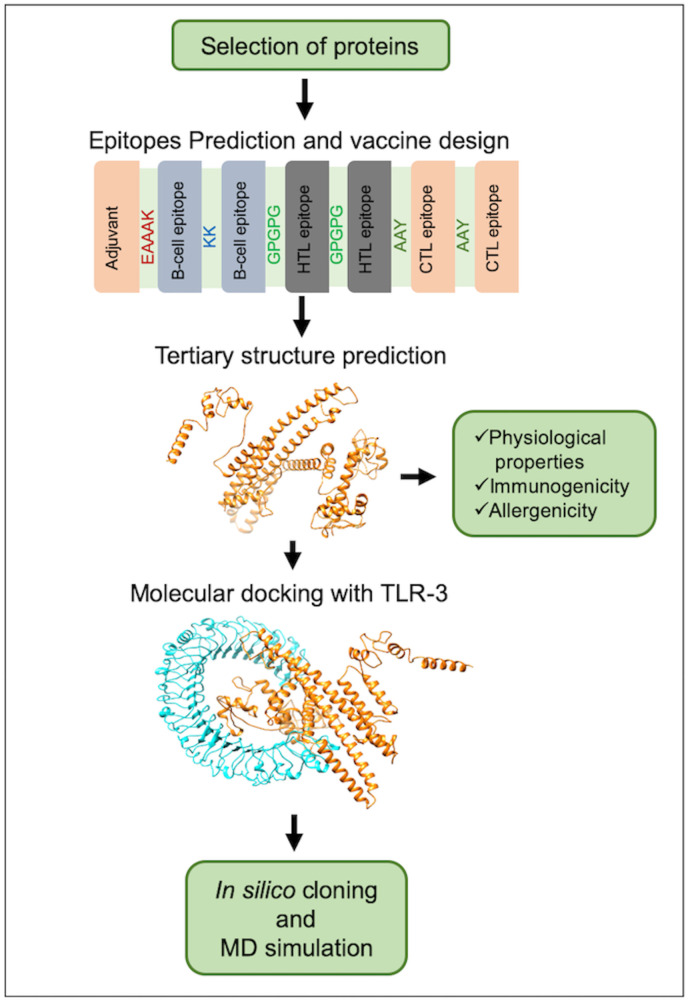
The amino acid sequence of the three SARS-CoV-2 proteins, namely, nucleocapsid protein, membrane glycoprotein, and surface spike glycoprotein, were retrieved from the NCBI database (Table 1). These proteins are known to have a prominent role in host receptor recognition, viral entry, and pathogenicity. The proteins with an antigenic score of greater than 0.8 (Table 1) were used further for the prediction of epitopes for subunit vaccine designing. A schematic representation of the methodology for the construction of the subunit vaccine candidate is shown in Fig. 1 .
Fig. 1.
Schematic representation of the multi-epitope subunit vaccine candidate designing using B-cell, CTL, and HTL epitopes.
3.2. Prediction of HTL, BCL and CTL epitopes
Helper T-lymphocytes are the key players of the adaptive immune response. They are involved in the activation of B-cells and cytotoxic T cells for antibody production and killing infected target cells, respectively. All three proteins were subjected to the IEDB MHC-II epitope prediction module for HTL prediction. A total of six highest immunogenic epitopes of 15-mer were selected based on their percentile rank and IC50 values. Also, all these epitopes showed positive scores on IFNepitope server output (Table 2 ). B-cells are the main components of humoral immunity during the adaptive immune response that produces antibodies, which recognize antigens. Therefore, it was necessary to predict B-cell epitopes before vaccine designing. ABCpred was performed for predicting B-cell epitope, and a total of 9 epitopes with top scores from the three proteins were considered for the vaccine (Table 3 ).
Table 2.
Predicted HTL specific epitopes and their percentile rank obtained from IEDB.
| S. No. | Protein name | Epitope | Percentile rank | SMM align IC50 (nM) | Allele | IFN-γ Inducer score |
|---|---|---|---|---|---|---|
| 1. | Nucleocapsid protein | GTWLTYTGAIKLDDK | 0.58 | 15 | HLA-DRB1*07:01 | 3 |
| AALALLLLDRLNQLE | 0.61 | 17 | HLA-DRB4*01:01 | 1 | ||
| 2. | Membrane glycoprotein | NRFLYIIKLIFLWLL | 0.12 | 31 | HLA-DRB4*01:01 | 1 |
| 3. | Surface spike glycoprotein | EFVFKNIDGYFKIYS | 0.17 | 10 | HLA-DRB5*01:01 | 2 |
| ITRFQTLLALHRSYL | 0.26 | 2 | HLA-DRB5*01:01 | 3 | ||
| ATRFASVYAWNRKRI | 0.49 | 10 | HLA-DRB5*01:01 | 0.56 |
Table 3.
Predicted B-cell binding epitopes with their probable score and start position.
| Sl. No. | Protein Name | Epitope | Position | Score |
|---|---|---|---|---|
| 1 | Nucleocapsid protein | TRRIRGGDGKMKDLSP | 91 | 0.94 |
| KSAAEASKKPRQKRTA | 249 | 0.93 | ||
| EGALNTPKDHIGTRNP | 136 | 0.93 | ||
| 2 | Membrane glycoprotein | RSMWSFNPETNILLNV | 107 | 0.89 |
| SFRLFARTRSMWSFNP | 99 | 0.88 | ||
| 3 | Surface spike glycoprotein | YACWHHSIGFDYVYNP | 6149 | 0.96 |
| VVKIYCPACHNSEVGP | 365 | 0.96 | ||
| TLKGGAPTKVTFGDDT | 814 | 0.95 | ||
| TSRYWEPEFYEAMYTP | 5304 | 0.94 |
CTL epitopes are essential for inducing MHC-I cellular immune response by neutralizing virus-infected cells and damaged cells via releasing cytotoxic proteins like granzymes, perforins, etc. The CTL epitopes were predicted for all selected proteins using the NetCTL 1.2 server. Here, A2, A3, and B7 supertypes were considered for prediction as they cover at least 88.3% of the total ethnic population. Eighteen epitopes with a combined score of >0.75 were finally considered for the vaccine (Table 4 ). All the selected HTL, CTL, and B-cell epitopes were subjected to the ToxinPred module to screen for their toxicity. Supplementary Table 1 shows that all epitopes chosen for the vaccine were non-toxic.
Table 4.
Predicted CTL epitopes for A2, A3, B7 super types.
| S. No. | Protein Name | Supertype | Epitopes | Score |
|---|---|---|---|---|
| 1 | Nucleocapsid protein | A2 | LLLDRLNQL | 1.2648 |
| GMSRIGMEV | 1.0266 | |||
| A3 | KSAAEASKK | 1.4421 | ||
| KTFPPTEPK | 1.4314 | |||
| B7 | FPRGQGVPI | 1.6470 | ||
| KPRQKRTAT | 1.6339 | |||
| 2 | Membrane glycoprotein | A2 | GLMWLSYFI | 1.3055 |
| FVLAAVYRI | 1.2094 | |||
| A3 | LSYFIASFR | 1.4994 | ||
| RIAGHHLGR | 1.2901 | |||
| B7 | LPKEITVAT | 1.1745 | ||
| RLFARTRSM | 0.9882 | |||
| 3 | Surface spike glycoprotein | A2 | YLQPRTFLL | 1.5152 |
| KIADYNYKL | 1.4347 | |||
| A3 | RLFRKSNLK | 1.7563 | ||
| GVYFASTEK | 1.4615 | |||
| B7 | SPRRARSVA | 1.5619 | ||
| IPTNFTISV | 1.5619 |
3.3. Subunit vaccine designing
A total of 6 HTLs, 18 CTLs, and 9 B-cell epitopes derived from the three proteins were used to design the subunit vaccine (566 amino acid residues) against SARS-CoV-2 (Supplementary Fig. 1). The human β-defensin 1(68 amino acid residues) sequence was added as an adjuvant followed by the HTL, CTL, and B-cell epitopes and linked by specific linkers.
3.4. Antigenicity and allergenicity prediction of constructed vaccine
An important parameter of vaccine designing is ensuring that the constructed vaccine is immunogenic to induce a humoral and/or cell-mediated immune response against the targeted virus. The computational data suggest that our vaccine is antigenic with a probability score of 0.513 and 0.732 predicted by VaxiJen v2.0 and ANTIGENPro servers, respectively. The allergenicity score was found to be −0.658 in AlgPRED prediction module. Additionally, the vaccine was also found to be non-allergic using AllerTOP v2.0.
3.5. Physiochemical characterization of designed vaccine
The designed vaccine construct is composed of 566 amino acids with a molecular weight of approximately 62.34 kDa. The theoretical pI was 10.21, suggesting the vaccine is significantly basic. The half-life of the vaccine was estimated to be 30 h in mammalian reticulocytes (in vitro), >20 h in yeast (in vivo), and >10 h in E. coli (in vivo), suggesting that the construct is stable in vivo. The instability index was estimated to be 24.76, suggesting a stable protein. The computed aliphatic index and grand average of hydropathicity were found to be 77.79 and −0.187, respectively, suggesting that the vaccine is thermostable and hydrophilic, respectively.
3.6. Structure prediction and validation
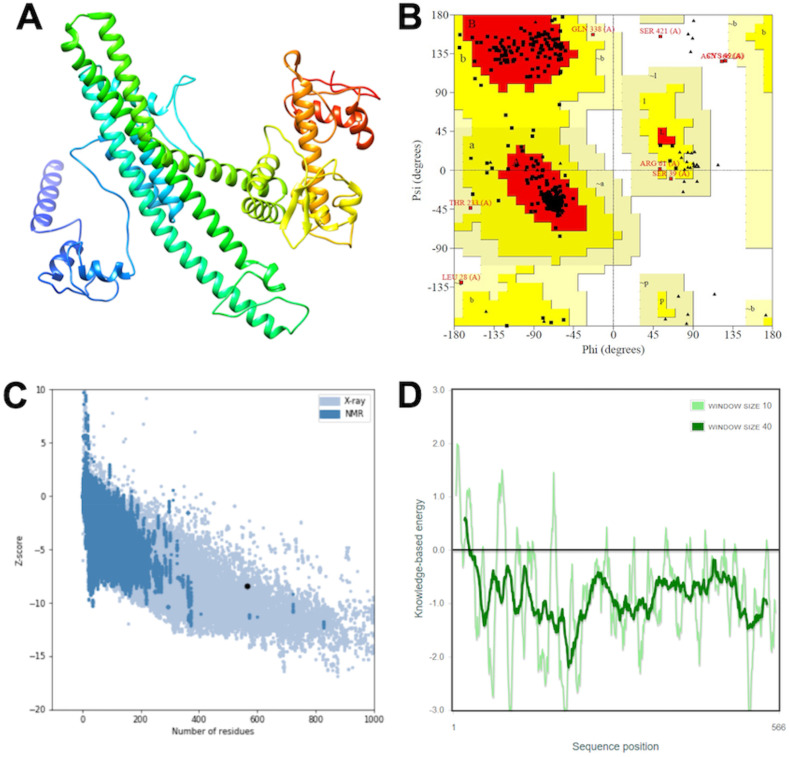
The secondary structure was predicted using the PSIPRED 4.0 server (Supplementary Fig. 2). The tertiary structure of the vaccine was predicted using the trRosetta modeling suite. The 3D model generated by trRosetta modeling was subjected to the PROCHECK server, where Ramachandran plot statistics were generated. The output showed 98.4% residues were present in the favored region, 1.0% residues in the generously allowed region, and 0.6% residues in disallowed regions. Further, the Z-score plot and energy plot was generated by the ProSA web server. The calculated Z-score (−8.46) lies within the X-ray crystal structure range. The energy plot suggested that all the residues have low energy value in the modeled structure (Fig. 2 ).
Fig. 2.
Tertiary structure model prediction and its validation (A) 3D model obtained for the multi-subunit vaccine protein. (B) Ramachandran plot showing the presence of amino acid residues in favored, allowed, and outlier regions. (C) ProSA-web z-score plot for predicted the 3D structure and (D) Energy plot for all residues in the predicted structure.
3.7. Vaccine-receptor docking
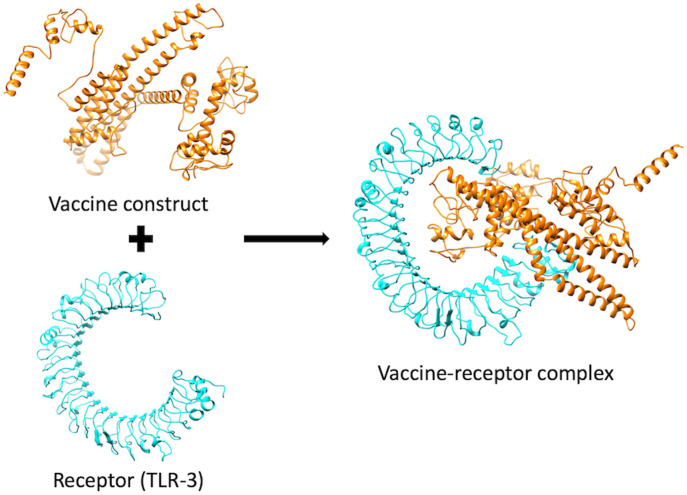
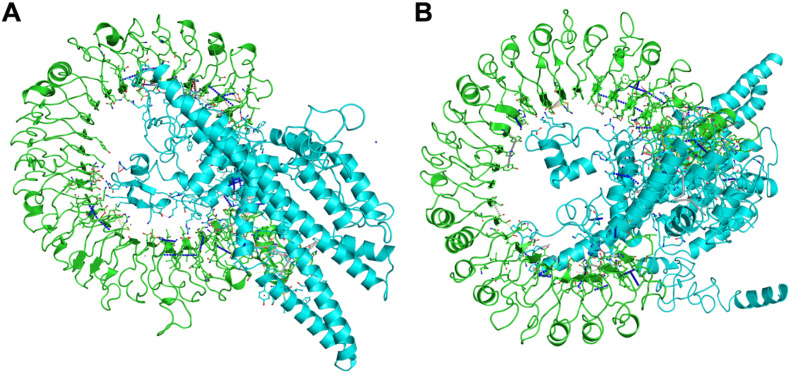
Vaccine-receptor docking was performed to evaluate the binding energy of the vaccine with its TLR3 receptor. ClusPro analysis provided 30 vaccine-receptor complexes with respective energy scores. The lowest energy complex with a binding energy of about −1491 kJ mol−1 was selected and subjected to MD simulation (Fig. 3 ).
Fig. 3.
Stable interaction between the vaccine construct and TLR3 after docking. The vaccine construct is shown in orange, while the TLR3 is shown in cyan.
3.8. MD simulations
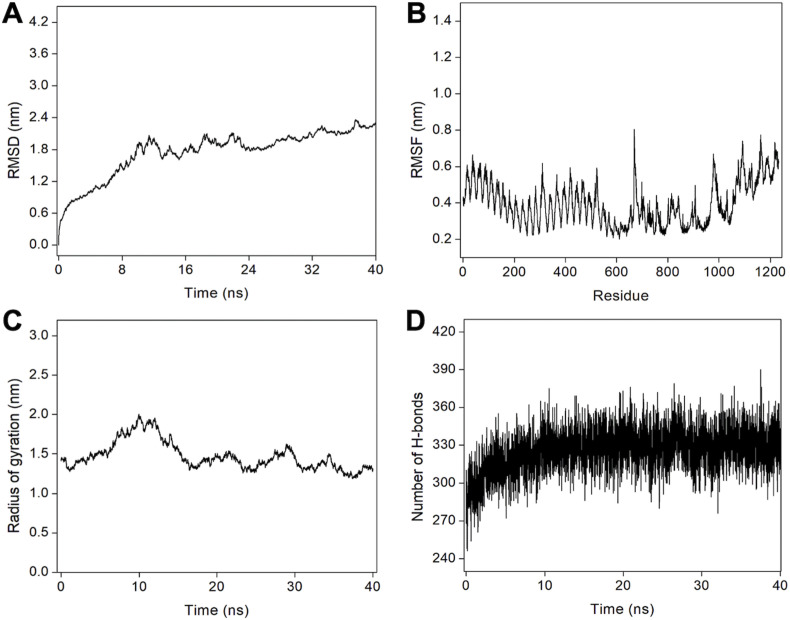
The binding modes, dynamics, and stability of the vaccine-TLR3 complex were evaluated using a 40 ns MD simulation study (Fig. 4 ). The atomic-level interaction between vaccine and TLR3 was determined, and root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg), hydrogen bond, and contact energy were calculated.
Fig. 4.
MD simulation of the vaccine-receptor complex. (A) RMSD for the amino acid backbone of the vaccine-receptor complex, (B) RMSF of amino acids side chain of the vaccine-TLR3 complex, (C) Rg as a function of simulation time and (D) Number of hydrogen bonds formed during MD simulation trajectory.
The RMSD data suggest that the receptor-vaccine complex was stabilized after about 20 ns until the end of the simulation (Fig. 4A). All the calculations were then done for the 20–40 ns MD simulation trajectory. Next, RMSF calculation, which gives information about the residue-wise dynamics of a protein with respect to its initial position, was done. An average RMSF value of 0.39 nm was observed for the complex (Fig. 4B). Our subsequent analysis of the changes in the Rg for the vaccine-TLR3 complex during the simulation was also determined, and the average Rg was found to be 1.47 nm, demonstrating the compactness of the TLR3 receptor with vaccine subunit during the simulations (Fig. 4C). We further analyzed the hydrogen bonds that play a vital role in stabilizing protein structure and the recognition of other protein partners in a complex. The vaccine-receptor complex formed an average of 327 hydrogen bonds, suggesting the favorable intermolecular interactions between the vaccine protein and the TLR3 receptor. The formation of a large number of hydrogen bonds and its stabilization during simulations reflect the specificity and selectivity of intermolecular interactions (Fig. 4D). In total, the complex formed about 5369 bonds during the 40 ns simulation run. Next, we further computed the contact energy for the vaccine-TLR3 complex, and it was found that while the starting complex structure exhibited total contact energy of −814.36 kcal mol−1, the stabilized complex exhibited total contact energy of −935.68 kcal mol−1, showing the increased stability during MD simulations. Next, we performed ED analysis where PCA is one of the important techniques that provide insight into the correlation of atomic movement of protein-protein complexes, raised from the collective motion of atoms that are controlled by the secondary structure of the proteins. Typically, the largest associated eigenvalues define the essential subspace in which most of the protein dynamics occur. For this, the clusters of stable states of PCA for the vaccine-TLR3 complex were visualized and analyzed. The trace value calculated from the covariance matrix of the vaccine-TLR3 was found to be 2.18 nm2, suggesting that the complex exhibited compact behavior during the simulation (Supplementary Fig. 3). Lastly, the detailed interactions between TLR3 and the vaccine protein were computed from the starting structure of MD simulations and the stabilized structure of the complex extracted from MD simulated trajectory (Fig. 5 and Table 5 ). The higher total number of interactions in the stabilized complex suggests the stability and tighter binding of the vaccine with TLR3.
Fig. 5.
Key interactions obtained from (A) the initial complex structure of TLR3 and vaccine complex, (B) the stabilized TLR3 vaccine complex obtained from MD simulation. TLR3 receptor is shown in green color, and the vaccine is shown in cyan color in both panels. van der Waals interactions, proximal interactions, polar contacts, hydrogen bonds, aromatic contacts, hydrophobic contacts, carbonyl interactions, and amide-amide interactions are shown in yellow, grey, red, white dashed, white long-dashed, green dashed, black-white dashed and in blue dashed lines respectively.
Table 5.
Details of the interactions occurred in the vaccine-receptor complex during MD simulation.
| Type of interactions | Initial structure before simulation | Stabilized structure after simulation |
|---|---|---|
| VdW interactions | 40 | 28 |
| Proximal interactions | 2136 | 2245 |
| Polar contacts | 69 | 69 |
| Hydrogen bonds | 47 | 45 |
| Aromatic contacts | 9 | 16 |
| Hydrophobic contacts | 77 | 108 |
| Carbonyl interactions | 7 | 5 |
| Total number of interactions | 2385 | 2516 |
3.9. Codon adaptation and in silico cloning
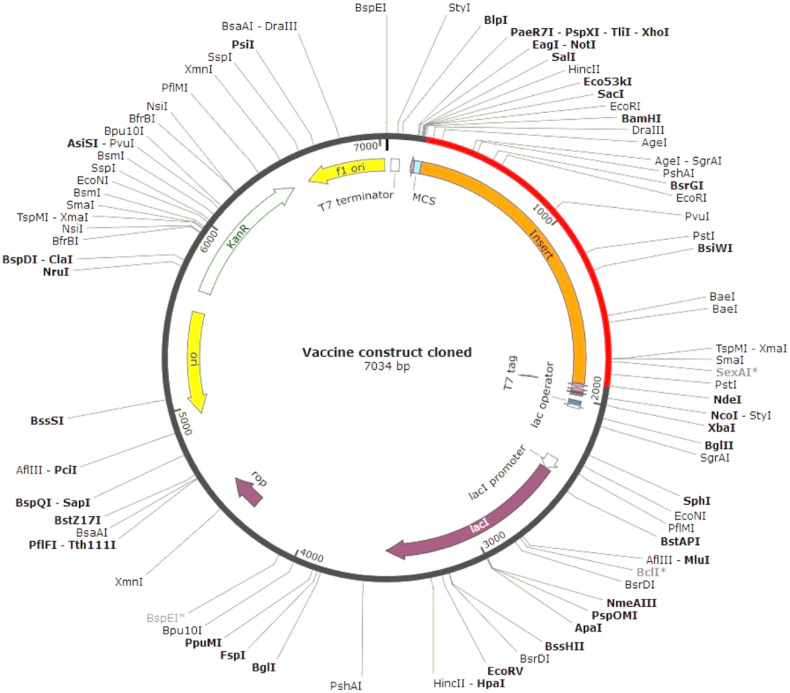
The codon optimization index ensures the relationship between codon usage and gene expression in a heterologous system. The JCAT output was further analyzed in NEBcutter, and at the N- and C-terminal ends of the optimized vaccine sequence, BamHI and NdeI restriction sites were added that are non-cutters for the vaccine construct but are present in the multiple cloning site of the selected expression vector pET28a(+). In silico clone was generated using the SnapGene 1.1.3 restriction cloning tool that resulted in a cloned product of 7034 bp (Fig. 6 ).
Fig. 6.
In silico cloning map showing the insert of vaccine protein-specific optimized codons (red) into the pET28a(+) expression vector.
4. Discussion
Some reports suggest that 5–10% of recovered patients in Wuhan test positive again; this indicates a possibility of a resurgence of COVID-19 in places where lockdowns have already worked. As a consequence, the spread can also be caused by asymptomatic carriers [[31], [32], [33]]. A positive re-test, however, may also be because the original test was false-negative, and the patient was not actually COVID-negative. Whatever may be the case, a vaccine is a better option for coronavirus management than drugs. The efforts to produce a vaccine against coronavirus are moving at a rapid pace. Two candidate vaccines are in Phase I clinical trials: i) An adenovirus type-5 vector-based vaccine, and ii) an LNP-encapsulated mRNA vaccine. Studies evaluating the safety and immunogenicity of these vaccines are underway. Additionally, several vaccine candidates are under preclinical evaluation [34]. Though these trials are underway, there are known situations that vaccines have failed. Recently few groups have tried designing subunit vaccines against SARS-CoV-2; however, their workflow involved either use of single protein for vaccine design [24,25] or used only CTL epitopes without considering the importance of B-cell or HTL epitopes [35]. We considered all of these points while designing the vaccine. Based on extensive bioinformatics analysis, we used three proteins to design a multi-epitope subunit vaccine against novel coronavirus SARS-CoV-2. These proteins are nucleocapsid protein (N), membrane glycoprotein (M), and the surface spike glycoprotein (S). The N protein is involved in packaging the viral genome into a helical ribonucleocapsid, and it plays a fundamental role during viral self-assembly [36]. The M protein is responsible for the assembly and immunogenicity of virus particles. The S protein mediates the entrance of the virus to human respiratory epithelial cells by interacting with cell surface receptor ACE2. The S protein has two regions: S1, for host cell receptor binding; and S2, for membrane fusion. The S protein is a key target for the development of vaccines, therapeutic antibodies, and diagnostics for coronavirus [15,37,38]. Although the S protein is a promising immunogen for protection, optimizing antigen design is critical to ensure an optimal immune response. Our vaccine contains a suitable adjuvant, HTL, CTL, and B-cell epitopes that are joined by suitable linkers. Furthermore, the epitopes were screened for their toxicity potential. The subunit vaccine was found to be thermostable, antigenic, and non-allergenic. Molecular docking and MD simulation provided insights about the interaction, stability, and dynamics of the vaccine-receptor complex. The data suggest constructive intermolecular interactions between the vaccine protein and the TLR3 receptor. Also, the in-silico cloning suggests the potential expression of the vaccine in a microbial expression system, thereby making it a potential vaccine against SARS-CoV-2 infection.
The development of an effective vaccine requires a detailed investigation of the immunological correlations with SARS-CoV-2. However, such approaches would not serve the urgency due to the emergency and severity of the disease outbreak. A computational prediction is, therefore, helpful for guiding scientists towards designing a vaccine and help control the disease. The development of a vaccine is a lengthy and expensive process, with high failure rates, and it typically takes multiple candidates and several years to produce a commercial vaccine. Upon optimization of the production process, the subunit vaccines can be rapidly tested and released in the market. They consist of only the antigenic portion of the pathogens that may directly elicit an immune response. Additionally, the vaccine does not utilize live pathogen, thus, reducing the risk of pathogenicity reversal. Hence, it can be used in immune-suppressed patients as well and elicit long-lived immunity. Computational studies suggest that our multi-epitope based subunit vaccine has a probability of showing good protective efficacy and safety against SARS-CoV-2 infection in humans. We suggest the synthesis and experimental evaluation of this vaccine to determine its immunogenic potency.
Authors contributions
PK and AKP carried out the experiments. PK and TT conceived the study and participated in its design and coordination. PK, AKP, KYJZ, and TT analyzed the data and drafted the manuscript. All authors read and approved the final manuscript.
CRediT authorship contribution statement
Parismita Kalita: Conceptualization, Methodology, Data curation, Writing - original draft. Aditya K. Padhi: Methodology, Data curation, Writing - original draft. Kam Y.J. Zhang: Writing - review & editing. Timir Tripathi: Conceptualization, Supervision, Writing - review & editing.
Declaration of competing interest
The authors have declared no conflict of interest.
Acknowledgements
The authors acknowledge RIKEN ACCC for the Hokusai supercomputing resources.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.micpath.2020.104236.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen K., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poon L.L., Chan Kh Fau - Wong O.K., Wong Ok Fau - Yam W.C., Yam Wc Fau - Yuen K.Y., Yuen Ky Fau - Guan Y., Guan Y., Fau - Lo Y.M.D., et al. Early diagnosis of SARS coronavirus infection by real time RT-PCR. J. Clin. Virol. 2003;28:233–238. doi: 10.1016/j.jcv.2003.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., et al. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 2020;6:315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lythgoe M.P., Middleton P. Ongoing clinical trials for the management of the COVID-19 pandemic. Trends Pharmacol. Sci. 2020 doi: 10.1016/j.tips.2020.03.006. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez M.A. Compounds with therapeutic potential against novel respiratory 2019 coronavirus. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00399-20. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., et al. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anonymous . 2020. Information for Clinicians on Therapeutic Options for COVID-19 Patients. USA. [Google Scholar]
- 11.Gautret P., Lagier J.-C., Parola P., Hoang V.T., Meddeb L., Mailhe M., et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020:105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., et al. A trial of lopinavir-ritonavir in adults hospitalized with severe covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001282. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W.-H., Strych U., Hotez P.J., Bottazzi M.E. The SARS-CoV-2 vaccine pipeline: an overview. Curr. Trop. Med. Rep. 2020 doi: 10.1007/s40475-020-00201-6. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roper R.L., Rehm K.E. SARS vaccines: where are we? Expert Rev. Vaccines. 2009;8:887–898. doi: 10.1586/erv.09.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020;38:1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 16.Thanh Le T., Andreadakis Z., Kumar A., Gomez Roman R., Tollefsen S., Saville M., et al. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020 doi: 10.1038/d41573-020-00073-5. In press. [DOI] [PubMed] [Google Scholar]
- 17.Anonymous . 2020. Moderna's work on a potential vaccine against COVID-19. USA. [Google Scholar]
- 18.Anonymous . 2020. NIH Clinical Trial of Investigational Vaccine for COVID-19 Begins. USA. [Google Scholar]
- 19.Hodgson J. The pandemic pipeline. Nat. Biotechnol. 2020 doi: 10.1038/d41587-020-00005-z. In press. [DOI] [PubMed] [Google Scholar]
- 20.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 22.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S., et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020 doi: 10.1038/s41423-020-0400-4. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abdelmageed M.I., Abdelmoneim A.H., Mustafa M.I., Elfadol N.M., Murshed N.S., Shantier S.W., et al. bioRxiv; 2020. Design of Multi Epitope-Based Peptide Vaccine against E Protein of Human 2019-nCoV: an Immunoinformatics Approach. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhattacharya M., Sharma A.R., Patra P., Ghosh P., Sharma G., Patra B.C., et al. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): immunoinformatics approach. J. Med. Virol. 2020;n/a:1–14. doi: 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mishra S. T Cell epitope-based vaccine design for pandemic novel coronavirus 2019-nCoV. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12029523.v2. [DOI] [Google Scholar]
- 27.Mukerjee S. 2020. The First Coronavirus Drug Candidate Is Set for Testing in China. [Google Scholar]
- 28.Clover Initiates Development of Recombinant Subunit-Trimer Vaccine for Wuhan Coronavirus. Clover Biopharmaceuticals; China: 2020. Chengdu. [Google Scholar]
- 29.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalita P., Lyngdoh D.L., Padhi A.K., Shukla H., Tripathi T. Development of multi-epitope driven subunit vaccine against Fasciola gigantica using immunoinformatics approach. Int. J. Biol. Macromol. 2019;138:224–233. doi: 10.1016/j.ijbiomac.2019.07.024. [DOI] [PubMed] [Google Scholar]
- 31.Feng E., Cheng A. In: National Public Radio: News and Analysis. Brumfiel G., editor. 2020. Mystery in Wuhan: recovered coronavirus patients test negative then positive. Washington, USA. [Google Scholar]
- 32.Beau H.D. SlashdotMedia; 2020. Some Recovered Coronavirus Patients in Wuhan Are Testing Positive Again. [Google Scholar]
- 33.Cyranoski D. We need to be alert: scientists fear second coronavirus wave as China’s lockdowns ease. Nature. 2020 doi: 10.1038/d41586-020-00938-0. In press. [DOI] [PubMed] [Google Scholar]
- 34.WHO . 2020. DRAFT Landscape of COVID-19 Candidate Vaccines – 21 March 2020. Geneva Switzerland. [Google Scholar]
- 35.Seema M. T cell epitope-based vaccine design for pandemic novel coronavirus 2019-nCoV. ChemRxiv. 2020 [Google Scholar]
- 36.Chang C.K., Chen Cm Fau - Chiang M-h, Chiang Mh Fau - Hsu Y-l, Hsu Yl Fau - Huang T-h, Huang T.H. Transient oligomerization of the SARS-CoV N protein--implication for virus ribonucleoprotein packaging. PloS One. 2013;8(5) doi: 10.1371/journal.pone.0065045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV--a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Al-Amri S.S., Abbas A.T., Siddiq L.A., Alghamdi A., Sanki M.A., Al-Muhanna M.K., et al. Immunogenicity of candidate MERS-CoV DNA vaccines based on the spike protein. Sci. Rep. 2017;7:44875. doi: 10.1038/srep44875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J., Anishchenko I., Park H., Peng Z., Ovchinnikov S., Baker D. Improved protein structure prediction using predicted interresidue orientations. Proc. Natl. Acad. Sci. U. S. A. 2020;117(3):1496–1503. doi: 10.1073/pnas.1914677117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.