Abstract
Continuous rise in the human population has resulted in an upsurge in food demand, which in turn demand grain yield enhancement of cereal crops, including rice. Rice yield is estimated via the number of tillers, grain number per panicles, and the number of spikes present per panicle. Marker-assisted selection (MAS) serve as one of the best ways to introduce QTLs/gene associated with yield in the rice plant. MAS has also been employed effectively in dissecting several other complex agricultural traits, for instance, drought, cold tolerance, salinity, etc. in rice plants. Thus, in this review, authors attempted to collect information about various genes/QTLs associated with high yield, including grain number, in rice and how different scheme of MAS can be employed to introduce them in rice (Oryza sativa L.) plant, which in turn will enhance rice yield. Information obtained to date suggest that, numerous QTLs, e.g., Gn1a, Dep1, associated with grain number and yield-related traits, have been identified either via mapping or cloning approaches. These QTLs have been successfully introduced into rice plants using various schemes of MAS for grain yield enhancement in rice. However, sometimes, MAS does not perform well in breeding, which might be due to lack of resources, skilled labors, reliable markers, and high costs associated with MAS. Thus, by overcoming these problems, we can enhance the application of MAS in plant breeding, which, in turn, may help us in increasing yield, which subsequently may help in bridging the gap between demand and supply of food for the continuously growing population.
Keywords: Marker-assisted selection, Rice, Gene pyramiding, Grain yield, QTLs mapping for grain yield, Marker-assisted backcross
Introduction
Rice (Oryza sativa L.) is one of the most important food crops across the world and is mainly grown in south Asian countries, specifically in India, China, Sri Lanka, Bangladesh Vietnam, Thailand, Philippines (Gupta et al. 2018, 2019; Donde et al. 2019b). Rice belongs to the family Poaceae. The genus Oryza comprised of twenty-two wild & two cultivated species, namely, Oryza glaberrima and O. sativa (Morishima 1984). Germplasm of all cultivated rice produced in America, Europe, Asia, the Middle East, and Africa mainly belong to O. sativa. Germplasm in West Africa belongs to O. glaberrima. Based on geographical distribution and morphological traits, Oryza sativa was further sub-classified as javanica (or tropical japonica), japonica, and indica (Takahashi 1984). Indica & Japonica are mainly grown in tropical/subtropical and temperate regions. The wild progenitors of Asian rice that are native to southern China are Oryza rufipogan and Oryza nivara, whereas O. glaberrima, known as African rice, mainly originated from the Niger river delta (Fuller 2011).
Though rice serves as a staple food crop across the world, continuous growth in the human population caused a concomitant upsurge in food demand (Wang et al. 2015). In addition to urbanization that is associated with a decrease in cropland, climate change, the occurrence of novel pests & infections are life-threating for stable crop production (Liu et al. 2016). Thus, there is an urgent requirement for the plant breeder to improvise the genetic make-up of crop plants, so it can tolerate biotic as well as abiotic stress condition (Donde et al. 2019a). This, in turn, will help in grain yield enhancement, which serves as one of the best solutions to aid the food demand of the ever-growing population. As rice is one the chief staple food crop for most of the human population, there is an urgent requirement to enhance its production, together with other cereal crops, in order to meet the continuous demand of food supply (Wang et al. 2015). In 2011, Varshney and his team suggested that global rice production needs to be doubled by 2050 for feeding the ever-growing population (Varshney et al. 2011).
Rice yield is generally estimated via the number of tillers, grain number per panicles and the number of spikes present per panicles. Earlier studies have also reported that grain number is directly associated with yield, and hence, it is the most essential trait for grain yield (Hua et al. 2002). The best possible way to increase grain number is by introducing high grain number genes or quantitative trait loci (QTL), like Gn1a, and APO1, into elite rice cultivars; which in turn will increase the food supply (Gouda et al. 2019). Several researchers have also proposed that narrow bottleneck and loss of useful alleles might be responsible for having several unproductive tillers and limited sink size, i.e., small panicles in rice (Khush 1999). Therefore, it is highly essential to identify and use various biomarkers or techniques like conventional breeding and molecular breeding approaches for producing new varieties with high and stable yield (Donde et al. 2019b). To date, numerous approaches, like, marker-assisted selection (MAS), have been developed to introduce these gene/QTLs into rice (Chukwu et al. 2019). Additionally, MAS has also been employed in dissecting several other complex agricultural traits, for instance, drought (Oladosu et al. 2019), cold tolerance (Shakiba et al. 2017), etc. in rice plant. In 2015, Suh and colleauges introduced five biotic stress resistance genes into the japonica rice cultivar employing MAS (Suh et al. 2015). Recently, several other studies have suggested that Hap 3 haplotypes (Ma et al. 2019), and GS3 (Nan et al. 2018) can be employed in MAS for enhancing yield in different rice species. Thus, in this review article, authors made an attempt to collect information about various biomarker associated with grain yield in rice and how MAS techniques can be employed to introduce them in rice (O. sativa L.) plant, which in turn will decrease the gap between food demand and supply of rapidly growing human population.
QTL mapping in rice
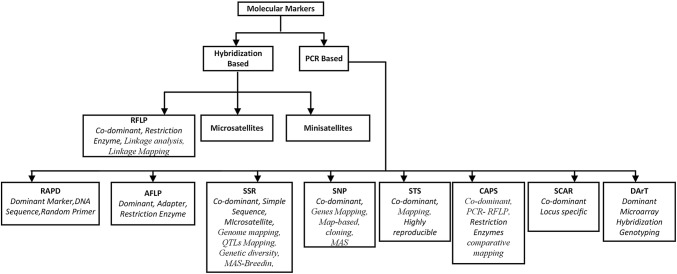
Molecular markers are the fragments of DNA that are associated with a certain location within the genome (Semagn et al. 2006). Molecular markers are either co-dominant or dominant identified through the sequence variation in the number of tandem repeats between nucleotides. Due to highly diverse and reproducible ability in the genome, the molecular markers are used as an ideal marker to study at the genetic level. Molecular markers are grouped by their mode of action to identify a gene and its action for the trait, hybridization-based detection, and transmission pattern of inheritance through the genome. Molecular markers are broadly classified as PCR based and hybridization-based markers (Joshi et al. 1999). The PCR based markers are comprised of amplified fragment length polymorphism (AFLP), single-nucleotide polymorphism (SNP), randomly amplified polymorphic DNA (RAPD), sequence characterized amplified region (SCAR), simple sequence repeats (SSR), cleaved amplified polymorphic sequences (CAPS), sequence-tagged site (STS), diversity array technology (DArT), whereas the hybridization-based markers consist of restriction fragment length polymorphism (RFLP), and mini-satellites (Fig. 1). Till date, several molecular markers associated with various biotic, abiotic stresses, qualitative and quantitative trait including plant height, heading date or other yield traits have been identified by mapping the corresponding QTL (Table 1) (Hittalmani et al. 2003). QTL is a genetic locus that denotes the character and action of several genes. The gene characters are either polygenic or quantitative trait that describes the function of a particular trait, for instance, grain number (Guo et al. 2015).
Fig. 1.
Molecular markers and their type that are used in plant breeding
Table 1.
Information about some important QTL Mapped for grain number and yield related traits in rice
| S.No | Gene/QTL | Trait | Parent | Chromosome no. | Marker interval | Marker type | References |
|---|---|---|---|---|---|---|---|
| 1 | EHD1 | Grain number | Nipponbare | 10 | C1369-C234 | CAPS | Doi et al. (2004) |
| 2 | SPP7 | Grain number | Minghui63 | 7 | R1440-C1023 | STS | Xing et al. (2008) |
| 3 | Ghd7 | Grain number | Minghui63 | 7 | RM5451, RM1135 | SSR | Xue et al. (2008) |
| 4 | Dep1 | Grain number | Shennong265 | 9 | RM3700-7424 | SSR | Huang et al. (2009) |
| 5 | EP3 | Grain number | Milyang23 | 2 | STS5803-5–TS5803-7 | STS | Piao et al. (2009) |
| 6 | Gnp4 | Grain number | Hanzhongxiangnuo | 4 | RM5586-5953 | SSR | Zhang et al. (2011) |
| 7 | LSCHL4 | Grain number | Nipponbare | 4 | STS4-5, RM349 | STS | Zhang et al. (2014) |
| 8 | NGP4 | Grain number | Nipponbare | 4 | RM6314- 5424 | SSR | Zhang et al. (2015) |
| 9 | GNS4 | Grain number | Zhonghua 11 | 4 | LYH54-LYH71 | STS | Zhou et al. (2017) |
| 10 | qTGW10-20.8 | Grain weight | Teqing/IRBB52 | 10 | Te20811-Te20882 | Indel | Zhu et al. (2019) |
QTL for grain number
To date, numerous QTLs, e.g. Gn1a, Dep1, OsSPL14, SCM2, associated with grain number have been identified and cloned (Gouda et al. 2019). Gn1a, which encodes a cytokinin oxidase/dehydrogenase (CKX2), is the first map-based grain number QTL cloned from Habataki utilizing near-isogenic lines (NIL) (Ashikari et al. 2005) and is associated with the depletion of the cytokinin. Reduced CKX2 expression causes cytokinin accretion in the inflorescence meristem, which in turn increases both grain number as well as branches, thereby enhancing grain yield (Ashikari et al. 2005). In 2012, Tsai and the team suggested that an external application of cytokinin may help in modulation gene expression of CKX in both shoots as well as roots of rice (Tsai et al. 2012). In 2013, Li and the team suggested that mutation in the promoter region of the CKX2 gene causes enhanced accretion of cytokinins nearby the apical meristem of rice, which subsequently enhances the production of grain number per panicle (Li et al. 2013). In another study, Yeh et al. (2015) suggested that the CKX2 suppression through RNAi enhances the tiller number as well as grain weight formation, which in turn enhances grain yield.
Huang et al. identified Dep1 for erect and dense panicle as well as high grain number in Shennong 265 (Huang et al. 2009). It controls erect panicle and has a positive effect on grain number via enhancing both primary as well as secondary branches via enhancing the meristematic activity, promoting cell proliferation and encodes an earlier unidentified phosphatidylethanolamine binding protein (PEBP)-like domain protein sharing some homology with the N-terminus of GS3. This QTL is located at chromosome 9 between RM3700 and RM7424. The grain yield is increased by up to 40.9%. Xu et al. (2016) reported that the γ subunit of G protein is involved in the modulation of grain number per panicle, erect panicle, nitrogen uptake, and stress tolerance. The expression study of Dep1 showed a dwarf and shorter panicle (Taguchi-Shiobara et al. 2011).
Miura et al. (2010) identified WFP (Wealthy Farmer’s Panicle) from ST12. It encodes Squamosa Promoter Binding Protein Like14 (a.k.a. OsSPL14) and is located on chromosome 8 mapped between RM223 and RM264. They reported that a higher expression of OsSPL14 is found at the early reproductive stages. OsSPL14 positively regulates panicle branching and grain number per panicle formation at the reproductive stage while negatively regulate the shoot branching in rice at the vegetative stage. Another study has also reported that the point mutation in OsSPL14 disturbs the normal OsmiR156-directed regulation of OsSPL14, which in turn causes high grain yield with reducing tiller number in rice (Jiao et al. 2010).
Another QTL, namely, SCM2, encodes Kelch-repeat containing F-box-containing protein, which helps in the large panicle formation via restricting the precocious conversion of inflorescence meristems into spikelet meristems (Ikeda et al. 2007). SCM2 is identical to APO1 that controls the panicle structure in rice. The panicles of APO1 loss-of-function mutants form fewer spikelets than the wild-type panicle. High expression of SCM2 in mutants increases the spikelet number, mainly via enhancing cell proliferation rate in both the meristem as well as enlarged vascular bundles. Earlier studies have also suggested that that the rice genotype Habataki carrying APO1 allele increases internode diameter, and its overexpression decreases panicle number in rice (Ookawa et al. 2010).
QTLs/genes for tiller number
Tiller number determines the panicle number, which is a key component of rice grain yield (Yan et al. 1998). Tillering traits are highly dependent on when and where lateral shoot branches develop. The development of tillers is affected by various environmental factors including planting density, and climatic conditions such as light, temperature, water supply etc. (Fukushima 2019). Tiller number per plant is a quantitative trait with a relatively low heritability of 29.8–49.6%. To date, many genes have been identified for tillers such as Dwarf 3 (d3), Dwarf 10 (D10), Dwarf 14(D14), Dwarf 17 (D17)/high-tillering Dwarf1 (HTD1), and Dwarf 27(d27).
The D10 was identified through map-based cloning by the backcrossing of Shiokari with Kikeibanshinriki (Ishikawa et al. 2005). This d10 mutant showed an increase in tiller as compare to the wild type. Arite et al. reported that d10 is an ortholog of MAX4, RMS1, and DAD1 in Arabidopsis, pea, and petunia, respectively (Arite et al. 2007). D10 gene plays essential role in strigolactone biosynthesis. Strigolactone hormones are responsible for shoot branching as well as tillering in rice. Zhang et al. reported that the strigolactone regulates the auxin level and helps in the biosynthesis of cytokinin in the shoot nodes and produce more tillers in rice. In Arabidopsis strigolactone have a positive effect on root hair elongation (Zhang et al. 2010).
The d3 gene was developed through map-based cloning and encodes an F-box leucine-rich repeats (LRR). It is an ortholog of MAX2/ORE9 in Arabidopsis that controls the axillary bud activity (Ishikawa et al. 2005). In the development stage of the rice plant, d3 helps hypocotyl elongation. A mutation in d3 produces a defected hypocotyl and decrease in shoot branching and senescence. Another genes, namely, d14 gene belongs to the α/β hydrolase superfamily having serine, histidine and aspartic acid at its hydrophobic sites (de Saint Germain et al. 2016). This d14 gene inhibits rice tillering and act as one key component in the strigolactone-dependent branching inhibition pathway (Arite et al. 2009). It is an orthologous of DAD2 & RMS3 in petunia & pea, respectively, and interacts with the F-box protein to regulate tillering through hydrolyzing strigolactone (Yao et al. 2018). On hydrolytic cleavage of strigolactone, it forms an intermediate D-ring, which enhance the strigolactone signalling in plant. This will increase the tiller number in rice. OsMADS57 gene along with D14 regulates tillering negatively. It regulates the expression of D14 to produce more tillers in rice (Guo et al. 2013). HTD1 is a mutant developed through map-based cloning by backcrossing of Nanjing 6 with Aataiyin 2 at chromosome 4. HTD1 is a recessive gene that controls tillering by producing more axillary buds in rice plants (Zou et al. 2005). It encodes MAX3, an ortholog of Arabidopsis, and is expressed in both root and shoot (Zou et al. 2006). A mutation in the HTD1 gene is reported to produce more tiller and reduces the plant height in rice, which in turn increases yield in rice plants (Zou et al. 2005).
QTLs/genes for heading date
The heading date is another important trait that determines the yield potential in rice. Several genes responsible for heading date have been identified to date. For instance, a QTL for heading date such as qDTH-5 has been identified from the BC2F2 population of Milyang 23 and O. rufipogan cross (Cho 2003). Yano et al. identified 14 QTLs responsible for flowering time and developed NILs by crossing Nipponbare and Kasalath as the donor for Hd3. They suggested that an epistatic interaction between Hd3 and Hd1 results in increased expression of photo periodicity in Nipponbare. Hd1 present in rice is an ortholog of CONSTANS (CO) gene in Arabidopsis and plays a significant role in flowering (Yano et al. 2000). Under short-day conditions, Hd1 promotes floral transition while suppressing under long-day conditions. Doi et al. reported that Ehd1 delayed flowering by activating the expression of Hd3a gene and RFT1 on short-day plants (Doi et al. 2004). It contains a B-type regulator which induces flowering under long and short-day condition. Florigen genes/QTLs were the main factors for flowering. Their expression occurs when it comes in contact with certain environmental signals to inducing flowering in the plant. Rice flowering genes, namely, Rice Flowering Locus T (RFT1) & Hd3a, are involved in the flowering pathway in rice (Komiya et al. 2008). Another QTL, namely, Ghd7, identified in rice, shows a pleiotropic effect on growth such as heading date, plant height and the number of grains per panicle (Xue et al. 2008). It directly affects the grain yield on rice by controlling the grain number. This may regulate the photoperiodic flowering time as well as light signalling on long-day conditions. Ghd7 increases grain number by producing more branches in rice (Brambilla and Fornara 2013). Chakraborty et al. mapped qHD-1-b at chromosome 1 between flanking markers RG109 to ME1014. This QTL is very sensitive to moisture stress found during flowering and results in high floret sterility (Chakraborty and Zeng 2011). A total of 32 QTLs were identified for flowering and grain yield-related traits (Huang et al. 2011).
QTL for other yield-related traits
In 2004, Lanceras and the team identified 77 QTLs for yield components, yield, panicle sterility after crossing IR62266 and CT9993 in rice (Lanceras et al. 2004). Xiao and the team detected 2, 3, 4, 4, 1, 2 and 2 and QTLs for grain yield, ear number per plant, kernel number per ear, 100 kernel weight, cob weight per ear, ear weight, and kernel weight per ear respectively, under irrigated condition (Xiao et al. 2005). In 2006, Yue and the team identified 39 QTLs for productivity, grain weight, leaf rolling, spikelet fertility, harvest index, and biomass, with a population of indica low land × tropical japonica upland. qGL3 has been mapped on chromosome 3 between RMw357- RMw353 (Yue et al. 2006). Gomez identified 24 QTLs associated with numerous plant production as well as physio-morphological traits during drought stress. QTLs number per trait under stress condition was: 1 for panicle length, 4 for leaf drying, 5 for leaf rolling, 5 for plant height, 3 for days to 50% flowering, 1 for straw yield, and 3 for grain yield (Gomez et al. 2006). Venuprasad and team identified DTY3.1 that have strong impact on grain yield under drought stress (Venuprasad et al. 2007). Bernier and the team investigated the impact of qtl12.1 on grain yield as well as its related traits in rice of eastern India as well as Philippines and suggested that the relative impacts of qtl12.1 on grain yield enhance with increasing drought stress intensity, from no effect under irrigated conditions to additive effect of > 40% of trait mean in the utmost harsh treatment (Bernier et al. 2009). Two QTLs, namely, qGYP-2-1 & qGYP-3-1, have been mapped from the backcrossing population BC3F1 of G52-9 and Yuexiangzhan (Jing et al. 2010). These QTLs were mapped on the chromosome 2 and 3 with a marker interval of RM282-RM49 which increases yield from 40.05 to 49.04%. Vikram and colleauges identified a major QTL, namely, qDTY1.1 for grain yield under reproductive-stage drought stress on rice chromosome 1 flanked by RM11943 and RM431 in all three populations. RILs were developed by crossing N22 with Swarna, IR64 and MTU1010. They recorded the phenotypic variation of 13.4%, 16.9% and 12.6% in 13.4%, Swarna, IR6, MTU1010, respectively. Continuous analysis over 2 years reveals that qDTY1.1 have an additive impact of 16.1%, 29.3% and 24.3%, and of mean yield in N22/MTU1010, N22/Swarna and N22/IR64, respectively, under drought stress. qDTY 1.1 also showed a positive impact on grain yield in non-stress situations in N22/Swarna, N22/IR64 and N22/MTU1010 (Vikram et al. 2011).
Cloning of QTLs for yield
Some of the rice QTLs have been identified through positional cloning. For example, Hd1, Hd6, Ghd7, DTH8, Ehd4, and DTH7 have been cloned for heading date in rice (Gao et al. 2014). Few genes controlling panicle length have also been cloned to date. For example, the gene short panicle1 (SP1) encodes a putative polypeptide transporter protein (PTR) that regulates the spike meristem activity, resulting in the short-panicle phenotype (Li et al. 2009). The gene DEP2 encodes a plant-specific protein deprived of any recognized functional domain and is highly essential for modulating panicle outgrowth as well as the elongation (Li et al. 2010). DEP3 encodes a patatin-like phospholipase A2, which plays key role in high grain yield (Qiao et al. 2011). LARGER PANICLE (LP) encodes a Kelch repeat-containing F-box protein, which is highly required for enhancement of both spikelet’s as well as branches and is also involved in regulating cytokinin concentration within plant tissues (Li et al. 2011). Detailed information about some yield-related QTLs identified through cloning is provided in Table 2. Further, the introduction of these QTLs in the rice plant via different approaches, like MAS, may increase yield.
Table 2.
Detailed information about some yield-related QTL identified through cloning
| S.No | Gene/QTL | Trait | Chromosome no. | Donor | Reference |
|---|---|---|---|---|---|
| 1 | Hd1 | Grain number | 6 | Kasalath | Yano et al. (2000) |
| 2 | LAX1 | Grain number | 1 | Zhonghua11 | Komatsu et al. (2001) |
| 3 | Ehd1 | Grain number | 10 | T65 | Doi et al. (2004) |
| 4 | Gn1a | Grain number | 1 | Habataki | Ashikari et al. (2005) |
| 5 | BRD2 | Grain number | 10 | Nipponbare | Hong et al. (2005) |
| 6 | Ghd7 | Grain number | 7 | Minghui63 | Xue et al. (2008) |
| 7 | DEP1 | Grain number | 9 | Shao314 | (Huang et al. 2009) |
| 8 | EP3 | Grain number | 2 | Hwasunchalbyeo | Piao et al. (2009) |
| 9 | DEP3 | Grain number | 6 | Hwacheongbyeo | Piao et al. (2009) |
| 10 | SP1 | Grain number | 11 | Zhonghua11 | Li et al. (2009) |
| 11 | SCM2/APO1 | Grain number | 6 | Habataki | Ookawa et al. (2010) |
| 12 | EP2/DEP2 | Grain number | 7 | Zhonghua11 | Li et al. (2010) |
| 13 | WFP/IPA1 | Grain number | 8 | Shaoniejing | Jiao et al. (2010) |
| 14 | PAP2 | Grain number | 3 | Nipponbare | Kobayashi et al. (2010) |
| 15 | Ghd8/DTH8 | Grain number | 8 | Zhenshan97 | Yan et al. (2011) |
| 16 | LAX2 | Grain number | 4 | Zhonghua11 | Tabuchi et al. (2011) |
| 17 | APO2 | Grain number | 4 | Habataki | Ikeda-Kawakatsu et al. (2012) |
| 18 | Nglf-1(OsARG) | Grain number | 4 | Kitaake | Ma et al. (2013) |
| 19 | FZP | Grain number | 7 | Zhonghua11 | Ren et al. (2018) |
| 20 | D1 | Tiller number | 5 | FL2 | Ashikari et al. (1999) |
| 21 | D2 | Tiller number | 1 | Kasalath | Hong et al. (2003) |
| 22 | D11 | Tiller number | 4 | Taichung65 | Tanabe et al. (2005) |
| 23 | D17/HTD1 | Tiller number | 4 | Nanjing6 | (Zou et al. 2006) |
| 24 | D88 | Tiller number | 3 | Lansheng | Gao et al. (2009) |
| 25 | D14 | Tiller number | 3 | Shiokari | Arite et al. (2009) |
| 26 | D10 | Tiller number | 1 | Indica9 | Yuan et al. (2013) |
| 27 | RGA1 | Panicle number | 4 | IR36 | Seo et al. (1995) |
| 28 | TLD1 | Panicle number | 11 | Hwayoung | Zhang et al. (2009) |
| 29 | ASP1 | Panicle number | 8 | TOS17 | Yoshida et al. (2012) |
| 30 | TAD1 | Panicle number | 3 | Minghui63 | Xu et al. (2012) |
| 31 | DLT | Panicle number | 6 | Zhonghua11 | Xiao et al. (2017) |
| 32 | OsPIN2 | Panicle number | 6 | Heijing2 | Wang et al. (2017) |
| 33 | GW2 | Grain weight | 2 | WY3 | Song et al. (2007) |
| 34 | GW5 | Grain weight | 5 | IR24 | Wan et al. (2008) |
| 35 | GW6a | Grain weight | 6 | WY3 | Qi et al. (2012) |
| 36 | GW8 | Grain weight | 8 | HJX74 | Wang et al. (2012b) |
| 37 | HGW | Grain weight | 6 | Zhonghua11 | Li et al. (2012) |
| 38 | TGW6 | Grain weight | 6 | Kasalath | Ishimaru et al. (2013) |
Marker-assisted selection (MAS)
MAS is a method of trait selection by using molecular markers (Lamont et al. 2014). For the first time, MAS was suggested by Smith and Simpson (1986) as well as by Soller and Beckmann (1990). In MAS, the initial step utilizes both phenotypes as well as genotypes for detecting genetic markers related to any trait(s) or phenotype(s). Subsequently, these markers are introduced into the genetic evaluation as well as selection techniques via computing their impact on the trait(s)/phenotype(s) (Lamont et al. 2014). Thus, it increases the efficiency of plant breeding by transferring the genes/QTLs of interest to the plant genome (Babu et al. 2004). The molecular markers could also help in increasing the efficiency of target gene introgression and recovery from the recurrent parent genome through backcrossing (Hospital 2005). Moreover, reliability, good quality of DNA, skilled marker assay, high level of parental polymorphism, and costs are considered as the five most important criteria for marker consideration (Mackill and Ni 2008). Successful implementation of MAS breeding in broad range of crops like barley, beans, cassava, chickpea, cowpea, groundnut, maize, potato, rice, sorghum, and wheat has been well documented by various researchers (Cobb et al. 2019).
MAS schemes
The two most important MAS schemes in plant breeding are (a) Marker-assisted backcross breeding (MABB) (b) Marker-assisted gene pyramiding. MABB is the most native form of MAS, and its main objective is to incorporate the most important gene from an agronomically inferior source, i.e. the donor parent into the elite cultivar, i.e. the recurrent parent. In this scheme, the final product is the line comprised of only the most essential gene from the donor parent, with the recurrent parent genotype present everywhere else in the genome (Hospital and Charcosset 1997; Hospital 2005). By employing markers specific for target loci could minimize the donor chromosome segment containing the target loci. This, in turn, will reduce the linkage drag and improve rice quality. MABB helps in the recovery of the desired gene by minimizing the chromosome size of the donor parent (Bishwas et al. 2016). MABB involves the successful removal of donor genetic background with the maximum recovery of recurrent parent’s genome. This approach is most widely used for the development of near-isogenic lines (NILs) by minimizing the donor segments flanking the target locus. NILs are the backcrossing mapping populations developed by crossing the donor containing the trait of interest with the recurrent parent. These populations (NILs) are first developed by Tanksley and Nelson through advance backcrossing of QTLs (Tanksley and Nelson 1996). The NILs are isolated through the screening of a large population of about 1000 individuals in BC2 or BC3 generation. Immortal populations were developed through repeated backcrossing through NILs. These are used as a tool for mapping of QTLs and markers linked to target genes in the breeding program. NILs are also used for pyramiding of QTLs and study the epistatic interaction between genes. QTL-NILs are constructed for the development of new varieties through the introgression of new genes in the elite genetic background.
Gene pyramiding is used as an essential approach for crop improvement, and it is more prevalent in cereals like wheat and rice by the introgressing genes of traits into elite varieties. The process of pyramiding involves the transfer of genes of the various traits into a single genotype from multiple parents by MAS. Different QTLs are used for pyramiding to increase yield (Ashikari and Matsuoka 2006). Gene pyramiding aims for (a) enhancing trait performance by combining two or more complementary genes, (b) remedying deficits by introgressing genes from other sources (c) increasing the durability of resistance, and (c) broadening the genetic base. Pyramiding of favourable genes for yield traits will make a valuable contribution to breeding programs to increase yield in plants. It is used to improve yield traits as well as other traits influenced by the environment. The method of gene pyramiding is carried out by using the MAS approach to minimize the breeding time, which in turn will reduce the population size and number of generations to identify the yield trait that has been transferred from an outer source. To date, a large number of genes/QTLs have been tagged with molecular markers to use in MAS for trait improvement (Arunakumari et al. 2016; Balachiranjeevi et al. 2018; Chukwu et al. 2019).
MAS for grain yield
As stated above, successful implementation of MAS breeding in broad range of crops like rice, barley, beans, cassava, chickpea, cowpea, groundnut, maize, potato, sorghum, and wheat has been well documented in different works of literature (Cobb et al. 2019). Both schemes of MAS, namely, MABB and MAP have been extensively employed for various conditions, including abiotic stress (Steele et al. 2006), and grain number (Ashikari et al. 2005).
MABB for grain yield
By using MABB, it possible to enhance yield by introgressing candidate genes in different crop plants. Septiningsih et al. developed BC2F2 populations by crossing IR64 with O. rufipogan (IRGC105491) for identifying QTL associated with different yield traits (Septiningsih et al. 2003). Zhang et al. (2006) developed near-isogenic lines (NILs) by crossing Zhenshan 97 with HR5 for spikelet per panicle (SPP), grains per panicle (GPP), plant height (PH), and heading date (HD). NILs were developed from IR64 and Azucena for four root trait QTLs through backcrossing (Shen et al. 2001). The Doubled haploid lines carrying four QTLs had greater than 50% of the recipient genome, i.e., IR64. Shen et al. (2001) found that the non-target trait association with some NILs results in increased plant height with reduced tiller number. All 9 NILs showed reduced tiller number, whereas three of the four NILs were significantly found taller than IR64. The small effects on QTLs could also be detected by NILs (Keurentjes et al. 2007). Ashikari et al. developed NILs for Gn1a in the background of Nipponbare from donor Habataki (Ashikari et al. 2005). He et al. (2006) developed qGY2-1, a grain yield QTL, by crossing Dongxiang (O. rufipogan) with Guichao2 (indica). Two QTLs, namely, gw8.1 and gw9.1, were developed from a cross between Hwaseongbyeo (Korean japonica cultivar) and IRGC105491 (O. rufipogan) at chromosome 8 and 9 (Xie et al. 2006, 2008). In another study, NILs were developed by transferring gw9.1 QTL from O. rufipogan to Hwaseongbyeo of temperate japonica background (Xie et al. 2008). Seven QTLs were identified in the BC3F4 population for thousand-grain weight, spikelet per panicle, grains per panicle, panicle length, spikelet density, heading date, and plant height. The grain yield increased up to 14–18% in NIL populations as compared to the Hwaseongbyeo parent (Xie et al. 2008).
Bernier et al. developed NILs for qDTY12.1 with average grain yield under drought conditions (Bernier et al. 2009). Zhang et al. (2009) developed 4 NILs in the background of Zhenshan 97 using a donor line of HR5. HR5 was selected as a donor for SPP and GPP QTLs to transfer into Zhenshan 97 background. The developed NILs in the BC4F2 population showed a 50% spikelet per panicle variation and 80% variation found in plant height and heading date. A backcrossing population BC4F2 was generated by crossing IR24 with Asominori (Wan et al. 2006). This showed a phenotypic variation of 33–35%. Wang et al. (2017) introgressed two yield genes secondary branch number (SBN) and spikelet per panicle (SPP) to a japonica cultivar Tainan 13 from the donor IR65598-112-2 a high yielding rice genotype. They found that the BC2F5 lines have an increase in SPP and SBN by 83.2% and 61%, respectively. Whereas NILs were developed for qSBN1 and qPBN6 from a cross between Sasanishiki, a japonica cultivar, with indica cultivar Habataki (Ando et al. 2008). The spikelet per panicle was reported 30% higher than Habataki. Luo et al. (2013) developed NILs by backcrossing of Hwayeongbyeo and O. rufipogan. They introgressed qSPP5 and qTGW5 into the Hwayeongbyeo background. An increase in 10-15 spikelets had been found in the NILs.
Imai et al. (2013) introgressed six yield QTLs yld1.1, yld2.1, yld3.2, yld6.1, yld8.1, yld9.1 to Jefferson background from O. rufipogan through MABB. All the lines showed a better yield as a comparison to control. The NILs carrying the yld2.1 and yld6.1 showed an increased yield percentage of 27.7 and 26.1, respectively. They also found that only the yld9.1 QTL containing ILs are showing lodging as compared to the wild parent. Singh et al. introgressed qGn4.1 for grain number per panicle into 12 rice mega varieties through marker-assisted backcross breeding (Singh et al. 2018). The BC2F3 plants showed an increase in grain number per panicle in all 12 backgrounds. Mishra et al. (2013) developed the backcross inbred line by crossing IR74371-46-1-1 and Sabitri for qDTY12.1 responsible for grain yield in the reproductive drought stress condition. A 45.3% grain yield found on drought, Henry et al. (2014) introgressed qDTY12.1 into the Vandana background. The NILs showed a higher yield under drought. Two QTLs qDTY3.2 and qDTY12.1vwere transferred to Sabitri, an upland variety of Nepal. The NILs developed through backcrossing showed early flowering up to 13 days and reduction of plant height up to 13 cm as compared to Sabitri (Dixit et al. 2017). Various other QTLs identified and mapped through MABB approach is depicted in Table 3.
Table 3.
Identification and mapping of QTLs through MABB approach
| S.No | QTLs | Cross | Population | Marker Interval | References |
|---|---|---|---|---|---|
| 1 | qGL-3a | Asominori/IR24 | BC4F2 | RMw357-RMw353 | Wan et al. (2006) |
| 2 | GW9.1 | Hwaseongbyeo/O.rufipogan | BC3F4 | RM24718.CNR11- RM30005CNR142 | Xie et al. (2008) |
| 3 | qSPP1 | Zhenshan97/HR5 | BC4F2 | MRG2746-RM490 | Zhang et al. (2009) |
| 4 | qGPP1 | Zhenshan97/HR5 | BC4F2 | MRG2746-RM490 | Zhang et al. (2009) |
| 5 | GW3 | Baodali/Zhongua | BC2F2 | WGW16-WGW19 | Gou et al. (2009) |
| 6 | qGY2-1 | Yuexiangzhan/G52-9 | BC3F3 | RM110-RM211 | Jing et al. (2010) |
| 7 | qGYP3-1 | Yuexiangzhan/G52-9 | BC3F3 | RM282-RM49 | Jing et al. (2010) |
| 8 | qDTH3.1, qDTH4.1, qDTH6.1 | Swarna/O. nivara | BC2F2 | RM22-RM517, RM185-RM204, RM314-RM241 | Swamy et al. (2011) |
| 9 | qNPB2.1, qNPB7.1, | Swarna/O. nivara | BC2F2 | RM3874-RM6, RM172-RM248 | Swamy et al. (2011) |
| 10 | qSPP5 | Hwayeongbyeo/CR6 | BC2F4 | RM413-RM194 | Luo et al. (2013) |
| 11 | qTGW5 | Hwayeongbyeo/CR7111-30 | BC4F3 | RM194 | Luo et al. (2013) |
Marker-assisted gene pyramiding for grain yield
Gene pyramiding is widely applicable in traits that are inherited but challenging to measure their phenotypic expression. Traits are improved by gene pyramiding if the gene affecting the particular traits are identified using functional markers. Genes that are difficult to transfer in a single genotype through conventional breeding are possible through marker-assisted gene pyramiding. Ashikari et al. (2006) pyramided Gn1a and Ph1 associated with grain number and plant height, respectively, by crossing between Koshihikari and Habataki to produce the NIL. The resultant NIL showed an increased grain number with reduced plant height. The pyramided lines showed an increased grain production of up to 23%, and plant height reduced to 20% as compared to Koshihikari. Ohsumi et al. (2011) developed NILs by backcrossing of Sasanishiki with high yielding parent Habataki. They pyramided two QTLs, namely, qSBN1 and qPBN6, responsible for primary and secondary branch formation in rice plant. The pyramiding lines carrying two QTLs showed a 62–65% increase in spikelet per panicle whereas single QTL qSBN1 produced 28–37% and qPBN6 9–16%. A yield recorded higher up to 4–12% due to carbohydrate translocation from stem to panicle. Zong et al. (2012) pyramided 4 QTLs for spikelet per panicle qSN-1a, qSN-1b, qSN-6, qSN-8 and 4 QTLs for 1000 grain weight qTGW5a, qTGW5b, qTGW8, qTGW10 by crossing RILS of AW35 × AW208 and AW51 × AW208. This shows an increase is grain number up to 10% with a grain weight of 8% in rice. Wang et al. pyramided QHD8 and GS3 in Zhenshan97 from donor parent 93–11 (Wang et al. 2012a). These pyramided lines show longer grains with higher yield per plant by 53%, spikelet number and flag leaf were increased by 14 and 30%. Yano et al. identified the strong culm QTL (SCM3) which is identical to OsTB1 control strigolactone signalling in rice (Yano et al. 2015). They developed NILs having two QTLs, namely, SCM2 and SCM3. The single QTLs showed strong culm as well as increased spikelet number, whereas two QTLs (SCM2 +SCM3) showed much stronger culm than single QTL carrying pyramided lines.
Shamsudin et al. pyramided three drought QTLs qDYT2.2, qDYT3.1, qDYT12.1 into MR219 background for yield. qDYT2.2and qDYT3.1 showed a positive effect for grain yield in low land areas (Shamsudin et al. 2016). Two QTLs (qDYT2.2 + qDYT3.1, qDYT2.2 + qDYT12.1, qDYT3.1 + qDYT12.1) showed an increase in grain yield under reproductive drought stress condition as compared to three QTLs combinations. Kumar et al. pyramided drought and submergence QTLs in MR219, Swarnasub1, Samba Mahsuri. The pyramided lines containing qDTY12.1 + qDYT3.1and qDYT2.2 + qDYT3.1 showed high yield under RS and NS condition (Kumar et al. 2018). Two drought QTLs qDTY12.1 and qDTY2.3 were introgressed into Funnabor 2 through the backcrossing method (Anyaoha et al. 2019). The pyramiding lines carrying these two QTLs show higher grain yield under drought and normal conditions. Thus, the scheme associated with MAS may serve as an important tool in plant breeding to increase yield by modulating various traits. However, as nothing is ideal or perfect in this world, MAS has certain limitations in plant breeding approaches.
Limitation and future perspectives
Irrespective of all advantages mentioned above, MAS still has certain limitations, which in turn may cause failure in breeding, for instance, lack of resources, skilled labors, reliable markers, and high costs associated with MAS. The most important expenditure involved prior to MAS is the generation of a genetic linkage map for the species of interest and recognition of linkage amongst genes/QTLs and economically important traits. Such expenditure could be significantly high for developing countries. In 2003, Dreher and the team suggested that these costs could be significantly reduced by increasing marker number considering the economies of scale as well as the absence of divisibility associated with numerous components of MAS (Dreher et al. 2003). This can be best achieved by establishing marker genotyping companies, which in turn will help marker genotyping to be outsourced. Presuming costs associated with outsourcing genotyping to be cheap, absence or presence of minimum logistic problems, this will enhance use of MAS in plant breeding (Collard and Mackill 2008). Thus, authors believe that formation of marker genotyping facilities as well as staff training in various rice breeding institutes across globe, identification of genes/QTLs controlling traits and tightly-linked markers, establishment as well as annotation of publicly available QTL/marker associated databases and generating new markers from DNA sequence data employing high throughput sequencing technology may enhance the application of MAS and, in turn, may help us in increasing grain yield.
Conclusion
In conclusion, it is well established that genetic diversity is the major factor in crop improvement programs, and MAS serves as the primary technique for bringing this diversity into breeding programs deprived of the associated linkage drag from otherwise poor-quality genomes of donor varieties. Molecular breeding plays an essential role in crop improvement by producing NILs, RILs and BILs populations and were used in QTLs and gene identifications. Therefore, MAS has a great role in discovering new QTLs, genes, and alleles responsible for increasing grain yield as well as biotic and abiotic stress tolerance of crops.
Abbreviations
- AFLP
Amplified fragment length polymorphism
- BC
Back cross
- BILs
Backcross inbred lines
- CAPS
Cleaved amplified polymorphic sequences
- GDP
Gross domestic product
- GPP
Grain number per panicle
- HD
Heading date
- MABB
Marker assisted backcross breeding
- MAP
Marker assisted gene pyramiding
- MAS
Marker assisted selection
- MDP
Marker data point
- mha
Million hector
- NILs
Near isogenic lines
- PH
Plant height
- QTL
Quantitative trait loci
- RAPD
Randomly amplified polymorphic DNA
- RFLP
Restriction fragment length polymorphism
- RILs
Recombinant inbred lines
- SCAR
Sequence characterized amplified region
- SNP
Single-nucleotide polymorphism
- SPP
Spikelet per panicle
- SSR
Simple sequence repeat
- STS
Sequence-tagged site
- UTR
Untranslated region
- VNTR
Variable number of tandem repeats
Funding
The authors acknowledge the Department of Science and Technology (DST), Government of India, New Delhi, for providing financial assistance through Grant number: DST/INSPIRE Fellowship/2013/992 to Inspire Fellow Miss. Gayatri Gouda
Compliance with ethical standards
Conflict of interest
None.
Ethical statement
This article does not contain any studies with human participants or animals.
Informed consent
This article does not require consent from any individual or organisation.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Manoj Kumar Gupta and Ravindra Donde have contributed equally to this work.
References
- Ando T, Yamamoto T, Shimizu T, et al. Genetic dissection and pyramiding of quantitative traits for panicle architecture by using chromosomal segment substitution lines in rice. Theor Appl Genet. 2008;116:881–890. doi: 10.1007/s00122-008-0722-6. [DOI] [PubMed] [Google Scholar]
- Anyaoha CO, Fofana M, Gracen V, et al. Introgression of two drought QTLs into FUNAABOR-2 early generation backcross progenies under drought stress at reproductive stage. Rice Sci. 2019;26:32–41. doi: 10.1016/j.rsci.2018.04.006. [DOI] [Google Scholar]
- Arite T, Iwata H, Ohshima K, et al. DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J. 2007;51:1019–1029. doi: 10.1111/j.1365-313X.2007.03210.x. [DOI] [PubMed] [Google Scholar]
- Arite T, Umehara M, Ishikawa S, et al. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 2009;50:1416–1424. doi: 10.1093/pcp/pcp091. [DOI] [PubMed] [Google Scholar]
- Arunakumari K, Durgarani CV, Satturu V, et al. Marker-assisted pyramiding of genes conferring resistance against bacterial blight and blast diseases into Indian rice variety MTU1010. Rice Sci. 2016;23:306–316. doi: 10.1016/j.rsci.2016.04.005. [DOI] [Google Scholar]
- Ashikari M, Matsuoka M. Identification, isolation and pyramiding of quantitative trait loci for rice breeding. Trends Plant Sci. 2006;11:344–350. doi: 10.1016/j.tplants.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Ashikari M, Sakakibara H, Lin S, et al. Cytokinin oxidase regulates rice grain production. Science. 2005;309:741–745. doi: 10.1126/science.1113373. [DOI] [PubMed] [Google Scholar]
- Ashikari M, Wu J, Yano M, et al. Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the α-subunit of GTP-binding protein. Proc Natl Acad Sci USA. 1999;96:10284–10289. doi: 10.1073/pnas.96.18.10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu R, Nair SK, Prasanna BM, Gupta HS. Integrating marker-assisted selection in crop breeding—prospects and challenges. Curr Sci. 2004;87:14. [Google Scholar]
- Balachiranjeevi CH, Naik B, Kumar A, et al. Marker-assisted pyramiding of two major, broad-spectrum bacterial blight resistance genes, Xa21 and Xa33 into an elite maintainer line of rice, DRR17B. PLoS ONE. 2018;13:e0201271. doi: 10.1371/journal.pone.0201271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier J, Kumar A, Venuprasad R, et al. Characterization of the effect of a QTL for drought resistance in rice, qtl12. 1, over a range of environments in the Philippines and eastern India. Euphytica. 2009;166:207–217. doi: 10.1007/s10681-008-9826-y. [DOI] [Google Scholar]
- Bishwas N, Sharma M, Hasan A, et al. Improvement of rice crop by marker-assisted backcross method. Magnesium. 2016;03:8. [Google Scholar]
- Brambilla V, Fornara F. Molecular control of flowering in response to day length in rice: control of flowering in rice. J Integr Plant Biol. 2013;55:410–418. doi: 10.1111/jipb.12033. [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Zeng ZB. QTL mapping for days to flowering under drought condition in rice (Oryza sativa L.) genome. Notulae Botanicae Horti Agrobotanici Cluj-Napoca. 2011;39:58–63. doi: 10.15835/nbha3915610. [DOI] [Google Scholar]
- Cho YC. QTLs analysis of yield and its related traits in wild rice relative Oryza rufipogon. Treat Crop Res. 2003;4:19–29. [Google Scholar]
- Chukwu SC, Rafii MY, Ramlee SI, et al. Marker-assisted selection and gene pyramiding for resistance to bacterial leaf blight disease of rice (Oryza sativa L.) Biotechnol Biotechnol Equip. 2019;33:440–455. doi: 10.1080/13102818.2019.1584054. [DOI] [Google Scholar]
- Cobb JN, Biswas PS, Platten JD. Back to the future: revisiting MAS as a tool for modern plant breeding. Theor Appl Genet. 2019;132:647–667. doi: 10.1007/s00122-018-3266-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collard BCY, Mackill DJ. Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans R Soc Lond B Biol Sci. 2008;363:557–572. doi: 10.1098/rstb.2007.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Germain A, Clavé G, Badet-Denisot M-A, et al. An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat Chem Biol. 2016;12:787–794. doi: 10.1038/nchembio.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit S, Yadaw RB, Mishra KK, Kumar A. Marker-assisted breeding to develop the drought-tolerant version of Sabitri, a popular variety from Nepal. Euphytica. 2017;213:184. doi: 10.1007/s10681-017-1976-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi K, Izawa T, Fuse T, et al. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004;18:926–936. doi: 10.1101/gad.1189604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donde R, Gupta MK, Gouda G, et al. Computational characterization of structural and functional roles of DREB1A, DREB1B and DREB1C in enhancing cold tolerance in rice plant. Amino Acids. 2019;51:839–853. doi: 10.1007/s00726-019-02727-0. [DOI] [PubMed] [Google Scholar]
- Donde R, Kumar J, Gouda G, et al. Assessment of genetic diversity of drought tolerant and susceptible rice genotypes using microsatellite markers. Rice Sci. 2019;26:239–247. doi: 10.1016/j.rsci.2019.01.004. [DOI] [Google Scholar]
- Dreher K, Khairallah M, Ribaut J-M, Morris M. Money matters (I): costs of field and laboratory procedures associated with conventional and marker-assisted maize breeding at CIMMYT. Mol Breed. 2003;11:221–234. doi: 10.1023/A:1022820520673. [DOI] [Google Scholar]
- Fukushima A. Varietal differences in tiller and panicle development determining the total number of spikelets per unit area in rice. Plant Prod Sci. 2019;22:192–201. doi: 10.1080/1343943X.2018.1562308. [DOI] [Google Scholar]
- Fuller DQ. Pathways to asian civilizations: tracing the origins and spread of rice and rice cultures. Rice. 2011;4:78–92. doi: 10.1007/s12284-011-9078-7. [DOI] [Google Scholar]
- Gao Z, Qian Q, Liu X, et al. Dwarf 88, a novel putative esterase gene affecting architecture of rice plant. Plant Mol Biol. 2009;71:265–276. doi: 10.1007/s11103-009-9522-x. [DOI] [PubMed] [Google Scholar]
- Gao H, Jin M, Zheng X-M, et al. Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. PNAS. 2014;111:16337–16342. doi: 10.1073/pnas.1418204111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez MS, Kumar SS, Jeyaprakash P, et al. Mapping QTLs linked to physio-morphological and plant production traits under drought stress in rice (Oryza sativa L.) in the target environment. Am J Biochem Biotechnol. 2006;2:161–169. doi: 10.3844/ajbbsp.2006.161.169. [DOI] [Google Scholar]
- Gouda G, Gupta MK, Donde R, et al. Computational approach towards understanding structural and functional role of cytokinin oxidase/dehydrogenase 2 (CKX2) in enhancing grain yield in rice plant. J Biomol Struct Dyn. 2019 doi: 10.1080/07391102.2019.1597771. [DOI] [PubMed] [Google Scholar]
- Guo S, Xu Y, Liu H, et al. The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat Commun. 2013;4:1–12. doi: 10.1038/ncomms2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Ku L, Qi J, et al. Genetic analysis and major quantitative trait locus mapping of leaf widths at different positions in multiple populations. PLoS ONE. 2015;10:e0119095. doi: 10.1371/journal.pone.0119095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta MK, Vadde R, Donde R, et al. Insights into the structure–function relationship of brown plant hopper resistance protein, Bph14 of rice plant: a computational structural biology approach. J Biomol Struct Dyn. 2018 doi: 10.1080/07391102.2018.1462737. [DOI] [PubMed] [Google Scholar]
- Gupta MK, Vadde R, Gouda G, et al. Computational approach to understand molecular mechanism involved in BPH resistance in Bt-rice plant. J Mol Graph Model. 2019;88:209–220. doi: 10.1016/j.jmgm.2019.01.018. [DOI] [PubMed] [Google Scholar]
- He G, Luo X, Tian F, et al. Haplotype variation in structure and expression of a gene cluster associated with a quantitative trait locus for improved yield in rice. Genome Res. 2006;16:618–626. doi: 10.1101/gr.4814006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry A, Dixit S, Mandal NP, et al. Grain yield and physiological traits of rice lines with the drought yield QTL qDTY12.1 showed different responses to drought and soil characteristics in upland environments. Funct Plant Biol. 2014;41:1066–1077. doi: 10.1071/FP13324. [DOI] [PubMed] [Google Scholar]
- Hittalmani S, Huang N, Courtois B, et al. Identification of QTL for growth—and grain yield-related traits in rice across nine locations of Asia. TAG Theor Appl Genet. 2003;107:679–690. doi: 10.1007/s00122-003-1269-1. [DOI] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Umemura K, et al. A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell. 2003;15:2900–2910. doi: 10.1105/tpc.014712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Ueguchi-Tanaka M, Fujioka S, et al. The rice brassinosteroid-deficient dwarf2 Mutant, defective in the rice homolog of arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell. 2005;17:2243–2254. doi: 10.1105/tpc.105.030973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospital F. Selection in backcross programmes. Philos Trans R Soc B Biol Sci. 2005;360:1503–1511. doi: 10.1098/rstb.2005.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospital F, Charcosset A. Marker-assisted Introgression of quantitative trait loci. Genetics. 1997;147:1469–1485. doi: 10.1093/genetics/147.3.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua JP, Xing YZ, Xu CG, et al. Genetic dissection of an elite rice hybrid revealed that heterozygotes are not always advantageous for performance. Genetics. 2002;162:1885–1895. doi: 10.1093/genetics/162.4.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Qian Q, Liu Z, et al. Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet. 2009;41:494–497. doi: 10.1038/ng.352. [DOI] [PubMed] [Google Scholar]
- Huang X, Zhao Y, Wei X, et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat Genet. 2011;44:32–39. doi: 10.1038/ng.1018. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Ito M, Nagasawa N, et al. Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate: APO1 regulates meristem fate in rice. Plant J. 2007;51:1030–1040. doi: 10.1111/j.1365-313X.2007.03200.x. [DOI] [PubMed] [Google Scholar]
- Ikeda-Kawakatsu K, Maekawa M, Izawa T, et al. ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1: Characterization of rice APO2/RFL gene. Plant J. 2012;69:168–180. doi: 10.1111/j.1365-313X.2011.04781.x. [DOI] [PubMed] [Google Scholar]
- Imai I, Kimball JA, Conway B, et al. Validation of yield-enhancing quantitative trait loci from a low-yielding wild ancestor of rice. Mol Breed. 2013;32:101–120. doi: 10.1007/s11032-013-9855-7. [DOI] [Google Scholar]
- Ishikawa S, Maekawa M, Arite T, et al. Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol. 2005;46:79–86. doi: 10.1093/pcp/pci022. [DOI] [PubMed] [Google Scholar]
- Ishimaru K, Hirotsu N, Madoka Y, et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat Genet. 2013;45:707–711. doi: 10.1038/ng.2612. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet. 2010;42:541–544. doi: 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- Jing Z, Qu Y, Yu C, et al. QTL analysis of yield-related traits using an advanced backcross population derived from common wild rice (Oryza rufipogon L) Mol Plant Breed. 2010 doi: 10.5376/mpb.2010.01.0001. [DOI] [Google Scholar]
- Joshi SP, Ranjekar PK, Gupta VS. Molecular markers in plant genome analysis. Curr Sci. 1999;77:230–240. [Google Scholar]
- Keurentjes JJB, Bentsink L, Alonso-Blanco C, et al. Development of a near-isogenic line population of arabidopsis thaliana and comparison of mapping power with a recombinant inbred line population. Genetics. 2007;175:891–905. doi: 10.1534/genetics.106.066423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khush GS. Green revolution: preparing for the 21st century. Genome. 1999;42:10. doi: 10.1139/g99-044. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Maekawa M, Miyao A, et al. PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol. 2010;51:47–57. doi: 10.1093/pcp/pcp166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Maekawa M, Shimamoto K, Kyozuka J. The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis-branch and spikelet development. Dev Biol. 2001;231:364–373. doi: 10.1006/dbio.2000.9988. [DOI] [PubMed] [Google Scholar]
- Komiya R, Ikegami A, Tamaki S, et al. Hd3a and RFT1 are essential for flowering in rice. Development. 2008;135:767–774. doi: 10.1242/dev.008631. [DOI] [PubMed] [Google Scholar]
- Kumar A, Sandhu N, Dixit S, et al. Marker-assisted selection strategy to pyramid two or more QTLs for quantitative trait-grain yield under drought. Rice. 2018;11:35. doi: 10.1186/s12284-018-0227-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont SJ, Dekkers JCM, Zhou H. Chapter 11—immunogenetics and the mapping of immunological functions. In: Schat KA, Kaspers B, Kaiser P, editors. Avian immunology. 2. Boston: Academic Press; 2014. pp. 205–221. [Google Scholar]
- Lanceras JC, Pantuwan G, Jongdee B, Toojinda T. Quantitative trait loci associated with drought tolerance at reproductive stage in rice. Plant Physiol. 2004;135:384–399. doi: 10.1104/pp.103.035527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Qian Q, Fu Z, et al. Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. Plant J. 2009;58:592–605. doi: 10.1111/j.1365-313X.2009.03799.x. [DOI] [PubMed] [Google Scholar]
- Li F, Liu W, Tang J, et al. Rice DENSE AND ERECT PANICLE 2 is essential for determining panicle outgrowth and elongation. Cell Res. 2010;20:838–849. doi: 10.1038/cr.2010.69. [DOI] [PubMed] [Google Scholar]
- Li M, Tang D, Wang K, et al. Mutations in the F-box gene LARGER PANICLE improve the panicle architecture and enhance the grain yield in rice. Plant Biotechnol J. 2011;9:1002–1013. doi: 10.1111/j.1467-7652.2011.00610.x. [DOI] [PubMed] [Google Scholar]
- Li J, Chu H, Zhang Y, et al. The rice HGW gene encodes a ubiquitin-associated (UBA) domain protein that regulates heading date and grain weight. PLoS ONE. 2012;7:e34231. doi: 10.1371/journal.pone.0034231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhao B, Yuan D, et al. Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression. Proc Natl Acad Sci USA. 2013;110:3167–3172. doi: 10.1073/pnas.1300359110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wei X, Sheng Z, et al. Polycomb protein OsFIE2 affects plant height and grain yield in rice. PLoS ONE. 2016;11:e0164748. doi: 10.1371/journal.pone.0164748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Ji S-D, Yuan P-R, et al. QTL mapping reveals a tight linkage between QTLs for grain weight and panicle spikelet number in rice. Rice (N Y) 2013;6:33. doi: 10.1186/1939-8433-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Cheng Z, Qin R, et al. OsARG encodes an arginase that plays critical roles in panicle development and grain production in rice. Plant J. 2013;73:190–200. doi: 10.1111/j.1365-313x.2012.05122.x. [DOI] [PubMed] [Google Scholar]
- Ma X, Feng F, Zhang Y, et al. A novel rice grain size gene OsSNB was identified by genome-wide association study in natural population. PLoS Genet. 2019;15:e1008191. doi: 10.1371/journal.pgen.1008191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackill DJ, Ni J. Molecular mapping and marker-assisted selection for major-gene traits in rice. In: Khush GS, Brar DS, Hardy B, editors. Rice genetics IV. USA: World Scientific Publishing Company; 2008. pp. 137–151. [Google Scholar]
- Mishra KK, Vikram P, Yadaw RB, et al. qDTY12.1: a locus with a consistent effect on grain yield under drought in rice. BMC Genet. 2013;14:12. doi: 10.1186/1471-2156-14-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Ikeda M, Matsubara A, et al. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet. 2010;42:545–549. doi: 10.1038/ng.592. [DOI] [PubMed] [Google Scholar]
- Morishima H (1984) Species relationships and the search for ancestor. Biol Rice 3–30
- Nan J, Feng X, Wang C, et al. Improving rice grain length through updating the GS3 locus of an elite variety Kongyu 131. Rice. 2018;11:21. doi: 10.1186/s12284-018-0217-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi A, Takai T, Ida M, et al. Evaluation of yield performance in rice near-isogenic lines with increased spikelet number. Field crops Res. 2011;120(1):68–75. doi: 10.1016/j.fcr.2010.08.013. [DOI] [Google Scholar]
- Oladosu Y, Rafii MY, Samuel C, et al. Drought resistance in rice from conventional to molecular breeding: a review. Int J Mol Sci. 2019 doi: 10.3390/ijms20143519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ookawa T, Hobo T, Yano M, et al. New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nat Commun. 2010;1:1–11. doi: 10.1038/ncomms1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao R, Jiang W, Ham T-H, et al. Map-based cloning of the ERECT PANICLE 3 gene in rice. Theor Appl Genet. 2009;119:1497–1506. doi: 10.1007/s00122-009-1151-x. [DOI] [PubMed] [Google Scholar]
- Qi P, Lin Y-S, Song X-J, et al. The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 2012;22:1666–1680. doi: 10.1038/cr.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Piao R, Shi J, et al. Fine mapping and candidate gene analysis of dense and erect panicle 3, DEP3, which confers high grain yield in rice (Oryza sativa L.) Theor Appl Genet. 2011;122:1439–1449. doi: 10.1007/s00122-011-1543-6. [DOI] [PubMed] [Google Scholar]
- Ren D, Hu J, Xu Q, et al. FZP determines grain size and sterile lemma fate in rice. J Exp Bot. 2018;69:4853–4866. doi: 10.1093/jxb/ery264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semagn K, Bjørnstad Å, Ndjiondjop MN. An overview of molecular marker methods for plants. Afr J Biotechnol. 2006;525(25):2540–2568. [Google Scholar]
- Seo H-S, Kim H-Y, Jeong J-Y, et al. Molecular cloning and characterization of RGA1 encoding a G protein a subunit from rice (Oryza sativa L. IR-36) Plant Mol Biol. 1995;27(6):1119–1131. doi: 10.1007/BF00020885. [DOI] [PubMed] [Google Scholar]
- Septiningsih EM, Prasetiyono J, Lubis E, et al. Identification of quantitative trait loci for yield and yield components in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon. Theor Appl Genet. 2003;107:1419–1432. doi: 10.1007/s00122-003-1373-2. [DOI] [PubMed] [Google Scholar]
- Shakiba E, Edwards JD, Jodari F, et al. Genetic architecture of cold tolerance in rice (Oryza sativa) determined through high resolution genome-wide analysis. PLoS ONE. 2017;12:e0172133. doi: 10.1371/journal.pone.0172133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamsudin NAA, Swamy BPM, Ratnam W, et al. Marker assisted pyramiding of drought yield QTLs into a popular Malaysian rice cultivar, MR219. BMC Genet. 2016;17:30. doi: 10.1186/s12863-016-0334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Courtois B, McNally KL, et al. Evaluation of near-isogenic lines of rice introgressed with QTLs for root depth through marker-aided selection. Theor Appl Genet. 2001;103:75–83. doi: 10.1007/s001220100538. [DOI] [Google Scholar]
- Singh VK, Ellur RK, Singh AK, et al. Effect of qGN4.1 QTL for grain number per panicle in genetic backgrounds of twelve different mega varieties of rice. Rice. 2018;11:8. doi: 10.1186/s12284-017-0195-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Simpson SP. The use of genetic polymorphisms in livestock improvement. J Anim Breed Genet. 1986;103:205–217. doi: 10.1111/j.1439-0388.1986.tb00083.x. [DOI] [Google Scholar]
- Soller M, Beckmann JS (1990) Molecular mapping of quantitative genes. In: Proceedings of the world congress on genetics applied to livestock production 96
- Song X-J, Huang W, Shi M, et al. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet. 2007;39:8. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- Steele KA, Price AH, Shashidhar HE, Witcombe JR. Marker-assisted selection to introgress rice QTLs controlling root traits into an Indian upland rice variety. Theor Appl Genet. 2006;112:208–221. doi: 10.1007/s00122-005-0110-4. [DOI] [PubMed] [Google Scholar]
- Suh J-P, Cho Y-C, Won Y-J, et al. Development of resistant gene-pyramided japonica rice for multiple biotic stresses using molecular marker-assisted selection. Plant Breed Biotech. 2015;3:333–345. doi: 10.9787/PBB.2015.3.4.333. [DOI] [Google Scholar]
- Swamy BPM, Kaladhar K, Ramesha MS, et al. Molecular mapping of QTLs for yield and yield-related traits in oryza sativa cv swarna × O. nivara (IRGC81848) backcross population. Rice Sci. 2011;18:178–186. doi: 10.1016/S1672-6308(11)60025-5. [DOI] [Google Scholar]
- Tabuchi H, Zhang Y, Hattori S, et al. LAX PANICLE2 of Rice Encodes a Novel Nuclear Protein and Regulates the Formation of AxillaryMeristems[W] Plant Cell. 2011;23:3276–3287. doi: 10.1105/tpc.111.088765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi-Shiobara F, Kawagoe Y, Kato H, et al. A loss-of-function mutation of rice DENSE PANICLE 1 causes semi-dwarfness and slightly increased number of spikelets. Breed Sci. 2011;61:17–25. doi: 10.1270/jsbbs.61.17. [DOI] [Google Scholar]
- Takahashi N. Differentiation of ecotypes in Oryza Sativa L. In: Tsunoda S, Takahashi N, editors. Developments in crop science. Amsterdam: Elsevier; 1984. pp. 31–67. [Google Scholar]
- Tanabe S, Ashikari M, Fujisawa S, Takatsuto S (2005) A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length | plant cell. http://www.plantcell.org/content/17/3/776. Accessed 13 Nov 2019 [DOI] [PMC free article] [PubMed]
- Tanksley SD, Nelson JC. Advanced backcross QTL analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor Appl Genet. 1996;92:191–203. doi: 10.1007/BF00223376. [DOI] [PubMed] [Google Scholar]
- Tsai Y-C, Weir NR, Hill K, et al. Characterization of genes involved in cytokinin signaling and metabolism from rice1[W][OA] Plant Physiol. 2012;158:1666–1684. doi: 10.1104/pp.111.192765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney RK, Bansal KC, Aggarwal PK, et al. Agricultural biotechnology for crop improvement in a variable climate: hope or hype? Trends Plant Sci. 2011;16:363–371. doi: 10.1016/j.tplants.2011.03.004. [DOI] [PubMed] [Google Scholar]
- Venuprasad R, Zenna N, Choi I-R, et al (2007) Identification of marker loci associated with tungro and drought tolerance in near-isogenic rice lines derived from IR64/Aday Sel. Int Rice Res Notes
- Vikram P, Swamy BM, Dixit S, et al. qDTY 1.1, a major QTL for rice grain yield under reproductive-stage drought stress with a consistent effect in multiple elite genetic backgrounds. BMC Genet. 2011;12:89. doi: 10.1186/1471-2156-12-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan XY, Wan JM, Jiang L, et al. QTL analysis for rice grain length and fine mapping of an identified QTL with stable and major effects. Theor Appl Genet. 2006;112:1258–1270. doi: 10.1007/s00122-006-0227-0. [DOI] [PubMed] [Google Scholar]
- Wan X, Weng J, Zhai H, et al. Quantitative trait loci (QTL) analysis for rice grain width and fine mapping of an identified QTL allele gw-5 in a recombination hotspot region on chromosome 5. Genetics. 2008;179(4):2239–2252. doi: 10.1534/genetics.108.089862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Xing Y, Li Z, Yu S. Improving rice yield and quality by QTL pyramiding. Mol Breed. 2012;29:903–913. doi: 10.1007/s11032-011-9679-2. [DOI] [Google Scholar]
- Wang S, Wu K, Yuan Q, Liu X, et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet. 2012;44(8):950–954. doi: 10.1038/ng.2327. [DOI] [PubMed] [Google Scholar]
- Wang J, Xu H, Li N, et al. Artificial selection of gn1a plays an important role in improving rice yields across different ecological regions. Rice. 2015;8:37. doi: 10.1186/s12284-015-0071-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S-S, Chen R-K, Chen K-Y, et al. Genetic mapping of the qSBN7 locus, a QTL controlling secondary branch number per panicle in rice. Breed Sci. 2017;67:340–347. doi: 10.1270/jsbbs.17007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao YN, Li XH, George ML, et al. Quantitative trait locus analysis of drought tolerance and yield in Maize in China. Plant Mol Biol Rep. 2005;23:155–165. doi: 10.1007/BF02772706. [DOI] [Google Scholar]
- Xiao Y, Liu D, Zhang G, et al. Brassinosteroids regulate OFP1, a DLT interacting protein, to modulate plant architecture and grain morphology in rice. Front Plant Sci. 2017 doi: 10.3389/fpls.2017.01698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Song M-H, Jin F, et al. Fine mapping of a grain weight quantitative trait locus on rice chromosome 8 using near-isogenic lines derived from a cross between Oryza sativa and Oryza rufipogon. Theor Appl Genet. 2006;113:885–894. doi: 10.1007/s00122-006-0348-5. [DOI] [PubMed] [Google Scholar]
- Xie X, Jin F, Song M-H, et al. Fine mapping of a yield-enhancing QTL cluster associated with transgressive variation in an Oryza sativa × O. rufipogon cross. Theor Appl Genet. 2008;116:613–622. doi: 10.1007/s00122-007-0695-x. [DOI] [PubMed] [Google Scholar]
- Xing YZ, Tang WJ, Xue WY, et al. Fine mapping of a major quantitative trait loci, qSSP7, controlling the number of spikelets per panicle as a single Mendelian factor in rice. Theor Appl Genet. 2008;116:789–796. doi: 10.1007/s00122-008-0711-9. [DOI] [PubMed] [Google Scholar]
- Xu C, Wang Y, Yu Y, et al. Degradation of MONOCULM 1 by APC/CTAD1 regulates rice tillering. Nat Commun. 2012;3:750. doi: 10.1038/ncomms1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Zhao M, Zhang Q, et al. The DENSE AND ERECT PANICLE 1 (DEP1) gene offering the potential in the breeding of high-yielding rice. Breed Sci. 2016;66:659–667. doi: 10.1270/jsbbs.16120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- Yan JQ, Zhu J, He CX, et al. Quantitative trait loci analysis for the developmental behavior of tiller number in rice (Oryza sativa L.) Theor Appl Genet. 1998;97:267–274. doi: 10.1007/s001220050895. [DOI] [Google Scholar]
- Yan W-H, Wang P, Chen H-X, et al. A Major QTL, Ghd8, Plays Pleiotropic Roles in Regulating Grain Productivity, Plant Height, and Heading Date in Rice. Mol Plant. 2011;4:319–330. doi: 10.1093/mp/ssq070. [DOI] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell. 2000;12:2473–2484. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Ookawa T, Aya K, et al. Isolation of a novel lodging resistance QTL gene involved in strigolactone signaling and its pyramiding with a QTL gene involved in another mechanism. Mol Plant. 2015;8:303–314. doi: 10.1016/j.molp.2014.10.009. [DOI] [PubMed] [Google Scholar]
- Yao R, Wang L, Li Y, et al. Rice DWARF14 acts as an unconventional hormone receptor for strigolactone. J Exp Bot. 2018;69:2355–2365. doi: 10.1093/jxb/ery014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh S-Y, Chen H-W, Ng C-Y, et al. Down-regulation of cytokinin oxidase 2 expression increases tiller number and improves rice yield. Rice (N Y) 2015 doi: 10.1186/s12284-015-0070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A, Ohmori Y, Kitano H, et al. ABERRANT SPIKELET AND PANICLE1, encoding a TOPLESS-related transcriptional co-repressor, is involved in the regulation of meristem fate in rice. Plant J. 2012;70:327–339. doi: 10.1111/j.1365-313X.2011.04872.x. [DOI] [PubMed] [Google Scholar]
- Yuan S, Wang T, Yin L, et al. Cloning and expression of gene responsible for high-tillering dwarf phenotype in indica rice mutant gsor23. Rice Sci. 2013;20:320–328. doi: 10.1016/S1672-6308(13)60134-1. [DOI] [Google Scholar]
- Yue B, Cui K, Yu S, et al. Molecular marker-assisted dissection of quantitative trait loci for seven morphological traits in rice (Oryza Sativa L.) Euphytica. 2006;150:131–139. doi: 10.1007/s10681-006-9101-z. [DOI] [Google Scholar]
- Zhang Y, Luo L, Xu C, et al. Quantitative trait loci for panicle size, heading date and plant height co-segregating in trait-performance derived near-isogenic lines of rice (Oryza sativa) Theor Appl Genet. 2006;113:361–368. doi: 10.1007/s00122-006-0305-3. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Luo L, Liu T, et al. Four rice QTL controlling number of spikelets per panicle expressed the characteristics of single Mendelian gene in near isogenic backgrounds. Theor Appl Genet. 2009;118:1035–1044. doi: 10.1007/s00122-008-0960-7. [DOI] [PubMed] [Google Scholar]
- Zhang S, Li G, Fang J, et al. The interactions among DWARF10, auxin and cytokinin underlie lateral bud outgrowth in rice. J Integr Plant Biol. 2010;52:626–638. doi: 10.1111/j.1744-7909.2010.00960.x. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li J, Yao G, et al. Fine mapping and cloning of the grain number per-panicle gene (Gnp4) on chromosome 4 in rice (Oryza sativa L.) Agric Sci China. 2011;10:1825–1833. doi: 10.1016/S1671-2927(11)60182-X. [DOI] [Google Scholar]
- Zhang G-H, Li S-Y, Wang L, et al. LSCHL4 from japonica cultivar, which Is allelic to NAL1, increases yield of indica super rice 93–11. Mol Plant. 2014;7:1350–1364. doi: 10.1093/mp/ssu055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Tang J, Zhou Y, et al. Characterization and fine mapping of NGP4c(t), a novel gene controlling the number of grains per panicle in rice. J Genet. 2015;94:513–517. doi: 10.1007/s12041-015-0553-6. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Tao Y, Zhu J, et al. GNS4, a novel allele of DWARF11, regulates grain number and grain size in a high-yield rice variety. Rice. 2017;10:34. doi: 10.1186/s12284-017-0171-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Zhang Z, Chen J, et al. Fine mapping of qTGW10-20.8, a QTL having important contribution to grain weight variation in rice. Crop J. 2019 doi: 10.1016/j.cj.2019.01.006. [DOI] [Google Scholar]
- Zong G, Wang A, Wang L, et al. A pyramid breeding of eight grain-yield related quantitative trait loci based on marker-assistant and phenotype selection in rice (Oryza sativa L.) J Genet Genomics. 2012;39:335–350. doi: 10.1016/j.jgg.2012.06.004. [DOI] [PubMed] [Google Scholar]
- Zou J, Chen Z, Zhang S, et al. Characterizations and fine mapping of a mutant gene for high tillering and dwarf in rice (Oryza sativa L.) Planta. 2005;222:604–612. doi: 10.1007/s00425-005-0007-0. [DOI] [PubMed] [Google Scholar]
- Zou J, Zhang S, Zhang W, et al. The rice HIGH-TILLERING DWARF1 encoding an ortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J. 2006;48:687–698. doi: 10.1111/j.1365-313X.2006.02916.x. [DOI] [PubMed] [Google Scholar]